Abstract
Myeloid-derived suppressor cells (MDSCs) promote tumor progression. The mechanisms of MDSC development during tumor growth remain unknown. Tumor exosomes (T-exosomes) have been implicated to play a role in immune regulation, however the role of exosomes in the induction of MDSCs is unclear. Our previous work demonstrated that exosomes isolated from tumor cells are taken up by bone marrow myeloid cells. Here, we extend those findings showing that exosomes isolated from T-exosomes switch the differentiation pathway of these myeloid cells to the MDSC pathway (CD11b+Gr-1+). The resulting cells exhibit MDSC phenotypic and functional characteristics including promotion of tumor growth. Furthermore, we demonstrated that in vivo MDSC mediated promotion of tumor progression is dependent on T-exosome prostaglandin E2 (PGE2) and TGF-β molecules. T-exosomes can induce the accumulation of MDSCs expressing Cox2, IL-6, VEGF, and arginase-1. Antibodies against exosomal PGE2 and TGF-β block the activity of these exosomes on MDSC induction and therefore attenuate MDSC-mediated tumor-promoting ability. Exosomal PGE2 and TGF-β are enriched in T-exosomes when compared with exosomes isolated from the supernatants of cultured tumor cells (C-exosomes). The tumor microenvironment has an effect on the potency of T-exosome mediated induction of MDSCs by regulating the sorting and the amount of exosomal PGE2 and TGF-β available. Together, these findings lend themselves to developing specific targetable therapeutic strategies to reduce or eliminate MDSC-induced immunosuppression and hence enhance host antitumor immunotherapy efficacy.
Keywords: Tumor exosomes, induction of myeloid-derived suppressor cells, PGE2, TGF-β, tumor growth
Introduction
Myeloid-derived suppressor cells (MDSCs) are expanded in large numbers during tumor growth, resulting in accumulation in secondary lymphoid organs, blood, and tumor tissue. These expanded myeloid cells contribute to tumor progression, providing supporting stroma and immune evasion1–4. In mice, removal of the primary tumor results in the reduction of the number of systemic MDSCs, revealing a causal role for MDSCs in tumor growth 5. Purified MDSCs have been shown to inhibit both CD4+ and CD8+ T cell responses in vitro6–8. Furthermore, studies imply that these cells can down-regulate T cell functions in vivo8 and promote tumor metastasis by releasing a number of chemokines9.
Expansion of MDSCs is likely dependent on tumor ability to secrete myeloid-influencing factors (ie, colony-stimulating factor-1, IL-6, vascular endothelial growth factor (VEGF), PGE2, and granulocyte-macrophage colony stimulating factor [GM-CSF])3, 10–12. However, which of these tumor-associated factors are critical for MDSC accumulation and how these tumor factors affect MDSC development have not been fully evaluated.
Many cells, including tumor cells, have the capacity to release exosomes. Recently, increased evidence has suggested that T-exosomes might act as a vehicle for transmitting signals for suppression thus having negative effects on antitumor immune responses. T-exosomes interfere directly with T cell effector functions to induce apoptosis in activated tumor-specific T cells 13–15 and suppress NK cell tumor cytotoxicity through the downmodulation of perforin expression 16. In addition T-exosomes also impair the capacity of bone marrow derived CD11b+ myeloid cells and CD14+ monocytes to differentiate into functional dendritic cells (DCs) 12, 15.
In the present study, we show that T-exosomes can induce the accumulation of MDSCs. Once T-exosomes are taken up, the MDSCs markedly increase production of inflammatory cytokines, including IL-6 and VEGF, and promote tumor growth. Blocking tumor exosomal PGE2 and TGF-β leads to the attenuation of T-exosome mediated induction of accumulation of MDSCs. PGE2 and TGF-β have been shown to suppress immune response by induction of MDSCs 17–20. We further show that tumor microenvironmental factors producing exosomal PGE2 and TGF-β are enriched or increased as the tumor progresses; further highlighting the importance of the tumor microenvironment in regulating T-exosome-mediated immune suppression and tumor growth.
Material and Methods
Mice
BALB/c mice were purchased from the Jackson Laboratory. Seven- to 10-week old female mice were used in this study. All animal studies were conducted within the guidelines established by the University of Alabama at Birmingham Institutional Animal Care and Use Committee.
Cell Culture
The TS/A cell line, 4T-1 cell line, murine mammary adenocarcinomas of spontaneous BALB/c origin, were maintained in vitro at 37°C in a humidified 5% CO2 atmosphere in air in complete medium (DMEM with 5% FBS) as described previously16. FBS used in cell cultures was exosome depleted by differential centrifugation using a method described previously 16.
Exosome preparation and electron microscopic examination
TS/A or 4T-1 tumor cells from 80% confluent cultures were injected subcutaneously into mice. Mice received 1.2 × 105 tumor cells in 50 μl of PBS per site at three sites in mammary fat pads. Tumor tissue was removed from BALB/c mice at day 7, 14, and 21 post injection, weighed and subsequently dissociated enzymatically for 1 h with 1 mg/ml type I collagenase (Sigma-Aldrich) in the presence of 50 units/ml of RNase and DNase (Sigma-Aldrich) ., Single cell suspensions of tumor tissue in PBS were washed by centrifugation at 600×g for 5 min. The cell supernatants were collected and used for exosome purification by differential centrifugation using a previously described method 16. The cell pellets were washed with PBS and total leukocytes were isolated using Percoll gradients as described 16. Total numbers of leukocytes isolated from tumor tissue were determined using a hemocytometer method. Isolated leukocytes (10,000/sample) were analyzed for the presence of CD11b, and Gr-1 molecules by flow cytometry. Percentages of double-positive CD11b+Gr-1+ myeloid cells were determined by FACS. Purity and integrity of sucrose gradient purified exosomes was analyzed using a Hitachi H7000 electron microscope (Electronic Instruments) as previously described 16. Care was taken to minimize potential contamination during T-exosome purification. The precautions taken included: 1) only using tumor tissues that had no necrosis; 2) bacterial cultures of collagenase digested tumor tissue were negative; 3) rerunning the exosomes isolated from the first run on another sucrose gradient; and 4) determining the presence of common exosome markers before using the T- and C-exosomes.
The TS/A and 4T-1 tumor cell derived exosomes (C-exosomes) were isolated from the supernatants of 36 h cell cultures using minor modifications of the method described above . In brief, the TS/A and 4T-1 cells were cultured in vitro at 37°C in a humidified 5% CO2 atmosphere in air in complete medium (DMEM with 5% exosome-depleted FCS). When the cells reached 60% confluence, the cells were washed two times with CD293 media (Invitrogen, sera free media originally developed for culturing 293 cells without FBS) and returned to culture in CD293 media until 90% confluency was achieved. The cells were cultured in fresh CD293 medium for an additional 36 h before the supernatants were harvested for exosome purification. The concentration of exosomes was determined by analyzing protein concentration using the Biorad protein quantitation assay kit (Biorad) with BSA as a standard.
Western blotting
Sucrose purified exosomes for western blots were prepared by lysing exosomes in radioimmunoprecipitation assay (RIPA) buffer containing 1 mM EDTA. Lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to polyvinylidene difluoride membranes. After transfer, blots were analyzed using an Odyssey infrared imaging system (LI-COR, Biosciences, NE, USA). Briefly, membranes were rinsed with PBS for several minutes and blocked with Odyssey blocking buffer for 1 h at 22°C. The membrane was then incubated for 1 h at 22°C in a dilution of a primary antibodies in blocking solution containing 0.1% Tween-20, after which the membrane was washed in PBS + 0.1% Tween-20. Subsequently the membrane was incubated in the dark with fluorescent secondary antibody at a dilution of 1:10,000 in blocking solution containing 0.1% Tween-20. Secondary antibodies were IRDye 800 anti-mouse (Molecular Probes, Rockland Immunochemicals, PA), and Alexa Fluor 700 anti-rabbit (Molecular Probes, OR) probes. Images were acquired with an Odyssey infrared imaging system and analyzed using software specified by Odyssey systems.
Protein analysis by MS/MS
C-exosomes and T-exosomes were lysed in protein lysis buffer and 100 μg of proteins were electrophoresed on 10% SDS-polyacrylamide gels. Coomassie-stained SDS-polyacrylamide gels were cut into 10 strips to correlate with the gel lanes and trypsinized. The digested peptides were loaded on a 100 nm × 10 cm capillary column packed in-house with C18 Monitor 100 A-spherical silica beads and eluted by a 1 h gradient of 10–100% acetonitrile, 0.1% TFA. Mass spectrometric analysis was performed and analyzed using an LTQ XL spectrometer (Thermo Finnigan) in the UAB Proteomic Core Facility.
UniProt protein IDs were enriched for GO terms using the Protein Information and Property Explorer (http://pipe.systemsbiology.net/pipe/#summary). Sorting of data on proteins based on cellular location and biological process were done using a spreadsheet.
In vivo induction of myeloid-derived suppressor cells by T-exosomes
Analysis of the production of T-exosomes revealed that ~100 ± 4.2 μg of T-exosomes are released from a tumor (1.1 ± 0.27 g of weight per tumor at 21 day after the injection). Based on the assumption that 100 μg of T-exosomes are released to the peripheral tissues continuously, we used 100 μg of exosomes for injection experiments. T-exosomes (100 μg) isolated from TS/A or 4T-1 tumor removed at day 21 post tumor cell injection were injected into BALB/c mice via the tail vein twice weekly for three weeks. Five mice received PBS and served as controls. One day after the final injection, mice were sacrificed and each organ was removed for isolation of CD11b+Gr-1+ cells using a protocol described below. The percentage of CD11b+Gr-1+ myeloid cells or others analyzed in the total number of leukocytes isolated from each organ were determined by FACS12.
To determine whether T-exosomes play a role in the accumulation of CD11b+Gr-1+ cells in the tumor, tumor cells (5×105) were co-injected with T-exosomes (50 μg). On day 5 and 10 post injection the tumors were removed. Tumor tissue was dissociated enzymatically for 1 h. with 1 mg/ml of type I collagenase. The resulting single cell suspensions of tumor tissue in PBS were washed by centrifugation at 1,000xg for 5 min. Cells (50,000/sample) were analyzed for the presence of CD11b and Gr-1+ by flow cytometry. The percentage of double positive myeloid cells, i.e., CD11b+Gr-1+, was determined by FACS analysis 12.
Bone marrow cell culture and spleen myeloid cell isolation
Bone marrow was isolated and cultured after RBC lysis as described previously12. Spleen cells were positively selected for CD11b using MACS beads (Miltenyi Biotec) and then stained with anti-Gr-1 Ab. CD11b+Gr-1+ populations were isolated by cell sorting. Purified spleen myeloid cells were used for co-injection with tumor cells. RBC-depleted bone marrow cells were cultured in RPMI 1640 medium containing 10% FBS, with the addition of glutamine, 2-ME, sodium pyruvate, nonessential amino acid, antibiotics (Invitrogen), and GM-CSF (20 ng/ml).
Labeling of exosomes and analysis of their target cells in vivo
C-exosomes and T-exosomes were labeled with the PKH67 green and PKH26 red fluorescent dyes, respectively, using a commercially available kit (Sigma-Aldrich) according to the manufacturer's instructions. The efficiency of labeling of the exosomes (>92%) with PKH dyes was determined by FACS analysis as described previously 12. Fifty μg mixtures of labeled C- and T- exosomes at a ratio of 1:1 were injected i.v. into BALB/c mice (5 mice). Twenty-four h post injection, the spleen, liver, and lung were resected and retained and the BM was collected. Single-cell suspensions of each tissue were prepared in RPMI 1640 medium and subjected to FACS analysis.
Flow cytometry
For cell surface marker staining, isolated cells were blocked at 4°C for 5 min with 10 μg/ml Mouse Fc Block (BD), and then reacted for 30 min at 4°C with various fluochrome-labeled Abs including appropriate isotype controls. After washing twice, cells were analyzed using a FACSCalibur (BD Biosciences).
Antibodies for flow cytometry and western blotting
The following antibodies were used for flow cytometry: fluorescein isothiocyanate (FITC) anti-mouse CD19, PE anti-mouse CD11c, PE anti-mouse Ly-6G and Ly-6C (Gr-1), and APC anti-mouse Mac-1 (CD11b) and PE anti-mouse F4/80 from e-Bioscience (San Diego, CA). Anti-mouse CD3, CD19, NK cell surface expression of DX5 and isotype-matched antibodies (all from BD Biosciences) were used as controls for nonspecific binding.
The following antibodies were used for western blotting: rabbit polyclonal anti-CD81, calnexin, and mouse monoclonal antibodies anti-CD9, anti-TSG101, and anti-CD63 all obtained from Santa Cruz Biotechnology (CA). Mouse monoclonal Ab anti-TGF-β was purchased from R&D Systems (Minneapolis, MN). Mouse monoclonal anti-PGE2 antibody was purchased from Cayman Chemical (Ann Arbor, MI). One μg of anti-TGF-β or 2 μg of anti-PGE2 antibody was used for blocking the effects of 100 μg TS/A T- exosomes on induction of CD11b+Gr-1+ bone marrow derived cells.
In vivo tumor growth assays
Tumor cells for injection were prepared from cultures grown to near confluency with 95% viability. Cells were enumerated, adjusted to the proper number, and mixed with sorted CD11b+Gr-1+ cells at the ratio of 3:1 (tumor cell:CD11b+Gr-1+). Tumor cells alone or mixed with CD11b+Gr-1+ cells were injected subcutaneously into the mammary fat pads of mice in 0.2ml injection volumes. Additional groups of mice were injected with tumor cells and tumor sized was measured once a week using calipers. Two independent measurements (length and width) were taken for each tumor weekly. Tumor size was calculated according to the formula V = L × W (L: length, mm. W: width, mm). Animals were sacrificed when the maximal allowable tumor size was reached or after observation for 50 days.
Reverse transcription-PCR
Total RNA from the CD11b+Gr-1+ cells prepusled for 12 h with T-exosomes (1 μg/ml) was extracted using TRIzol reagent (Invitrogen) and reverse transcription-PCR (RT-PCR) analysis was done as previously described 21. Specific primers used in the RT-PCR were mouse Cox2, Arginase-1, IL-10 and GAPDH. 21. Three replicates from each cDNA were analyzed. The primers for mouse Cox2 were: forward primer 5'--TGAGTACCGCAAACGCTT CT -3' and reverse primer 5'-CTCCCCAAAGATAGCATCTGG -3'. The primers for mouse arginase-1 were: forward primer 5'- CAGAAGAATGGAAGAGTCAG-3' and reverse primer 5'-CAGATATGCAGGGAGTCACC-3'. The primers for mouse IL-10 were: forward primer 5'- CCAGTTTTACCTGGTAGAAGTGATG-3'. The primers for mouse GAPDH were: forward primer 5'-AGGTCATCCCAGAGCTGAACG-3' and reverse primer 5'- ACC CTGTTGCTGTAGCCGTAT-3'.
TaqMan RT-PCR
An ABI PRISM 7700 sequence detection system (Applied Biosystems) was used for amplification and detection of the gene transcripts of interest. In each TaqMan run, serial 5-fold dilutions of a single-stranded cDNA derived from a commercially available mouse positive control RNA (Applied Biosystems) were amplified to create a standard curve, and values of unknown samples were estimated relative to this standard curve. Standard curves showed a linear relationship between the copy number (defined as 1 ng of 1000-bp DNA = 9.1 × 1011 molecules) of the original internal standard and the number of PCR cycles that were required to exceed a preset threshold, according to the method described previously 21. PCRs for each sample were run in duplicate in three 5-fold serial dilutions. From these standard curves, the relative amount of cDNA for the 18S ribosomal RNA, GAPDH, and each gene was determined and expressed as a ratio of the amount of this material in cells treated or untreated with TS/A exosomes after samples were standardized to the relative expression of the 18S ribosomal RNA and GAPDH.
ELISA for VEGF, IL-6, TNF-α, and PGE2
Mouse VEGF produced in cultures was determined using a VEGF ELISA (R&D Systems, Minneapolis, MN). Mouse TNF-α and IL-6 produced in culture supernatants was also quantified using an ELISA (e-Bioscience). PGE2 levels were determined using the prostaglandin E2 monoclonal EIA kit (Cayman Chemical). In each case the assay was run according to the manufacturer's instructions.
Neutralization of exosome PGE2 and TGF-β
One hundred μg of exosomes (based on protein content) were incubated for 1 h at 22°C with mouse anti-TGF-β antibody (1μg per ml, R&D System), mouse anti-PGE2 antibody (Cayman Chemical), or mouse IgG at 2μg per ml final concentration. (This concentration was chosen because more than 1 μg/ml of anti-TGF-β antibody or 2 μg/ml of anti-PGE2 antibody did not further reverse T-exosome mediated induction of CD11b+Gr-1+ cells in an in vitro assay 12). The exosomes were then washed once at 100,000 × g for 1 h, and resuspended in 200 ul of PBS for use in induction assays on bone marrow derived CD11b+Gr-1+ cells.
Data analysis
All data represents the mean ± SEM, and the error bars in figures also represent SEM. Unpaired two-tailed t tests were used to compare significant differences between two groups. One-way ANOVA followed by Bonferroni tests were used to analyze data for more than two groups.
Results
Myeloid-derived suppressor cells induced by murine breast carcinoma T-exosomes promote tumor growth
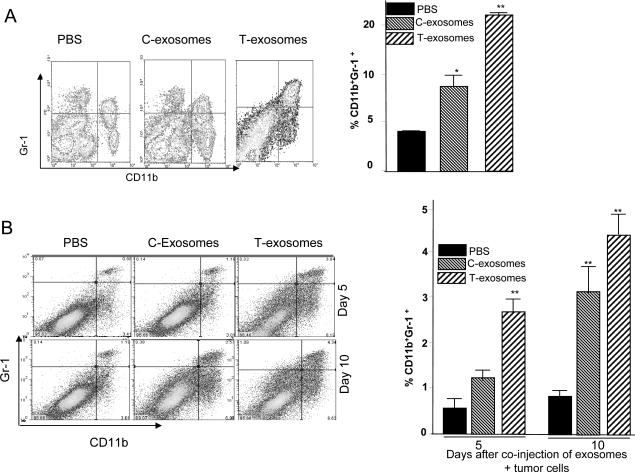
Our previous data suggest that T-exosomes are taken up by bone marrow precursor cells (Gr-1+CD11b+) in vivo 12. Experiments were conducted in a mouse model using intravenously injected exosomes isolated from TS/A tumor removed at day 21 post tumor cell injection to examine the effects of T-exosomes or C-exosomes on the induction of accumulation of Gr-1+CD11b+ populations. After twice weekly injections over a 3 week period, FACS analysis of the splenocytes of mice (Figure 1A) demonstrated that the apparent splenomegaly (data not shown) was associated with marked accumulation of cells expressing Gr-1 and CD11b markers, but did not reveal an increase in cells expressing CD3, CD19, DX5 or CD11c (data not shown). A less dramatic increase in the percentage of CD11b+Gr-1+ cells occurred when mice were treated with C-exosomes (Figure 1A, right panel). This result suggested that tumor derived factors enhance T-exosome mediated induction of CD11b+Gr-1+ cells. We also looked for CD11b+Gr-1+cells in other tissues and secondary lymphoid organs. No significant increases in CD11b+Gr-1+cells were observed in the mesenteric lymph nodes or bone marrow (data not shown). However, in lung tissue a marked increase in the percent of CD11b+Gr-1+ cells was noted 21 d post injection where C-exosome and T-exosome injected mice had 8.2% and 14.2% CD11b+Gr-1+ cells, respectively, and PBS injected mice had 3.2% CD11b+Gr-1+ cells. Similar results were observed when exosomes isolated from 4T-1 tumor bearing mice were used for the injection. The limited induction in CD11b+Gr-1+cells in the spleen of mice treated with C-exosomes is most likely not due to preferential up take of T-exosomes because CD11b+Gr-1+cells took up the co-injected PKH67 labeled C-exosomes and PKH26 labeled T-exosomes with equal efficacy as determined by FACS analysis (data not shown). The data published by other groups indicate that the majority of MDSCs accumulate in both spleen and tumor, and we further determined whether T-exosomes played a role in the accumulation of CD11b+Gr-1+ cells in the tumor. Co-injection of tumor and T-exosomes resulted in an increase of CD11b+Gr-1+cell accumulation in the tumor when evaluated over a 10 day period (Figure 1B). Collectively, these results suggest that T-exosomes play a role in MDSCs accumulation in the spleen and tumor.
Figure 1. CD11b+Gr-1+ cells induced by T-exosomes promote tumor growth and progression.
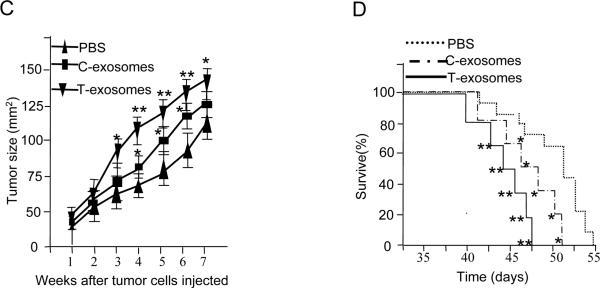
7-week old female BALB/c mice (n = 10) were injected intravenously twice weekly for three weeks with exosomes isolated from TS/A tumor removed at day 21 post tumor cell injection (T-exosomes) or exosomes isolated from the supernatants of 36 h cultured TS/A tumor cells (C-exosomes) (100 μg/mouse) . One day after the final injection, the expression of CD11b+Gr-1+ markers on spleen cells was determined using FACS (A, left panel) and the percent of spleen CD11b+Gr-1+ cells was calculated as described in the Material and Methods. Data represent the mean ± SEM of five replicates (A, right panel), * p < 0.05, **p < 0.01. 7-week old female BALB/c mice (n = 5/group) were subcutaneously co-injected with TS/A cells (5×105) and T-exosomes (50 μg). At day 5 and 10 post injection tumors were removed. Collagnase digested single cell suspensions of tumor tissue were stained with anti-CD11b and anti-Gr-1 antibodies, and the percent CD11b+Gr-1+ cells was determined using FACS (B left panel). Data represent the mean ± SEM of five replicates (B, right panel), **p < 0.01. CD11b+Gr-1+ cells were FACS sorted from spleen of naïve BALB/c mice treated with PBS or from naïve BALB/c mice treated with T- and C- exosomes as described in figure legend 1A. The sorted cells were then mixed with TS/A tumor cells (CD11b+Gr-1+ cells : tumor cells = 1:3). Seven-week old female BALB/c mice were injected subcutaneously in the mammary fat pads with TS/A tumor cells alone (1.2×105) or premixed with spleen CD11b+Gr-1+ cells. The growth of the implanted tumor was measured throughout a 50-day period (C). Each point represents the mean tumor size ± SD of 5 mice from each group. The mortality of mice was calculated through day 55 (D). *p < 0.05; **p < 0.01.
To examine whether CD11b+Gr-1+ cells induced by T-exosomes affect tumor growth directly, we sorted CD11b+Gr-1+ cells from the spleen of naïve mice or mice having been treated with C-exosomes or T-exosomes and coinjected BALB/c mice subcutaneously with TS/A cells (1.2 × 105) and the isolated CD11b+Gr-1+ cells (0.4 × 105). Tumor size was measured using calipers. We found that tumor growth occurred significantly faster when TS/A cells were coinjected with CD11b+Gr-1+ cells from mice treated with T-exosomes than tumor cells coinjected with naïve mouse CD11b+Gr-1+ cells (p < 0.01) (Figure 1C). Results of tumor growth experiments also indicated that spleen CD11b+Gr-1+ cells isolated from mice pretreated with C-exosomes promote tumor growth but to a slower degree than CD11b+Gr-1+ cells from T-exosome treated mice (p < 0.05). However, this difference in tumor growth was reduced approximately 42 days after tumor inoculation (Figure 1C). This is likely due to the significant induction of endogenous CD11b+Gr-1+ cells in host mice bearing large tumors and having higher amounts of exosomes released. The co-injection of T-exosome or C-exosomes –pretreated CD11b+Gr-1+ cells also led to early deaths in increased numbers of mice (Figure 1D). These data support the idea that T-exosomes induce the accumulation of CD11b+Gr-1+ cells which directly promotes tumor growth.
The tumor microenvironment has effects on T-exosome-mediated induction of a number of tumor growth factors
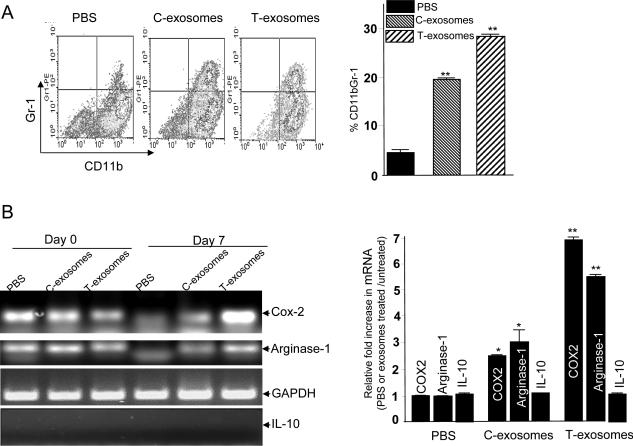
To examine whether the tumor microenvironment has an effect on tumor derived exosome mediated induction of CD11b+Gr-1+ population, bone marrow derived cells were cultured ex vivo in 24-well plates in the presence of C-exosomes or T-exosomes isolated from TS/A tumor removed from day 21 post tumor cell injected mice. In the absence of T-exosomes the majority of these cells differentiate into myeloid dendritic cells in the presence of GM-CSF 12. In contrast, culturing bone marrow derived cells for 7 d with C-exosomes or T-exosomes led to ~19% and ~29%, respectively, of these cells retaining CD11b+Gr-1+ cells phenotypically compared with ~3% of PBS control treated cells (Figure 2A). To determine the potential effects of T-exosomes on induction of genes/cytokines associated with promoting tumor growth, cytokine profiles in the 7-d cultured supernatants were determined using an ELISA and the expression of genes associated with tumor growth was quantified by RT-PCR and TaqMan PCR analysis of RNA isolated from the 7-d cultured CD11b+Gr-1+ cells. Treatment with T-exosomes most effectively up-regulated Cox2 and arginase-1 expression in 7 day cultured bone marrow cells than did C-exosome treatment (Figure 2B, top panel). Similar results were obtained from additional T-exosomes (n=5) isolated from different tumor-bearing mice when the expression of Cox2 and arginase-1 mRNA was quantified by TaqMan PCR analysis. Cox2 and arginase-1 mRNA was most up-regulated after bone marrow-derived CD11b+Gr-1+ cells were treated with T-exosomes (Fig. 2B, bottom panel). This result was not due to the amount of mRNA used for the TaqMan PCR analysis as there were no differences in the amount of IL-10 gene amplified (Fig. 2B, bottom panel).
Figure 2. The tumor microenvironment has a significant effect on T-exosome mediated induction of proinflammation cytokines, tumor growth factors of MDSCs and tumor progression.
Bone marrow cells were cultured in RPMI 1640 supplemented with 10% FBS and 20 ng/ml recombinant mouse GM-CSF. C- and T- exosomes (1 μg/ml) were added to the cultured cells after the addition of the GM-CSF to the cultures. After 7 days in culture, the cells were analyzed for the expression of CD11b+Gr-1+ cells using FACS. One representative of three independent experiments is shown (A, left panel). All FACS analysis results are presented as the mean ± SEM obtained for three samples in three independent experiments (A, right panel). A portion of 7-day cultured cells treated as described above was sorted for CD11b+Gr-1+ by FACS and used for RT-PCR analysis of mouse Cox2, arginase-1, and IL-10 expression. GAPDH mRNA levels are included as a loading control. The results are representative of three independent experiments of RT-PCR results (B, left panel). The same RNA used for RT-PCR was also used for real-time PCR to quantitate Cox2, Arginase-1, and IL-10 (B, roght panel) using the protocol as described in the Materials and Methods. Data were first normalized from nontreated cells and then presented as ratios of PBS, C-, and T- exosome-treated/untreated CD11b+Gr-1+ cells (n = 5) in triplicate, mean ± SEM; *, p < 0.005; **, p < 0.01.
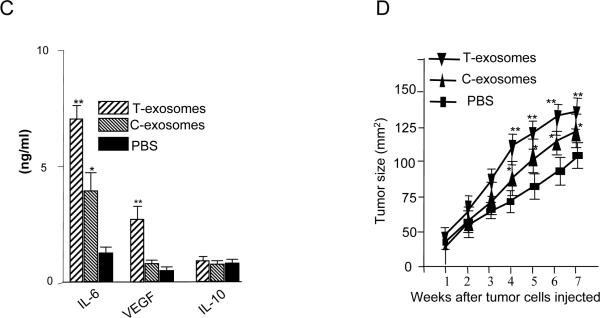
The supernatants of 7-day cultures were collected and induction of IL-6, VEGF, and IL-10 was determined using an ELISA as described in the Material and Methods section. The results represent the mean ± SEM of triplicate cultures (C). After 7-days in culture, the FACS sorted CD11b+Gr-1+ cells were mixed with TS/A tumor cells at the ratio of 1:3 and the cell suspension was injected into the mammary fat pad of 7-week-old female BALB/c mice. The size of the implanted tumors was measured weekly. Each point represents the mean size ±SEM of the 5 mice from each group (D), *, p < 0.05; **, p < 0.01.
The quantity of VEGF and IL-6 in the culture supernatants was significantly higher when T-exosomes were added (P < 0.01, Figure 2C). The response appeared selective as the induction of IL-10 was not observed (Figure 2C). Interestingly, when C-exosomes were added to bone marrow cells, there was no induction of VEGF (Figure 2C). Consistent with the above findings, the coinjection of T-exosome pretreated CD11b+Gr-1+ cells with TS/A tumor cells resulted in the most rapid tumor growth among the three groups of mice (Figure 2D). Taken together, these results suggest that in taking up T-exosomes, bone marrow derived immature myeloid cells gain an ability to induce inflammatory cytokine IL-6 and tumor growth factors that promote tumor growth. Although both C- and T- exosomes can induce myeloid-derived suppressor cells, T-exosomes are far more potent than C-exosomes at inducing MDSCs.
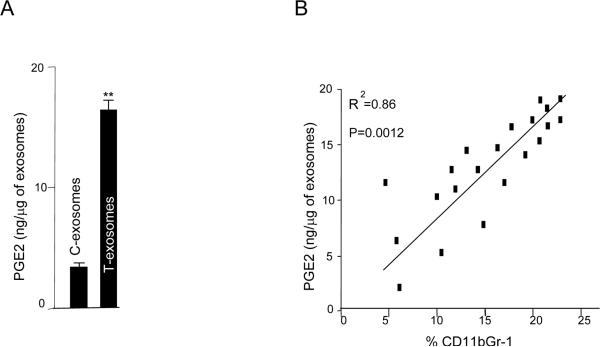
T-exosomes are enriched with PGE2 as a tumor progresses resulting in induction of MDSCs
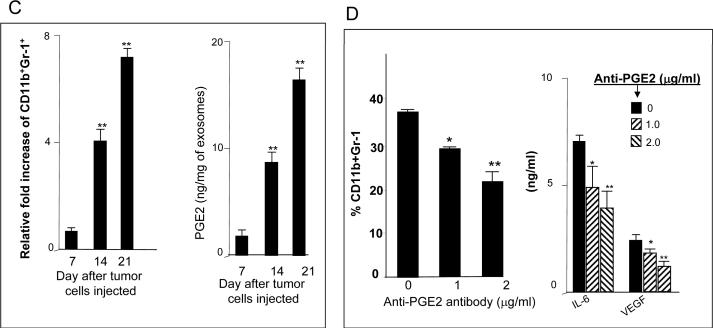
Given the results showing the majority of products induced by T-exosomes are found to be proinflammatory associated, in particular Cox2, we speculated that T-exosomes may contain sufficient quantities of PGE2 to induce a number of proinflammatory cytokines including Cox2 that facilitates tumor growth. The ELISA results showed that PGE2 is highly enriched in the T-exosomes when compared with C-exosomes (Figure 3A). In addition, we also found that higher percentages of CD11b+Gr-1+ cells infiltrating a tumor are correlated with increased amounts of PGE2 (Figure 3B). To determine if T-exosomes with increased quantities of PGE2 are capable of inducing more CD11b+Gr-1+ cells, the induction of bone marrow derived CD11b+Gr-1+ cells was determined in the presence of T-exosomes purified at different times after tumor cell injection. FACS analysis of 7-day bone marrow T-exosome cultures indicated that higher percentages of CD11b+Gr-1+ cells were induced with T-exosomes isolated from later stages of tumor growth (Figure 3C, left panel) where the T-exosomes contained higher quantities of PGE2 (Figure 3C, right panel). To further confirm the role of exosomal PGE2 in the induction of CD11b+Gr-1+ myeloid cells and proinflammatory cytokine expression, anti-PGE2 antibody was used to block exosomal PGE2 effects. Pre-incubation of T-exosomes with anti-PGE2 antibody led to a partial inhibition of tumor exosomal mediated induction of MDSCs (Figure 3D, left panel) with a concomitant reduction of proinflammatory cytokines (Figure 3D, right panel) when 7-day cultures of bone marrow derived cells were analyzed.
Figure 3. The tumor microenvironment regulates the enrichment of tumor exosomal PGE2 which in turn promotes the induction of CD11b+Gr-1+ cells.
One μg of C-exosomes and T-exosomes was diluted in 50 μl of RPMI1640 and used to quantify PGE2 amounts using an ELISA. All data represent the mean ± SEM obtained for four samples in three independent experiments (A). The mean PGE2 levels of T-exosomes prepared from TS/A tumor removed from BALB/c mice at day 7, 14 and 21 post injection were calculated based on PGE2 ELISA data. PGE2 ELISA data represent the mean ± SEM obtained for four samples in three independent experiments and were plotted against the mean percents of CD11b+Gr-1+ cells isolated from corresponding tumor samples (%CD11b+Gr-1+/10,000 leukocytes cells isolated from tumor tissue). Each point represents the mean value of each tumor. The correlation between the two variables was calculated using a linear regression analysis_(B). Total bone marrow cells were isolated from the BM of 2-mo-old female BALB/c mice and stimulated with GM-CSF in the presence of TS/A T-exosomes isolated from tumor removed at different days after injecting tumor cells or BSA as a control (1 μg/ml for 7 days). The expression of CD11b+Gr-1+ markers on cell surfaces was determined using FACS. Data are presented as the n-fold increase of CD11b+Gr-1+ cells caused by addition of T-exosomes compared with BSA treatment (C, left panel). T-exosomes were prepared from TS/A tumor removed from BALB/c mice at day 7, 14 and 21 post injection of tumor cells. One μg of supernatant from the T-exosomes was diluted in 50 μl of RPMI 1640 and used to quantify PGE2 amounts using an ELISA. All data represent the mean ± SEM obtained for four samples in three independent experiments (C, right panel). Bone marrow derived cells were cultured as described in Figure 3C with the addition of T-exosomes-d21 preincubated with anti-PGE2 antibody. At day 7 in culture, the percent of double positive myeloid cells for Gr-1 and CD11b were determined using FACS (D, left panel). The supernatants of the 7-day cultures were collected and the amounts of IL-6, and VEGF (D, right panel) were determined using an ELISA. Data are given as the mean ± SEM obtained for five samples in two independent experiments, *, p < 0.05; **, p < 0.01.
Neutralization of tumor exosomal PGE2 and TGF-β attenuates the induction of MDSCs and reverses T-exosome mediated promotion of tumor growth
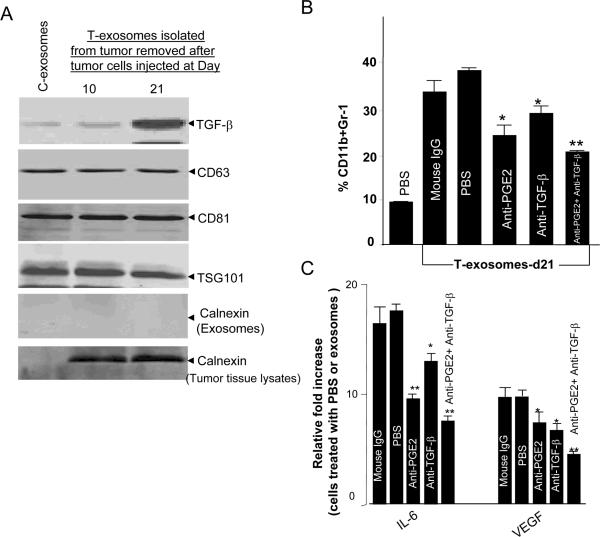
Since neutralization of exosomal PGE2 only partially reverses the induction of CD11b+Gr-1+ myeloid cells, we searched for additional molecules that might promote the induction of MDSCs. Another group has reported that TGF-β, which causes immune suppression, is packed into exosomes13. Using western blots our data also shows that TGF-β is packed into both C- and T- exosomes and is enriched as a tumor progresses (Figure 4A). The results of analyzing other exosomal protein markers, including CD63, CD81, and Tsg101, (Figure 4A) suggests that the tumor microenvironment regulates enrichment of TGF-β in a selective fashion as the increase of other proteins was not observed. Calnexin was also detected in the lysates of tumor tissue, but not in the purified exosome preparations (Figure 4A), indicating that our exosomal preparations were free of contamination with non-exosomal membrane proteins. To further determine if the tumor microenvironment only regulates sorting of TGF-β into exosomes, we did a comparison of protein profiles of T-exosomes isolated from TS/A tumor with C-exosomes. Specific proteins were at undetectable levels in C-exosomes when compared with protein amounts in T-exosomes (Table 1). This analysis indicated a protein composition of T-exosomes typical of exosomes derived from other cell types 22–27. The proteins are not detected in the C-exosomes (Table 1) but in both cases the exosomes contained proteins known to be involved in cell metabolism, signal transduction, cell endocytosis, protein transport, and several class E vacuolar protein-sorting (VPS) proteins (associated with MVB biogenesis) (Table 2), with 36 of the proteins identified in this study having been described previously in exosomes derived from other cell types (Table 3) 22–27.
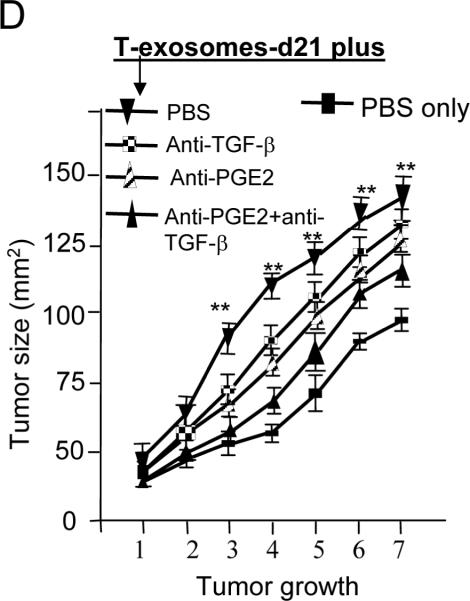
Figure 4. T-exosomal TGF-β has an additive effect with PGE2 on the induction of tumor growth factors of MDSCs and tumor growth.
(A) Fifty μg T-exosomes isolated from TS/A tumor removed at day 10 and 21 after the initial tumor cells were injected or 50 μg of C-exosomes were lysed using a protein lysing buffer, electrophoresed, and immunoblotted with antibodies as indicated on Figure 4A. Results are representative of 2 separate experiments. (B) 2 × 106 bone marrow cells were pulsed with T-exosomes-d21 pretreated with combinations of different antibodies or PBS as a control and then cultured in the presence of GM-CSF (20 ng/ml). After 7-days in culture, the expression of CD11bGr-1 on cell surfaces was determined using FACS (B), and the amount of IL-6, and VEGF quantified using an ELISA (C). Data are given as the means ± SEM obtained for five samples in two independent experiments, *, p < 0.05; **, p < 0.01. The effect of exosomal PGE2 and TGF-β on TS/A tumor cell growth was determined by co-injection of bone marrow derived CD11b cells pretreated, as indicated in Figure 4D, with 1.2 × 105 of TS/A tumor cells (CD11b : TS/A tumor cells = 1:3). The growth of the implanted tumor was measured weekly through a 50-day period. Each point represents the mean of average areas of tumors of 5 mice.
Table 1.
Proteins detected in T-exosomes but not in C-exosomes using LTQ-LC/MS
| Ion transport | |
|
| |
| UniRef100_Q3THL7 | Voltage-dependent anion channel 1 |
|
| |
| UniRef100_Q3TTN3 | Voltage-dependent anion channel 3 |
|
| |
| UniRef100_Q99JR1 | Sideroflexin 1 |
|
| |
| Apoptosis | |
|
| |
| UniRef100_Q3UDJ2 | Sphingosine phosphate lyase 1 |
|
| |
| UniRef100_P27773 | Protein disulfide isomerase associated 3 |
|
| |
| UniRef100_O35295 | Purine rich element binding protein B |
|
| |
| UniRef100_Q3TIN2 | Glutaminyl-tRNA synthetase |
|
| |
| Protein transport | |
|
| |
| UniRef100_P24668 | Mannose-6-phosphate receptor, cation dependent |
|
| |
| UniRef100_Q8BJL4 | Lectin, mannose-binding 2 |
|
| |
| Metabolic process | |
|
| |
| UniRef100_Q3UDJ2 | Sphingosine phosphate lyase 1 |
|
| |
| UniRef100_Q8VBT6 | Apolipoprotein B48 receptor |
|
| |
| UniRef100_A8XUV0 | t-complex protein 1 |
|
| |
| UniRef100_P11352 | Glutathione peroxidase 1 |
|
| |
| Immune | |
|
| |
| UniRef100_A2ALM8 | B-cell receptor-associated protein 31 |
|
| |
| UniRef100_P28063 | Proteasome (prosome, macropain) subunit, beta type 8 (large multifunctional peptidase 7) |
|
| |
| Vesicle-mediated transport | |
|
| |
| UniRef100_A2ALM8 | B-cell receptor-associated protein 31 |
|
| |
| Differentiation | |
|
| |
| UniRef100_P16045 | Lectin, galactose binding, soluble 1 |
|
| |
| UniRef100_Q4ADG5 | PRP19/PSO4 pre-mRNA processing factor 19 homolog (S. cerevisiae) |
|
| |
| UniRef100_O70551 | Serine/arginine-rich protein specific kinase 1 |
|
| |
| UniRef100_P11352 | Glutathione peroxidase 1 |
|
| |
| UniRef100_O35295 | Purine rich element binding protein B |
|
| |
| UniRef100_Q99JR1 | Sideroflexin 1 |
Table 2.
Identities of proteins found in exosomes
| Ion transport | |
| UniRef100_Q3TIC2 | Ras homolog gene family, member B |
|
| |
| UniRef100_Q8K314 | ATPase, Ca++ transporting, plasma membrane 1 |
|
| |
| UniRef100_P14824 | Annexin A6 |
|
| |
| UniRef100_Q3UMR9 | Solute carrier family 5 (inositol transporters), member 3 |
|
| |
| UniRef100_P09528 | Ferritin heavy chain 1 |
|
| |
| UniRef100_P49817 | Caveolin, caveolae protein 1 |
|
| |
| UniRef100_Q8VDN2 | ATPase, Na+/K+ transporting, alpha 1 polypeptide |
|
| |
| UniRef100_Q3TIP8 | Chloride intracellular channel 1 |
|
| |
| UniRef100_P97370 | ATPase, Na+/K+ transporting, beta 3 polypeptide |
|
| |
| UniRef100_Q8CFE6 | Solute carrier family 38, member 2 |
|
| |
| UniRef100_A2ALM4 | Solute carrier family 6 (neurotransmitter transporter, creatine), member 8 |
|
| |
| UniRef100_P53986 | Solute carrier family 16 (monocarboxylic acid transporters), member 1 |
|
| |
| UniRef100_P46467 | Vacuolar protein sorting 4b (yeast |
| Apoptosis | |
| UniRef100_Q8BH78 | Reticulon 4 |
|
| |
| UniRef100_Q3TIC2 | Ras homolog gene family, member B |
|
| |
| UniRef100_P12815 | Programmed cell death 6 |
|
| |
| UniRef100_P49769 | Presenilin 1 |
|
| |
| UniRef100_P26231 | Catenin (cadherin associated protein), alpha 1 |
|
| |
| UniRef100_P62960 | Y box protein 1 |
|
| |
| UniRef100_P68510 | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide |
|
| |
| UniRef100_Q3U2W7 | v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog |
|
| |
| UniRef100_A1E2B8 | Heat shock protein 1A |
|
| |
| UniRef100_P10126 | Eukaryotic translation elongation factor 1 alpha 1 |
|
| |
| UniRef100_Q3TED2 | Programmed cell death 6 interacting protein |
| Endocytosis | |
| UniRef100_Q62351 | Transferrin receptor |
|
| |
| UniRef100_Q8BH64 | EH-domain containing 2 |
|
| |
| UniRef100_Q3TLP8 | RAS-related C3 botulinum substrate 1 |
|
| |
| UniRef100_Q91ZX7 | Low density lipoprotein receptor-related protein 1 |
|
| |
| UniRef100_P49817 | Caveolin, caveolae protein 1 |
|
| |
| UniRef100_P35278 | RAB5C, member RAS oncogene family |
| Protein transport | |
| UniRef100_Q3TIC2 | Ras homolog gene family, member B |
|
| |
| UniRef100_P49769 | Presenilin 1 |
|
| |
| UniRef100_Q14AA6 | RIKEN cDNA 1700009N14 gene |
|
| |
| UniRef100_P68510 | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide |
|
| |
| UniRef100_Q5SXR6 | Clathrin, heavy polypeptide (Hc) |
|
| |
| UniRef100_Q3UK08 | Tumor susceptibility gene 101 |
|
| |
| UniRef100_Q3UUX9 | guanosine diphosphate (GDP) dissociation inhibitor 2 |
|
| |
| UniRef100_Q3TED2 | programmed cell death 6 interacting protein |
|
| |
| UniRef100_P35278 | RAB5C, member RAS oncogene family |
|
| |
| UniRef100_O09044 | synaptosomal-associated protein 23 |
|
| |
| UniRef100_P61028 | RAB8B, member RAS oncogene family |
|
| |
| UniRef100_P53994 | RAB2A, member RAS oncogene family |
|
| |
| UniRef100_P46467 | vacuolar protein sorting 4b (yeast) |
|
| |
| UniRef100_Q8R0J7 | vacuolar protein sorting 37B (yeast) |
|
| |
| UniRef100_Q8R105 | vacuolar protein sorting 37C (yeast) |
|
| |
| UniRef100_Q9D1C8 | vacuolar protein sorting 28 (yeast) |
|
| |
| UniRef100_Q8BHH2 | RAB9B, member RAS oncogene family |
|
| |
| UniRef100_P55258 | RAB8A, member RAS oncogene family |
|
| |
| UniRef100_P46638 | RAB11B, member RAS oncogene family |
|
| |
| UniRef100_Q50HW9 | RAB14, member RAS oncogene family |
| Metabolic process | |
| UniRef100_Q3TED3 | ATP citrate lyase |
|
| |
| UniRef100_P49769 | presenilin 1 |
|
| |
| UniRef100_P06151 | lactate dehydrogenase A |
|
| |
| UniRef100_P14152 | malate dehydrogenase 1, NAD (soluble) |
|
| |
| UniRef100_P10852 | solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 |
|
| |
| UniRef100_Q14AA6 | RIKEN cDNA 1700009N14 gene |
|
| |
| UniRef100_P19096 | fatty acid synthase |
|
| |
| UniRef100_P17751 | triosephosphate isomerase 1 |
|
| |
| UniRef100_Q05816 | fatty acid binding protein 5, epidermal |
|
| |
| UniRef100_A6ZI47 | aldolase 1, A isoform, retrogene 2 |
|
| |
| UniRef100_Q9DBJ1 | phosphoglycerate mutase 1 |
|
| |
| UniRef100_P15532 | similar to Nucleoside diphosphate kinase A (NDK A) |
|
| |
| UniRef100_Q01768 | non-metastatic cells 2, protein (NM23B) expressed in |
|
| |
| UniRef100_Q3UMR9 | solute carrier family 5 (inositol transporters), member 3 |
|
| |
| UniRef100_P68510 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide |
|
| |
| UniRef100_P59108 | copine II |
|
| |
| UniRef100_Q61753 | 3-phosphoglycerate dehydrogenase |
|
| |
| UniRef100_P49817 | caveolin, caveolae protein 1 |
|
| |
| UniRef100_P58242 | sphingomyelin phosphodiesterase, acid-like 3B |
|
| |
| UniRef100_Q3TXD3 | vesicle amine transport protein 1 homolog (T californica) |
|
| |
| UniRef100_P62774 | myotrophin |
|
| |
| UniRef100_P50247 | S-adenosylhomocysteine hydrolase |
|
| |
| UniRef100_P80316 | chaperonin subunit 5 (epsilon) |
|
| |
| UniRef100_P42932 | chaperonin subunit 8 (theta) |
|
| |
| UniRef100_P80317 | chaperonin subunit 6a (zeta) |
|
| |
| UniRef100_P08030 | adenine phosphoribosyl transferase |
|
| |
| UniRef100_Q3UK08 | tumor susceptibility gene 101 |
|
| |
| UniRef100_Q8VDN2 | ATPase, Na+/K+ transporting, alpha 1 polypeptide |
|
| |
| UniRef100_A2A607 | GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
| Immune | |
| UniRef100_O19476 | histocompatibility 2, D region locus 1 |
|
| |
| UniRef100_P49769 | presenilin 1 |
|
| |
| UniRef100_P04202 | Transforming growth factor beta-1 |
| Cell adhesion | |
| UniRef100_A2AKI5 | integrin alpha V |
|
| |
| UniRef100_Q3TIC2 | ras homolog gene family, member B |
|
| |
| UniRef100_P49769 | presenilin 1 |
|
| |
| UniRef100_Q3UNG0 | laminin, gamma 1 |
|
| |
| UniRef100_Q3UEG9 | flotillin 2 |
|
| |
| UniRef100_P10493 | nidogen 1 |
|
| |
| UniRef100_A2AU04 | integrin alpha 6 |
|
| |
| UniRef100_P02469 | laminin B1 subunit 1 |
|
| |
| UniRef100_O88792 | F11 receptor |
|
| |
| UniRef100_P26231 | catenin (cadherin associated protein), alpha 1 |
|
| |
| UniRef100_Q64727 | vinculin |
|
| |
| UniRef100_Q05793 | perlecan (heparan sulfate proteoglycan 2) |
|
| |
| UniRef100_P40240 | CD9 antigen |
|
| |
| UniRef100_P11688 | integrin alpha 5 (fibronectin receptor alpha) |
|
| |
| UniRef100_Q3TLP8 | RAS-related C3 botulinum substrate 1 |
|
| |
| UniRef100_P09055 | integrin beta 1 (fibronectin receptor beta) |
|
| |
| UniRef100_Q01768 | non-metastatic cells 2, protein (NM23B) expressed in |
|
| |
| UniRef100_P16045 | lectin, galactose binding, soluble 1 |
|
| |
| UniRef100_P35441 | thrombospondin 1 |
|
| |
| UniRef100_Q3UHT9 | myosin, heavy polypeptide 9, non-muscle |
|
| |
| UniRef100_P21956 | milk fat globule-EGF factor 8 protein |
|
| |
| UniRef100_A2APM1 | CD44 antigen |
|
| |
| UniRef100_ A2ABW7 | laminin, alpha 5 |
| Signal transduction | |
| UniRef100_Q3TIC2 | ras homolog gene family, member B |
|
| |
| UniRef100_P10107 | annexin A1 |
|
| |
| UniRef100_Q3U7U8 | RAS-related protein-1a |
|
| |
| UniRef100_Q14AA6 | RIKEN cDNA 1700009N14 gene |
|
| |
| UniRef100_Q3TLP8 | RAS-related C3 botulinum substrate 1 |
|
| |
| UniRef100_P68510 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide |
|
| |
| UniRef100_P49817 | caveolin, caveolae protein 1 |
|
| |
| UniRef100_P68254 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide |
|
| |
| UniRef100_Q99PT1 | Rho GDP dissociation inhibitor (GDI) alpha; Rho GDP dissociation inhibitor (GDI) alpha |
|
| |
| UniRef100_Q9DAS9 | guanine nucleotide binding protein (G protein), gamma 12 |
|
| |
| UniRef100_P08752 | guanine nucleotide binding protein (G protein), alpha inhibiting 2 |
|
| |
| UniRef100_Q3U1B1 | guanine nucleotide binding protein (G protein), beta 1 |
|
| |
| UniRef100_Q3TJH1 | guanine nucleotide binding protein (G protein), alpha inhibiting 3 |
|
| |
| UniRef100_A2A607 | GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
|
| |
| UniRef100_P21278 | guanine nucleotide binding protein, alpha 11 |
|
| |
| UniRef100_P10833 | Harvey rat sarcoma oncogene, subgroup R |
|
| |
| UniRef100_Q3UUX9 | guanosine diphosphate (GDP) dissociation inhibitor 2 |
|
| |
| UniRef100_Q3U2W7 | v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog |
|
| |
| UniRef100_Q6ZQK2 | IQ motif containing GTPase activating protein 1 |
|
| |
| UniRef100_Q8CCG5 | v-ral simian leukemia viral oncogene homolog B (ras related) |
|
| |
| UniRef100_P63321 | v-ral simian leukemia viral oncogene homolog A (ras related) |
|
| |
| UniRef100_Q3TED2 | programmed cell death 6 interacting protein |
|
| |
| UniRef100_P35278 | RAB5C, member RAS oncogene family |
|
| |
| UniRef100_P61028 | RAB8B, member RAS oncogene family; RAB8B, member RAS oncogene family |
|
| |
| UniRef100_P53994 | RAB2A, member RAS oncogene family |
|
| |
| UniRef100_Q8BHH2 | RAB9B, member RAS oncogene family |
|
| |
| UniRef100_P55258 | RAB8A, member RAS oncogene family |
|
| |
| UniRef100_P46638 | RAB11B, member RAS oncogene family |
|
| |
| UniRef100_Q50HW9 | RAB14, member RAS oncogene family |
| Vesicle-mediated transport | |
| UniRef100_Q5SXR6 | clathrin, heavy polypeptide (Hc) |
|
| |
| UniRef100_P59108 | copine II |
|
| |
| UniRef100_Q3UUX9 | guanosine diphosphate (GDP) dissociation inhibitor 2 |
|
| |
| UniRef100_O09044 | synaptosomal-associated protein 23 |
|
| |
| UniRef100_P53994 | RAB2A, member RAS oncogene family |
|
| |
| UniRef100_P13020 | gelsolin |
|
| |
| UniRef100_P63024 | vesicle-associated membrane protein 3 |
| Differentiation | |
| UniRef100_A2AKI5 | integrin alpha V |
|
| |
| UniRef100_Q3TIC2 | ras homolog gene family, member B |
|
| |
| UniRef100_P49769 | presenilin 1 |
|
| |
| UniRef100_O88792 | F11 receptor |
|
| |
| UniRef100_P09055 | integrin beta 1 (fibronectin receptor beta) |
|
| |
| UniRef100_P16045 | lectin, galactose binding, soluble 1 |
|
| |
| UniRef100_P49817 | caveolin, caveolae protein 1 |
|
| |
| UniRef100_Q3UHT9 | myosin, heavy polypeptide 9, non-muscle |
|
| |
| UniRef100_P62774 | myotrophin; myotrophin |
|
| |
| UniRef100_P53783 | SRY-box containing gene 1 |
|
| |
| UniRef100_P17742 | peptidylprolyl isomerase A (cyclophilin A) |
|
| |
| UniRef100_A0PJ91 | heat shock protein 90, alpha (cytosolic), class A member 1 |
|
| |
| UniRef100_Q3UK08 | tumor susceptibility gene 101 |
|
| |
| UniRef100_A2A607 | GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
|
| |
| UniRef100_P46935 | E3 ubiquitin-protein ligase NEDD4 |
|
| |
| UniRef100_P21278 | guanine nucleotide binding protein, alpha 11 |
|
| |
| UniRef100_Q3U2W7 | v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog |
|
| |
| UniRef100_Q5M9P3 | ribosomal protein S19 |
| Cell cycle | |
| UniRef100_Q3TIC2 | Ras homolog gene family, member B |
|
| |
| UniRef100_P14069 | S100 calcium binding protein A6 (calcyclin) |
|
| |
| UniRef100_Q3UKW2 | Calmodulin 1 |
|
| |
| UniRef100_P10107 | Annexin A1 |
|
| |
| UniRef100_Q3U7U8 | RAS-related protein-1a |
|
| |
| UniRef100_P42208 | Septin 2 |
|
| |
| UniRef100_P27661 | H2A histone family, member X |
|
| |
| UniRef100_Q14AA6 | RIKEN cDNA 1700009N14 gene |
|
| |
| UniRef100_P09055 | Integrin beta 1 (fibronectin receptor beta) |
|
| |
| UniRef100_Q3V4A1 | Serine/threonine kinase 11 |
|
| |
| UniRef100_P60122 | RuvB-like protein 1 |
|
| |
| UniRef100_Q3UK08 | Tumor susceptibility gene 101 |
|
| |
| UniRef100_Q3UXQ6 | Ribosomal protein S4, X-linked |
Table 3.
Identities of exosome proteins found in T-exosomes and C-exosomes
| Accession no. | Identified protein |
|---|---|
| UniRef100_Q3TIC2 | Ras homolog gene family, member B |
| UniRef100_Q8K314 | ATPase, Ca++ transporting, plasma membrane 1 |
| UniRef100_P14824 | Annexin A6 |
| UniRef100_Q3UMR9 | Solute carrier family 5 (inositol transporters), member 3 |
| UniRef100_P09528 | Ferritin heavy chain 1 |
| UniRef100_P49817 | Caveolin, caveolae protein 1 |
| UniRef100_Q8VDN2 | ATPase, Na+/K+ transporting, alpha 1 polypeptide |
| UniRef100_P97370 | ATPase, Na+/K+ transporting, beta 3 polypeptide |
| UniRef100_Q8CFE6 | Solute carrier family 38, member 2 |
| Solute carrier family 6 (neurotransmitter transporter, | |
| UniRef100_A2ALM4 | creatine), member 8 |
| Solute carrier family 16 (monocarboxylic acid | |
| UniRef100_P53986 | transporters), member 1 |
| UniRef100_P46467 | Vacuolar protein sorting 4b (yeast |
| UniRef100_P61028 | RAB8B, member RAS oncogene family |
| UniRef100_ P53994 | RAB2A, member RAS oncogene family |
| UniRef100_P04202 | Transforming growth factor beta-1 |
| UniRef100_Q8R0J7 | vacuolar protein sorting 37B (yeast) |
| UniRef100_Q3TIC2 | ras homolog gene family, member B |
| UniRef100_P49769 | presenilin 1 |
| UniRef100_Q3UNG0 | laminin, gamma 1 |
| UniRef100_Q3UEG9 | flotillin 2 |
| UniRef 100_Q02053 | ubiquitin-like modifier activating enzyme 1 |
| UniRef 100_Q921I1 | transferrin |
| UniRef100_P35278 | RAB5C, member RAS oncogene family |
| RAB8B, member RAS oncogene family; RAB8B, member | |
| UniRef100_P61028 | RAS oncogene family |
| UniRef100_P53994 | RAB2A, member RAS oncogene family |
| UniRef100_Q8BHH2 | RAB9B, member RAS oncogene family |
| UniRef100_P55258 | RAB8A, member RAS oncogene family |
| UniRef100_P46638 | RAB11B, member RAS oncogene family |
| UniRef100_Q50HW9 | RAB14, member RAS oncogene family |
| UniRef100_Q5SXR6 | clathrin, heavy polypeptide (Hc) |
| UniRef 100_P41731 | CD63 antigen |
| UniRef100_P40240 | CD9 antigen |
| heat shock protein 90, alpha (cytosolic), class A member | |
| UniRef100_A0PJ91 | 1 |
| UniRef100_Q3UK08 | tumor susceptibility gene 101 |
| UniRef 100_P19096 | fatty acid synthase |
| UniRef 100_P35762 | CD 81 antigen |
We determined the effect of anti-PGE2 and anti-TGF-β antibodies on induction of bone marrow CD11b+Gr-1+ cells. and found that neutralization of tumor exosomal PGE2 and TGF-β led to a more significant reduction in the induction CD11b+Gr-1+ cells (P < 0.01, Figure 4B) than anti-PGE2 or anti-TGF-β alone (Figure 4B). In addition, neutralization of PGE2 and TGF-β together also resulted in a decreased induction of expression of IL-6 and VEGF (Figure 4C). More importantly, blocking exosomal PGE2 and TGF-β by pre-incubation of T-exosomes with anti-PGE2 and anti-TGF-β antibodies reduced tumor growth when tumor cells were co-injected with bone marrow derived CD11b+Gr-1+ cells pulsed with T-exosomes (Figure 4D).
Discussion
The present study revealed that T-exosomes can cause the accumulation of myeloid-derived suppressor cells in a tumor. This is supported by the finding that 1) mice injected with T-exosomes developed enlarged spleens containing accumulations of CD11b+Gr-1+ MDSCs; 2) these exosome-induced CD11b+Gr-1+ cells also released proinflammatory cytokine IL-6 and tumor growth factor VEGF; and 3) the co-injection of T-exosomes taken up by bone marrow derived CD11b+Gr-1+ cells with tumor cells led to the promotion of tumor growth. We further identified that tumor exosomal PGE2 and TGF-β play a role in the induction of the accumulation of MDSCs. Together, these data provide evidence of the immune suppression properties of exosomal PGE2 and TGF-β in cancer.
PGE2, a product of inflammation, is thought to promote tumor growth by inducing neoangiogenesis 28–33 and by inducing the accumulation of MDSCs 19. In addition to PGE2 regulating production of a number of cytokines, including IL-6 34–38 and VEGF 29, 31, 39–41, PGE2 also regulates arginase 1 42–44. In the late stages of cancer, TGF-β promotes tumor spreading by enhancing the expression of VEGF 17, 20, 45–50. The current study shows that both PGE2 and TGF-β are enriched in exosomes isolated from late stages of tumor growth. Blocking exosomal PGE2 and TGF-β pathways (with anti-PGE2 and TGF-β antibodies) results in the attenuation of tumor exosomal induced expression of VEGF, and delays tumor growth in an in vivo transplantation tumor model.
Further characterization of the T-exosomes was performed by examining the effects of the tumor microenvironment on T-exosome mediated induction of MDSCs. We found that the tumor microenvironment can dramatically effect the recruitment of PGE2 and TGF-β into the exosomes. The more advanced the tumor stage, the more enriched T-exosomes are with PGE2 and TGF-β. It is conceivable that the tumor microenvironment during the early stages of tumor growth or establishment may regulate the sorting of exosomal proteins in/out differently from during later stages of tumor growth. During the early stages of tumor development, the host immune response against tumor growth may be dominant in the tumor microenvironment when compared to tumor derived immune suppression factors. The exosomes released during the early stages of tumor growth and establishment may actually stimulate host immune responses 51. However, as the tumor becomes established new factors regulated by the tumor microenvironment are expressed 52–57 and these factors could drive the sorting of tumor exosomal molecules toward those molecules that tend to cause immune suppression. This study identifies potential markers for further identification of the tumor microenvironment derived master regulator(s) that preferentially sort immune suppressor molecules such as exosomal PGE2 and TGF-β into exosomes.
Exosomes packed with multiple immune suppression molecules, including PGE2 and TGF-β, could act on a variety of levels and pathways to suppress immune function in multiple targets simultaneously. Therefore, given the broad array of bioactive molecules incorporated into tumor-produced exosomes, the possibility exists that other factors might be involved in the induction of MDSCs (besides exosomal PGE2 and TGF-β). As a matter of fact, our mass spectrometry analysis results suggest that other proteins might be selectively sorted in or out of T-exosomes. These proteins could also play a role in exosome-mediated induction of MDSCs. This assumption is supported with the evidence that neutralization of the activities of exosomal PGE2 and TGF-β does not lead to complete inhibition of T-exosome mediated induction of MDSCs. Therefore, the higher amounts of exosomal PGE2 or TGF-β may be one reason why these molecules are more effective but not necessarily the master regulators of induction of MDSCs and tumor promoting activity of these cells. One important factor to explore is the regulation of protein sorting in and out of exosomes. Data published by other groups indicate that ceramid may play a role in exosomal protein sorting58. Ceramide regulates cell apoptosis. Our MS/LC data suggest that a relatively large group of proteins regulating cell apoptosis is preferentially sorted into T-exosomes. Whether these proteins are regulated by ceramide warrants further investigated.
Regulators within cells are tightly controlled, but may be affected by microenvironment changes. Therefore, difference in the induction of MDSCs and the tumor promoting activity of cells induced by C-exosomes and T-exosomes might be due to tumor microenvironment factors. Tumor microenvironment factors could have effects on the T-exosome mediated accumulation of MDSCs in the tumor and spleen through multiple steps, from controlling quality and quantity of the exosomes released from the tumor to in vivo trafficking routs and targeted cells of the exosomes. These issues are important and require further study.
Finally, caution should be exercised so as to not over interpret our results in regard to the resource of T-exosome producer cells, although our data indicate that the exosomes released by tumor cells can induce MDSCs. Other types of cells rather than tumor cells including infiltrating MDSCs, tumor stroma cells and regulatory T cells should not be excluded for their potential release of exosomes. Future experiments of this nature will be fundamental to understanding how the tumor cells and stroma cells in the tumor microenvironment communicate with infiltrating immune cells via exosomes to regulate immune suppression in a host.
Acknowledgments
This work was supported by National Institutes of Health (NIH) grant no. RO1CA116092, RO1CA107181, R01AT004294; Birmingham Veterans Administration Medical Center (VAMC) Merit Review Grants (H.-G.Z.); and a grant from the Susan G. Komen Breast Cancer Foundation. We thank Dr. Jerald Ainsworth for editorial assistance.
References
- 1.Mareel M, Madani I. Tumour-associated host cells participating at invasion and metastasis : targets for therapy? Acta Chir Belg. 2006;106:635–40. doi: 10.1080/00015458.2006.11679971. [DOI] [PubMed] [Google Scholar]
- 2.Nagaraj S, Gabrilovich DI. Myeloid-derived suppressor cells. Adv Exp Med Biol. 2007;601:213–23. doi: 10.1007/978-0-387-72005-0_22. [DOI] [PubMed] [Google Scholar]
- 3.Serafini P, Borrello I, Bronte V. Myeloid suppressor cells in cancer: recruitment, phenotype, properties, and mechanisms of immune suppression. Semin Cancer Biol. 2006;16:53–65. doi: 10.1016/j.semcancer.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 4.Yang L, DeBusk LM, Fukuda K, Fingleton B, Green-Jarvis B, Shyr Y, Matrisian LM, Carbone DP, Lin PC. Expansion of myeloid immune suppressor Gr+CD11b+ cells in tumor-bearing host directly promotes tumor angiogenesis. Cancer Cell. 2004;6:409–21. doi: 10.1016/j.ccr.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 5.Liu C, Yu S, Kappes J, Wang J, Grizzle WE, Zinn KR, Zhang HG. Expansion of spleen myeloid suppressor cells represses NK cell cytotoxicity in tumor-bearing host. Blood. 2007;109:4336–42. doi: 10.1182/blood-2006-09-046201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kusmartsev S, Nagaraj S, Gabrilovich DI. Tumor-associated CD8+ T cell tolerance induced by bone marrow-derived immature myeloid cells. J Immunol. 2005;175:4583–92. doi: 10.4049/jimmunol.175.7.4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mirza N, Fishman M, Fricke I, Dunn M, Neuger AM, Frost TJ, Lush RM, Antonia S, Gabrilovich DI. All-trans-retinoic acid improves differentiation of myeloid cells and immune response in cancer patients. Cancer Res. 2006;66:9299–307. doi: 10.1158/0008-5472.CAN-06-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagaraj S, Gupta K, Pisarev V, Kinarsky L, Sherman S, Kang L, Herber DL, Schneck J, Gabrilovich DI. Altered recognition of antigen is a mechanism of CD8+ T cell tolerance in cancer. Nat Med. 2007;13:828–35. doi: 10.1038/nm1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hiratsuka S, Watanabe A, Aburatani H, Maru Y. Tumour-mediated upregulation of chemoattractants and recruitment of myeloid cells predetermines lung metastasis. Nat Cell Biol. 2006;8:1369–75. doi: 10.1038/ncb1507. [DOI] [PubMed] [Google Scholar]
- 10.Bartz H, Avalos NM, Baetz A, Heeg K, Dalpke AH. Involvement of suppressors of cytokine signaling in toll-like receptor-mediated block of dendritic cell differentiation. Blood. 2006;108:4102–8. doi: 10.1182/blood-2006-03-008946. [DOI] [PubMed] [Google Scholar]
- 11.Isomoto H, Mott JL, Kobayashi S, Werneburg NW, Bronk SF, Haan S, Gores GJ. Sustained IL-6/STAT-3 signaling in cholangiocarcinoma cells due to SOCS-3 epigenetic silencing. Gastroenterology. 2007;132:384–96. doi: 10.1053/j.gastro.2006.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu S, Liu C, Su K, Wang J, Liu Y, Zhang L, Li C, Cong Y, Kimberly R, Grizzle WE, Falkson C, Zhang HG. Tumor exosomes inhibit differentiation of bone marrow dendritic cells. J Immunol. 2007;178:6867–75. doi: 10.4049/jimmunol.178.11.6867. [DOI] [PubMed] [Google Scholar]
- 13.Clayton A, Mitchell JP, Court J, Mason MD, Tabi Z. Human tumor-derived exosomes selectively impair lymphocyte responses to interleukin-2. Cancer Res. 2007;67:7458–66. doi: 10.1158/0008-5472.CAN-06-3456. [DOI] [PubMed] [Google Scholar]
- 14.Taylor DD, Akyol S, Gercel-Taylor C. Pregnancy-associated exosomes and their modulation of T cell signaling. J Immunol. 2006;176:1534–42. doi: 10.4049/jimmunol.176.3.1534. [DOI] [PubMed] [Google Scholar]
- 15.Valenti R, Huber V, Iero M, Filipazzi P, Parmiani G, Rivoltini L. Tumor-released microvesicles as vehicles of immunosuppression. Cancer Res. 2007;67:2912–5. doi: 10.1158/0008-5472.CAN-07-0520. [DOI] [PubMed] [Google Scholar]
- 16.Liu C, Yu S, Zinn K, Wang J, Zhang L, Jia Y, Kappes JC, Barnes S, Kimberly RP, Grizzle WE, Zhang HG. Murine mammary carcinoma exosomes promote tumor growth by suppression of NK cell function. J Immunol. 2006;176:1375–85. doi: 10.4049/jimmunol.176.3.1375. [DOI] [PubMed] [Google Scholar]
- 17.Donovan D, Harmey JH, Toomey D, Osborne DH, Redmond HP, Bouchier-Hayes DJ. TGF beta-1 regulation of VEGF production by breast cancer cells. Ann Surg Oncol. 1997;4:621–7. doi: 10.1007/BF02303745. [DOI] [PubMed] [Google Scholar]
- 18.Kaminska B, Wesolowska A, Danilkiewicz M. TGF beta signalling and its role in tumour pathogenesis. Acta Biochim Pol. 2005;52:329–37. [PubMed] [Google Scholar]
- 19.Sinha P, Clements VK, Fulton AM, Ostrand-Rosenberg S. Prostaglandin E2 promotes tumor progression by inducing myeloid-derived suppressor cells. Cancer Res. 2007;67:4507–13. doi: 10.1158/0008-5472.CAN-06-4174. [DOI] [PubMed] [Google Scholar]
- 20.Xiong B, Gong LL, Zhang F, Hu MB, Yuan HY. TGF beta1 expression and angiogenesis in colorectal cancer tissue. World J Gastroenterol. 2002;8:496–8. doi: 10.3748/wjg.v8.i3.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang HG, Hyde K, Page GP, Brand JP, Zhou J, Yu S, Allison DB, Hsu HC, Mountz JD. Novel tumor necrosis factor alpha-regulated genes in rheumatoid arthritis. Arthritis Rheum. 2004;50:420–31. doi: 10.1002/art.20037. [DOI] [PubMed] [Google Scholar]
- 22.Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci U S A. 2004;101:13368–73. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skokos D, Botros HG, Demeure C, Morin J, Peronet R, Birkenmeier G, Boudaly S, Mecheri S. Mast cell-derived exosomes induce phenotypic and functional maturation of dendritic cells and elicit specific immune responses in vivo. J Immunol. 2003;170:3037–45. doi: 10.4049/jimmunol.170.6.3037. [DOI] [PubMed] [Google Scholar]
- 24.Wolfers J, Lozier A, Raposo G, Regnault A, Thery C, Masurier C, Flament C, Pouzieux S, Faure F, Tursz T, Angevin E, Amigorena S, et al. Tumor-derived exosomes are a source of shared tumor rejection antigens for CTL cross-priming. Nat Med. 2001;7:297–303. doi: 10.1038/85438. [DOI] [PubMed] [Google Scholar]
- 25.Thery C, Regnault A, Garin J, Wolfers J, Zitvogel L, Ricciardi-Castagnoli P, Raposo G, Amigorena S. Molecular characterization of dendritic cell-derived exosomes. Selective accumulation of the heat shock protein hsc73. J Cell Biol. 1999;147:599–610. doi: 10.1083/jcb.147.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mears R, Craven RA, Hanrahan S, Totty N, Upton C, Young SL, Patel P, Selby PJ, Banks RE. Proteomic analysis of melanoma-derived exosomes by two-dimensional polyacrylamide gel electrophoresis and mass spectrometry. Proteomics. 2004;4:4019–31. doi: 10.1002/pmic.200400876. [DOI] [PubMed] [Google Scholar]
- 27.Wubbolts R, Leckie RS, Veenhuizen PT, Schwarzmann G, Mobius W, Hoernschemeyer J, Slot JW, Geuze HJ, Stoorvogel W. Proteomic and biochemical analyses of human B cell-derived exosomes. Potential implications for their function and multivesicular body formation. J Biol Chem. 2003;278:10963–72. doi: 10.1074/jbc.M207550200. [DOI] [PubMed] [Google Scholar]
- 28.Ding YB, Shi RH, Tong JD, Li XY, Zhang GX, Xiao WM, Yang JG, Bao Y, Wu J, Yan ZG, Wang XH. PGE2 up-regulates vascular endothelial growth factor expression in MKN28 gastric cancer cells via epidermal growth factor receptor signaling system. Exp Oncol. 2005;27:108–13. [PubMed] [Google Scholar]
- 29.Mauritz I, Westermayer S, Marian B, Erlach N, Grusch M, Holzmann K. Prostaglandin E(2) stimulates progression-related gene expression in early colorectal adenoma cells. Br J Cancer. 2006;94:1718–25. doi: 10.1038/sj.bjc.6603146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Owen JL, Iragavarapu-Charyulu V, Gunja-Smith Z, Herbert LM, Grosso JF, Lopez DM. Up-regulation of matrix metalloproteinase-9 in T lymphocytes of mammary tumor bearers: role of vascular endothelial growth factor. J Immunol. 2003;171:4340–51. doi: 10.4049/jimmunol.171.8.4340. [DOI] [PubMed] [Google Scholar]
- 31.Shao J, Jung C, Liu C, Sheng H. Prostaglandin E2 Stimulates the beta-catenin/T cell factor-dependent transcription in colon cancer. J Biol Chem. 2005;280:26565–72. doi: 10.1074/jbc.M413056200. [DOI] [PubMed] [Google Scholar]
- 32.Takahashi A, Kono K, Ichihara F, Sugai H, Fujii H, Matsumoto Y. Vascular endothelial growth factor inhibits maturation of dendritic cells induced by lipopolysaccharide, but not by proinflammatory cytokines. Cancer Immunol Immunother. 2004;53:543–50. doi: 10.1007/s00262-003-0466-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X, Klein RD. Prostaglandin E2 induces vascular endothelial growth factor secretion in prostate cancer cells through EP2 receptor-mediated cAMP pathway. Mol Carcinog. 2007;46:912–23. doi: 10.1002/mc.20320. [DOI] [PubMed] [Google Scholar]
- 34.Eisengart CA, Mestre JR, Naama HA, Mackrell PJ, Rivadeneira DE, Murphy EM, Stapleton PP, Daly JM. Prostaglandins regulate melanoma-induced cytokine production in macrophages. Cell Immunol. 2000;204:143–9. doi: 10.1006/cimm.2000.1686. [DOI] [PubMed] [Google Scholar]
- 35.Elgert KD, Alleva DG, Mullins DW. Tumor-induced immune dysfunction: the macrophage connection. J Leukoc Biol. 1998;64:275–90. doi: 10.1002/jlb.64.3.275. [DOI] [PubMed] [Google Scholar]
- 36.Fiebich BL, Hull M, Lieb K, Gyufko K, Berger M, Bauer J. Prostaglandin E2 induces interleukin-6 synthesis in human astrocytoma cells. J Neurochem. 1997;68:704–9. doi: 10.1046/j.1471-4159.1997.68020704.x. [DOI] [PubMed] [Google Scholar]
- 37.Fiebich BL, Schleicher S, Spleiss O, Czygan M, Hull M. Mechanisms of prostaglandin E2-induced interleukin-6 release in astrocytes: possible involvement of EP4-like receptors, p38 mitogen-activated protein kinase and protein kinase C. J Neurochem. 2001;79:950–8. doi: 10.1046/j.1471-4159.2001.00652.x. [DOI] [PubMed] [Google Scholar]
- 38.Liu XH, Kirschenbaum A, Lu M, Yao S, Klausner A, Preston C, Holland JF, Levine AC. Prostaglandin E(2) stimulates prostatic intraepithelial neoplasia cell growth through activation of the interleukin-6/GP130/STAT-3 signaling pathway. Biochem Biophys Res Commun. 2002;290:249–55. doi: 10.1006/bbrc.2001.6188. [DOI] [PubMed] [Google Scholar]
- 39.Liu XH, Kirschenbaum A, Lu M, Yao S, Dosoretz A, Holland JF, Levine AC. Prostaglandin E2 induces hypoxia-inducible factor-1alpha stabilization and nuclear localization in a human prostate cancer cell line. J Biol Chem. 2002;277:50081–6. doi: 10.1074/jbc.M201095200. [DOI] [PubMed] [Google Scholar]
- 40.Pai R, Szabo IL, Soreghan BA, Atay S, Kawanaka H, Tarnawski AS. PGE(2) stimulates VEGF expression in endothelial cells via ERK2/JNK1 signaling pathways. Biochem Biophys Res Commun. 2001;286:923–8. doi: 10.1006/bbrc.2001.5494. [DOI] [PubMed] [Google Scholar]
- 41.Timoshenko AV, Chakraborty C, Wagner GF, Lala PK. COX-2-mediated stimulation of the lymphangiogenic factor VEGF-C in human breast cancer. Br J Cancer. 2006;94:1154–63. doi: 10.1038/sj.bjc.6603067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Corraliza IM, Soler G, Eichmann K, Modolell M. Arginase induction by suppressors of nitric oxide synthesis (IL-4, IL-10 and PGE2) in murine bone-marrow-derived macrophages. Biochem Biophys Res Commun. 1995;206:667–73. doi: 10.1006/bbrc.1995.1094. [DOI] [PubMed] [Google Scholar]
- 43.Ochoa AC, Zea AH, Hernandez C, Rodriguez PC. Arginase, prostaglandins, and myeloid-derived suppressor cells in renal cell carcinoma. Clin Cancer Res. 2007;13:721s–6s. doi: 10.1158/1078-0432.CCR-06-2197. [DOI] [PubMed] [Google Scholar]
- 44.Rodriguez PC, Hernandez CP, Quiceno D, Dubinett SM, Zabaleta J, Ochoa JB, Gilbert J, Ochoa AC. Arginase I in myeloid suppressor cells is induced by COX-2 in lung carcinoma. J Exp Med. 2005;202:931–9. doi: 10.1084/jem.20050715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gaspar NJ, Li L, Kapoun AM, Medicherla S, Reddy M, Li G, O'Young G, Quon D, Henson M, Damm DL, Muiru GT, Murphy A, et al. Inhibition of transforming growth factor beta signaling reduces pancreatic adenocarcinoma growth and invasiveness. Mol Pharmacol. 2007;72:152–61. doi: 10.1124/mol.106.029025. [DOI] [PubMed] [Google Scholar]
- 46.Schwarte-Waldhoff I, Schmiegel W. Smad4 transcriptional pathways and angiogenesis. Int J Gastrointest Cancer. 2002;31:47–59. doi: 10.1385/IJGC:31:1-3:47. [DOI] [PubMed] [Google Scholar]
- 47.Benckert C, Jonas S, Cramer T, Von Marschall Z, Schafer G, Peters M, Wagner K, Radke C, Wiedenmann B, Neuhaus P, Hocker M, Rosewicz S. Transforming growth factor beta 1 stimulates vascular endothelial growth factor gene transcription in human cholangiocellular carcinoma cells. Cancer Res. 2003;63:1083–92. [PubMed] [Google Scholar]
- 48.Teraoka H, Sawada T, Nishihara T, Yashiro M, Ohira M, Ishikawa T, Nishino H, Hirakawa K. Enhanced VEGF production and decreased immunogenicity induced by TGF-beta 1 promote liver metastasis of pancreatic cancer. Br J Cancer. 2001;85:612–7. doi: 10.1054/bjoc.2001.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheng D, Lee YC, Rogers JT, Perkett EA, Moyers JP, Rodriguez RM, Light RW. Vascular endothelial growth factor level correlates with transforming growth factor-beta isoform levels in pleural effusions. Chest. 2000;118:1747–53. doi: 10.1378/chest.118.6.1747. [DOI] [PubMed] [Google Scholar]
- 50.Li Z, Shimada Y, Uchida S, Maeda M, Kawabe A, Mori A, Itami A, Kano M, Watanabe G, Imamura M. TGF-alpha as well as VEGF, PD-ECGF and bFGF contribute to angiogenesis of esophageal squamous cell carcinoma. Int J Oncol. 2000;17:453–60. doi: 10.3892/ijo.17.3.453. [DOI] [PubMed] [Google Scholar]
- 51.Zeelenberg IS, Ostrowski M, Krumeich S, Bobrie A, Jancic C, Boissonnas A, Delcayre A, Le Pecq JB, Combadiere B, Amigorena S, Thery C. Targeting tumor antigens to secreted membrane vesicles in vivo induces efficient antitumor immune responses. Cancer Res. 2008;68:1228–35. doi: 10.1158/0008-5472.CAN-07-3163. [DOI] [PubMed] [Google Scholar]
- 52.Johrer K, Pleyer L, Olivier A, Maizner E, Zelle-Rieser C, Greil R. Tumour-immune cell interactions modulated by chemokines. Expert Opin Biol Ther. 2008;8:269–90. doi: 10.1517/14712598.8.3.269. [DOI] [PubMed] [Google Scholar]
- 53.Hu M, Polyak K. Microenvironmental regulation of cancer development. Curr Opin Genet Dev. 2008 doi: 10.1016/j.gde.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Witz IP. Yin-yang activities and vicious cycles in the tumor microenvironment. Cancer Res. 2008;68:9–13. doi: 10.1158/0008-5472.CAN-07-2917. [DOI] [PubMed] [Google Scholar]
- 55.Noonan DM, De Lerma Barbaro A, Vannini N, Mortara L, Albini A. Inflammation, inflammatory cells and angiogenesis: decisions and indecisions. Cancer Metastasis Rev. 2008;27:31–40. doi: 10.1007/s10555-007-9108-5. [DOI] [PubMed] [Google Scholar]
- 56.Stetler-Stevenson WG. The tumor microenvironment: regulation by MMP-independent effects of tissue inhibitor of metalloproteinases-2. Cancer Metastasis Rev. 2008;27:57–66. doi: 10.1007/s10555-007-9105-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Albini A, Mirisola V, Pfeffer U. Metastasis signatures: genes regulating tumor-microenvironment interactions predict metastatic behavior. Cancer Metastasis Rev. 2008;27:75–83. doi: 10.1007/s10555-007-9111-x. [DOI] [PubMed] [Google Scholar]
- 58.Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brugger B, Simons M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319:1244–7. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]