Abstract
The MAGE antigens are frequently expressed cancer vaccine targets. However, quantitative analysis of MAGE expression in upper aero-digestive tract (UADT) tumor cells and its association with T cell recognition has not been performed, hindering the selection of appropriate candidates for MAGE specific immunotherapy. Using quantitative RT-PCR (QRT-PCR), we evaluated the expression of MAGE-3/6 in 65 UADT cancers, 48 normal samples from tumor matched sites and 7 HLA-A*0201+squamous cell carcinoma of the head and neck (SCCHN) cell lines. Expression results were confirmed using western blot. HLA-A*0201:MAGE-3(271–279) specific cytotoxic T lymphocytes (MAGE-CTL) from SCCHN patients and healthy donors showed that MAGE-3/6 expression was highly associated with CTL recognition in vitro. Based on MAGE-3/6 expression we could identify 31 (47%) of the 65 UADT tumors which appeared to express MAGE-3/6 at levels that correlated with efficient CTL recognition. To confirm that the level of MAGE-3 expression was responsible for CTL recognition, two MAGE-3/6 mRNAhigh SCCHN cell lines, PCI-13 and PCI-30, were subjected to MAGE-3/6 specific knockdown. RNAi–transfected cells showed that MAGE expression, and MAGE-CTL recognition, were significantly reduced. Furthermore, treatment of cells expressing low MAGE-3/6 mRNA with a demethylating agent, 5-aza-2'-deoxycytidine (DAC), increased the expression of MAGE-3/6 and CTL recognition. Thus, using QRT-PCR UADT cancers frequently express MAGE-3/6 at levels sufficient for CTL recognition, supporting the use of a QRT-PCR based assay for the selection of candidates likely to respond to MAGE-3/6 immunotherapy. Demethylating agents could increase the number of patients amenable for targeting epigenetically modified tumor antigens in vaccine trials.
Keywords: MAGE, Head and neck cancer, T cells, QRT-PCR
Introduction
Upper aero-digestive tract (UADT) malignancies include tumors of the head and neck, esophagus and lung. These tumors are often grouped due to their similar carcinogenic etiology and the fact that patients suffering from UADT primary tumors are at high risk of developing a second UADT later in life1. Thus, immunotherapy against commonly expressed tumor antigens (TA) in UADT tumors is appealing as secondary prevention to improve the survival and prognosis of these often aggressive tumors.
Of the many TA found in epithelial malignancies, the MAGE family of cancer-testis antigens are attractive targets for immunotherapy due to their relatively frequent expression in a variety of tumor types as compared to non-neoplastic tissues2. The MAGE-3 protein contains a 9 amino acid peptide (FLWGPRALV, residues 271–279) that is also encoded in the MAGE-6 and-12 genes in the exact same position, differing by a single, relatively conservative residue at position nine (V to I). This epitope appears to be cross-reactive between MAGE-3, -6 and -123. CTL specific for this epitope have been found in patients with MAGE-3+ tumors4, 5 and have been generated in vitro 6 and in vivo following vaccination7.
Previous reports that UADT cancers express genes of the MAGE family used standard semi-quantitative RT-PCR techniques or immunohistochemistry8–11. These techniques are at least semi-quantitative and provide little information on whether sufficient levels of this TA are expressed and processed to permit MAGE-specific CTL recognition. Furthermore, the correlation between quantitative levels of MAGE gene expression and CTL recognition of tumor cells has not been established, hindering the estimate of actual cancer patients suitable for MAGE targeted immunotherapy. Due to the large subset of tumors with little or not detectable MAGE expression, the use of demethylating agents such as 2'-Deoxy-5-azacytidine (DAC) that upregulate the expression of MAGE-3, might improve the clinical responses to these immunotherapies.
Thus, for the first time quantitative MAGE-3/6 expression has been determined using a rapid QRT-PCR assay we developed in a series of UADT tumors. Our studies suggest that level of antigen expression is an important factor in CTL recognition of malignant cells. Quantitative MAGE-3/6 specific expression in UADT cancers should be investigated to determine the levels sufficient to permit HLA-A*0201:MAGE-3271–279 specific CTL recognition, which could be applied to clinical vaccine trial cohorts.
Materials and methods
Tissues and Pathological Evaluation
Tumor and normal tissue specimens were obtained from the University of Pittsburgh Medical Center through IRB approved protocols. Primary tumor or normal tissue was snap frozen in liquid nitrogen and later embedded in OCT for frozen sectioning and RNA isolation. Twenty 5-micron sections were cut from each tissue for RNA isolation. In addition, sections were cut and placed on slides for H&E analysis at the beginning, middle (between the tenth and eleventh sections for RNA), and end of the sections for RNA isolation. All three H&E slides from each specimen underwent pathological review to confirm presence of tumor, percentage of tumor, and to identify the presence of any contaminating tissues. All of the unstained slides were stored at −20°C.
Cell lines
The HLA-A*0201+ SCCHN cell lines: SCC-4, SCC-90, PCI-13, PCI-30, JHU-011, -012 (gifts of Dr James Rocco, Massachusetts, Eye and Ear Infirmary) and UD-SCC-6 were used; their characteristics and derivation have been published elsewhere12. Cells lines were kept in culture using DMEM with 8% FBS, 2% L-Glut and 1% P/S and checked for mycoplasma every 30 days. HLA-A*0201 status was determined using a combination of flow cytometry and SSCP-based PCR analysis as described13. The T2 mutant cells that lack expression of the antigen presenting machinery genes LMP2/7 and TAP1/214 were grown in AIM-V serum free media and cleaned using a ficoll gradient every 30 days.
Peptide and Tetramer
The University of Pittsburgh Peptide Synthesis facility produced the MAGE-3271–279 (FLWGPRALV) and HIV-1 POL476–484 (ILKEPVHGV) peptides, using F-MOC technology. These peptides were purified to >90% purity as confirmed by HPLC and mass spectrometry. The lyophilized peptides were re-suspended at 1mg/ml in DMSO and used at the concentrations noted. Lyophilized MAGE-3271–279 peptide was used by the NIH Tetramer Core Facility at Emory University, to produce the HLA-A* 0201:FLWGPRALV tetramer linked the phycoerythrin (PE) fluorophore.
Antibodies
The following antibodies were used in the ELISPOT assays: HLA-A, -B, -C-specific mAb W6/3215, HLA-A2 and -A68-specific mAb BB7.216, and HLA-DR, specific mAb L24317. The MAGE-A3/6 antibody (goat polyclonal IgG mAb (F-14): sc-33233 (Santa Cruz biotechnology, Inc.) and a donkey anti-goat antibody IgG-HRP: sc-2020 (Santa Cruz biotechnology,) were used for protein detection in Western blots.
ELISPOT assays
IFN-γ ELISPOT assays were performed, as described previously13. All experiments used negative and positive controls to confirm CTL specificity, consisting of T2 cells without peptide or pulsed with TA peptide (1µM for 2 hours at RT) or PMA/ionomycin-treated CTL for maximum spots, respectively. T2 cells loaded with TA peptide were incubated with eitherW6/32, BB7.2 or L243 as an irrelevant Ab. These mAb were incubated at 10 µg/ml for 30 min to confirm HLA class I and HLA A*0201 antigen restricted, but not HLA class II antigen-restricted T cell activity. Spot numbers and sizes were determined with computer-assisted video image analysis (Cellular Technologies).
Flow Cytometry
For tetramer staining, PBMC were incubated in 1% BSA/PBS for 2.5 hours in the dark at room temperature (RT) with tetramer at the concentrations noted. After this, cells were washed of unbound tetramer, chilled on ice and a cocktail of anti-CD8 CyChrome, anti-CD3-ECD (Becton Dickinson) was added and incubated for 30 minutes after which cells were washed and fixed with 2% paraformaldehyde. Flow cytometry was performed on a Beckman Coulter EPICS XL machine, and data analyzed using EXPO32 software.
RNA extraction
Total RNA from all frozen tumor samples and normal mucosal tissues was isolated using the RNeasy Mini Kit (Qiagen Inc.) according to the manufacturer’s protocol. The mucosa was carefully dissected from any subjacent muscle or lymphoid tissue before RNA extraction, so that only normal squamous epithelium was studied. The RNA pellet was dried under a vacuum and resuspended in 30–50 µl of DEPC treated water. The integrity of RNA samples was confirmed by analyzing 1 µg of total RNA on 1.2% (w/v) denaturing formaldehyde-agarose gels. Before DNA synthesis, RNA was treated with deoxyribonuclease I, Amplification Grade (Invitrogen) for 15 min at room temperature to avoid DNA contamination. DNase I was inactivated by incubation with 25 mM EDTA at 65 °C for 10 min. To extract RNA from the SCCHN cell lines used in the study, one to two million cells were plated on a T-75 flask and the media replaced with fresh media a day after. Cells were harvested the using Trypsin EDTA (Invitrogen) and high quality RNA was extracted using Trizol (Gibco, San Diego CA) and purified with the RNeasy isolation kit (Qiagen, Valencia, CA). RNA was cleaned from DNA using a DNAse (Ambion, Austin, TX) and resuspended in RNA secure solution and quantified.
Detection and quantitation of MAGE-3/6 by QRT-PCR
Expression of MAGE-3 and MAGE-6 was determined by quantitative reverse transcription polymerase chain reaction (QRT-PCR), using a previously validated assay specific for the quantification of both MAGE genes18. The following common primer and probe sequences were used to co-amplify and detect both genes: Reverse primer: GGA GAC CCA CTG GCA GAT CT, Forward primer: AGG AGG CAA GGT TCT GAG GG, Probe 5’ 6-FAM-ACA GGC TGA CCT GGA CCA GAG G/3’ 6-TAMTp. A mixture of the Universal Human Reference RNA (Stratagene, La Jolla, CA) and RNAs from human placenta, thyroid, heart, colon, PCI-13 cell line and SKBR3 cell line served as a universal positive expression control for all the genes. Reverse transcription was performed using 2 μg of RNA paired with random hexamer primers as described previously19. QRT-PCR was then carried out using the Applied Biosystems 7700 Sequence Detection Instrument. Relative expression of the MAGE gene to the endogenous control gene, β-glucuronidase (GUS), was calculated using the ΔΔCT method described previously20. All of the experiments were performed in triplicates or quadruplicates.
RNA interference (RNAi)
RNAi oligonucleotides specific for human MAGE-3/6 were purchased from Santa Cruz Biotechnology, Inc. (product sc-45284). RNAi knockdown was performed by transfection of these RNAi oligonucleotides, according to the manufacturer's protocol. Scrambled control oligonucleotides (sc-37007, Santa Cruz) were also used. Briefly, cells were seeded in 6-well plates without antibiotics and transiently transfected with control or MAGE-3/6 specific RNAi at a final concentration of 100 nmol/L by using Lipofectamine (Invitrogen).
Western Blot Analysis
HLA-A*0201+ SCCHN cell lines were homogenized and insoluble cell debris was removed by centrifugation at 12000 g for 30 minutes. Protein concentration of samples and bovine serum albumin standard were determined using the BCA™ protein assay kit (Thermo Scientific) by measurement the absorbance at or near 562 nm on a plate reader. Equal amounts of protein (50 µg) solubilized in sample buffer were separated on 10% SDS polyacrylamide gels and transferred electrophoretically to polyvinylidene difluoride membranes. Membranes were blocked in TBS containing 0.5% Tween 20 plus 5% nonfat dried milk for 1 h at room temperature and probed with primary antibodies at 4°C overnight. A 1:200 dilution of the MAGE-A3/6 antibody (goat polyclonal IgG mAb, F-14: sc-33233 and a 1:2,000 dilution of donkey anti-goat antibody IgG-HRP: sc-2020 were used for protein detection. The F-14 antibody detects a common epitope specific to the MAGE-A3 and MAGE-A6 protein. β-Actin expression was determined with a 1:4,000 dilution of an anti-β-actin mAb (Sigma-Aldrich) and used as a loading control. Quantitation was performed after densitometry of repeated experiments.
Generation of TA specific CTL by in vitro stimulation (IVS)
Specific CTL against MAGE-3271–279 were generated from HLA-A*0201+ PBMC obtained from two SCCHN patients and five healthy donors after informed consent was obtained through an IRB approved protocol. CTL were generated as described elsewhere21 with the following modifications. Monocytes were selected by plastic adherence for 2 hours at 37°C, and cultured in 10% FBS serum/IMDM with rhIL-4 and rhGM-CSF at 1000 IU/ml each for six days. Six days later, the culture medium was removed, and the immature DC were cultured in AIM-V supplemented with 1000 IU/ml rhGM-CSF and 1000 IU/ml rhIL-4, 10ng/ml rhIL-6, 10 ng/ml TNF-α, and 10 ng/ml IL-1β and 1ug/ml PGE2. Mature DC were harvested on day 8 and pulsed with MAGE-3271–279 (FLWGPRALV) peptide at 10µM for six hours at RT and then irradiated (50 Gy). Autologous CD8+ T cells were negatively isolated from PBMC with immunomagnetic beads (Miltenyi Biotech, Germany) and added (2×106) to the peptide-pulsed DC (2×105) in a final volume of 2 ml of culture medium (24-well tissue culture plate) along with 5ng/ml IL-7 and 20 units/ml IL-2. On day 7 and weekly thereafter, lymphocytes were restimulated with autologous irradiated DC pulsed with peptide in culture medium supplemented with 20 units/ml IL-2 and 5 ng/ml IL-7. The stimulated CD8+ T cells were analyzed for specificity in IFN-γ ELISPOT and 51Cr release assays at day 21 and then every 7 days after the most recent stimulation. Specific binding was also confirmed with tetramer staining as described.
Treatment with DAC (5-Aza-2'-deoxycytidine)
Treatment of the cell lines 5-Aza-2'-deoxycytidine (Sigma Corp. St. Louis Mo) was done as previously described22 with the following modifications: stocks of 1mg/ml DAC in 50% acetic acid were kept in the dark at −80C. Cells were plated in T75 flasks at a 40% confluence a day before starting the treatment. Culture media was replaced with fresh media containing 1µM DAC every 12 hours for the first 48 hours, after which the cells were left to rest for two days and then used in molecular and functional assays.
Statistical Analysis
T cell reactivity as measured by the ELISPOT assay was considered positive if test wells were significantly greater than background wells when using a two tailed permutation test at α ≤ 0.05. For comparison of MAGE–3/6 expression between cell lines, a two tailed Student’s t test was employed.
Results
MAGE-3271–279 specific CTL generated from healthy donors and SCCHN patients
CTL lines specific for HLA-A*0201:MAGE-3271–279 were generated from two SCCHN patients and from five healthy donors, as described previously6. After IVS, CTL were characterized for MAGE-3271–279 specificity using IFN-γ ELISPOT and tetramer staining, as shown in Figure 1.
Figure 1. Analysis of HLA-A*0201:MAGE-3271–279 specific CTL, cross-reactivity between MAGE-3 and MAGE-6 and avidity generated by IVS.
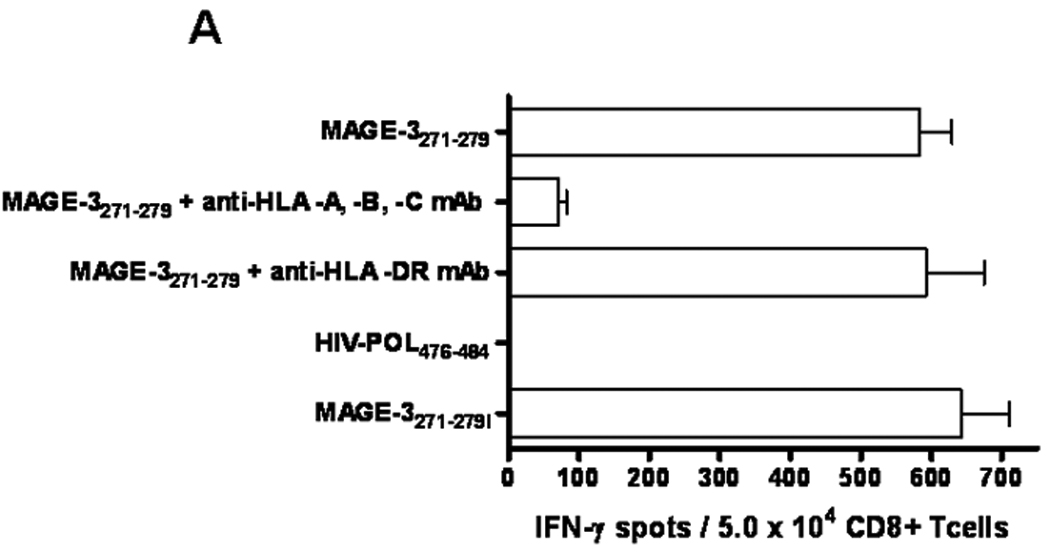
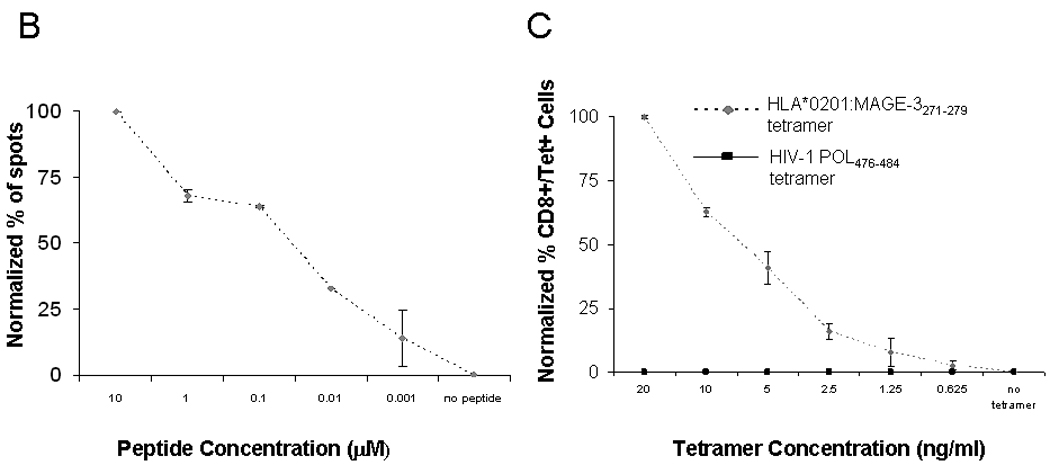
A) Reactivity of MAGE-3271–279 peptide-specific CD8+ T cells against peptide-pulsed target cells and also that recognizes the MAGE-3271–279I in ELISPOT IFN-Γ. This recognition was blocked by HLA class I and HLA-A*0201 antigen restricted, but not HLA class II antigen-restricted T cell activity and no reactivity with irrelevant peptide. Data represent the mean spot number of triplicates with 104 MAGE-3271–279 peptide-specific CD8+ T cells initially seeded per well. One representative experiment of three performed is depicted. B) IFN-γ ELISPOT assay were performed, as described previously13. T2 cells without peptide or pulsed with tenfold dilutions of the MAGE-3271–279 peptide (10µM to 0.001µM). ELISPOT IFN-γ release was normalized to the highest number of spots from each assay and plotted for comparison of CTL generated from each of the healthy donors or SCCHN patients. C) HLA-A*0201:MAGE-3271–279 tetramer binding to MAGE-3271–279 specific CTL after serial dilutions of tetramer. MAGE-3271–279 specific CTL were incubated with 6 different HLA-A*0201:MAGE-3271–279 tetramer concentrations (20 ng/ml to 0.625 ng/ml) and compared to unstimulated HLA-matched PBMC from a healthy donor using flow cytometry. The results were standardized to the percentage of cells bound to tetramer at the highest tetramer concentration.
CTL specificity and sensitivity was demonstrated by IFN-γ ELISPOT assays using T2 cells loaded with MAGE-3271–279 peptide or irrelevant, control peptide HIV-1 POL476–484. HLA class I restricted activity was determined by blockade of recognition of the MAGE-3271–279 peptide pulsed T2 cells incubated with the HLA-A, -B and –C specific mAb W6/32, but not with the HLA-DR specific blocking antibody L243 (Figure 1A). MAGE-3271–279 specific CTL recognition of T2 cells pulsed with MAGE-3271–279I (10µg/ml at 37°C, Figure 1A) indicated that MAGE-3271–279 specific CTL were cross-reactive with the MAGE-3271–279I (FLWGPRALI) peptide that is present in MAGE-6 and MAGE-12 protein, as described previously3. CTL reactivity was quantified as the number of IFN-γ spots above background (defined by reactivity to irrelevant peptide pulsed T2 cells), and confirmed by HLA-A*0201 MAGE-3271–279 loaded loaded tetramer staining, which was not observed with an HIV-1 POL476–484 loaded tetramer as a negative control or using unstimulated HLA-A*0201+ PBMC (data not shown).
To determine the sensitivity of MAGE-3271–279 specific CTL recognition, peptide titration experiments using IFN-γ ELISPOT were performed with MAGE-3271–279 pulsed T2 cells (Figure 1B). Sequential, tenfold dilutions of MAGE-3271–279 from 10µM to 0.001µM were incubated with T2 cultured in serum free media. IFN-γ ELISPOT activity of MAGE-CTL was significantly higher (p≤0.05, two tailed permutation test) than recognition of T2 cells incubated with media alone or with the control peptide, POL476–484. Reactivity of MAGE-CTL cultures was compared (at the same E:T ratio), and results were standardized using 100% as the maximal number of spots observed for that CTL culture and 0% as the number of spots obtained using T2 cells pulsed with POL476–484 peptide to allow comparisons between each cell line. Figure 1B shows a representative experiment of this titration, indicating half-maximal CTL recognition at approximately 50nM (Figure 1B).
To characterize the avidity of MAGE-CTL, these cells were incubated with decreasing tetramer concentrations, ranging from 20 ng/ml to 0.625 ng/ml, and analyzed by flow cytometry. The results of these assays were standardized to the maximal percentage of CTL bound at the highest tetramer concentration. As shown in Figure 1C, MAGE-CTL showed comparable titration curves suggesting consistency of the TCR avidity between CTL used for recognition of tumor cells.
MAGE-3271–279 specific CTL recognition does not correlate with MAGE-3 expression by RT-PCR
After the specificity and sensitivity of the MAGE-CTL lines had been determined, a panel of HLA-A*0201+ SCCHN cell lines12 were tested for CTL reactivity using IFN-γ ELISPOT or 51Cr release assays (not shown). Interestingly, only PCI-13, PCI-30, JHU-012, and UD-SCC-6 cell lines were recognized by MAGE-CTL (Figure 2). HLA class I specificity is demonstrated by blocking of IFN-γ release after pre-treatment an HLA-A,-B,-C specific mAb (W6/32), and HLA-A specific mAb (LGIII.1.47,23) but not by an HLA-DR specific mAb (Figure 2, p≤0.05, two tailed permutation test).
Figure 2. Recognition of SCCHN cell lines by HLA-A*0201:MAGE-3271–279 specific CTL.
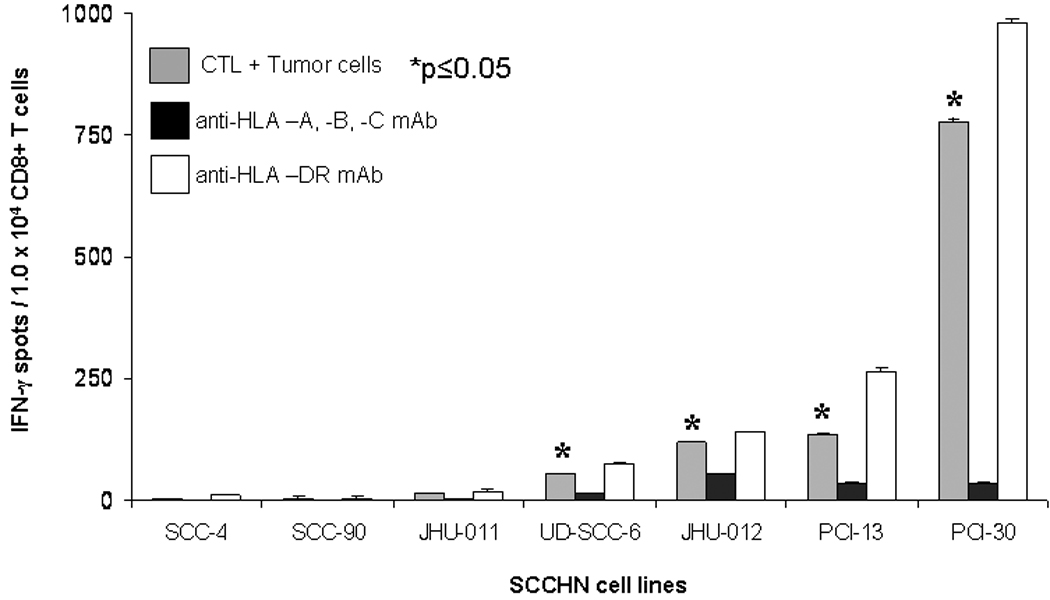
Seven HLA-A*0201+ SCCHN cell lines were incubated with HLA-A* 0201:MAGE-3271–279 specific CTL in ELISPOT IFN-γ assay, as described in Material and Methods. The HLA-A, -B, -C specific mAb W6/32 or the HLA –DR specific mAb L243 were used to determine class I restricted activity. Positive responses were scored for significant CTL recognition using the two tailed permutation test.
Due to the differential recognition of some SCCHN cells, we sought to determine if MAGE-CTL recognition was correlated with detection of MAGE-3 expression by standard RT-PCR. MAGE-CTL recognition in the panel of 7 SCCHN cells lines compared with MAGE-3 expression by standard RT-PCR, as described8. Interestingly, this experiment demonstrated a clear 90bp DNA band (Figure 3A), as predicted for the MAGE-3 gene amplification product, in the SCCHN cell lines SCC-4, SCC-90, and JHU-011, despite the lack of recognition by MAGE-3271–279 specific CTL (Figure 2). This experiment demonstrated that MAGE-3 specific CTL recognition does not reliably correlate with the expression of MAGE-3 by standard RT-PCR.
Figure 3. MAGE-3 expression in HLA-A*0201+ SCCHN cell lines.
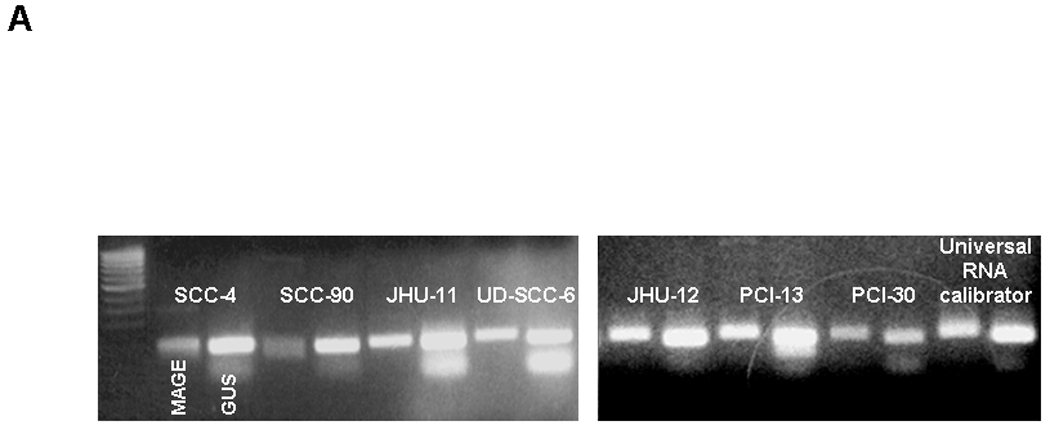
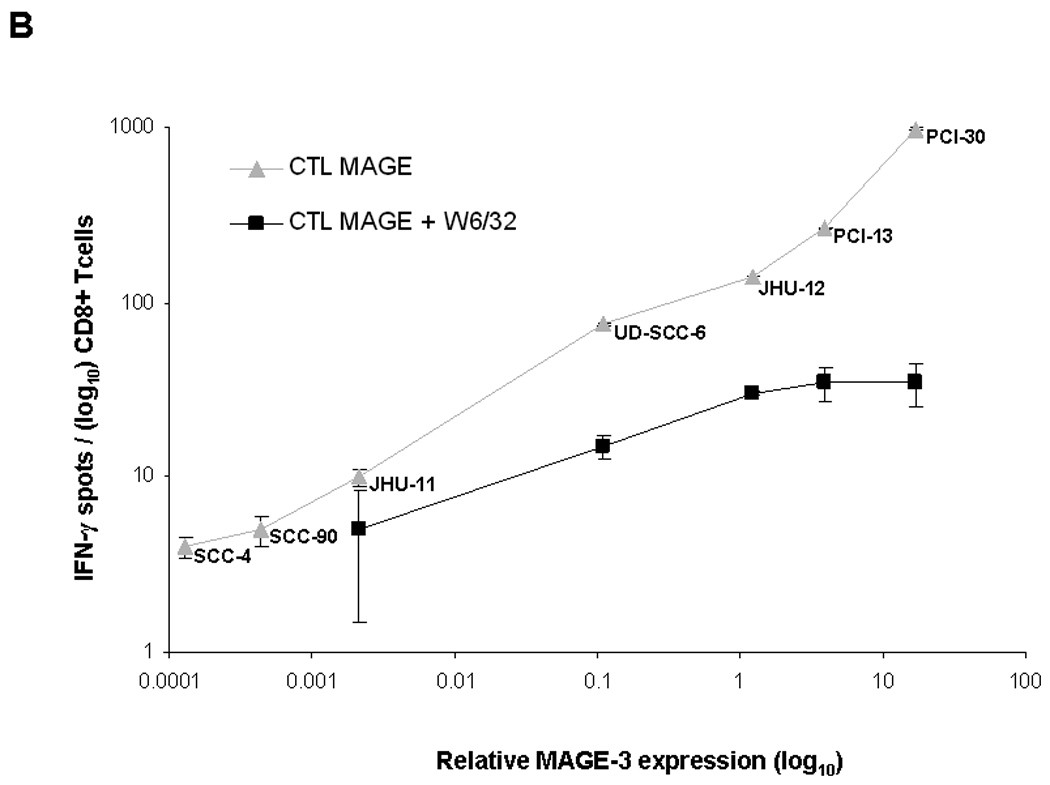
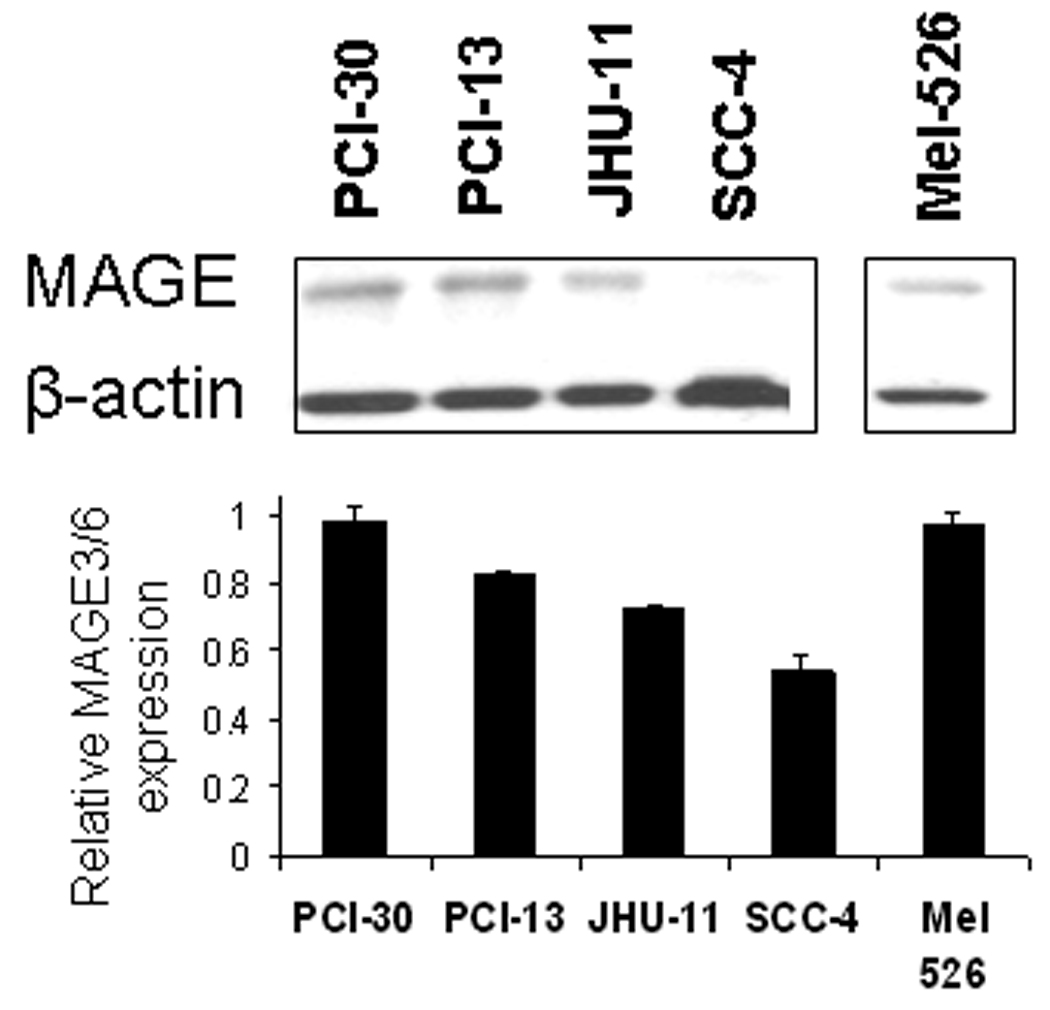
A) Standard RT-PCR was performed after RNA extraction from 7 SCCHN cell lines. Each cell line demonstrates two bands, one corresponding to the MAGE-3 gene (90 bp amplicon, right) and a second one corresponding to the control gene β-glucoronidase (81 bp amplicon GUS, left). Each cell line showed MAGE-3 bands of varying intensity, despite similar levels of GUS expression. B) MAGE-3 expression was determined in the same seven SCCHN cell lines by QRT-PCR, relative to the expression of control gene β-glucoronidase (GUS). Relative MAGE-3 expression between cell lines was determined using the 2−ΔCt method. All experiments were performed twice from individual RNA isolations and using the UR+ control RNA to compare between assays. All these seven HLA-A*0201+ SCCHN cell lines were incubated with HLA-A*0201:MAGE-3271–279 specific CTL in ELISPOT IFN-γ assay, as described in Materials and Methods. The HLA-A, -B, -C specific mAb W6/32 were used to determine class I restricted activity. C) Western blot for MAGE-3/6 expression in four SCCHN cell lines and one melanoma cell line (Mel 526). For these experiments, 50µg of total protein lysate per well from SCCHN cell lines were separated on 10% SDS polyacrylamide gels and transferred electrophoretically to polyvinylidene difluoride membranes. Membranes were probed with primary antibodies at 4°C overnight. A 1:200 dilution of the MAGE-A3/6 antibody (goat polyclonal IgG mAb, F-14: sc-33233 and a 1:2,000 dilution of donkey anti-goat antibody IgG-HRP: sc-2020 were used for protein detection. β-Actin expression was determined with a 1:4,000 dilution of an anti-β-actin mAb (Sigma-Aldrich) and used as a loading control. Quantitation was performed after densitometry of repeated experiments.
Level of MAGE-3 expression by QRT-PCR correlates with in vitro recognition of SCCHN cell lines by MAGE-3271–279 specific CTL
As shown above, we found that subset of SCCHN cell lines were not recognized by MAGE-3271–279 specific CTL despite their expression of MAGE-3 by RT-PCR. Therefore, we developed a novel QRT-PCR assay for MAGE-3/6 expression24. In order to correlate the level of MAGE-3 expression with MAGE-3271–279 specific CTL recognition, each of these seven SCCHN cell lines was analyzed by QRT-PCR for MAGE-3/6 expression relative to the control gene, GUS (Figure 3B). Quantitative level of MAGE expression was plotted against CTL recognition using IFN-γ ELISPOT, as shown in Figure 3B. QRT-PCR analysis of each cell line was repeated at least three times from separate RNA extractions to ensure reproducibility of results. This analysis indicated that relative level of MAGE-3:GUS expression correlated with recognition by MAGE-3271–279 specific CTL. Recognition was HLA class I restricted, as shown by the black curve (Figure 3B), indicating blocking of CTL recognition by incubation of tumor cells with anti-HLA-A, -B, -C specific mAb, W6/32. These data suggested that level of MAGE-3/6 expression is crucial for CTL recognition and that quantitative levels must be tested to determine whether sufficient antigen is likely to be processed and presented to CTL.
To determine whether protein expression correlated with level of MAGE-3/6 expression, western blot analysis of SCCHN cell lines was performed with anti-MAGE goat polyclonal IgG mAb (Figure 3C). Western blotting showed strong expression of MAGE-3 in MAGE-3 mRNAhigh SCCHN cells, PCI-30 and PCI-13; as determined using QRT-PCR, and confirmed very low expression in MAGE-3 mRNAlow SCCHN cells, SCC-4. Quantification by densitometry indicated the range of protein expression of approximately two-fold between high and low-expressing cell lines, as compared to roughly 200-fold variation in MAGE-3/6 expression using QRT-PCR (Figure 3B).
Knockdown of MAGE-3 reduces recognition of SCCHN cells by MAGE-3 specific CTL
To determine that the level of MAGE-3 expression was responsible for CTL recognition of tumor cells, we transfected two MAGE-3high SCCHN cell lines, PCI-13 and PCI-30, with MAGE-3/6 specific RNAi or control, scrambled oligonucleotides. These SCCHN cells were analyzed for level of MAGE-3/6 knockdown using QRT-PCR and CTL recognition, using IFN-γ ELISPOT assay. RNAi–transfected PCI-13 and PCI-30 cells showed substantially decreased MAGE expression, by 85–90% (Figure 4A), respectively, when compared with PCI-30 cells transfected with control, scrambled RNAi oligonucleotides. After RNAi-mediated MAGE-3 knockdown, we observed significantly reduced MAGE-CTL recognition of PCI-30 cells (p≤0.05, permutation test, Figure 4B).
Figure 4. Knockdown of MAGE-3/6 expression in HLA-A*0201+ SCCHN cell line PCI-30 and recognition by HLA-A*0201:MAGE-3271–279 specific CTL.
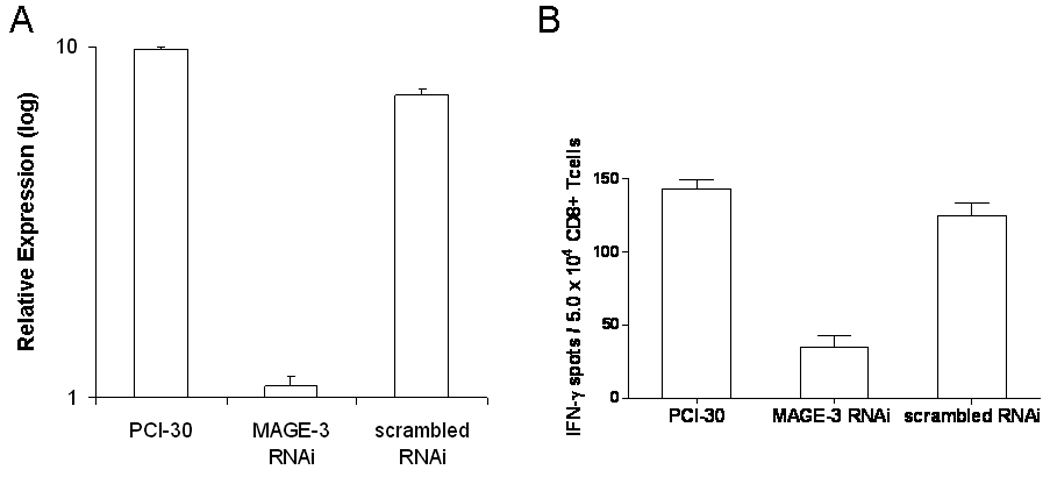
A) PCI-30 were transfected with MAGE-3/6 specific RNAi oligonucleotides to knock down MAGE-3/6 gene expression, and were analyzed using QRT-PCR. B) ELISPOT assays were performed using PCI-30 cells after MAGE-3/6 knockdown by RNAi, as described in panel A. All experiments were performed twice, and scrambled RNAi primer-transfected PCI-30 cells were used as controls.
Expression of MAGE-3 in lung, esophagus and head and neck cancers by QRT-PCR
Having determined that SCCHN cell lines with relative MAGE-3 expression >0.1 exhibited MAGE-3271–279 specific CTL recognition in vitro, we evaluated a large series of UADT cancer specimens to estimate a frequency of UADT cancer patients who would be suitable candidates for MAGE-3 targeted immunotherapy. MAGE-3 expression was quantified in carcinomas of the head and neck (n=26), esophagus (n=19) and lung (n=20) and in 48 site matched normal tissues (Figure 5A–C). A universal RNA calibrator was used to normalize QRT-PCR results between cell lines and tissues from each site and to calculate relative MAGE-3 expression values, as described24. Using the relative MAGE-3:GUS expression to endogenous ratio >0.1, we predict that sufficient MAGE-3 expression for CTL recognition would be present in approximately 54% of SCCHN, 32% of esophageal and 55% of lung cancer specimens. Overall, approximately 41% of the UADT patients would be classified as suitable MAGE-3 specific vaccine candidates.
Figure 5. MAGE-3 expression in UADT cancers.
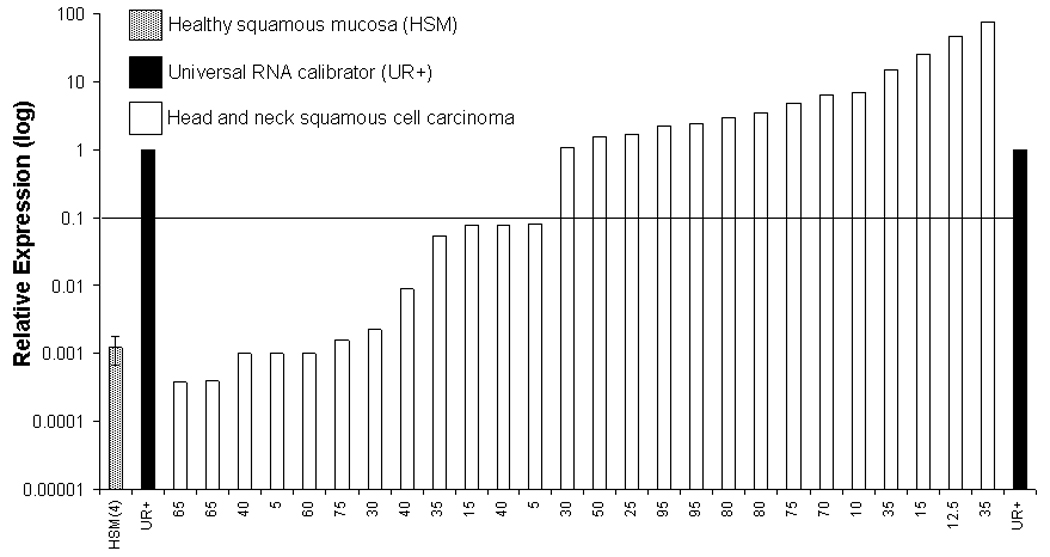
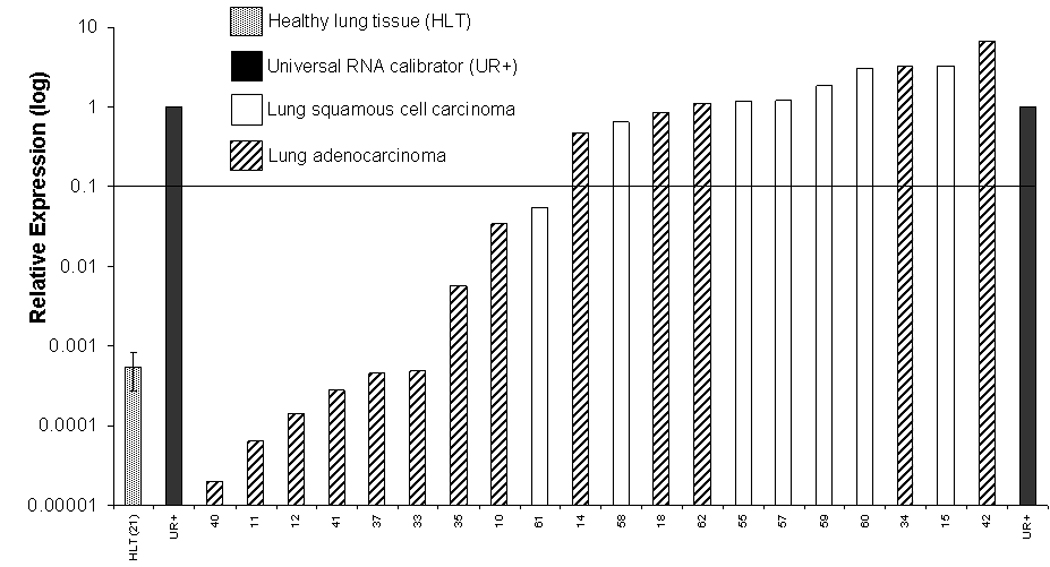
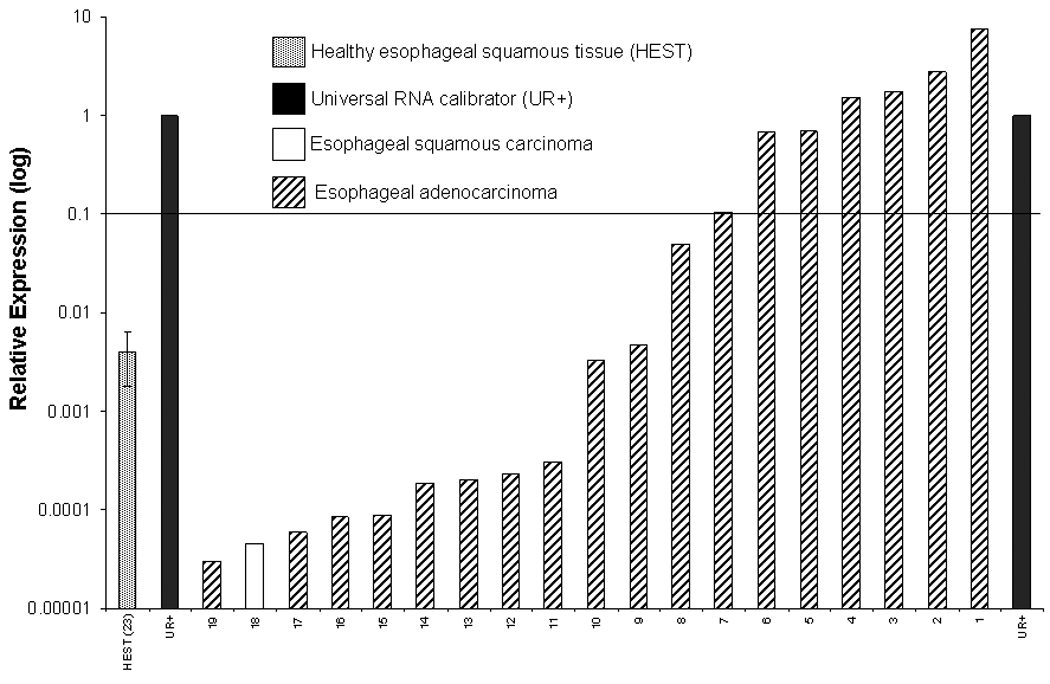
A) Relative expression of MAGE-3 by QRT-PCR in 26 SCCHN and four healthy squamous mucosal samples (HSM). B) Relative expression of MAGE-3 determined by QRT-PCR in 20 squamous or adenocarcinomas of the lung and 21 healthy lung tissues (HLT). C) Relative expression of MAGE-3 determined by QRT-PCR in 19 squamous or adenocarcinomas of the esophagus and 23 healthy esophageal squamous tissues (HEST).
Induced MAGE-3 gene expression by demethylation results in MAGE-3271–279 specific CTL recognition
MAGE-3low SCCHN cells, SCC-4 and SCC-90, demonstrated nearly undetectable MAGE-3 expression, based on QRT-PCR and were not recognized in vitro by MAGE-3271–279 specific CTL (Figure 2). Treatment of these cells with the demethylating agent 5-Aza-2'-deoxycytidine (DAC,25) upregulated MAGE-3 expression to a level sufficient for MAGE-3271–279 specific CTL recognition. DAC treatment also increased the relative expression of MAGE-3 100–1000 fold as determined by QRT-PCR, and resulted in MAGE-3271–279 specific CTL recognition in both SCCHN cells tested (p≤0.05, permutation test, Figure 6). These results are consistent with a level of MAGE-3 relative expression necessary for specific recognition by CTL in vitro.
Figure 6. MAGE-3 expression and HLA-A*0201:MAGE-3271–279 specific CTL recognition of the SCCHN cell line SCC4 and SCC-90 after treatment with demethylating agent, DAC.
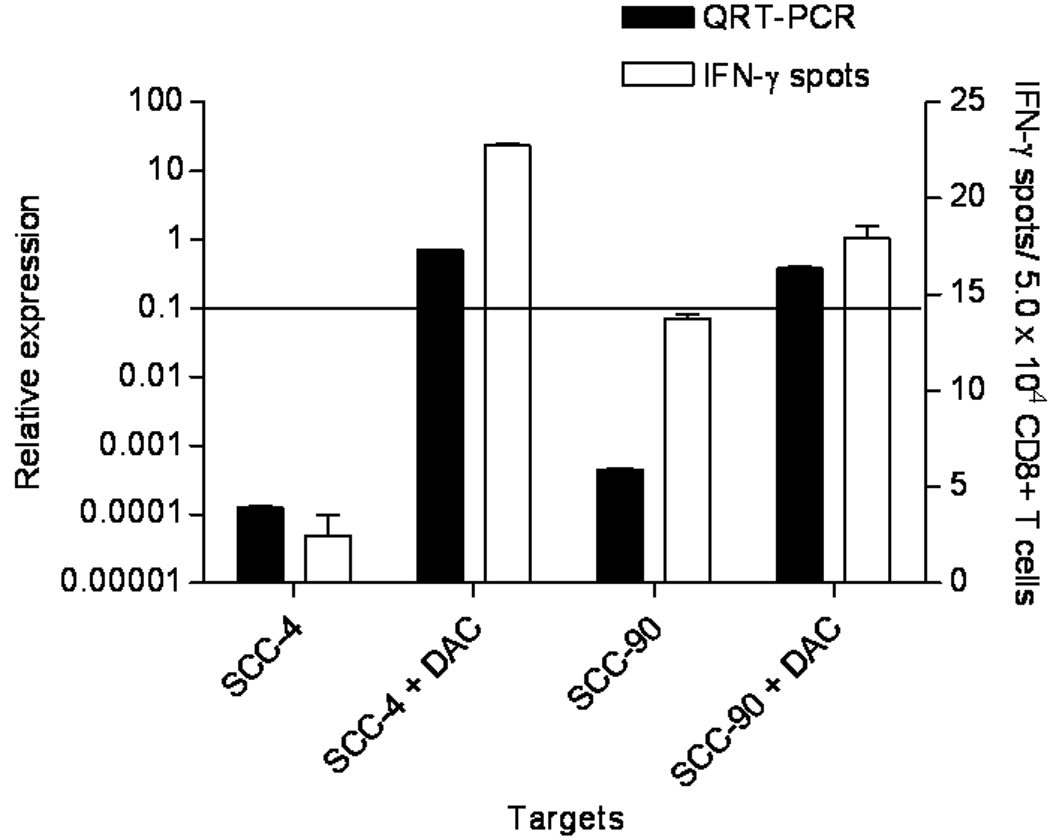
The MAGE-3low SCCHN cell line SCC-4 and SCC-90 were treated with 5-AZA as described in Materials and Methods and analyzed for expression of MAGE-3 by QRT-PCR. Under these conditions we also tested HLA-A*0201:MAGE-3271–279 specific CTL recognition of the same SCC-4 and SCC-90 cells in an ELISPOT IFN-γ assay. Dark bars represent the relative MAGE-3 expression and white bars represent number of IFN-γ spots in ELISPOT. Results were analyzed for statistical significance after DAC treatment.
Discussion
MAGE gene products appear to be promising targets for T cell based immunotherapy of UADT and phase III clinical trial evaluation is ongoing in lung cancer patients. The selection of appropriate candidates for TA specific vaccination requires an understanding of the determinants of MAGE-3/6 expression. We sought use a novel QRT-PCR assay, in which MAGE-3/6 expression was strongly associated with likelihood of recognition by MAGE-specific CTL. Thus, selection of appropriate candidates for MAGE immunotherapy may be facilitated based on the quantitative level of MAGE expression. In addition, those tumor cells with low levels of MAGE-3 relative expression and not recognized by MAGE-3271–279 specific CTL recognition, might become suitable candidates for MAGE specific immunotherapy after treatment with demethylating agents such as DAC. These results need to be validated in larger cohorts and in clinical trial specimens.
MAGE-3 was found at highly variable levels in different UADT cancer specimens (Figure 6) and 40–50% of the tumors and cell lines included in our study. Interestingly, while Western blot analysis generally indicated correlation of mRNA levels with protein expression, the range of variation between SCCHN cells was much lower using the immunoblotting technique than QRT-PCR. The analysis presented here confirms that UADT cancers are good targets for MAGE-specific immunotherapy. The clinical value of MAGE-3 as a TA in UADT cancer patients is also supported by our findings of the retention of moderate to high avidity CTL in circulating PBMC from SCCHN patients. The MAGE-3271–279 peptide serial dilutions and HLA-A*0201-MAGE–3271–279 tetramer titration studies indicate recognition of this HLA:TA peptide complexes at peptide concentrations of 10−8 M using CTL expanded from HLA-A*0201 healthy donors or SCCHN PBMC. Thus, the theoretical advantage of cancer-testis TA such as MAGE-3 gene products probably due to lack of deletion of high avidity T cells is supported by our studies. Likewise using DC-based IVS, we were able to generate cytolytic phenotype effector T cells based on their CD56 expression by flow cytometry (data not shown). Measurement of expression and reactivity of MAGE-3271–279 specific CTL in vivo should include such TCR avidity considerations as well as level of antigen expression and clinical vaccine responses.
While others have shown that HLA-A2+-MAGE-3/6+ melanoma cell lines are not always recognized by HLA-A2-MAGE-3271–279 specific CTL in vitro, immune responses against this epitope have been found in MAGE-3 vaccinated melanoma patients in vivo3, 26. Furthermore, we have previously shown that the MAGE-3271–279 epitope is processed and presented in SCCHN cell lines6. In this study, HLA-A*0201-MAGE-3271–279 specific CTL have been isolated from SCCHN patient PBMC for the first time, characterized and used against several HLA-A*0201+-MAGE-3+ cell lines, showing that the MAGE-3271–279 epitope is widely processed and presented in SCCHN cells. While we have not identified a specific cutoff level of expression, we would suggest that QRT-PCR may be used to identify those with very low levels of mRNA for MAGE-3/6 who are unlikely to benefit from MAGE-specific immunotherapy if it is not appreciably expressed in their tumors.
Our study differs with the reports by others using QRT-PCR in melanoma cells with correlation to CTL responses27, 28 besides the etiologic differences, UADT malignancies often differ from more immunogenic cancers such as melanoma in TA expression, antigen processing29 and CTL recognition, and mechanisms of immune escape. Vaccine approaches must take such variation between cancer types into account. Since MAGE gene products are not expressed in non-neoplastic tissues (except for germinal cells or the thymus), it has been proposed that T cell tolerance would be less imprinted as with other wild type “self” antigens such as HER2 30 or wild type p53 peptides 31. Our data comparing frequency and avidity of CTL from healthy and SCCHN donor PBMC are consistent with this notion.
MAGE-3 expression was measured relative to GUS in each experiment and normalized to a standard RNA sample (the calibrator) in order to compare data from different PCR runs on different days. Absolute quantification on a per cell level may not be possible or necessary in general for any diagnostic test. The important feature is that MAGE-3/6 expression should be measured in a reliable, standardized assay that correlates with CTL recognition. The precise nature of this cut-off (relative copy number, absolute copy number, delta Ct etc) will depend on the qPCR approach used (including the primers, instrumentation, detection methodology, analysis settings etc.) and will likely be refined by future experiments on larger UADT sample sets.
Despite relatively low expression of MAGE-3 as determined by QRT-PCR HLAA* 0201 MAGE271–279 specific CTL recognition was observed. This finding implies that a wider group of SCCHN patients could be vaccinated than might be initially considered. We propose that MAGE–3 expression be determined in vivo with regard to its quantitative level of expression, since we found a strong correlation between level of MAGE-3/6 and magnitude of CTL recognition of individual SCCHN cell lines. A limitation of our study was that amplification of MAGE-3 and -6 can occur due to the high degree of homology between these genes, and the peptide encoded by MAGE-2, -3, -6, and -12 may be recognized by our cross-reactive CTL. However, RNAi-mediated knockdown of MAGE-3/6 using specific oligonucleotide primers led to significant reduction in CTL recognition of the SCCHN cells, suggesting these genes as the source of the processed peptide. A future study will be necessary to correlate specific peptide epitopes expressed in cells due top expression of each MAGE gene, perhaps using mass spectrometric techniques. Nonetheless, our findings reinforce the potential value of MAGE-specific immunotherapeutic strategies for UADT malignancies. With the use of 5-AZA-Cdr agent (DAC), the transcriptional silencing of a MAGE-3 gene could be partially reversed in cancer cells, which were subsequently and recognized by CTL. This is a potential approach to enhance the effectiveness of immunotherapy.
MAGE expression may be regulated both by transcriptional epigenetic mechanisms as well as post-transcriptionally. Hypermethylated regions leading to transcriptional silencing of tumor suppressor and DNA repair genes have often been detected in malignant cells 32–34. In general human cancer have high levels of CpG islands hypermethylation and it is one of the complex process of carcinogenesis, and acts directly. Loriot et al. reported that during the neoplastic process cancerous cells undergo a transient phase of epigenetic instability leading to demethylation of the MAGE-A1 gene 35. The MAGE-A3 promoter region from −198 to +131 after DNA sequencing following bisulfite treatment includes 19 CpG islands, which may become methylated directly 32–34, 36. MAGE-A6 shares a common promoter with MAGE-A3 and there for is likely regulated together to a large degree with demythylating agents 37. A large CpG island is located in the promoter region surrounding the noncoding exon 1 of human MAGE-A3 DNA 38. Various primers designed for the MAGE-A3 promoter often coamplify the MAGE-A6 promoter, which is indistinguishable from MAGE-A3 promoter CpG islands 37. We cannot exclude that the beneficial effect of DAC on these cell lines is partially the resulting effect of upregulation of HLA-encoded gene products, leading to enhanced CTL recognition.
As a hypo-methylating agent, DAC has been reported to induce re-expression of methylated gene 39, suppressing the tumor proliferation, but others have shown that DAC causes DNA damage due to structural instability at its incorporation sites. It may also induce chromatin modifications elading to altered gene expression. DNA methyltransferase inhibitors can induce cytotoxicity in human cancer cell lines by genotoxicity and demethylation of genomic DNA. It is difficult to easily determine whether DAC acts directly and primarily through effects on DNA methylation or through other means. In any case, the basic mechanisms of the actions of DAC are complex, and the details of the molecular mechanism and pathway involved in the interaction of DAC leading to enhanced gene expression is the topic of another study. Thus, DAC may act directly as well as indirectly, but the ultimate outcome of MAGE upregulation would in theory result in enhancement of MAGE peptides presented and MAGE-CTL recognition of tumor cells.
While upregulation of HLA class I antigens may occur, we determined that a strong correlation existed between MAGE-3 upregulation and CTL recognition after DAC treatment. If this is the case with normal tissues, treatment with this agent could worsen autoimmunity of normal tissues induced by vaccination. However, autoimmunity has not been observed clinically in DAC treated patients40. Furthermore, some of the toxicities pertaining to vaccination might be tolerable if anti-tumor activity is observed. The high rate of recurrence in these patients and second primary tumors of the UADT merit novel approaches such as vaccines, which may induce anti-tumor activity and long term memory. Clinical trials testing this hypothesis are necessary to definitively address this potential issue. For this reason, we suggest the incorporation of a QRT-PCR assay or similar quantitative assay in any current or future MAGE vaccine trials.
Acknowledgements
This study was funded by R01 CA111806.
References
- 1.Vikram B, Strong EW, Shah JP, Spiro R. Second malignant neoplasms in patients successfully treated with multimodality treatment for advanced head and neck cancer. Head Neck Surg. 1984;6:734–737. doi: 10.1002/hed.2890060306. [DOI] [PubMed] [Google Scholar]
- 2.De Plaen E, Arden K, Traversari C, Gaforio JJ, Szikora JP, De Smet C, Brasseur F, van der Bruggen P, Lethe B, Lurquin C, et al. Structure, chromosomal localization, and expression of 12 genes of the MAGE family. Immunogenetics. 1994;40:360–369. doi: 10.1007/BF01246677. [DOI] [PubMed] [Google Scholar]
- 3.Gajewski TF, Fallarino F, Ashikari A, Sherman M. Immunization of HLA-A2+ melanoma patients with MAGE-3 or MelanA peptide-pulsed autologous peripheral blood mononuclear cells plus recombinant human interleukin 12. Clin Cancer Res. 2001;7:895s–901s. [PubMed] [Google Scholar]
- 4.Valmori D, Lienard D, Waanders G, Rimoldi D, Cerottini JC, Romero P. Analysis of MAGE-3-specific cytolytic T lymphocytes in human leukocyte antigen-A2 melanoma patients. Cancer Res. 1997;57:735–741. [PubMed] [Google Scholar]
- 5.Zerbini A, Pilli M, Soliani P, Ziegler S, Pelosi G, Orlandini A, Cavallo C, Uggeri J, Scandroglio R, Crafa P, Spagnoli GC, Ferrari C, et al. Ex vivo characterization of tumor-derived melanoma antigen encoding gene-specific CD8+cells in patients with hepatocellular carcinoma. J Hepatol. 2004;40:102–109. doi: 10.1016/s0168-8278(03)00484-7. [DOI] [PubMed] [Google Scholar]
- 6.Lopez-Albaitero A, Nayak JV, Ogino T, Machandia A, Gooding W, DeLeo AB, Ferrone S, Ferris RL. Role of antigen-processing machinery in the in vitro resistance of squamous cell carcinoma of the head and neck cells to recognition by CTL. J Immunol. 2006;176:3402–3409. doi: 10.4049/jimmunol.176.6.3402. [DOI] [PubMed] [Google Scholar]
- 7.Atanackovic D, Altorki NK, Stockert E, Williamson B, Jungbluth AA, Ritter E, Santiago D, Ferrara CA, Matsuo M, Selvakumar A, Dupont B, Chen YT, et al. Vaccine-induced CD4+ T cell responses to MAGE-3 protein in lung cancer patients. J Immunol. 2004;172:3289–3296. doi: 10.4049/jimmunol.172.5.3289. [DOI] [PubMed] [Google Scholar]
- 8.Kienstra MA, Neel HB, Strome SE, Roche P. Identification of NY-ESO-1, MAGE-1, and MAGE-3 in head and neck squamous cell carcinoma. Head Neck. 2003;25:457–463. doi: 10.1002/hed.10223. [DOI] [PubMed] [Google Scholar]
- 9.Eura M, Ogi K, Chikamatsu K, Lee KD, Nakano K, Masuyama K, Itoh K, Ishikawa T. Expression of the MAGE gene family in human head-and-neck squamous-cell carcinomas. Int J Cancer. 1995;64:304–308. doi: 10.1002/ijc.2910640504. [DOI] [PubMed] [Google Scholar]
- 10.Lee KD, Eura M, Ogi K, Nakano K, Chikamatsu K, Masuyama K, Ishikawa T. Expression of the MAGE-1, -2, -3, -4, and -6 genes in non-squamous cell carcinoma lesions of the head and neck. Acta Otolaryngol. 1996;116:633–639. doi: 10.3109/00016489609137901. [DOI] [PubMed] [Google Scholar]
- 11.Park JW, Kwon TK, Kim IH, Sohn SS, Kim YS, Kim CI, Bae OS, Lee KS, Lee KD, Lee CS, Chang HK, Choe BK, et al. A new strategy for the diagnosis of MAGE-expressing cancers. J Immunol Methods. 2002;266:79–86. doi: 10.1016/s0022-1759(02)00105-9. [DOI] [PubMed] [Google Scholar]
- 12.Lin CJ, Grandis JR, Carey TE, Gollin SM, Whiteside TL, Koch WM, Ferris RL, Lai SY. Head and neck squamous cell carcinoma cell lines: established models and rationale for selection. Head Neck. 2007;29:163–188. doi: 10.1002/hed.20478. [DOI] [PubMed] [Google Scholar]
- 13.Sirianni N, Ha PK, Oelke M, Califano J, Gooding W, Westra W, Whiteside TL, Koch WM, Schneck JP, DeLeo A, Ferris RL. Effect of human papillomavirus-16 infection on CD8+ T-cell recognition of a wild-type sequence p53264-272 peptide in patients with squamous cell carcinoma of the head and neck. Clin Cancer Res. 2004;10:6929–6937. doi: 10.1158/1078-0432.CCR-04-0672. [DOI] [PubMed] [Google Scholar]
- 14.Salter RD, Cresswell P. Impaired assembly and transport of HLA-A and -B antigens in a mutant TxB cell hybrid. Embo J. 1986;5:943–949. doi: 10.1002/j.1460-2075.1986.tb04307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parham PBC, Bodmer WF. Use of a monoclonal antibody (W6/32) in structural studies of HLA-A,B,C, antigens. J Immunol. 1979;123:342–349. [PubMed] [Google Scholar]
- 16.Parham PBW. Monoclonal antibody to a human histocompatibility alloantigen, HLA-A2. Nature. 1978;276:397–399. doi: 10.1038/276397a0. [DOI] [PubMed] [Google Scholar]
- 17.Goldman JM, Hibbin J, Kearney L, Orchard K, Th'ng KH. HLA-DR monoclonal antibodies inhibit the proliferation of normal and chronic granulocytic leukaemia myeloid progenitor cells. Br J Haematol. 1982;52:411–420. doi: 10.1111/j.1365-2141.1982.tb03910.x. [DOI] [PubMed] [Google Scholar]
- 18.Ferris RL, Xi L, Raja S, J.L H, Wang J, Gooding W, Kelly L, Ching J, Godfrey TE. Application of novel tumor markers for molecular staging of cervical lymph nodes in squamous cell carcinoma of the head and neck (SCCHN) Cancer Research. 2005 doi: 10.1158/0008-5472.CAN-04-3717. In press. [DOI] [PubMed] [Google Scholar]
- 19.Wang J, Xi L, Hunt JL, Gooding W, Whiteside TL, Chen Z, Godfrey TE, Ferris RL. Expression pattern of chemokine receptor 6 (CCR6) and CCR7 in squamous cell carcinoma of the head and neck identifies a novel metastatic phenotype. Cancer Res. 2004;64:1861–1866. doi: 10.1158/0008-5472.can-03-2968. [DOI] [PubMed] [Google Scholar]
- 20.ABI Prism 7700 Sequence Detection System Technical Bulletin 2. Relative Quantitaion of Gene expression: Applied Biosciences. 1997 [Google Scholar]
- 21.Mailliard RBW-KA, Cai Q, Wesa A, Hilkens CM, Kapsenberg ML, Kirkwood JM, Storkus WJ, Kalinski P. alpha-type-1 polarized dendritic cells: a novel immunization tool with optimized CTL-inducing activity. Cancer Research. 2004;64:5934–5937. doi: 10.1158/0008-5472.CAN-04-1261. [DOI] [PubMed] [Google Scholar]
- 22.Coral S, Sigalotti L, Altomonte M, Engelsberg A, Colizzi F, Cattarossi I, Maraskovsky E, Jager E, Seliger B, Maio M. 5-aza-2'-deoxycytidine-induced expression of functional cancer testis antigens in human renal cell carcinoma: immunotherapeutic implications. Clin Cancer Res. 2002;8:2690–2695. [PubMed] [Google Scholar]
- 23.Wang X, Liang B, Rebmann V, Lu J, Celis E, Kageshita T, Grosse-Wilde H, Ferrone S. Specificity and functional characteristics of anti-HLA-A mAbs LGIII-147.4.1 and LGIII-220.6.2. Tissue Antigens. 2003;62:139–148. doi: 10.1034/j.1399-0039.2003.00087.x. [DOI] [PubMed] [Google Scholar]
- 24.Ferris RL, Xi L, Raja S, Hunt JL, Wang J, Gooding WE, Kelly L, Ching J, Luketich JD, Godfrey TE. Molecular staging of cervical lymph nodes in squamous cell carcinoma of the head and neck. Cancer Res. 2005;65:2147–2156. doi: 10.1158/0008-5472.CAN-04-3717. [DOI] [PubMed] [Google Scholar]
- 25.Serrano A, Tanzarella S, Lionello I, Mendez R, Traversari C, Ruiz-Cabello F, Garrido F. Rexpression of HLA class I antigens and restoration of antigen-specific CTL response in melanoma cells following 5-aza-2'-deoxycytidine treatment. Int J Cancer. 2001;94:243–251. doi: 10.1002/ijc.1452. [DOI] [PubMed] [Google Scholar]
- 26.Paczesny S, Banchereau J, Wittkowski KM, Saracino G, Fay J, Palucka AK. Expansion of melanoma-specific cytolytic CD8+ T cell precursors in patients with metastatic melanoma vaccinated with CD34+ progenitor-derived dendritic cells. J Exp Med. 2004;199:1503–1511. doi: 10.1084/jem.20032118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lethe B, van der Bruggen P, Brasseur F, Boon T. MAGE-1 expression threshold for the lysis of melanoma cell lines by a specific cytotoxic T lymphocyte. Melanoma research. 1997;7 Suppl 2:S83–S88. [PubMed] [Google Scholar]
- 28.Riker AI, Kammula US, Panelli MC, Wang E, Ohnmacht GA, Steinberg SM, Rosenberg SA, Marincola FM. Threshold levels of gene expression of the melanoma antigen gp100 correlate with tumor cell recognition by cytotoxic T lymphocytes. Int J Cancer. 2000;86:818–826. doi: 10.1002/(sici)1097-0215(20000615)86:6<818::aid-ijc10>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 29.Campoli M, Chang CC, Ferrone S. HLA class I antigen loss, tumor immune escape and immune selection. Vaccine. 2002;20 Suppl 4:A40–A45. doi: 10.1016/s0264-410x(02)00386-9. [DOI] [PubMed] [Google Scholar]
- 30.Kobayashi H, Wood M, Song Y, Appella E, Celis E. Defining promiscuous MHC class II helper T-cell epitopes for the HER2/neu tumor antigen. Cancer Res. 2000;60:5228–5236. [PubMed] [Google Scholar]
- 31.Theobald M, Biggs J, Dittmer D, Levine AJ, Sherman LA. Targeting p53 as a general tumor antigen. Proc Natl Acad Sci U S A. 1995;92:11993–11997. doi: 10.1073/pnas.92.26.11993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qiu G, Fang J, He Y. 5' CpG island methylation analysis identifies the MAGE-A1 and MAGE-A3 genes as potential markers of HCC. Clin Biochem. 2006;39:259–266. doi: 10.1016/j.clinbiochem.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 33.Wischnewski F, Friese O, Pantel K, Schwarzenbach H. Methyl-CpG binding domain proteins and their involvement in the regulation of the MAGE-A1, MAGE-A2, MAGE-A3, and MAGE-A12 gene promoters. Mol Cancer Res. 2007;5:749–759. doi: 10.1158/1541-7786.MCR-06-0364. [DOI] [PubMed] [Google Scholar]
- 34.Wischnewski F, Pantel K, Schwarzenbach H. Promoter demethylation and histone acetylation mediate gene expression of MAGE-A1, -A2, -A3, and -A12 in human cancer cells. Mol Cancer Res. 2006;4:339–349. doi: 10.1158/1541-7786.MCR-05-0229. [DOI] [PubMed] [Google Scholar]
- 35.Loriot A, Boon T, De Smet C. Five new human cancer-germline genes identified among 12 genes expressed in spermatogonia. Int J Cancer. 2003;105:371–376. doi: 10.1002/ijc.11104. [DOI] [PubMed] [Google Scholar]
- 36.Zhu X, Asa SL, Ezzat S. Fibroblast growth factor 2 and estrogen control the balance of histone 3 modifications targeting MAGE-A3 in pituitary neoplasia. Clin Cancer Res. 2008;14:1984–1996. doi: 10.1158/1078-0432.CCR-07-2003. [DOI] [PubMed] [Google Scholar]
- 37.Kondo T, Zhu X, Asa SL, Ezzat S. The cancer/testis antigen melanoma-associated antigen-A3/A6 is a novel target of fibroblast growth factor receptor 2-IIIb through histone H3 modifications in thyroid cancer. Clin Cancer Res. 2007;13:4713–4720. doi: 10.1158/1078-0432.CCR-07-0618. [DOI] [PubMed] [Google Scholar]
- 38.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 39.Juttermann R, Li E, Jaenisch R. Toxicity of 5-aza-2'-deoxycytidine to mammalian cells is mediated primarily by covalent trapping of DNA methyltransferase rather than DNA demethylation. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:11797–11801. doi: 10.1073/pnas.91.25.11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richardson BC. Role of DNA methylation in the regulation of cell function: autoimmunity, aging and cancer. J Nutr. 2002;132:2401S–2405S. doi: 10.1093/jn/132.8.2401S. [DOI] [PubMed] [Google Scholar]


