Abstract
In response to small molecule signals such as retinoids or steroids, nuclear receptors activate gene expression to regulate development in different tissues. microRNAs turn off target gene expression within cells by binding complementary regions in mRNA transcripts, and they have been broadly implicated in development and disease. Here we show that the C. elegans nuclear receptor DAF-12 and its steroidal ligand directly activate promoters of let-7 microRNA family members to downregulate the microRNA target hbl-1 and drive progression of epidermal stem cells from second to third larval stage patterns of cell division. Conversely, the unliganded receptor represses microRNA expression during developmental arrest. These findings identify microRNAs as components of a hormone-coupled molecular switch that shuts off earlier developmental programs to allow for later ones.
Lipophilic hormones coordinate organism-wide developmental progression in metazoans by binding to nuclear hormone receptors (NHR), converting the presence or absence of ligand into changes in gene expression patterns(1). This regulation is conserved in the nematode C. elegans, where the nuclear hormone receptor (NHR) DAF-12, a homolog of vertebrate liver-X and vitamin D receptors, regulates developmental progression or arrest in response to the environment(2, 3). In favorable environments, activation of TGF-β and insulin/IGF signaling cascades result in production of the DAF-12 steroidal ligands, the dafachronic acids (e.g. Δ4–DA), which promote rapid progression through four larval stages (L1–L4) to reproductive adults(4). In unfavorable environments, endocrine systems are suppressed and the unliganded DAF-12 causes arrest at a stress-resistant, long-lived alternative third larval stage, called the dauer diapause (L3d) (5).
A more cell-intrinsic level of developmental control is exerted by microRNAs (miRs). miRs are ~20–22nt long RNA molecules that bind to the 3’UTR of target mRNAs and decrease their expression (6–8). Null mutants for several miR genes show tissue-selective failure of progression from one stage-specific program to the next, generally described as heterochronic phenotypes. These phenotypes are most visible in the hypodermis, where hypodermal seam cells undergo invariant asymmetric stem cell division patterns, in which one daughter cell fuses to the hypodermal syncitium whereas the other retains stem cell character and its capacity to divide (9). Only during L2, do seam cells undergo one proliferative division prior to stem cell division, and repetition or loss of this program leads to changes in overall seam cell number in later stages. Finally, seam cell division ceases altogether by adulthood. Seam cells in animals with mutation of the microRNA lin-4 repeat L1 programs of asymmetic cell division during L2 stage, a triple deletion of the let-7 microRNA homologs mir-48, -84, -241 (referred to as let-7s) repeat L2 programs of cell proliferation during L3 stage, and let-7 null mutants repeat larval stage divisions and molting behavior in adults(10–12). daf-12(rh61rh411) null mutants exhibit a heterochronic phenotype similar to triple deletion of let-7 family members mir-48,mir-241(nDf51);mir-84(n4037), resulting in extra seam cells at the L3 stage (Table S1) (2, 11). This observation suggested that daf-12 might directly activate the let-7s miRs. To test this hypothesis, we fused the miR promoters (mir-241p and mir-84p) to the luciferase gene and co-transfected the reporters with DAF-12 into human cells. DAF-12 and Δ4–DA strongly activated mir-241p and mir-84p, whereas other promoters gave little or no signal (Fig. 1A). Deletion analysis of the mir-241 promoter revealed that whereas several deletion mutations retained transcriptional activity in the presence of DAF-12 and Δ4–DA, fragment #4 produced the highest fold induction and its removal substantially reduced activity (Fig. 1B). This fragment contained two pairs of DAF-12-response-elements (REs) as described by Shostak(13). In the full-length promoter context, mutation of one RE-pair in mir-241p and two REs in mir-84p led to a ~7 and ~3 fold decrease in activation, respectively. Gel mobility shift assays confirmed in vitro binding of DAF-12 to these REs whereas mutated versions abolished the interaction (Fig. 1C).
Figure 1. DAF-12 and dafachronic acid (Δ4–DA) activate microRNA promoters in vitro.
A. Activation of miR promoters in HEK293T cells. Promoters of let-7 homologs, mir-84 and mir-241, are strongly activated in the presence of DAF-12 and 400nM Δ4–DA, whereas other microRNAs are relatively unaffected. Luciferase assays were measured in triplicate, (with SD). EtOH, ethanol vehicle control; ptk-luc, empty luciferase vector.
B. Mutation analysis of mir-241 and mir-84 promoters reveals DAF-12/Δ4–DA activating elements. Deletion analysis of the mir-241p shows that the highest relative induction occurs with fragment #4, which contains four DAF-12 REs, 241abcd. Deletion or point mutation of 241ab elements (in red) abolishes activation (blue bars). Similarly, point mutation of DAF-12 REs in the mir-84 promoter, mir-84a and mir-84b reduce expression (red bars).
C. Gel mobility shift assay of DAF-12 and mir-241p. 32P-radiolabeled oligos containing the WT 241b element are shifted (s) by nuclear extracts expressing DAF-12::FLAG and supershifted (ss) in the presence of anti-FLAG antibody. Unlabeled WT 241b-oligo competes away the shift but addition of point-mutated oligo does not.
To examine miR expression in vivo, we generated transgenic worms containing the miR promoters fused to GFP. Wild type worms containing mir-241p::GFP gave a broad expression pattern as described(14). However, daf-12 nulls showed decreased expression most noticeably in the excretory cells (exc), as well as muscles, pharynx and intestine whereas neuronal expression seemed less affected, revealing tissue selectivity of daf-12 regulation (Fig. 2A–D). Null mutants for cytochrome P450 daf-9(dh6) fail to produce Δ4–DA, and the unliganded DAF-12 interacts with its co-repressor DIN-1/SHARP to repress its transcriptional targets, causing constitutive developmental arrest and dauer larvae formation(15–17). In these hormone deficient larvae, mir-241p::GFP expression was tightly repressed in most tissues (Fig 2E). Supplementation with Δ4–DA rescued this arrest and brought mir-241p::GFP expression back to WT levels (Fig 2F). Tight repression was also relieved in daf-9(dh6);din-1(dh149) double null mutants lacking the corepressor, with mir-241p::GFP expression levels similar to daf-12 nulls (Fig. 2G,H). Unlike daf-12 nulls, however, Δ4–DA supplementation of daf-9;din-1 worms restored mir-241p::GFP expression back to WT levels. Point mutation of all four daf-12-REs in mir-241p resulted in the same weak expression level in WT, daf-12 null with ligand, and daf-9 null, and daf-9;din-1 doubles ±ligand (Fig 2I–K, Fig. S1), revealing that these REs mediate both activation and repression. Thus, liganded DAF-12 works through its REs to mildly activate mir-241p in some tissues, whereas unliganded DAF-12 together with DIN-1 tightly represses expression in nearly all tissues.
Figure 2. DAF-12 and Δ4–DA regulate microRNA promoters in vivo.
Panels A–K, mir-241p::GFP. Images show representative L3 animals, with indicated cell types (white arrowhead, excretory cell, exc; outlined arrowheads, neuron, neu; muscle, mus; intestine, int,; pharynx, ph). Bar graphs quantify the percentage of worms with excretory cell GFP expression as either strong (green), weak (yellow), or off (red), (two independent experiments, left and right, n=10 animals each). mir-241p::GFP expression level in WT (N2) grown without ligand (EtOH) (A) is decreased in daf-12(rh61rh411)/NHR null (C), strongly repressed in daf-9(dh6)/CYP450 null (E) or not activated in daf-9(dh6)/CYP450;din-1(dh149)/SHARP double null (G) but rescued nearly to WT level by growth on 250 nM Δ4–DA (F, H). Δ4–DA has no effect in WT (B) or daf-12 null (D) animals. Point mutation of all four DAF-12 REs abolishes differences of tested genetic backgrounds or ligand (I–K, and Fig S1).
Panels L–O, mir-84p::GFP. Epidermal seam cells (arrowheads) express mir-84p::GFP in WT N2 (L), but not in daf-12 nulls (M). Seam cell expression is absent in hormone deficient daf-9;din-1 animals (N), but restored to nearly WT levels by Δ4–DA supplementation (O). Left, percentage of L3 animals showing weak or no seam cell expression (2 independent experiments, n=20 animals each).
(P) Relative quantification of microRNAs by Q-PCR. miR expression is decreased in daf-12, daf-9;din-1, and repressed in daf-9 mutants. In daf-9 genotypes, expression was Δ4–DA dependent. Q-PCR was carried out using the TaqMan system (see Fig S5 and SOM for data analysis).
As reported, mir-84p::GFP was expressed in pharynx, somatic gonad, seam cells, vulva cells, and occasionally in the intestine(14, 18). Seam cell expression was absent in daf-12 null and daf-9;din-1 null worms, and expression was increased almost to WT levels in daf-9;din-1 animals by addition of Δ4–DA (Fig. 2L–O), explaining daf-12 heterochronic phenotypes as a failure to activate the miRs in temporally patterned tissues. By contrast, expression in other tissues such as the pharynx, were less affected, again showing tissue-specific daf-12 regulation (Fig. S2). The transcriptional regulation of the let-7s is also reflected in the abundance of total mature microRNAs as measured by Taqman Q-PCR. daf-12 mutants and daf-9;din-1 animals showed decreased levels compared to WT, whereas daf-9 nulls showed tight repression of let-7 family of miRs (Fig. 2P, Fig. S3–Fig. S5). As expected, expression in daf-9, daf-9;din1, but not daf-12 mutants was rescued by Δ4–DA.
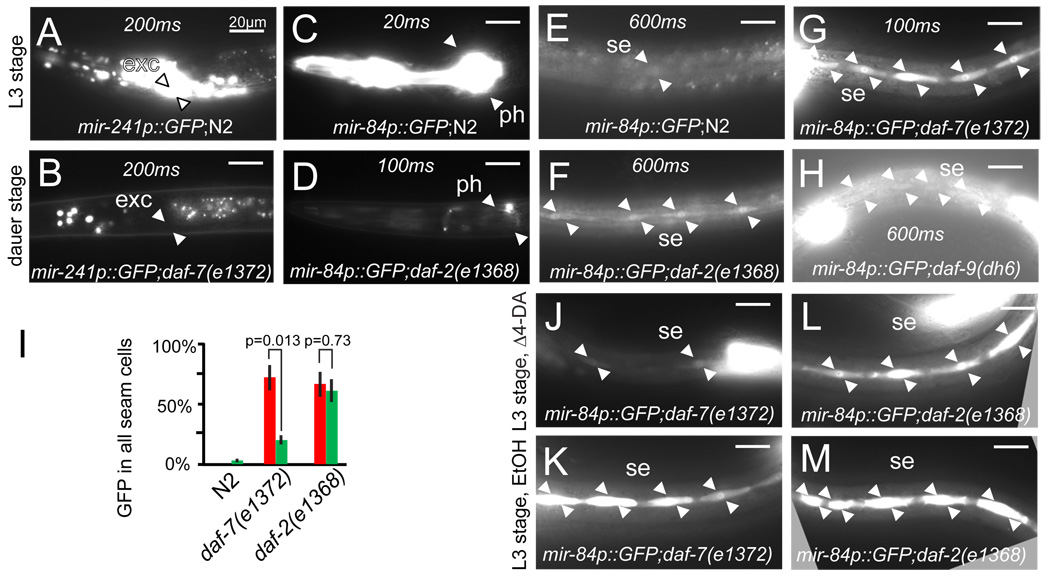
The dauer signaling pathways work upstream of daf-12 to govern organismal developmental progression and Δ4–DA production. We therefore sought to see if let-7s expression was affected by dauer constitutive mutant backgrounds of daf-7/TGF-β, daf-2/Insulin/IGF-I Receptor, and daf-9/CYP450. mir-241p::GFP expression was nearly completely repressed in these dauer larvae, whereas mir-84p::GFP was down regulated in some tissues such as the pharynx (Fig. 3A–D), but consistently upregulated and more penetrant in others such as the seam (Fig 3E–H). Even in reproductively growing L3 larvae, seam cell expression was more penetrant in daf-2(e1368) and daf-7(e1372) mutants than WT (Fig. 3G). Δ4–DA supplementation largely reversed this effect in daf-7, but not in the daf-2 background (Fig. 3I–M), suggesting that insulin/IGF and TGF-β signaling distinctly regulate mir-84p expression in a Δ4–DA-independent and Δ4–DA-dependent manner.
Figure 3. microRNA regulation by dauer signaling pathways.
mir-241p::GFP shows high expression in continuously growing WT (A), but low expression in daf-7(e1372) dauer larvae (B). mir-84p::GFP shows high expression in the pharynx of continuously growing WT (C) but low expression in daf-2(e1368) dauers (D). mir-84p::GFP seam cell expression (E) is elevated in daf-2 and daf-9 mutants during dauer stage (F, H) and is even higher in daf-7 mutants during reproductive growth at 20°C (G). Penetrant seam expression is reversed by 500 nM Δ4–DA in daf-7 but not daf-2 during reproductive growth (I–M). Animals were assayed during L3/L3d stages, n>20. Red bars, ethanol vehicle, green bars, Δ4–DA (SEM).
The zinc finger protein hbl-1/hunchback is responsible for L2 proliferative programs of seam cells (11). hbl-1 loss leads to a loss of proliferative programs, and hence a decreased seam cell number (Table S1). let-7s is proposed to inhibit hbl-1, through microRNA-mediated repression of its 3’UTR because let-7s mutants result in both increased seam number and hbl-1::GFP expression at L3 (11, 19). Given that DAF-12 activates let-7s, we sought to examine interactions with hbl-1. Consistent with NHR signaling promoting hbl-1 inhibition, daf-12 mutants and daf-9;din-1 double mutants have extra seam cells, the latter reversed by Δ4–DA (Table S1). Moreover, the penetrant extra seam phenotype of daf-12(rh61) ligand binding domain mutant was dependent on functional hbl-1(+), placing daf-12 upstream of hbl-1 by genetic epistasis. Accordingly, we observed consistent upregulated hypodermal expression of hbl-1p::GFP::hbl-1-3’UTR during L3 in daf-12(rh61), but not in WT (Fig. 4AB). This upregulation was likely due to loss of UTR-mediated repression, as exchange of the hbl-1-3’UTR for a non-repressed unc-54-3’UTR led to equal hypodermal expression levels in both WT and daf-12(rh61) background (Fig. 4CD).
Figure 4. let-7s repression target, hbl-1, is regulated by DAF-12.
A GFP-fusion to hbl-1 promoter and 3’-UTR is repressed in hypodermis at mid L3 (28h) in WT (A). In the daf-12(rh61) mutant, reporter signal is upregulated in the hypodermis (arrows) and other tissues (B) (exposure 250ms). A GFP-fusion to the hbl-1 promoter, containing the unc-54-3’-UTR lacks substantial up-regulation in the hypodermis (C, D), though body muscles show modest reporter upregulation (D) (exposure time 50ms)
Model for nuclear receptor/microRNA signaling cascades (E). In response to favorable environmental signals, activated insulin/IGF and TGF-β pathways induce Δ4–DA biosynthesis through DAF-9/CYP450. Right: Liganded DAF-12 activates L3 programs and expression of let-7s and thereby inhibits HBL-1 and genes of L2 programs, resulting in developmental progression (E). Left: During unfavorable conditions, unliganded DAF-12 together with DIN-1, repress L3 programs and let-7s, allowing derepression of L2 programs and/or developmental arrest. Dauer signaling also has Δ4–DA-independent outputs onto miRs.
In this work, we show that the nuclear hormone receptor DAF-12 directly regulates the let-7 relatives, mir-84 and mir-241, connecting organism-wide commitments to cell intrinsic programs (Fig. 4E). These studies suggest a model whereby in unfavorable environments, downregulated insulin/IGF-1 and TGF-β pathways suppress dafachronic acid production and unliganded DAF-12 together with corepressor DIN-1 repress miR expression in most tissues and specify developmental arrest. Conversely, in favorable environments, stimulation of insulin/IGF-1 and TGF-β growth signaling pathways results in dafachronic acid production. Liganded DAF-12 activates let-7 homologs, which in turn downregulate their target, hbl-1, allowing L2/L3 transitions in the hypodermis. The use of a NHR-miR coupled molecular switch to turn off earlier programs to allow for later ones is a function likely to be conserved, and may be a paradigm for understanding hormone- dependent developmental progression, stem cell differentiation, maturation or tumor formation in metazoans. In particular, activation of new programs, and inhibition of earlier ones is critical for the fidelity of distinct developmental states, which may be less apparent in more complex animals whose cell lineages are unknown. In fact, DAF-12 itself is downregulated by let-7 at later stages, suggesting that both feedforward and feedback loops drive transitions (20). In D. melanogaster, the steroid hormone ecdysone and its cognate receptor regulate developmental progression in part via mir-14, but it is unknown whether regulation is direct or indirect (21). Our studies also reveal the intricacy of NHR signaling in an organismal context. They give visible evidence that liganded receptors can activate their targets, while unliganded receptors can repress them, with vastly different outcomes for progression or arrest. This hormone dependent modulation of target gene expression around basal transcription mirrors that seen with the vertebrate homolog LXR (22). Finally, DAF-12/NHR regulation of the miRs is highly tissue- and stage-specific, implicating other transcription factors, coregulators, and chromatin factors in the control of miR expression.
Supplementary Material
References and Notes
- 1.Mangelsdorf DJ, et al. Cell. 1995;83:835. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antebi A, Culotti JG, Hedgecock EM. Development. 1998;125:1191. doi: 10.1242/dev.125.7.1191. [DOI] [PubMed] [Google Scholar]
- 3.Antebi A, Yeh WH, Tait D, Hedgecock EM, Riddle DL. Genes Dev. 2000;14:1512. [PMC free article] [PubMed] [Google Scholar]
- 4.Motola DL, et al. Cell. 2006;124:1209. doi: 10.1016/j.cell.2006.01.037. [DOI] [PubMed] [Google Scholar]
- 5.Fielenbach N, Antebi A. Genes Dev. 2008 Aug 15;22:2149. doi: 10.1101/gad.1701508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel DP. Cell. 2004 Jan 23;116:281. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Lee RC, Feinbaum RL, Ambros V. Cell. 1993 Dec 3;75:843. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 8.Wightman B, Ha I, Ruvkun G. Cell. 1993 Dec 3;75:855. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 9.Rougvie AE. Development. 2005 Sep;132:3787. doi: 10.1242/dev.01972. [DOI] [PubMed] [Google Scholar]
- 10.Chalfie M, Horvitz HR, Sulston JE. Cell. 1981 Apr;24:59. doi: 10.1016/0092-8674(81)90501-8. [DOI] [PubMed] [Google Scholar]
- 11.Abbott AL, et al. Dev Cell. 2005 Sep;9:403. doi: 10.1016/j.devcel.2005.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reinhart BJ, et al. Nature. 2000 Feb 24;403:901. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 13.Shostak Y, Van Gilst MR, Antebi A, Yamamoto KR. Genes Dev. 2004 Oct 15;18:2529. doi: 10.1101/gad.1218504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esquela-Kerscher A, et al. Dev Dyn. 2005 Dec;234:868. doi: 10.1002/dvdy.20572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerisch B, Weitzel C, Kober-Eisermann C, Rottiers V, Antebi A. Dev Cell. 2001 Dec;1:841. doi: 10.1016/s1534-5807(01)00085-5. [DOI] [PubMed] [Google Scholar]
- 16.Jia K, Albert PS, Riddle DL. Development. 2002 Jan;129:221. doi: 10.1242/dev.129.1.221. [DOI] [PubMed] [Google Scholar]
- 17.Ludewig AH, et al. Genes Dev. 2004 Sep 1;18:2120. doi: 10.1101/gad.312604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayes GD, Frand AR, Ruvkun G. Development. 2006 Dec;133:4631. doi: 10.1242/dev.02655. [DOI] [PubMed] [Google Scholar]
- 19.Li M, Jones-Rhoades MW, Lau NC, Bartel DP, Rougvie AE. Dev Cell. 2005 Sep;9:415. doi: 10.1016/j.devcel.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 20.Grosshans H, Johnson T, Reinert KL, Gerstein M, Slack FJ. Dev Cell. 2005 Mar;8:321. doi: 10.1016/j.devcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 21.Varghese J, Cohen SM. Genes Dev. 2007 Sep 15;21:2277. doi: 10.1101/gad.439807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Repa JJ, et al. Science. 2000 Sep 1;289:1524. doi: 10.1126/science.289.5484.1524. [DOI] [PubMed] [Google Scholar]
- 23.Our thanks go to G. Hayes, G. Ruvkun, V. Ambros, A. Rougvie for strains, N. Timchenko for gel shift support, D. Magner, S. Greene, F. Schroeder for manuscript comments. This work was supported by NIH grant GM077201 and the Ellison Medical Foundation (AA), and the Howard Hughes Medical Institute and the Robert A. Welch Foundation (DJM).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.