Abstract
Genetic studies of human susceptibility to Schistosoma (bloodfluke) infections have previously identified a genetic locus determining infection intensity with the African species, S. mansoni, in the 5q31-33 region of the human genome which is known to contain the Th2 immune response cluster including the genes encoding the IL-4, IL-5 and IL-13 cytokines. These cytokines are key players in inflammatory immune responses, and have previously been implicated in human susceptibility to infection with the Asian species, S. japonicum. In a nested case-control study, we genotyped 30 HapMap tagging SNPs across these three genes in 159 individuals identified as putatively susceptible to re-infection with S. japonicum and 133 putatively resistant individuals. A third group comprising 113 individuals susceptible to symptomatic infection was also included. The results provided no significant association at a global level between re-infection predisposition and any of the individual SNPs or haplotype blocks. However, two tagging SNPs in IL-5 demonstrated globally significant association with susceptibility to symptomatic infection. They were in strong linkage disequilibrium with each other and were found to belong to the same haplotype block which also provided a significant association after permutation testing. This haplotype was located in the 3’-UTR region of IL-5 suggesting that variants in this region of IL-5 may modulate the immune response in these individuals with symptomatic infection.
Keywords: Human, Parasitic-Helminth, Cytokines
Introduction
The digenetic trematode (blood fluke) Schistosoma japonicum, transmitted by the amphibious fresh water snail Oncomelania, causes intestinal and hepatosplenic schistosomiasis in China, the Philippines, and Indonesia. Currently, approximately 880,000 individuals are estimated to be infected in China, predominantly in the Yangtze River basin and the lake regions of Hunan and Jiangxi provinces (1). The over-distribution of faecal egg counts commonly observed in endemic populations is indicative of an innate susceptibility to S. japonicum infection in a minority of individuals. Individuals have been identified as being putatively susceptible or resistant to re-infection in both Chinese (2) and Filipino (3) populations. Variance components analyses have demonstrated significant clustering of infection and infection intensities in families for both S. japonicum (4) and the African species, S. mansoni (5, 6), and strong heritable factors were estimated to contribute to the control of human infection. Genetic studies have investigated infection intensity levels in S. mansoni-infected families and identified a co-dominant major gene playing a role in human susceptibility (7). The SM1 locus was subsequently linked to the 5q31-q33 region of the human genome (8-10), a region known to contain the Th2 gene cluster encoding the IL-4, IL-5 and IL-13 cytokines. Many immunological studies have demonstrated the role of these cytokines in the immunomodulation of several helminth, including schistosome, infections in murine models (11, 12) and in humans (13-17). Other studies have shown a marked increase in the levels of IL-5 and IL-13 in individuals identified as being resistant to schistosome infection (18, 19). Further, two polymorphisms in the IL-13 gene promoter region have been shown to be associated with with S. haematobium infections (20). These findings led us to undertake a nested case-control study to investigate human susceptibility to S. japonicum re-infection targeting IL-4, IL-5 and IL-13 as candidate genes for SNP typing and association analysis.
Materials and Methods
Study location and population
The participants from this study were selected from 8 administrative villages: Aiguo, Dingshan, Fuqian, Xindong, Yufeng, Caojia, Hexi and Tangmei from the Poyang Lake region, Jiangxi province, China which is highly endemic for S. japonicum. The total number of subjects from all eight villages who undertook an interview was 5794. Each administrative village was comprised of 4-8 ‘natural villages’. Each individual was allocated a personal identification code (PID) comprised of their administrative village code, natural village code, household and household member code. Demographic data were collected as well as water contact and schistosomiasis history using a validated questionnaire (21). Children under the age of 5 were not included in this study.
Questionnaire and sample selection
A cohort of 779 individuals was selected from the study based on exposure to infection and S. japonicum infection history. To ensure that all cohort members had similar water exposure, only full time residents of the village were included who were over the age of 25 and had regular water contact. A questionnaire was constructed to assess schistosomiasis history with respect to previous diagnosed infection, previous treatment (following diagnosis) with the highly effective drug praziquantel, previous control treatment and symptomatic infection (see below for definition). All individuals were also interviewed regarding their occupation, whether the occupation involved water contact, the length of time in the current occupation and previous water contact. At the time of interview, participants were asked to provide a saliva sample using a commercially available collection kit (Oragene, DNAgenotek, Inc., Ottawa, Ontario, Canada). In accordance with the protocol, individuals were given a cup of water to rinse their mouths. Donors were then instructed to provide at least 2 ml of saliva in a vial labelled with their PID, name and sex. Of the 779 cohort individuals, 192 were lost to follow up and 587 individuals were interviewed, of which two refused to provide a saliva sample.
Phenotype classification
All individuals included in the phenotype groups had to have significant occupational water exposure. Susceptibility/resistance to infection was examined using the questionnaire data. Susceptibility to infection/re-infection was defined as an individual having had ten or more diagnosed schistosome infections in his/her life time. Individuals susceptible to symptomatic infection were defined as those who had been diagnosed (by faecal egg count) with S. japonicum infection which was accompanied by three of the following symptoms: high fever, weakness, loss of appetite, headaches and dizziness. Resistant individuals were categorised as having had fewer than five known infections and fewer than five treatments. All resistant subjects had to have been in their occupation for at least 15 years to ensure adequate exposure. The length of time in occupation, age and average numbers of infections per year for each group are shown in Table 1. There were 159 individuals identified as susceptible to infection/re-infection, 113 were susceptible to acute infection, and 133 were categorised as resistant to infection/re-infection. The remainder were omitted from the analysis as they could not be definitively classified into one of the three defined phenotypes.
Table I.
Characteristics of the different phenotype groups
| Phenotype Group | N | No of infections (Range) |
Age (Range) |
Length of time in occupation (Range) |
No of infections/year in occupation (95% CI) |
|---|---|---|---|---|---|
| Resistant | 133 | 1.71 (0-5) |
48.81 (30-76) |
28.88 (15-60) |
0.067 (0.056-0.077) |
| Susceptible to re-infection | 159 | 30.63 (10-70) |
50.69 (27-86) |
29.27 (5-60) |
1.234 (1.071-1.398) |
| Susceptible to symptomatic infection | 113 | 17.71 (1-70) |
49.63 (29-82) |
29.28 (2-58) |
0.649 (0.491-0.808) |
DNA purification
DNA was purified from the saliva samples according to the manufacturer’s instructions (Oragene, DNAgenotek, Inc., Ottawa, Ontario, Canada). DNA concentration was measured by spectrophotometry. Aliquots of all samples were taken and subsequently adjusted to provide standard stock solutions of 20ng/μl. The A280/A260 ratio was estimated to provide an indication of the quality of the sample. Low ratios (<1.6) may indicate possible salt or ethanol contaminants whereas high values may suggest the presence of fragmented DNA, RNA or bacterial DNA. Only samples which provided a yield of greater than 20ng/ul and a A280/A260 ratio of >1.6 and <1.95 were included for genotyping analysis.
SNP selection
We selected tagging SNPs across the IL-4/IL-13 and IL-5 genes based on data from the public databases including the International HapMap Project (http://www.hapmap.org/) and NCBI (http://www.ncbi.nlm.nih.gov/). SNPs were selected beginning 20kb upstream and extending 20kb downstream for each gene. IL-4 and IL-13 were only 12.5 kb apart and thus were treated as one region. SNPs identified in the region which had frequency information in Hapmap (for Chinese populations) were used to identify tagSNPs. A total of 20 tagSNPs were selected in these three genes, selecting from phase I and II HapMap data so other SNPs within the interval were in strong linkage disequilibrium (r2 co-efficient of ≥ 0.8) with one of the tagSNPs. An additional 10 SNPs were selected from the literature with evidence of previous associations with atopic conditions, malaria and S. haematobium infections and included in the study (20, 22-26).
Genotyping
Assays were designed to type 30 SNPs across the 5q31-33 locus using the Sequenom MassARRAY Assay Design software (version 3.0). A MALDI-TOF Mass Spectrometer (Sequenom Inc, San Diego) was used to type the SNPs using iPLEX™ chemistry. PCR reactions were carried out in 2.5 μl reactions in standard 384-well plates. PCR was performed using approximately 12.5ng genomic DNA for each sample, 0.5 unit of Taq polymerase (HotStarTaq, Qiagen, Valencia, CA), 500umol of each dNTP, and 100 nmol of each PCR primer. PCR thermal cycling in an ABI-9700 instrument was 15 min at 94°C, followed by 45 cycles of 20 sec at 94°C, 30 sec at 56°C, 60 sec at 72°C. To the completed PCR reaction, 1 μl of Shrimp Alkaline Phosphatase (SAP) master mix containing 1.4 units of ASP in final concentration was added and incubated for 30 min at 37°C followed by inactivation for 5 min at 85°C. After adjusting the concentrations of extension primers to equilibrate signal-to-noise ratios, the post-PCR primer extension reaction of the iPLEX assay was performed in a final 5 μl of extension reaction containing 0.1 μl termination mix, 0.02 μl DNA polymerase (Sequenom, San Diego, CA), and 600 nM to 1200 nM extension primers. A two-step 200 short cycles program was used for the iPLEX reaction: initial denaturation was 30 sec at 94°C followed by 5 cycles of 5 sec at 52°C and 5 sec at 80°C. Additional 40 annealing and extension cycles were then looped back to 5 sec at 94°C, 5 sec at 52°C and 5 sec at 80°C. A final extension was done at 72°C for three min and then the sample was cooled to 20°C. The iPLEX reaction products were desalted by diluting samples with 15 μl water and 3 μl resin to optimize mass spectrometric analysis and then spotted on a SpectroChip (Sequenom), processed and analysed in a Compact Mass Spectrometer by MassARRAY Workstation (version 3.3) software (Sequenom).
Statistical analysis
Genotype frequencies for all SNP variants were examined and the results were tested for departures from Hardy-Weinberg equilibrium (HWE) separately for both the susceptible groups and the resistant group. This was achieved using the Haploview program (27). Genotypes for all but one marker (rs2243250) were consistent with HWE and no obvious genotyping errors were apparent thus the data for this marker were excluded from further analysis. Three markers were found to have no allelic variation in this population (rs2069743, rs2243231 and rs2243240).
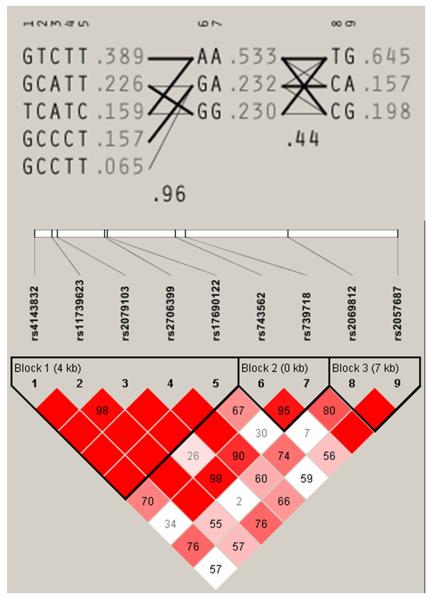
Haplotype blocks were determined using the default method of Gabriel et al. (28) and are displayed schematically with their population frequencies and inter-block connections. Linkage disequilibrium (LD) plots were also obtained and illustrate the estimated LD between SNPs, where red regions depict strong LD (1.0) with strong confidence (LOD>2.0); blue squares depict strong LD with weak confidence (LOD<2.0); and pale-red and white regions represent low LD (<1.0) and state the LD value within the box.
The haploview program was also used to test for association between the phenotypes and individual markers or combinations of markers (haplotypes). P-values were then corrected for multiple testing of all the SNPs and haplotypes within the region through 10000 permutations. This adjusts p-values derived from multiple statistical tests to correct for the occurrence of false positives and provides region-wide empirical p-values for each marker. ‘Local’ significance therefore can be described as the probability that an association for a particular SNP marker is due to chance and concerns a single test of the null hypothesis of no linkage; whereas a ‘global’ significance level is the probability that an association is detected within a given set of SNPs and involves sampling over a large number of tests to find the most significant result. This is achieved in Haploview by maintaining the individual genotype as a whole while the individual’s status is shuffled. The method preserves the correlation between SNPs (linkage disequilibrium) while breaking the relation between status and the genotypes. For each replicate or permutation, each SNP was tested for association and the most significant p-value was stored. To assess the potential effect of each significant SNP on the risk of disease, logistic regression was performed to obtain odds ratios and 95% confidence intervals (C.I.). These compared the carriers of the minor allele to homozygous carriers of the major allele for each significant SNP.
Ethical approval
Written ethical approval for this study was obtained at the national, provincial and village levels within China, and approval for the study was granted by the ethics committees of Jiangxi Provincial Institute of Parasitic Diseases, Chinese Center for Disease Control and Prevention and the Queensland Institute of Medical Research, prior to commencement. Study participants identified as stool egg-positive for schistosomiasis were treated with 40 mg/kg of praziquantel, the current dosage recommended by the WHO. Oral informed consent was obtained from all adults and from parents or guardians of minors aged 5 or above who were involved in the project.
Results
In total, thirty SNPs were genotyped spanning a region of 20 kb across the IL5 locus and a 24kb region across the IL13 and IL4 genes (an average spacing of one SNP every 3.03 kb and 1.51kb, respectively; Figure 1). Genotypes were obtained for most individuals with a mean completion rate of 95.45%. The minor allele frequencies of the SNPs ranged from 0.011 to 0.477 (Tables 1 and 2). In our study, we have investigated the 5q31-33 region of the human genome, more specifically, the IL-4, IL-5 and IL-13 genes. Across these loci we found little evidence of linkage disequilibrium between markers at different ends of the genes (Figure 2). We chose SNPs across the 5q31-33 region using a SNP tagging strategy where representative SNPs were genotyped that had a high correlation (r2 > 0.8) with other known SNPs in these genes. Common variants increasing risk of S. japonicum infection or acute infection would be expected to show evidence of association with one or more of the tagging SNPs genotyped. We found evidence for association between S. japonicum infection and individual tagging SNPs in both IL-5 and IL-13 for allelic association tests. Tests of association between S. japonicum infection and haplotypes or combinations of SNPs also showed evidence for IL-5 variation contributing to risk of S. japonicum infection. However, all associations found with S. japonicum infection were not found to be significant, when tested for global significance.
Figure 1.
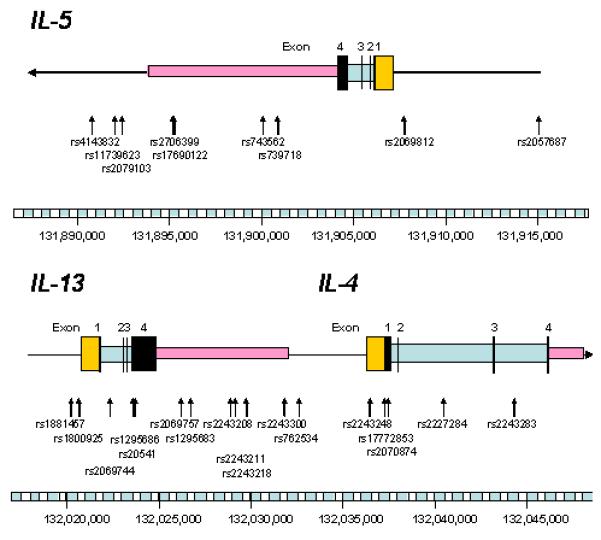
Human IL5, IL-4 and IL-13 and the genomic structure of each gene showing the location of the tag SNPs genotyped
Table II.
SNPs genotyped across the 5q31-33 region and associations with infection with S. japonicum
| # | dbSNP ID | Position | Gene | Role | Alleles | Frequency | Association (X2) | p-value |
|---|---|---|---|---|---|---|---|---|
| 1 | rs4143832 | 131890876 | IL-5 | A/C | 0.151 | 0.224 | 0.0926 | |
| 2 | rs11739623 | 131892051 | IL-5 | C/T | 0.38 | 0.426 | 0.2934 | |
| 3 | rs2079103 | 131892405 | IL-5 | G/T | 0.403 | 2.824 | 0.0197 | |
| 4 | rs2706399 | 131895601 | IL-5 | 3’-UTR | A/G | 0.153 | 4.251 | 0.0281 |
| 5 | rs17690122 | 131895734 | IL-5 | 3’-UTR | A/G | 0.151 | 0.132 | 0.1156 |
| 6 | rs743562 | 131900282 | IL-5 | 3’-UTR | C/T | 0.477 | 0.122 | 0.9351 |
| 7 | rs739718 | 131900972 | IL-5 | 3’-UTR | A/G | 0.249 | 2.706 | 0.109 |
| 8 | rs2069812 | 131907815 | IL-5 | Promoter | C/T | 0.332 | 1.74 | 0.3275 |
| 9 | rs2057687 | 131915144 | IL-5 | Promoter | A/G | 0.144 | 2.611 | 0.0911 |
| 10 | rs1881457 | 132020308 | IL-13 | Promoter | A/C | 0.231 | 0.307 | 0.3118 |
| 11 | rs1800925 | 132020708 | IL-13 | Promoter | C/T | 0.167 | 0.047 | 0.4883 |
| 12 | rs2069744 | 132022568 | IL-13 | Intron | C/T | 0.096 | 0.641 | 0.1945 |
| 13 | rs1295686 | 132023742 | IL-13 | Intron (Boundary) |
A/G | 0.191 | 4.793 | 0.035 |
| 14 | rs20541 | 132023863 | IL-13 | Exon | C/T | 0.339 | 2.23 | 0.2745 |
| 15 | rs2069757 | 132026312 | IL-13 | Intron | A/G | 0.127 | 1.337 | 0.109 |
| 16 | rs1295683 | 132026775 | IL-13 | 3’-UTR | C/T | 0.338 | 1.136 | 0.4566 |
| 17 | rs2243208 | 132029050 | IL-13 | 3’-UTR | A/G | 0.141 | 1.438 | 0.0678 |
| 18 | rs2243211 | 132029321 | IL-13 | 3’-UTR | A/C | 0.036 | 0.116 | 0.4634 |
| 19 | rs2243218 | 132029923 | IL-13 | 3’-UTR | A/G | 0.041 | 0.263 | 0.4025 |
| 20 | rs2243300 | 132031985 | IL-13 | 3’-UTR | G/T | 0.039 | 0.493 | 0.2784 |
| 21 | rs762534 | 132032655 | IL-4 | Promoter | A/C/T | 0.051 | 2.51 | 0.0667 |
| 22 | rs2243248 | 132036543 | IL-4 | Promoter | G/T | 0.057 | 0.064 | 0.6994 |
| 23 | rs17772853 | 132037489 | IL-4 | Exon | C/T | 0.012 | 0.107 | 0.6223 |
| 24 | rs2070874 | 132037609 | IL-4 | Exon | C/T | 0.188 | 0.245 | 0.899 |
| 25 | rs2227284 | 132040624 | IL-4 | Intron | A/C | 0.134 | 0.026 | 0.6849 |
| 26 | rs2243283 | 132044492 | IL-4 | Intron | C/G | 0.244 | 0.382 | 0.8302 |
Figure 2.
Haplotype blocks identified in each gene and linkage disequilibrium plot of single nucleotide polymorphism (SNP) estimated as r2 using Haploview for IL-5. SNP codes are provided in order of location along each gene; red squares depict strong LD (1.0) with strong confidence (LOD>2.0); blue boxes depict strong LD (1.0) with weak confidence (LOD<2.0); pale-red and white regions represent low LD (<1.0); the LD value is provided within each box. Plots based on Haploview output.
Investigation of allelic associations between symptomatic infection and individual tagging SNPs detected five significant SNPs; one in IL-13 and four in IL-5 (Table 2) all of which belonged to the same haplotype block (Figure 2). Of these, two SNPs in IL-5 (rs4143832 and rs17690122) remained significant after permutation tests (p = 0.025 and p = 0.019, respectively) and were in strong LD with each other (Figure 2). Logistic regression provided an odds ratio of 2.3 (p = 0.004; C.I. = 1.30-4.01) and 2.25 (p = 0.005; C.I. = 1.27-4.00) for each SNP marker, respectively. Haplotype association tests also detected two significant haplotypes associated with symptomatic infection (Table 3). Permutation tests showed a p-value of 0.019 for one haplotype.
Table III.
SNPs genotyped across the 5q31-33 region and associations with symptomatic S. japonicum infection
| # | dbSNP ID | Position | Gene | Role | Alleles | Frequency | Association (X2 value) |
p-value |
|---|---|---|---|---|---|---|---|---|
| 1 | rs4143832 | 131890876 | IL-5 | A/C | 0.159 | 7.107 | 0.0014* | |
| 2 | rs11739623 | 131892051 | IL-5 | C/T | 0.388 | 0.432 | 0.3621 | |
| 3 | rs2079103 | 131892405 | IL-5 | G/T | 0.402 | 2.292 | 0.0176 | |
| 4 | rs2706399 | 131895601 | IL-5 | 3’-UTR | A/G | 0.141 | 1.597 | 0.0237 |
| 5 | rs17690122 | 131895734 | IL-5 | 3’-UTR | A/G | 0.16 | 8.073 | 0.0011* |
| 6 | rs743562 | 131900282 | IL-5 | 3’-UTR | C/T | 0.463 | 0.563 | 0.3908 |
| 7 | rs739718 | 131900972 | IL-5 | 3’-UTR | A/G | 0.239 | 0.358 | 0.3587 |
| 8 | rs2069812 | 131907815 | IL-5 | Promoter | C/T | 0.328 | 1.744 | 0.6754 |
| 9 | rs2057687 | 131915144 | IL-5 | Promoter | A/G | 0.13 | 0.225 | 0.3338 |
| 10 | rs1881457 | 132020308 | IL-13 | Promoter | A/C | 0.251 | 0.569 | 0.0926 |
| 11 | rs1800925 | 132020708 | IL-13 | Promoter | C/T | 0.178 | 1.257 | 0.1183 |
| 12 | rs2069744 | 132022568 | IL-13 | Intron | C/T | 0.1 | 0.968 | 0.1092 |
| 13 | rs1295686 | 132023742 | IL-13 | Intron (Boundary) |
A/G | 0.337 | 0.019 | 0.1677 |
| 14 | rs20541 | 132023863 | IL-13 | Exon | C/T | 0.336 | 0.004 | 0.706 |
| 15 | rs2069757 | 132026312 | IL-13 | Intron | A/G | 0.14 | 0 | 0.1546 |
| 16 | rs1295683 | 132026775 | IL-13 | 3’-UTR | C/T | 0.187 | 0.227 | 0.7437 |
| 17 | rs2243208 | 132029050 | IL-13 | 3’-UTR | A/G | 0.151 | 0.935 | 0.0366 |
| 18 | rs2243211 | 132029321 | IL-13 | 3’-UTR | A/C | 0.038 | 0.023 | 0.5867 |
| 19 | rs2243218 | 132029923 | IL-13 | 3’-UTR | A/G | 0.042 | 0.07 | 0.4294 |
| 20 | rs2243300 | 132031985 | IL-13 | 3’-UTR | G/T | 0.04 | 0.215 | 0.2961 |
| 21 | rs762534 | 132032655 | IL-4 | Promoter | A/C/T | 0.044 | 2.355 | 0.0978 |
| 22 | rs2243248 | 132036543 | IL-4 | Promoter | G/T | 0.059 | 0.119 | 0.5717 |
| 23 | rs17772853 | 132037489 | IL-4 | Exon | C/T | 0.011 | 1.015 | 0.3123 |
| 24 | rs2070874 | 132037609 | IL-4 | Exon | C/T | 0.193 | 1.135 | 0.3878 |
| 25 | rs2227284 | 132040624 | IL-4 | Intron | A/C | 0.136 | 1.13 | 0.3334 |
| 26 | rs2243283 | 132044492 | IL-4 | Intron | C/G | 0.232 | 2.401 | 0.5494 |
Still significant after permutation test at a p<0.05 level
Discussion
The 5q31-q33 region of the human genome contains genes encoding the IL-4, IL-5 and IL-13 cytokines which are known players in the Th2 type immune response. These have been shown to have prominent roles in human susceptibility to infection with several parasitic helminths including Ascaris, Trichuris and hookworm (13-17). Moreover, combined segregation and linkage analysis identified this region to contain a susceptibility locus (SM1) controlling S. mansoni infection intensity, and polymorphisms in IL-13 have also been associated with infection susceptibility to S. haematobium (20) and susceptibility to severe malaria in Thailand (24). These cytokines act together to produce a pro-inflammatory response to schistosome infection characterised by increased eosinophil and IgE production (29-31) Such responses are typical of reactions to allergens and, as such, are commonly observed with asthma and allergy (32-34). Polymorphisms in these genes have also been associated with asthma and atopic dermatitis (34-38).
In this study, we investigated the relationship between this region of the genome and infection with S. japonicum. We detected two SNPs, one in IL-5 and one in IL-13, with allelic associations to infection susceptibility, but both associations did not withstand permutation testing, thus indicating that the associations could be due to chance. A major challenge underlying this study was that of precise phenotype definition; given the extensive levels of schistosomiasis control in China, it is becoming increasingly difficult to accurately determine the number of previous praziquantel drug treatments and infection diagnoses in schistosome-endemic communities. Consequently, we applied highly stringent criteria in order to identify putatively susceptible and resistant individuals as accurately as possible. This resulted in a relatively low number of susceptible and resistant individuals which limited the power of the study to detect discrete genetic associations with infection. The SNP identified in IL-5, however, belongs to the same haplotype block as the two SNPs associated with symptomatic S. japonicum infection and are in strong linkage disequilibrium with each other. The SNP detected in IL-13 has also been implicated in asthma and associated elevated IgE levels (39, 40). Further work to increase the sample size and investigate the potential biological implications of these SNPs on IL-5 and IL-13 expression would provide more insight into the potential role of these mutations in susceptibility to schistosome infection.
Symptomatic infection was also investigated in this study. This outcome of infection is typically seen in individuals experiencing their first schistosome infection though symptoms can be present in those who have a sudden heavy exposure to the parasite and amongst chronically infected individuals. There are many symptoms and variations in the clinical presentation of schistosome infection and disease ranging from fever and eosinophilia to diffuse abdominal pain and hepatomegaly (2). The difference between infection and disease is frequently confused and often requires clarification. In this study we investigated symptomatic infection which we define as the manifestation of a hyper-allergenic response to infection with S. japonicum, diagnosed by a local doctor (by parasitological examination), and characterised by at least three of the following symptoms: high fever, weakness, loss of appetite, headache and dizziness. We identified 113 individuals who had previously experienced symptoms associated with schistosome infection and were diagnosed by a local doctor at that time. Two SNPs within 15kb downstream from IL-5 (one in the 3’-UTR region) were found to be globally significant (P<0.05). They were in strong linkage disequilibrium with each other and belonged to the same haplotype block identified in Haploview (Figure 2). The haplotype also showed an association after testing for global significance (p= 0.019) suggesting that variants in IL-5 could be contributing to risk of symptomatic infection. Odds ratios and C.I.s obtained for both SNPs were similar, reflecting the high level of LD between the two markers and indicating that homozygous genotypes of the major allele are associated with a 2-fold decrease in the risk of having a symptomatic reaction if infected with S. japonicum. It has been shown previously in this population that symptomatic infection is strongly associated with re-infection (data not shown) indicating that those who had previously experienced a symptomatic reaction in response to infection were twice as likely to be re-infected (4).
Marked elevated levels of circulating serum IL-5 have been observed in individuals previously identified as putatively resistant to schistosome infection/re-infection (19, 41, 42) and IL-5-induced eosinophilia has shown to be a common symptom in acute schistosomiasis patients (2, 43, 44). Further, IL-5 contributes to the immune response against various pathogens and infectious agents including gastro-intestinal helminths such as hookworms (16) and Trichuris (45). It is important to note, however, that the phenotype in this study could also be reflecting a genetic predisposition to hyper-allergenic reactions and inflammatory responses that are associated with acute and chronic infection rather than susceptibility to schistosome infection per se. Other allergies, asthma and other atopic syndromes may also be associated with these individuals predisposed to a symptomatic response to infection and this merits further investigation.
It is noteworthy also that the two significant SNPs detected in this study may not be the cause of the association and possible neighbouring SNPs that are in strong LD could be accounting for the association observed with symptomatic infection. However, the strong LD of these SNPs with each other and their location in the same haplotype block would suggest that the true infection-predisposing variant does lie in the 3’-UTR region of IL-5. Previous functional studies have shown that mutations in the 3’-UTR region can have an effect on mRNA stability or translational efficiency by interfering with the mRNA-binding protein interaction (46-48).
This is the first study to investigate the genetics underlying susceptibility to S. japonicum infection. Despite the restricted sample size of this investigation, a significant association was observed implicating IL-5 with symptomatic infection with S. japonicum. Further work to increase the sample size and to replicate the study in populations in neighbouring endemic provinces in China would provide further information on the role of IL-5 variants in determining risk of S. japonicum infection. Rapid identification of those more pre-disposed to infection and/or a symptomatic reaction to infection would assist in minimising the morbidity associated with S. japonicum infection, allow for more targeted treatment of susceptible individuals and reduce overall costs to the individual and the health care system in China.
Table IV.
Haplotype blocks in the 5q31-33 region and associations with symptomatic S. japonicum infection
| Block | Haplotype | Case, Control Ratios | X2 value | p-value |
|---|---|---|---|---|
| Block 1 | ||||
| GTCTT | 0.389 | 81.0 : 141.0, 105.0 : 151.0 | 1.028 | 0.3105 |
| GCATT | 0.226 | 49.0 : 173.0, 59.0 : 197.0 | 0.065 | 0.7983 |
| TCATC | 0.159 | 48.0 : 174.0, 28.0 : 228.0 | 10.15 | 0.0014* |
| GCCCT | 0.157 | 25.0 : 197.0, 50.1 : 205.9 | 6.208 | 0.0127 |
| GCCTT | 0.065 | 17.0 : 205.0, 13.9 : 242.1 | 0.986 | 0.3206 |
| Block 2 | ||||
| AA | 0.533 | 121.4 : 100.6, 133.1 : 122.9 | 0.345 | 0.5569 |
| GA | 0.232 | 44.1 : 177.9, 66.9 : 189.1 | 2.62 | 0.1055 |
| GG | 0.23 | 54.2 : 167.8, 55.8 : 200.2 | 0.469 | 0.4933 |
| Block 3 | ||||
| TG | 0.645 | 141.0 : 81.0, 167.5 : 88.5 | 0.193 | 0.6603 |
| CG | 0.198 | 50.0 : 172.0, 44.5 : 211.5 | 1.99 | 0.1583 |
| CA | 0.157 | 31.0 : 191.0, 44.0 : 212.0 | 0.934 | 0.3338 |
| Block 4 | ||||
| AC | 0.763 | 162.0 : 60.0, 202.9 : 53.1 | 2.608 | 0.1063 |
| CT | 0.174 | 45.0 : 177.0, 38.0 : 218.0 | 2.44 | 0.1183 |
| CC | 0.063 | 15.0 : 207.0, 15.1 : 240.9 | 0.152 | 0.6968 |
| Block 5 | ||||
| GG | 0.652 | 147.0 : 75.0, 164.7 : 91.3 | 0.182 | 0.6694 |
| AG | 0.231 | 44.0 : 178.0, 66.3 : 189.7 | 2.463 | 0.1165 |
| AA | 0.117 | 31.0 : 191.0, 25.0 : 231.0 | 2.026 | 0.1546 |
| Block 6 | ||||
| TTTA | 0.795 | 172.0 : 50.0, 208.0 : 48.0 | 1.037 | 0.3085 |
| CCGG | 0.145 | 33.7 : 188.3, 35.7 : 220.3 | 0.149 | 0.6996 |
| CCTG | 0.049 | 13.3 : 208.7, 10.2 : 245.8 | 0.985 | 0.321 |
Still significant after permutation test at a p<0.05 level
Abbreviations
- SNP
Single nucleotide polymorphism
- 3’-UTR
Three prime-untranslated region
Footnotes
This study was supported by the National Institute of Allergy and Infectious Diseases (NIAID) (Tropical Medicine Research Center grant 1 P 50AI-39461) and a National Health and Medical Research Council of Australia and Wellcome Trust (UK) International Collaborative Research Grants Scheme Award.
References
- 1.Zhou XN, Jiang QW, Sun LP, Wang TP, Hong QB, Zhao GM, Wen LY, Ying ZC, Wu XH, Lin DD. Control and surveillance of schistosomiasis in China. Chin J Schisto Contr. 2005;17:161–165. [Google Scholar]
- 2.Ross AG, Vickers D, Olds GR, Shah SM, McManus DP. Katayama syndrome. Lancet Infect Dis. 2007;7:218–224. doi: 10.1016/S1473-3099(07)70053-1. [DOI] [PubMed] [Google Scholar]
- 3.Acosta LP, Aligui GD, Tiu WU, McManus DP, Olveda RM. Immune correlate study on human Schistosoma japonicum in a well-defined population in Leyte, Philippines: I. Assessment of ‘resistance’ versus ‘susceptibility’ to S. japonicum infection. Acta Trop. 2002;84:127–136. doi: 10.1016/s0001-706x(02)00176-6. [DOI] [PubMed] [Google Scholar]
- 4.Ellis MK, Li Y, Rong Z, Chen H, McManus DP. Familial aggregation of human infection with Schistosoma japonicum in the Poyang Lake region, China. Int J Parasitol. 2006;36:71–77. doi: 10.1016/j.ijpara.2005.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bethony J, Gazzinelli A, Lopes A, Pereira W, Alves-Oliveira L, Williams-Blangero S, Blangero J, Loverde P, Correa-Oliveira R. Genetic epidemiology of fecal egg excretion during Schistosoma mansoni infection in an endemic area in Minas Gerais, Brazil. Mem Inst Oswaldo Cruz. 2001;96(Suppl):49–55. doi: 10.1590/s0074-02762001000900007. [DOI] [PubMed] [Google Scholar]
- 6.Bethony J, Williams JT, Blangero J, Kloos H, Gazzinelli A, Soares-Filho B, Coelho L, Alves-Fraga L, Williams-Blangero S, Loverde PT, Correa-Oliveira R. Additive host genetic factors influence fecal egg excretion rates during Schistosoma mansoni infection in a rural area in Brazil. Am J Trop Med Hyg. 2002;67:336–343. doi: 10.4269/ajtmh.2002.67.336. [DOI] [PubMed] [Google Scholar]
- 7.Abel L, Demenais F, Prata A, Souza AE, Dessein A. Evidence for the segregation of a major gene in human susceptibility/resistance to infection by Schistosoma mansoni. Am J Hum Genet. 1991;48:959–970. [PMC free article] [PubMed] [Google Scholar]
- 8.Marquet S, Abel L, Hillaire D, Dessein A. Full results of the genome-wide scan which localises a locus controlling the intensity of infection by Schistosoma mansoni on chromosome 5q31-q33. Eur J Hum Genet. 1999;7:88–97. doi: 10.1038/sj.ejhg.5200243. [DOI] [PubMed] [Google Scholar]
- 9.Marquet S, Abel L, Hillaire D, Dessein H, Kalil J, Feingold J, Weissenbach J, Dessein AJ. Genetic localization of a locus controlling the intensity of infection by Schistosoma mansoni on chromosome 5q31-q33. Nat Genet. 1996;14:181–184. doi: 10.1038/ng1096-181. [DOI] [PubMed] [Google Scholar]
- 10.Muller-Myhsok B, Stelma FF, Guisse-Sow F, Muntau B, Thye T, Burchard GD, Gryseels B, Horstmann RD. Further evidence suggesting the presence of a locus, on human chromosome 5q31-q33, influencing the intensity of infection with Schistosoma mansoni. Am J Hum Genet. 1997;61:452–454. doi: 10.1016/S0002-9297(07)64073-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finkelman FD, Shea-Donohue T, Goldhill J, Sullivan CA, Morris SC, Madden KB, Gause WC, Urban JF., Jr. Cytokine regulation of host defense against parasitic gastrointestinal nematodes: lessons from studies with rodent models. Annu Rev Immunol. 1997;15:505–533. doi: 10.1146/annurev.immunol.15.1.505. [DOI] [PubMed] [Google Scholar]
- 12.Gause WC, Urban JF, Jr., Stadecker MJ. The immune response to parasitic helminths: insights from murine models. Trends Immunol. 2003;24:269–277. doi: 10.1016/s1471-4906(03)00101-7. [DOI] [PubMed] [Google Scholar]
- 13.Cooper PJ, Chico ME, Sandoval C, Espinel I, Guevara A, Kennedy MW, Urban JF, Jr, Griffin GE, Nutman TB. Human infection with Ascaris lumbricoides is associated with a polarized cytokine response. J Infect Dis. 2000;182:1207–1213. doi: 10.1086/315830. [DOI] [PubMed] [Google Scholar]
- 14.Jackson JA, Turner JD, Rentoul L, Faulkner H, Behnke JM, Hoyle M, Grencis RK, Else KJ, Kamgno J, Boussinesq M, Bradley JE. T helper cell type 2 responsiveness predicts future susceptibility to gastrointestinal nematodes in humans. J Infect Dis. 2004;190:1804–1811. doi: 10.1086/425014. [DOI] [PubMed] [Google Scholar]
- 15.Jackson JA, Turner JD, Rentoul L, Faulkner H, Behnke JM, Hoyle M, Grencis RK, Else KJ, Kamgno J, Bradley JE, Boussinesq M. Cytokine response profiles predict species-specific infection patterns in human GI nematodes. Int J Parasitol. 2004;34:1237–1244. doi: 10.1016/j.ijpara.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 16.Quinnell RJ, Pritchard DI, Raiko A, Brown AP, Shaw MA. Immune responses in human necatoriasis: association between interleukin-5 responses and resistance to reinfection. J Infect Dis. 2004;190:430–438. doi: 10.1086/422256. [DOI] [PubMed] [Google Scholar]
- 17.Turner JD, Faulkner H, Kamgno J, Cormont F, Van Snick J, Else KJ, Grencis RK, Behnke JM, Boussinesq M, Bradley JE. Th2 cytokines are associated with reduced worm burdens in a human intestinal helminth infection. J Infect Dis. 2003;188:1768–1775. doi: 10.1086/379370. [DOI] [PubMed] [Google Scholar]
- 18.Al-Sherbiny M, Osman A, Barakat R, El Morshedy H, Bergquist R, Olds R. In vitro cellular and humoral responses to Schistosoma mansoni vaccine candidate antigens. Acta Trop. 2003;88:117–130. doi: 10.1016/s0001-706x(03)00195-5. [DOI] [PubMed] [Google Scholar]
- 19.Leenstra T, Acosta LP, Wu HW, Langdon GC, Solomon JS, Manalo DL, Su L, Jiz M, Jarilla B, Pablo AO, McGarvey ST, Olveda RM, Friedman JF, Kurtis JD. T-helper-2 cytokine responses to Sj97 predict resistance to reinfection with Schistosoma japonicum. Infect Immun. 2006;74:370–381. doi: 10.1128/IAI.74.1.370-381.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kouriba B, Chevillard C, Bream JH, Argiro L, Dessein H, Arnaud V, Sangare L, Dabo A, Beavogui AH, Arama C, Traore HA, Doumbo O, Dessein A. Analysis of the 5q31-q33 locus shows an association between IL13-1055C/T IL-13-591A/G polymorphisms and Schistosoma haematobium infections. J Immunol. 2005;174:6274–6281. doi: 10.4049/jimmunol.174.10.6274. [DOI] [PubMed] [Google Scholar]
- 21.Ross AG, Yuesheng L, Sleigh AS, Yi L, Williams GM, Wu WZ, Xinsong L, Yongkang H, McManus DP. Epidemiologic features of Schistosoma japonicum among fishermen and other occupational groups in the Dongting Lake region (Hunan Province) of China. Am J Trop Med Hyg. 1997;57:302–308. doi: 10.4269/ajtmh.1997.57.302. [DOI] [PubMed] [Google Scholar]
- 22.Bugawan TL, Mirel DB, Valdes AM, Panelo A, Pozzilli P, Erlich HA. Association and interaction of the IL4R, IL4, and IL13 loci with type 1 diabetes among Filipinos. Am J Hum Genet. 2003;72:1505–1514. doi: 10.1086/375655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He JQ, Chan-Yeung M, Becker AB, Dimich-Ward H, Ferguson AC, Manfreda J, Watson WT, Sandford AJ. Genetic variants of the IL13 and IL4 genes and atopic diseases in at-risk children. Genes Immun. 2003;4:385–389. doi: 10.1038/sj.gene.6363985. [DOI] [PubMed] [Google Scholar]
- 24.Ohashi J, Naka I, Patarapotikul J, Hananantachai H, Looareesuwan S, Tokunaga K. A single-nucleotide substitution from C to T at position -1055 in the IL-13 promoter is associated with protection from severe malaria in Thailand. Genes Immun. 2003;4:528–531. doi: 10.1038/sj.gene.6364010. [DOI] [PubMed] [Google Scholar]
- 25.Wang M, Xing ZM, Lu C, Ma YX, Yu DL, Yan Z, Wang SW, Yu LS. A common IL-13 Arg130Gln single nucleotide polymorphism among Chinese atopy patients with allergic rhinitis. Hum Genet. 2003;113:387–390. doi: 10.1007/s00439-003-1001-x. [DOI] [PubMed] [Google Scholar]
- 26.Wei CL, Cheung W, Heng CK, Arty N, Chong SS, Lee BW, Puah KL, Yap HK. Interleukin-13 genetic polymorphisms in Singapore Chinese children correlate with long-term outcome of minimal-change disease. Nephrol Dial Transplant. 2005;20:728–734. doi: 10.1093/ndt/gfh648. [DOI] [PubMed] [Google Scholar]
- 27.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 28.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 29.Dutra WO, Correa-Oliveira R, Dunne D, Cecchini LF, Fraga L, Roberts M, Soares-Silveira AM, Webster M, Yssel H, Gollob KJ. Polarized Th2 like cells, in the absence of Th0 cells, are responsible for lymphocyte produced IL-4 in high IgE-producer schistosomiasis patients. BMC Immunol. 2002;3:8. doi: 10.1186/1471-2172-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fitzsimmons CM, Stewart TJ, Hoffmann KF, Grogan JL, Yazdanbakhsh M, Dunne DW. Human IgE response to the Schistosoma haematobium 22.6 kDa antigen. Parasite Immunol. 2004;26:371–376. doi: 10.1111/j.0141-9838.2004.00721.x. [DOI] [PubMed] [Google Scholar]
- 31.McKenzie GJ, Fallon PG, Emson CL, Grencis RK, McKenzie AN. Simultaneous disruption of interleukin (IL)-4 and IL-13 defines individual roles in T helper cell type 2-mediated responses. J Exp Med. 1999;189:1565–1572. doi: 10.1084/jem.189.10.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balzar S, Strand M, Rhodes D, Wenzel SE. IgE expression pattern in lung: relation to systemic IgE and asthma phenotypes. J Allergy Clin Immunol. 2007;119:855–862. doi: 10.1016/j.jaci.2006.12.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holgate ST. The epidemic of allergy and asthma. Nature. 1999;402:B2–4. doi: 10.1038/35037000. [DOI] [PubMed] [Google Scholar]
- 34.Kabesch M, Depner M, Dahmen I, Weiland SK, Vogelberg C, Niggemann B, Lau S, Illig T, Klopp N, Wahn U, Reinhardt D, von Mutius E, Nickel R. Polymorphisms in eosinophil pathway genes, asthma and atopy. Allergy. 2007;62:423–428. doi: 10.1111/j.1398-9995.2006.01300.x. [DOI] [PubMed] [Google Scholar]
- 35.Chiang CH, Tang YC, Lin MW, Chung MY. Association between the IL-4 promoter polymorphisms and asthma or severity of hyperresponsiveness in Taiwanese. Respirology. 2007;12:42–48. doi: 10.1111/j.1440-1843.2006.00960.x. [DOI] [PubMed] [Google Scholar]
- 36.Hosseini-Farahabadi S, Tavakkol-Afshari J, Rafatpanah H, Farid Hosseini R, Khaje Daluei M. Association between the Polymorphisms of IL-4 Gene Promoter (−590C>T), IL-13 Coding Region (R130Q) and IL-16 Gene Promoter (−295T>C) and Allergic Asthma. Iran J Allergy Asthma Immunol. 2007;6:9–14. [PubMed] [Google Scholar]
- 37.Nagarkatti R, Kumar R, Sharma SK, Ghosh B. Association of IL4 gene polymorphisms with asthma in North Indians. Int Arch Allergy Immunol. 2004;134:206–212. doi: 10.1159/000078767. [DOI] [PubMed] [Google Scholar]
- 38.Noguchi E, Shibasaki M, Arinami T, Takeda K, Yokouchi Y, Kawashima T, Yanagi H, Matsui A, Hamaguchi H. Association of asthma and the interleukin-4 promoter gene in Japanese. Clin Exp Allergy. 1998;28:449–453. doi: 10.1046/j.1365-2222.1998.00256.x. [DOI] [PubMed] [Google Scholar]
- 39.Maier LM, Howson JM, Walker N, Spickett GP, Jones RW, Ring SM, McArdle WL, Lowe CE, Bailey R, Payne F, Todd JA, Strachan DP. Association of IL13 with total IgE: evidence against an inverse association of atopy and diabetes. J Allergy Clin Immunol. 2006;117:1306–1313. doi: 10.1016/j.jaci.2005.12.1354. [DOI] [PubMed] [Google Scholar]
- 40.Sadeghnejad A, Karmaus W, Arshad S. Hasan, Ewart S. IL13 gene polymorphism association with cord serum immunoglobulin E. Pediatr Allergy Immunol. 2007 doi: 10.1111/j.1399-3038.2006.00524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Medhat A, Shehata M, Bucci K, Mohamed S, Dief AD, Badary S, Galal H, Nafeh M, King CL. Increased interleukin-4 and interleukin-5 production in response to Schistosoma haematobium adult worm antigens correlates with lack of reinfection after treatment. J Infect Dis. 1998;178:512–519. doi: 10.1086/515630. [DOI] [PubMed] [Google Scholar]
- 42.Rodrigues V, Jr., Piper K, Couissinier-Paris P, Bacelar O, Dessein H, Dessein AJ. Genetic control of schistosome infections by the SM1 locus of the 5q31-q33 region is linked to differentiation of type 2 helper T lymphocytes. Infect Immun. 1999;67:4689–4692. doi: 10.1128/iai.67.9.4689-4692.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bierman WF, Wetsteyn JC, van Gool T. Presentation and diagnosis of imported schistosomiasis: relevance of eosinophilia, microscopy for ova, and serology. J Travel Med. 2005;12:9–13. doi: 10.2310/7060.2005.00003. [DOI] [PubMed] [Google Scholar]
- 44.de Jesus AR, Silva A, Santana LB, Magalhaes A, de Jesus AA, de Almeida RP, Rego MA, Burattini MN, Pearce EJ, Carvalho EM. Clinical and immunologic evaluation of 31 patients with acute schistosomiasis mansoni. J Infect Dis. 2002;185:98–105. doi: 10.1086/324668. [DOI] [PubMed] [Google Scholar]
- 45.Bradley JE, Jackson JA. Immunity, immunoregulation and the ecology of trichuriasis and ascariasis. Parasite Immunol. 2004;26:429–441. doi: 10.1111/j.0141-9838.2004.00730.x. [DOI] [PubMed] [Google Scholar]
- 46.Akagawa H, Tajima A, Sakamoto Y, Krischek B, Yoneyama T, Kasuya H, Onda H, Hori T, Kubota M, Machida T, Saeki N, Hata A, Hashiguchi K, Kimura E, Kim CJ, Yang TK, Lee JY, Kimm K, Inoue I. A haplotype spanning two genes, ELN and LIMK1, decreases their transcripts and confers susceptibility to intracranial aneurysms. Hum Mol Genet. 2006;15:1722–1734. doi: 10.1093/hmg/ddl096. [DOI] [PubMed] [Google Scholar]
- 47.Bertram L, Parkinson M, McQueen MB, Mullin K, Hsiao M, Menon R, Moscarillo TJ, Blacker D, Tanzi RE. Further evidence for LBP-1c/CP2/LSF association in Alzheimer’s disease families. J Med Genet. 2005;42:857–862. doi: 10.1136/jmg.2004.024596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Day DA, Tuite MF. Post-transcriptional gene regulatory mechanisms in eukaryotes: an overview. J Endocrinol. 1998;157:361–371. doi: 10.1677/joe.0.1570361. [DOI] [PubMed] [Google Scholar]