Abstract
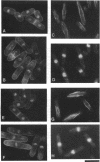
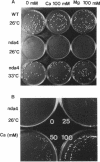
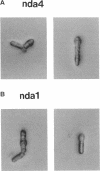
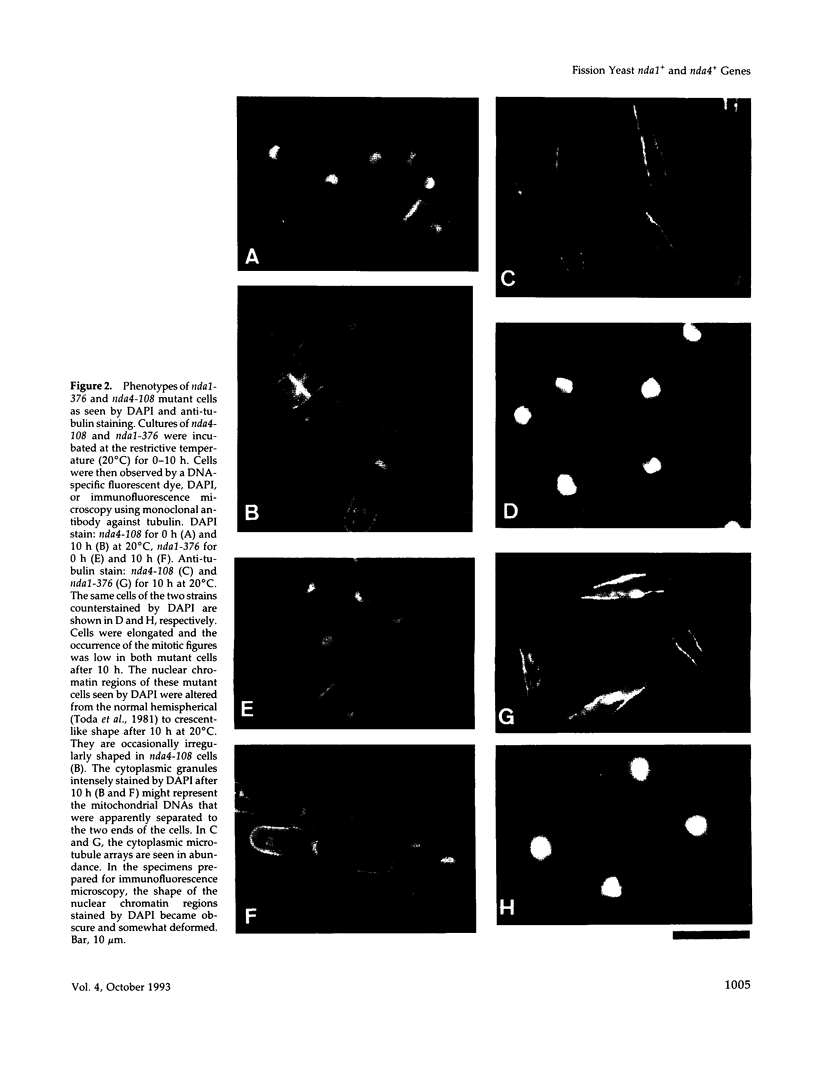
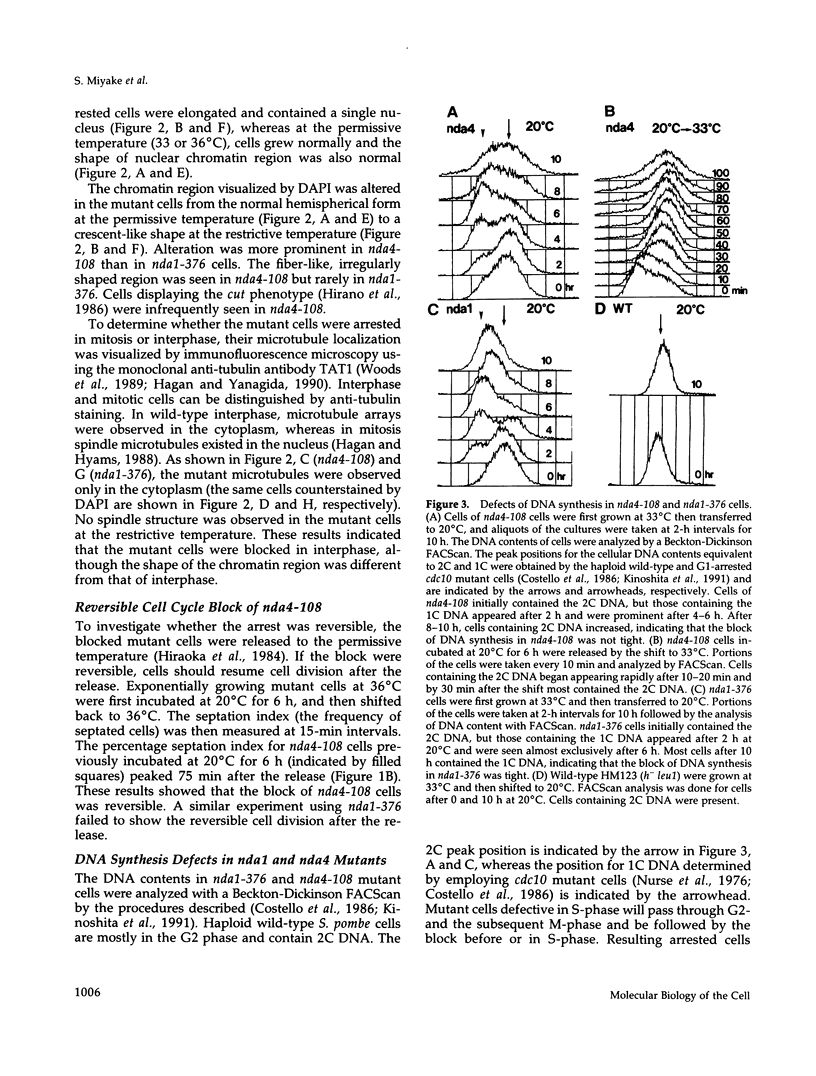
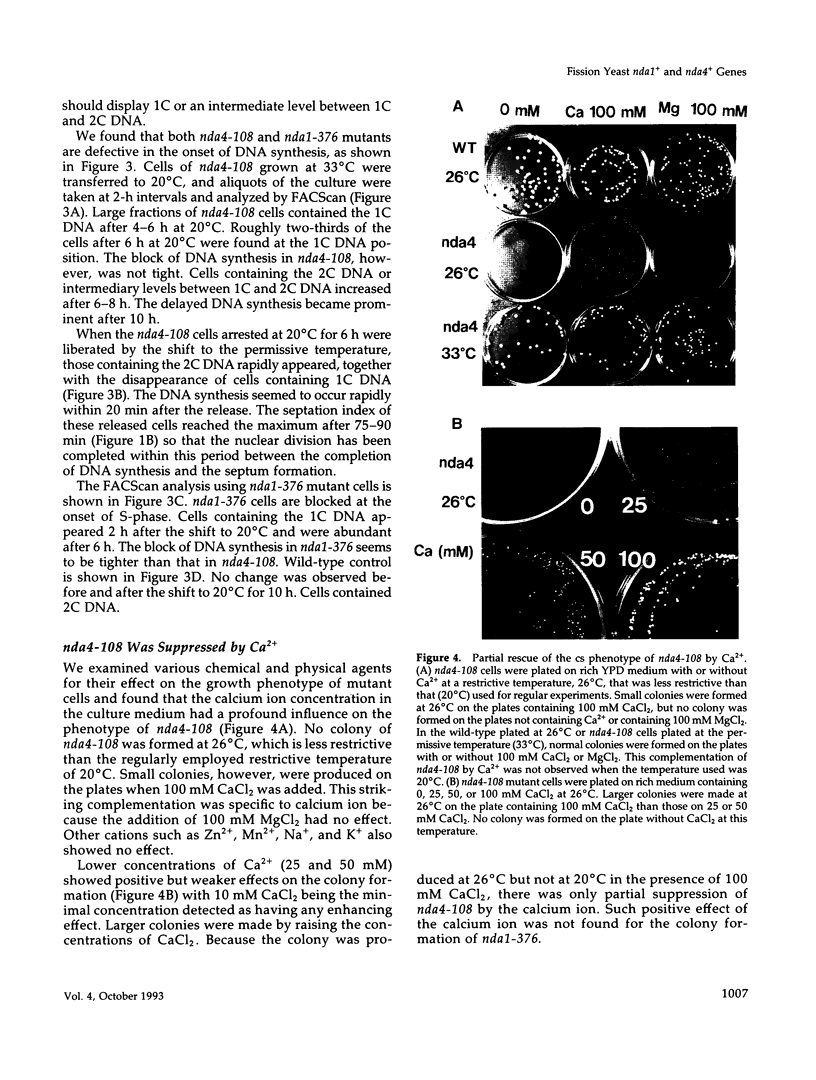
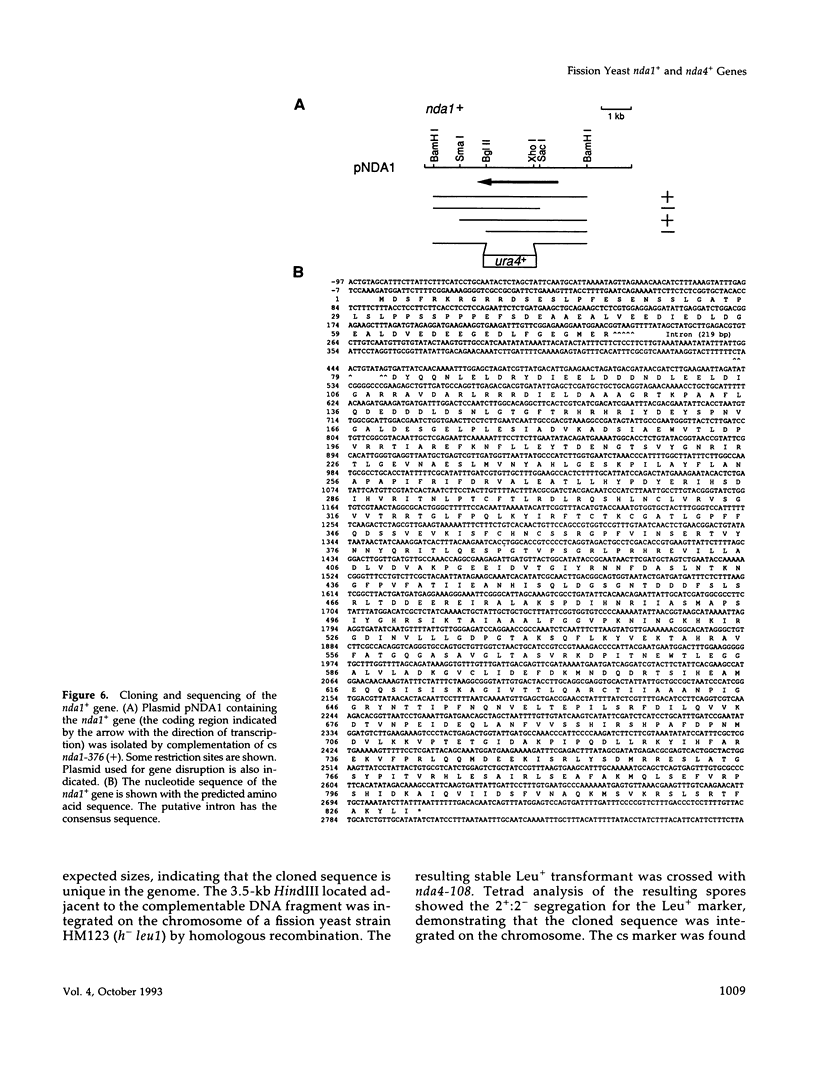
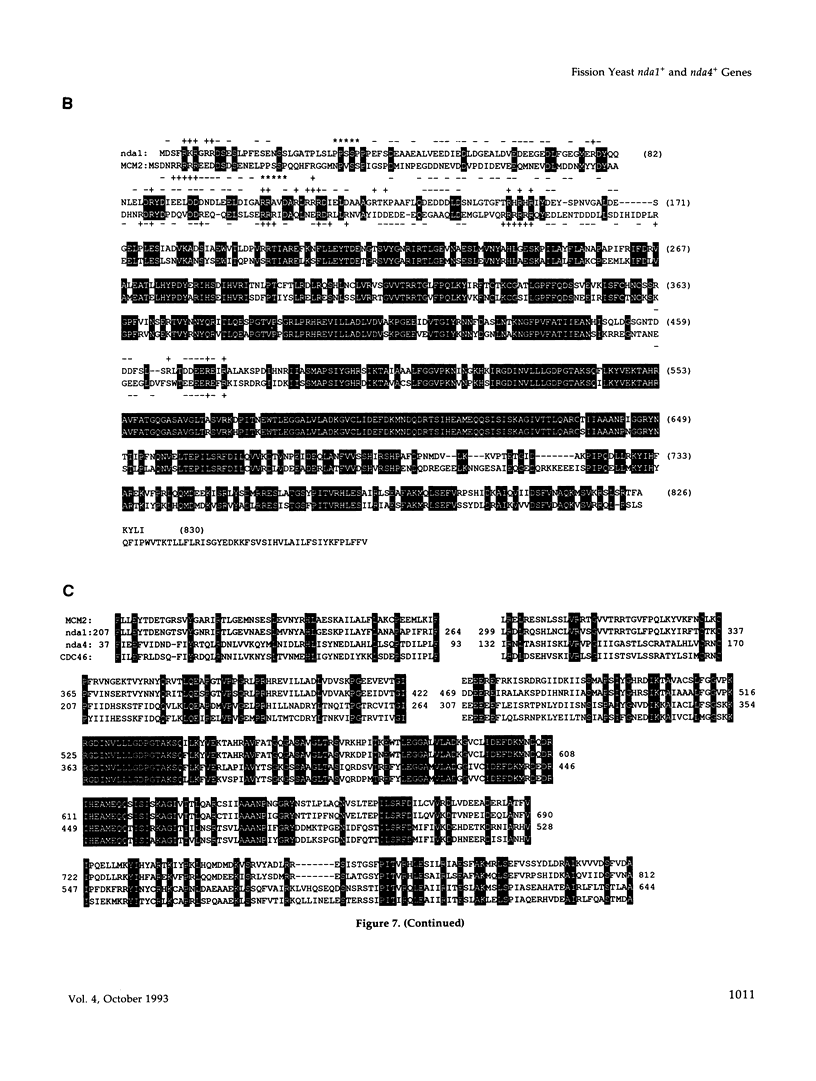
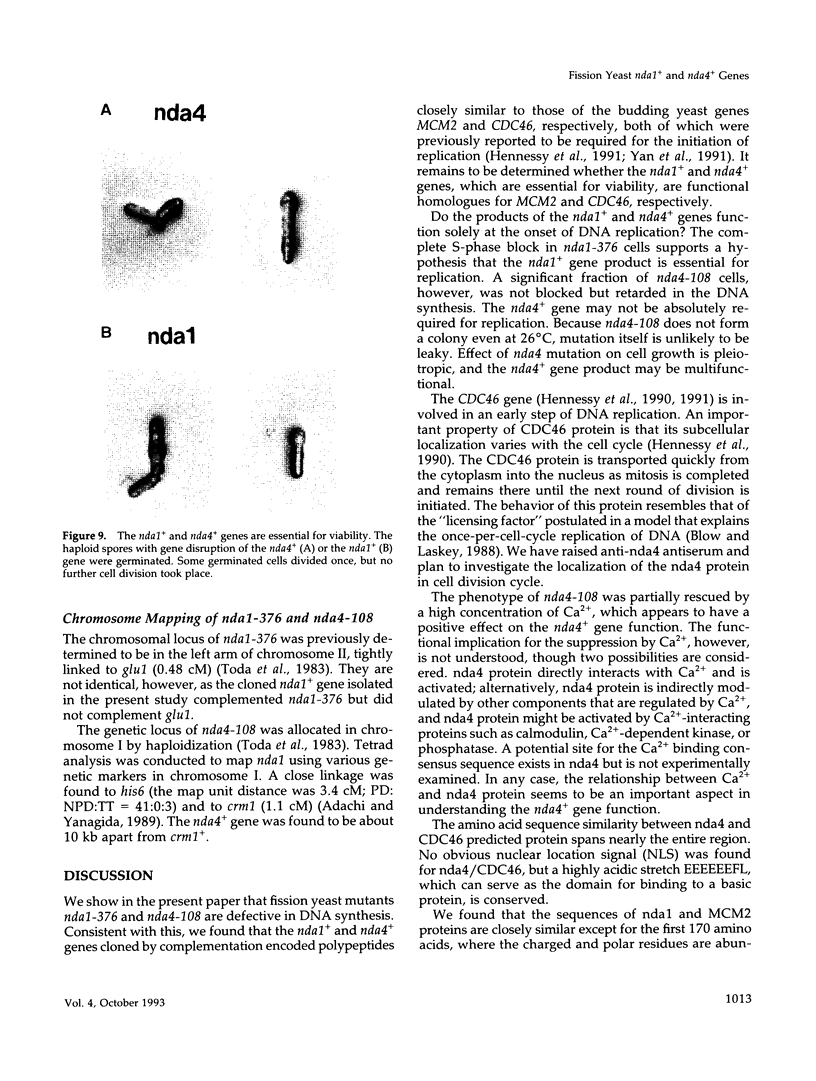
Fission yeast cold-sensitive mutants nda1-376 and nda4-108 display a cell cycle block phenotype at the restrictive temperature (cell elongation with the single nucleus) accompanied by an alteration in the nuclear chromatin region. DNA content analysis shows that the onset of DNA synthesis is blocked or greatly delayed in both mutant cells, the block being reversible in nda4-108. Upon release to the permissive temperature, nda4-108 cells resumed replicating DNA, followed by mitosis and cytokinesis. The nda4 phenotype was partly rescued by the addition of Ca2+ to the medium; Ca2+ plays a positive role in the nda4+ function. The predicted protein sequences of nda1+ and nda4+ isolated by complementation are similar to each other and also, respectively, to those of the budding yeast, MCM2 and CDC46, both of which are members of the gene family required for the initiation of DNA replication. The central domains of these proteins are conserved, whereas the NH2- and COOH- domains are distinct. Results of the disruption of the nda1+ and nda4+ genes demonstrates that they are essential for viability.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adachi Y., Toda T., Niwa O., Yanagida M. Differential expressions of essential and nonessential alpha-tubulin genes in Schizosaccharomyces pombe. Mol Cell Biol. 1986 Jun;6(6):2168–2178. doi: 10.1128/mcb.6.6.2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi Y., Yanagida M. Higher order chromosome structure is affected by cold-sensitive mutations in a Schizosaccharomyces pombe gene crm1+ which encodes a 115-kD protein preferentially localized in the nucleus and its periphery. J Cell Biol. 1989 Apr;108(4):1195–1207. doi: 10.1083/jcb.108.4.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow J. J., Laskey R. A. A role for the nuclear envelope in controlling DNA replication within the cell cycle. Nature. 1988 Apr 7;332(6164):546–548. doi: 10.1038/332546a0. [DOI] [PubMed] [Google Scholar]
- Coxon A., Maundrell K., Kearsey S. E. Fission yeast cdc21+ belongs to a family of proteins involved in an early step of chromosome replication. Nucleic Acids Res. 1992 Nov 11;20(21):5571–5577. doi: 10.1093/nar/20.21.5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson S. I., Surosky R. T., Tye B. K. The phenotype of the minichromosome maintenance mutant mcm3 is characteristic of mutants defective in DNA replication. Mol Cell Biol. 1990 Nov;10(11):5707–5720. doi: 10.1128/mcb.10.11.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan I. M., Hyams J. S. The use of cell division cycle mutants to investigate the control of microtubule distribution in the fission yeast Schizosaccharomyces pombe. J Cell Sci. 1988 Mar;89(Pt 3):343–357. doi: 10.1242/jcs.89.3.343. [DOI] [PubMed] [Google Scholar]
- Hagan I., Yanagida M. Novel potential mitotic motor protein encoded by the fission yeast cut7+ gene. Nature. 1990 Oct 11;347(6293):563–566. doi: 10.1038/347563a0. [DOI] [PubMed] [Google Scholar]
- Hennessy K. M., Clark C. D., Botstein D. Subcellular localization of yeast CDC46 varies with the cell cycle. Genes Dev. 1990 Dec;4(12B):2252–2263. doi: 10.1101/gad.4.12b.2252. [DOI] [PubMed] [Google Scholar]
- Hennessy K. M., Lee A., Chen E., Botstein D. A group of interacting yeast DNA replication genes. Genes Dev. 1991 Jun;5(6):958–969. doi: 10.1101/gad.5.6.958. [DOI] [PubMed] [Google Scholar]
- Hirano T., Funahashi S., Uemura T., Yanagida M. Isolation and characterization of Schizosaccharomyces pombe cutmutants that block nuclear division but not cytokinesis. EMBO J. 1986 Nov;5(11):2973–2979. doi: 10.1002/j.1460-2075.1986.tb04594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka Y., Toda T., Yanagida M. The NDA3 gene of fission yeast encodes beta-tubulin: a cold-sensitive nda3 mutation reversibly blocks spindle formation and chromosome movement in mitosis. Cell. 1984 Dec;39(2 Pt 1):349–358. doi: 10.1016/0092-8674(84)90013-8. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanbe T., Hiraoka Y., Tanaka K., Yanagida M. The transition of cells of the fission yeast beta-tubulin mutant nda3-311 as seen by freeze-substitution electron microscopy. Requirement of functional tubulin for spindle pole body duplication. J Cell Sci. 1990 Jun;96(Pt 2):275–282. doi: 10.1242/jcs.96.2.275. [DOI] [PubMed] [Google Scholar]
- Kinoshita N., Goebl M., Yanagida M. The fission yeast dis3+ gene encodes a 110-kDa essential protein implicated in mitotic control. Mol Cell Biol. 1991 Dec;11(12):5839–5847. doi: 10.1128/mcb.11.12.5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maine G. T., Sinha P., Tye B. K. Mutants of S. cerevisiae defective in the maintenance of minichromosomes. Genetics. 1984 Mar;106(3):365–385. doi: 10.1093/genetics/106.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K., Nurse P. Cell division cycle mutants altered in DNA replication and mitosis in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet. 1981;182(1):119–124. doi: 10.1007/BF00422777. [DOI] [PubMed] [Google Scholar]
- Nurse P., Thuriaux P., Nasmyth K. Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet. 1976 Jul 23;146(2):167–178. doi: 10.1007/BF00268085. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha P., Chang V., Tye B. K. A mutant that affects the function of autonomously replicating sequences in yeast. J Mol Biol. 1986 Dec 20;192(4):805–814. doi: 10.1016/0022-2836(86)90030-6. [DOI] [PubMed] [Google Scholar]
- Thömmes P., Fett R., Schray B., Burkhart R., Barnes M., Kennedy C., Brown N. C., Knippers R. Properties of the nuclear P1 protein, a mammalian homologue of the yeast Mcm3 replication protein. Nucleic Acids Res. 1992 Mar 11;20(5):1069–1074. doi: 10.1093/nar/20.5.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T., Adachi Y., Hiraoka Y., Yanagida M. Identification of the pleiotropic cell division cycle gene NDA2 as one of two different alpha-tubulin genes in Schizosaccharomyces pombe. Cell. 1984 May;37(1):233–242. doi: 10.1016/0092-8674(84)90319-2. [DOI] [PubMed] [Google Scholar]
- Toda T., Umesono K., Hirata A., Yanagida M. Cold-sensitive nuclear division arrest mutants of the fission yeast Schizosaccharomyces pombe. J Mol Biol. 1983 Aug 5;168(2):251–270. doi: 10.1016/s0022-2836(83)80017-5. [DOI] [PubMed] [Google Scholar]
- Toda T., Yamamoto M., Yanagida M. Sequential alterations in the nuclear chromatin region during mitosis of the fission yeast Schizosaccharomyces pombe: video fluorescence microscopy of synchronously growing wild-type and cold-sensitive cdc mutants by using a DNA-binding fluorescent probe. J Cell Sci. 1981 Dec;52:271–287. doi: 10.1242/jcs.52.1.271. [DOI] [PubMed] [Google Scholar]
- Woods A., Sherwin T., Sasse R., MacRae T. H., Baines A. J., Gull K. Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J Cell Sci. 1989 Jul;93(Pt 3):491–500. doi: 10.1242/jcs.93.3.491. [DOI] [PubMed] [Google Scholar]
- Yan H., Gibson S., Tye B. K. Mcm2 and Mcm3, two proteins important for ARS activity, are related in structure and function. Genes Dev. 1991 Jun;5(6):944–957. doi: 10.1101/gad.5.6.944. [DOI] [PubMed] [Google Scholar]