Abstract
MicroRNAS (miRNAs) have been suggested to play important roles in the central nervous system during development as well as disease. miRNAs appear to be dysregulated in a number of neurodegenerative diseases, developmental disorders and as a result of stroke. Each miRNA has the ability to regulate hundreds of messenger RNA transcripts, both by causing degradation of the mRNA and by inhibition of protein translation. Recent findings suggest that it may eventually be possible to treat some neurological disorders by restoring or inhibiting miRNAs altered by disease pathology. Both viral delivery and administration of modified oligonucleotides mimicking or inhibiting specific miRNAs have been effective in model systems. Artificial miRNAs have also been generated for the repression of specific transcripts. Alteration of miRNA expression by disease and insult also holds the potential for improved diagnostic tools. Lastly, miRNAs have been shown to control cellular proliferation and specification, suggesting that manipulation of miRNAs in cultured cells could result in more convenient generation of pure cell populations for transplantation.
Introduction
MicroRNAs (miRNAs) are short (20-23 base pair) single-stranded RNAs, with sequences partially complementary to one or more mRNAs, that play important roles in the post-transcriptional regulation of gene expression (Carthew and Sontheimer, 2009). The molecular mechanisms by which cells produce miRNAs, and how those miRNAs target specific mRNAs to either cause degradation of the mRNA or inhibit protein translation are reviewed in articles in this issue of Neuromolecular Medicine. Increasing evidence suggests that miRNAs are dysregulated in several neurological disorders. These small non-coding regulatory RNAs may therefore provide opportunities for the diagnosis and treatment of damaged and diseased nervous systems. The developing and mature central nervous system (CNS) expresses one of the richest diversities of miRNAs of any tissue (Krichevsky et al. 2003; Miska et al. 2004; Sempere et al. 2004; Smirnova et al. 2005; Bak et al. 2008; Kapsimali et al. 2007). These miRNAs have already been shown to contribute to cellular proliferation and specification (Schwamborn et al., 2009; Zhao et al., 2009), and to disease-relevant processes such as inflammation (Lukiw et al., 2008; Dahrap et al., 2009), in mammalian nervous systems.
A potential drawback in the therapeutic use of miRNAs in the nervous system is alteration of normal physiology in addition to disease pathology due to the many potential targets of a given miRNA. Endogenous miRNAs regulate hundreds of targets by mRNA degradation or translational repression of protein production (Lim et al. 2005; Baek et al., 2008; Selbach et al., 2008). Since not all targets of a particular miRNA are likely to be disease-related, any therapeutic perturbation of miRNA expression will likely have side-effects unrelated to the disease process. Endogenous miRNAs also appear to regulate different transcripts with varying efficiency, and the effects of expressing these miRNAs therapeutically will likely have differential effects depending on the transcriptome of a given cell population (Lim et al. 2005; Baek et al. 2008; Selbach et al. 2008). Very few miRNA:mRNA target pairings have been validated to date and only two studies have made an attempt to examine the entire proteome and transcriptome influenced by specific miRNAs in a cell population (Baek et al., 2008; Selbach et al., 2008). Before miRNAs are used therapeutically, the effects of over- or under-expressing the miRNA should be examined in all cell populations manipulated to avoid potentially unwanted effects from the hundreds of target transcripts. Caution should be used when using over-expression of miRNAs, as an above physiological abundance of a miRNA could result in binding to seed regions not normally targeted. Despite this complexity, at least one study reported that miRNAs can be manipulated in vivo over a long- time period with no apparent negative side effects (Elmén et al., 2008a)
Mechanisms of Manipulation and Delivery
Currently, one of the most promising methods of miRNA manipulation has been the use and systemic delivery of modified oligonucleotides. Both sense and antisense miRNAs and artificial miRNAs have all been employed in-vivo in mammalian systems with varying effects on target mRNAs and cell and tissue functions. The most common oligonucleotide modifications used in the manipulation of miRNAs are Locked Nucleic Acids (LNA), 2′-O-methyl–modification, and phosphorothioate backbones. LNA and 2′-O-methyl modified oligonucleotides contain modifications (sugar modifications or the addition of a methylene bridge) which result in increased stability of RNA duplexes. LNA-modified oligonucleotides appear to be more effective at binding miRNAs than oligonucleotides containing 2′-O-methyl–modification (Elmén et al., 2008a). LNA-modified oligonucleotides also dramatically improve detection of miRNAs by both in-situ hybridization and northern blot (Várallyay et al., 2008; Silahtaroglu et al., 2007). Additional modification of oligonucleotides to include a phosphorothioate backbone results in decreased degradation by nucleases and increased membrane permeability. These modifications improve the effectiveness of any oligonucleotide-based treatment and provide a means to systemically express or inhibit miRNAs.
The approach of using antisense oligonucleotides to bind and disrupt endogenous miRNAs, in some cases dubbed ‘antagomirs’ or ‘antiMirs’, has been used in vivo in several systems (Krützfeldt et al., 2005). Effective in vivo knockdown using LNA and phosphorothioate modified oligonucleotides has been demonstrated in mice and non-human primates (Elmén et al., 2008a, 2008b). Elmen and colleagues delivered LNA-modified oligonucleotides targeting miR-122 intravenously and demonstrated a decrease in circulating cholesterol levels lasting seven weeks. Importantly, no apparent toxicity was observed as a result of this treatment. Systemic administration of modified oligonucleotides through either intravenous injection or cerebrospinal fluid (CSF) infusion provides a relatively non-invasive and apparently non-toxic means of harnessing miRNAs for therapeutic benefit. Recently, “miRNA sponges”, custom-designed transcripts targeted to a specific miRNA, have been developed as a stable alternative to antisense oligonucleotides and transfected in vitro (Ebert et al., 2007). These sponges could also be delivered using viral vectors to allow for stable inhibition of specific, disease-related miRNAs, and RNA sponges also have the potential to inhibit entire families of miRNAs. The blood-brain-barrier (BBB) represents a problem to any therapy involving systemic delivery of oligonucleotides. One solution involves the addition of a rabies-virus peptide that allows oligonucleotides to cross the BBB and target neurons (Kumar et al. 2007), however other solutions maybe required due to possible acquired immune responses to this peptide.
More invasive, viral delivery of miRNAs, antisense miRNAs and shRNAs (short hairpin RNAs) has been used extensively in the study of miRNAs. Viral methods allow for more precise spatially targeted therapies through tissue-specific viral injections, potentially avoiding some of the pitfalls of systemically delivered oligonucleotides. A shRNA is a sequence of RNA that makes a tight hairpin turn and is expressed from a DNA template and processed by cellular enzymatic processing machinery into small double-stranded RNAs that mediate RNA interference. However, reports of toxicity resulting from viral shRNA delivery have recently emerged (Grimm et al., 2008; McBride et al., 2008). This toxicity has been observed to cause cell death in both the striatum and cerebellum when delivered using adeno-associated virus (AAV) (McBride et al., 2008; Boudreau et al., 2009). Another group demonstrated that nearly 50% of the AAV-shRNA vectors (23/49) tested and delivered intravenously resulted in death (Grimm et al., 2006). This morbidity appears to be dependent on the design of shRNA, as other studies delivering AAV-shRNA resulted in tissue damage, but not death (McBride et al., 2008). The negative effects of using shRNAs as a therapeutic tool appear to result from the abundance of shRNAs interfering with endogenous RNAi processes by competing for components of the RISC pathway (Castanotto et al., 2007; McBride et al., 2008). By placing a miRNA or siRNA under a strong promoter, the resulting abundance of the small interfering RNA overwhelms components of the RISC complex and disrupts endogenous processes using the RISC complex. Although shRNAs are effective at knockdown of specific genes, the potential hazards and variability associated with shRNA induced toxicity likely preclude its widespread therapeutic use.
In three recent studies the investigators created artificial miRNAs by placing the inhibitory sequence within a 60-70bp pre-miRNA hairpin structure, also known as a “miRNA scaffold” (Castanotto et al., 2007; Boudreau et al., 2009; McBride et al. 2008). This approach resulted in effective suppression of the target gene (huntingtin) with no apparent toxicity (McBride et al., 2008). Artificial miRNAs were found expressed in less abundance than shRNAs and did not compete with endogenous miRNAs. These studies demonstrate the use of endogenous miRNA scaffolds in the construction of artificial miRNAs targeted to specific genes is both safe and effective in animal models.
Alteration of miRNAs in CNS Disease and Dysfunction
A number of reports have emerged showing neurological disease-related perturbation of miRNAs (Wang et al., 2008; Hébert et al., 2008; Kim et al., 2007; Wang et al., 2008b; Johnson et al. 2008; Packer et al. 2008). These alterations in miRNA expression may be a direct consequence of disease or may occur as a result of the loss of a specific cell population (Wang et al., 2008; Kim et al., 2007). Altered expression of miRNAs prior to the onset of or during the course of disease pathology raises the possibility that expressing or inhibiting specific miRNAs might ameliorate the disease process and provide an effective treatment. Altered expression of miRNAs has been observed in Alzheimer's disease (AD), Parkinson's disease (PD), Huntington's disease (HD), Tourette's syndrome, and prion-induced neurodegeneration (Cogswell et al., 2008; Faghihi et al., 2008; Hebert et al., 2008; Packer et al., 2008; Saba et al., 2008; Wang et al., 2008a, 2008b) (Fig. 1). Other disease-related processes, such as inflammation, also alter miRNA expression and therapy might be mediated by manipulation of these miRNAs as well (see Urbich et al., 2008 and Sheedy et al., 2008 for reviews). Additionally, depression and mental health disorders such as schizophrenia also appear to involve changes in miRNA expression (Beveridge et al., 2008).
Figure 1. Involvement of miRNAs in neurodegenerative pathologies.
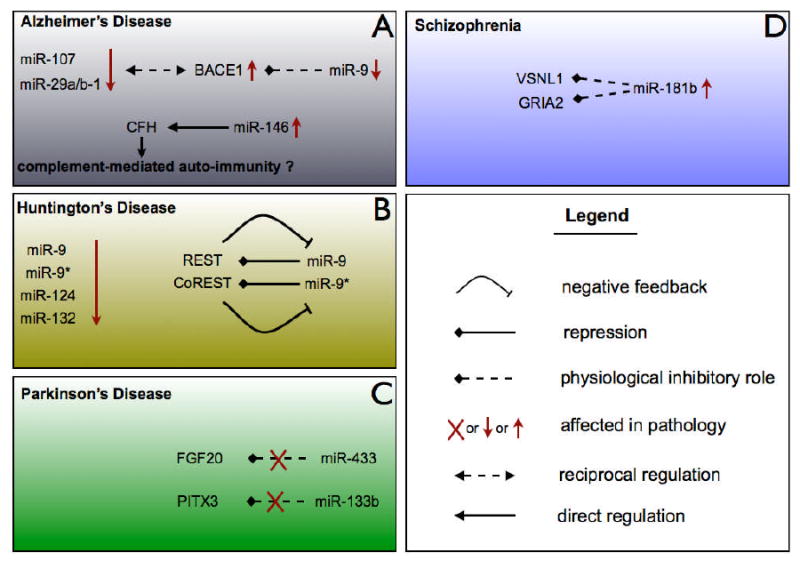
A. miR-9 is known to physiologically repress BACE1; miR-107 and miR-29a/b-1 reciprocally repress BACE1. In Alzheimer's disease, levels of miR-9, miR-107 and miR-19a/b-1 are downregulated, which results in increased BACE1 levels. In addition, miR-146 is increased in Alzheimer's disease, which causes its target, complement factor H to be downregulated which could result in complement-mediated autoimmunity. B. In Huntington disease patients, miR-9, miR-9*, miR-124 and miR-132 are down regulated; miR-9 and miR-9* were shown to repress REST and CoREST respectively. The latter repress miR-9 and miR-9* in a positive feedback loop. The loss of REST and CoREST repression results in neuronal cell loss and further neurodegeneration in this disease. C. In Parkinson's disease, mutations in the 3′ UTR of two genes were found to prevent the binding of two microRNAs: miR-433 for the FGF20 gene which was correlated with increased alpha-synuclein, and miR-133b for the PITX3 gene which is important for midbrain dopaminergic maturation and function. D. In schizophrenia patients, miR-181b is upregulated and results in increased repression of VSNL1 and GRIA2.
Increasing evidence suggests roles for miRNAs in the pathology of AD, particularly with respect to the regulation of beta-amyloid precursor protein-converting enzyme 1 (BACE1). Mouse models and human samples have all implicated altered miRNA expression in the AD process (Boissonneault et al., 2009; Wang et al., 2008; Hébert et al., 2008). Evidence suggests that dysregulation of miRNAs plays a role in both inherited and sporadic forms of AD (Hébert et al., 2008; Wang et al., 2008). These studies have largely found miRNAs involved in the regulation of BACE1, a protein that cleaves APP resulting in an intermediate APP fragment that can then be cleaved by gamma-secretase to generate the amyloid beta-peptide (Mattson, 2004a). Additionally, in vivo inhibition of BACE1 by siRNAs in a transgenic mouse model resulted in improved pathology, suggesting a promising therapeutic role for proper regulation of BACE1 in AD (Singer et al., 2005). In mouse models of AD, both miR-298 and miR-328 repress BACE1 expression (Boissonneault et al., 2009).
Three recent papers examined miRNA expression in human AD tissue and found a number of miRNAs with altered expression (Wang et al., 2008; Hebert et al., 2008; Lukiw et al., 2008). Wang and colleagues found an early decrease in expression of miR-107 that correlates with increased BACE1 expression and also validated this interaction. Hebert and colleagues found miR-29a/b-1 expression is decreased in sporadic AD patients, and demonstrated a decrease in BACE1 activity as a result of miR-29a/b-1 expression. They also demonstrated that miR-9 can regulate BACE1 expression and is altered in tissue samples from brain regions affected by pathology in sporadic AD patients. Lastly, Lukiw et al. found upregulation of miR-146a in human AD brain tissue and validated an interaction between miR-146a and complement factor H (CFH) (Lukiw et al., 2008). This interaction suggests a role for miR-146a in AD pathology-associated inflammation, revealing the possibility that inhibition of miR-146a in-vivo could lead to a reduction in disease-related inflammation.
Screening for miRNAs in HD brain tissue samples has demonstrated that expression of several miRNAs change with disease progression in both mouse models of HD and in human patients (Johnson et al. 2008; Packer et al., 2008). These miRNAs include miR-9/9*, 29b, 124a and 132. The role of miR-124 has been explored by several groups and has a strong role in regulating neural fate (Conaco et al. 2006, Makeyev et al. 2007). Although miR-124 expression results in a neuronal splicing pattern through the repression of PTBP1, the overexpression and disruption of miR-124 in the developing neural tube produced little perturbation of neuronal fate and suggested a role in maintenance of neuronal fate (Cao et al. 2007). The function of miR-9/9* was further investigated using in-vitro tools and shown to regulate the REST complex through miR-9 repression of REST and miR-9* repression of CoREST. It should be noted that while Packer et al. demonstrated repression of REST by miR-9 and Co-REST by miR-9*, this interaction was not shown in HD tissue. In addition to the decrease observed in HD tissue, these miRNAs are downstream of RE1 elements, suggesting that miR-9/9* and REST/CoREST function in a positive feedback loop. This is similar to a feedback loop observed between miR-133b and Pitx3 (Kim et al. 2007) and may represent a general theme in miRNA regulation that should be taken into consideration when planning miRNA-based therapies (Tsang et al. 2007).
Disruption of miRNA binding sites could also result in disease pathology, as is the case in PD. Wang and colleagues screened 729 families with PD patients and found a disruption in the binding site of miR-433 in the 3′UTR of the FGF20 gene (Wang et al. 2008). Increased expression of FGF20 has been correlated with increased alpha-synuclein and this could result in cytotoxicity associated with PD. This is strikingly similar to a reported case of a mutation observed in Tourette's Syndrome, where the 3′ UTR of the SLITRK1 gene resulting in lack of miR-189 being able to repress SLITRK1 (Abelson et al. 2005). In these cases it is possible that therapeutic repression of the target gene, either with artificial miRNAs or with modified oligonucleotides could have a therapeutic benefit. Another report found a lack of miR-133b expression in PD patient tissue when compared to controls (Kim et al. 2007). Further investigation of miR-133b demonstrated that it represses the transcription factor Pitx3 in a feedback loop and is necessary for midbrain dopaminergic maturation and function.
Mental health also appears to be influenced by miRNA expression. Tissue from the temporal lobe of schizophrenia patients has increased levels of miR-181b relative to control tissue (Beveridge et al. 2008). In the latter study, VSNL1 and GRIA2 were also confirmed as repressed targets of miR-181b in schizophrenia. In another study lithium and valproate, which are used to treat bipolar disorder, were administered to adult rats through food followed by miRNA expression profiling in the hippocampus. Expression of several miRNAs, including miR-34a, 30c, 30c, 136 and two members of the let-7 family, let-7b and let-7c, were found altered by these treatments. (Zhou et al. 2008). This study also demonstrated that miR-34a inhibits translation of metabotropic glutamate receptor 7 (GRM7), suggesting that miR-34a might mediate some of the effects of lithium and valproate in the CNS. Additionally, miR-30a-5p has been shown to inhibit BDNF in the prefrontal cortex (Mellios et al. 2008). BDNF is believed to mediate the antidepressant effects of treatments such as exercise and electroconvulsive shock, and has other beneficial effects including neuroprotection and stimulation of hippocampal neurogenesis (Mattson et al. 2004b). Thus, increasing evidence suggests that misregulation of miRNAs might be involved in mental health disorders including depression, bipolar disorder and schizophrenia, raising the possibility that miRNA-based treatments such as modified oligonucleotides might provide effective treatment.
Taken together, these findings described above provide evidence for the involvement of miRNAs in neurodegeneration and psychiatric disorders. Both alterations in miRNA expression and disruption of miRNA mediated repression by mutations in 3′UTRs have roles in neurological dysfunction. Manipulation of endogenous miRNAs altered as a result of disease pathology or introduction of artificial miRNAs, either through administration of oligonucleotides or viral vector-based technology could provide effective treatment for these conditions. Additionally, the evidence that miRNAs mediate the effects of antidepressants could provide new and more effective means for treating depression and related illnesses.
miRNAs in Transplantation and Regenerative Medicine
In addition to neurodegenerative disease and psychiatric disorders, miRNA-mediated therapy also has potential benefit for ischemia and traumatic insults to the CNS. Two studies have profiled miRNA expression using a middle cerebral artery occlusion – reperfusion rat model of transient focal ischemic stroke. Jeyaseelan and colleagues performed a profiling study of miRNAs at two time-points post occlusion (24 and 48 hours) and examined miRNA levels in both brain and blood (Jeyaseelan et al. 2008). Unique miRNAs were observed at both 24 and 48 hours post occlusion and common miRNAs were observed between blood and brain samples. These changes could play a role in the infiltration of immune cells post-occlusion. A large number of miRNAs assayed (20%) were observed to change at time points between 3 hours and 3 days, and expression of miRNAs was also observed to change with reperfusion time (Dharap et al. 2009). Dharap and colleagues also attempted to reduce miR-145 expression using antisense oligonucleotides and demonstrated an increase in SOD2, a neuroprotective antioxidant enzyme, but this increase resulted in little therapeutic benefit. These two studies both reveal a temporal component to post-ischemic miRNA expression that was previously unknown. Another study demonstrated the ability of oligonucleotide antisense delivery to manipulate post-ischemia angiogenesis, presenting another potential therapeutic target following CNS ischemia (van Soligen et al. 2008).
Traumatic head and spinal cord injury are major causes of morbidity and mortality, particularly in children and young adults. In one study a microarray platform was used to examine changes in the expression of miRNAs in the hippocampus at 3 and 24 hr after controlled cortical impact injury in a rat model of traumatic brain injury (Redell et al., 2009). Bioinformatic analysis of miRNAs significantly altered by traumatic brain injury, and their predicted targets, revealed proteins involved in signal transduction, transcriptional regulation, and cell proliferation and differentiation. Traumatic injury may involve excitotoxic damage to neurons and the potential use of miRNA technology to treat excitotoxic neuronal injury is also being pursued. For example, lentiviral transduction of human mesenchymal stem cells with anti-adenylate kinases cassettes resulted in increased production of adenosine by the cells (Ren et al., 2007). The latter cells were then transplanted into the hippocampus of mice 1 week prior to injection of the excitotoxin kainic acid into the amygdala. Whereas epileptic seizures and death of hippocampal CA3 neurons occurred in mice transplanted with control cells, seizures were suppressed and death of CA3 neurons prevented in mice transplanted with the cells in which adenylate kinase was inhibited by miRNA expression.
In addition to reducing inflammation and damage resulting from ischemia or trauma, in vitro transfection of miRNAs could lead to improved generation of cells for transplantation. miRNAs also present a tool for a more indirect therapeutic benefit if used to improve the generation of cells for transplantation. miRNAs function both at a general level, such as regional specification and at a more precise cell population-restricted level. Expression of miR-124, part of a family of miRNAs regulated by REST, regulates an entire neuronal alternative splicing pattern through the repression of PTBP1 (Conaco et al. 2006, Makeyev et al. 2007). Where expression of miR-124 results in a more general neuronal identity, miR-133b shows limited expression to midbrain dopaminergic neurons and is necessary for midbrain dopaminergic fate (Kim et al. 2007). Manipulation of the latter two and possibly other relevant miRNAs in vitro could lead to improved generation of cells for transplantation.
Developing endogenous repair strategies through manipulation of miRNAs in-vivo in neurogenic regions is also a possibility. A large number of miRNAs have already been identified as important for the regulation of cellular proliferation in cancer cells (Croce et al. 2005), and some of those miRNAs may be relevant to neural stem cells. In the adult CNS two neurogenic niches with the potential for endogenous cellular replacement exist, the sub-granular zone (SGZ) of the hippocampal dentate gyrus and the sub-ventricular zone (SVZ) (see Zhao et al. 2008 for a review). It is likely that miRNAs play important roles in the regulation of proliferation, migration, specification and integration of adult-born neurons and glial cells that arise from neural stem cells. Recent findings have indeed demonstrated roles for miRNAs in the behavior of neural stem cells. Targeted ablation of Dicer in neural progenitor cells in the embryonic mouse cerebral cortex had little effect on the proliferation and survival of the progenitor cells, and instead caused a dramatic suppression of neuronal differentiation and apoptosis of newly generated neurons (De Pietri et al., 2008). By knocking down and overexpressing specific miRNAs in mouse embryonic stem cells, Krichevsky et al. (2006) showed that miR-124 and miR-9 regulate neural lineage differentiation. Further understanding of the miRNAs involved in these processes and advances in delivery methods for viruses or modified oligonucleotides could lead to endogenous repair strategies for neurodegenerative diseases such as AD, as well as head and spinal cord injury and stroke.
miRNAs As Diagnostic Tools
Although largely unexplored, a possible use for miRNAs in a clinical setting is the improvement of diagnosis. Studies have examined the possibility of using blood and cerebrospinal fluid (CSF) to screen for miRNA biomarkers of disease and injury. In one case, multiple commonly altered miRNAs were found between brain regions and blood in a rat model of transient focal ischemic stroke (Jeyaseelan et al. 2009). Further work needs to be done to examine the usefulness of these miRNAs as biomarkers for ischemia. This observation does raise the possibility that miRNAs in blood could be used to screen for other disease pathology.
In one study, samples from brain regions and CSF from AD and control patients were compared (Cogswell et al. 2008). The miRNAs observed altered in AD patients CSF did not correspond to miRNAs altered in the brain regions examined, raising the possibility that the altered expression might be related to an immune response or changes in endothelial cell expression related to disease pathology. As evidenced by miR-107, in some cases miRNA levels are altered before pathology or symptoms of AD present, raising the possibility that immune-related responses might also be present early in the disease process. Even if these miRNAs serve as an indirect indication of AD pathology, screening for these miRNAs in patient CSF could represent a substantial improvement in the diagnosis of AD.
Altered Gene Expression as a Cause of Neurodegenerative Disorders
Emerging evidence suggests that the most common sporadic forms of AD and PD may result from increased expression of genes encoding the beta-amyloid precursor protein (APP), tau (AD) and alpha-synuclein (PD) (Singleton et al., 2004). Together with age-related impairment of proteasomal degradation (Keller et al., 2000; Hol et al., 2005), increased production of APP, tau and synuclein may result in the accumulation of these pathogenic proteins within vulnerable neuronal populations. An early clue that pointed to increased production of a pathogenic protein as being pivotal for a neurological disorder came from studies of Down Syndrome, a genetic disorder resulting from triplication of chromosome 21. The gene encoding APP is on chromosome 21, and essentially all people with Down Syndrome develop Abeta and tau pathology that is virtually identical to that of AD. Data from studies of trisomy 16 mice, a model of Down syndrome, suggest that increased APP expression is sufficient to cause neuropathological changes similar to those of AD (Cataldo et al., 2003; Salehi et al., 2006). The mutant Huntingtin protein (Htt) also serves as another example of gene disruption resulting in altered transcriptional profiles. Mutant Htt results in decreased levels of BDNF and other RE1 containing transcripts (Zuccato et al. 2007). Perhaps the strongest evidence that increased expression of a disease-associated protein can cause a neurodegenerative disorder comes from a study showing that triplication of the alpha-synuclein gene locus causes PD in a family with inherited PD (Singleton et al., 2003).
In light of the kinds of evidence described in the previous paragraph, therapeutic approaches aimed at reducing the expression of alpha-synuclein, APP and tau in neurons are being increasingly pursued. Recent studies using cell culture and animal models have demonstrated the ability to express miRNAs in neurons using lentiviral vectors configured to produce short hairpin RNAs. For example, the DYT1 dystonia disease-linked protein torsinA was knocked down in primary neuronal cultures using a recombinant feline immunodeficiency virus or lentiviral vectors expressing shRNAs (Gonzalez-Alegre et al., 2005; Harper and Gonzalez-Alegre, 2008). Lentiviral-mediated RNA interference has also been used to suppress alpha-synuclein levels in neurons in culture and in vivo in rat brain (Sapru et al., 2006). Lentivirus-based gene therapies for a range of diseases are at various stages of clinical development (Cockrell and Kafri, 2007), and we expect that RNA interference-based therapies for neurodegenerative disorders will eventually be tested.
Conclusion
Although miRNAs were first discovered in 1991 in invertebrates, the study of CNS miRNAs in injury, disease disease and normal function is still in its infancy (Lee et al. 1993). miRNAs appear altered in many neurodegenerative diseases and play important roles in many disease-related processes, such as inflammation and oncogenesis. Further understanding of the role of these miRNAs in normal and disease physiology may lead to improved treatment. Changes in miRNA expression that precede disease pathology, such as the decrease in miR-107 that precedes plaque formation in AD patients could provide improved detection of difficult to diagnose diseases (Wang et al. 2008). Manipulation of miRNAs in vitro could lead to more efficient and effective transplantation therapies for PD, HD and other diseases.
References
- Abelson JF, Kwan KY, O'Roak BJ, Baek DY, Stillman AA, Morgan TM, Mathews CA, Pauls DL, Rasin MR, Gunel M, Davis NR, Ercan-Sencicek AG, Guez DH, Spertus JA, Leckman JF, Dure LS, 4th, Kurlan R, Singer HS, Gilbert DL, Farhi A, Louvi A, Lifton RP, Sestan N, State MW. Sequence variants in SLITRK1 are associated with Tourette's syndrome. Science. 2005 Oct 14;310(5746):317–20. doi: 10.1126/science.1116502. [DOI] [PubMed] [Google Scholar]
- Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008 Sep 4;455(7209):64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge NJ, Tooney PA, Carroll AP, Gardiner E, Bowden N, Scott RJ, Tran N, Dedova I, Cairns MJ. Dysregulation of miRNA 181b in the temporal cortex in schizophrenia. Hum Mol Genet. 2008 Apr 15;17(8):1156–68. doi: 10.1093/hmg/ddn005. [DOI] [PubMed] [Google Scholar]
- Boissonneault V, Plante I, Rivest S, Provost P. MicroRNA-298 and microRNA-328 regulate expression of mouse beta-amyloid precursor protein-converting enzyme 1. J Biol Chem. 2009 Jan 23;284(4):1971–81. doi: 10.1074/jbc.M807530200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmistrova OA, Goltsov AY, Abramova LI, Kaleda VG, Orlova VA, Rogaev EI. MicroRNA in schizophrenia: genetic and expression analysis of miR-130b (22q11) Biochemistry (Mosc) 2007 May;72(5):578–82. doi: 10.1134/s0006297907050161. [DOI] [PubMed] [Google Scholar]
- Boudreau RL, Martins I, Davidson BL. Artificial microRNAs as siRNA shuttles: improved safety as compared to shRNAs in vitro and in vivo. Mol Ther. 2009 Jan;17(1):169–75. doi: 10.1038/mt.2008.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Pfaff SL, Gage FH. A functional study of miR-124 in the developing neural tube. Genes Dev. 2007 Mar 1;21(5):531–6. doi: 10.1101/gad.1519207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanotto D, Sakurai K, Lingeman R, Li H, Shively L, Aagaard L, Soifer H, Gatignol A, Riggs A, Rossi JJ. Combinatorial delivery of small interfering RNAs reduces RNAi efficacy by selective incorporation into RISC. Nucleic Acids Res. 2007;35(15):5154–64. doi: 10.1093/nar/gkm543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataldo AM, Petanceska S, Peterhoff CM, Terio NB, Epstein CJ, Villar A, Carlson EJ, Staufenbiel M, Nixon RA. App gene dosage modulates endosomal abnormalities of Alzheimer's disease in a segmental trisomy 16 mouse model of down syndrome. J Neurosci. 2003;23:6788–6792. doi: 10.1523/JNEUROSCI.23-17-06788.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockrell AS, Kafri T. Gene delivery by lentivirus vectors. Mol Biotechnol. 2007;36:184–204. doi: 10.1007/s12033-007-0010-8. [DOI] [PubMed] [Google Scholar]
- Cogswell JP, Ward J, Taylor IA, Waters M, Shi Y, Cannon B, Kelnar K, Kemppainen J, Brown D, Chen C, Prinjha RK, Richardson JC, Saunders AM, Roses AD, Richards CA. Identification of miRNA changes in Alzheimer's disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis. 2008 May;14(1):27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]
- Conaco C, Otto S, Han JJ, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci U S A. 2006 Feb 14;103(7):2422–7. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005 Jul 15;122(1):6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- De Pietri Tonelli D, Pulvers JN, Haffner C, Murchison EP, Hannon GJ, Huttner WB. miRNAs are essential for survival and differentiation of newborn neurons but not for expansion of neural progenitors during early neurogenesis in the mouse embryonic neocortex. Development. 2008;135:3911–3921. doi: 10.1242/dev.025080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharap A, Bowen K, Place R, Li LC, Vemuganti R. Transient focal ischemia induces extensive temporal changes in rat cerebral microRNAome. J Cereb Blood Flow Metab. 2009;29(4):675–687. doi: 10.1038/jcbfm.2008.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007 Sep;4(9):721–6. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmén J, Lindow M, Schütz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjärn M, Hansen HF, Berger U, Gullans S, Kearney P, Sarnow P, Straarup EM, Kauppinen S. LNA-mediated microRNA silencing in non-human primates. Nature. 2008b;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- Elmén J, Lindow M, Silahtaroglu A, Bak M, Christensen M, Lind-Thomsen A, Hedtjärn M, Hansen JB, Hansen HF, Straarup EM, McCullagh K, Kearney P, Kauppinen S. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008a;36:1153–1162. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G, 3rd, Kenny PJ, Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008 Jul;14(7):723–30. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998 Feb 19;391(6669):806–11. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Alegre P, Bode N, Davidson BL, Paulson HL. Silencing primary dystonia: lentiviral-mediated RNA interference therapy for DYT1 dystonia. J Neurosci. 2005;25:10502–10509. doi: 10.1523/JNEUROSCI.3016-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006 May 25;441(7092):537–41. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- Harper SQ, Staber PD, He X, Eliason SL, Martins IH, Mao Q, Yang L, Kotin RM, Paulson HL, Davidson BL. RNA interference improves motor and neuropathological abnormalities in a Huntington's disease mouse model. Proc Natl Acad Sci U S A. 2005 Apr 19;102(16):5820–5. doi: 10.1073/pnas.0501507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper SQ, Gonzalez-Alegre P. Lentivirus-mediated RNA interference in mammalian neurons. Methods Mol Biol. 2008;442:95–112. doi: 10.1007/978-1-59745-191-8_8. [DOI] [PubMed] [Google Scholar]
- Hébert SS, Horré K, Nicolaï L, Papadopoulou AS, Mandemakers W, Silahtaroglu AN, Kauppinen S, Delacourte A, De Strooper B. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer's disease correlates with increased BACE1/beta-secretase expression. Proc Natl Acad Sci U S A. 2008 Apr 29;105(17):6415–20. doi: 10.1073/pnas.0710263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hol EM, van Leeuwen FW, Fischer DF. The proteasome in Alzheimer's disease and Parkinson's disease: lessons from ubiquitin B+1. Trends Mol Med. 2005;11:488–495. doi: 10.1016/j.molmed.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Jeyaseelan K, Lim KY, Armugam A. MicroRNA expression in the blood and brain of rats subjected to transient focal ischemia by middle cerebral artery occlusion. Stroke. 2008 Mar;39(3):959–66. doi: 10.1161/STROKEAHA.107.500736. [DOI] [PubMed] [Google Scholar]
- Johnson R, Zuccato C, Belyaev ND, Guest DJ, Cattaneo E, Buckley NJ. A microRNA-based gene dysregulation pathway in Huntington's disease. Neurobiol Dis. 2008 Mar;29(3):438–45. doi: 10.1016/j.nbd.2007.11.001. [DOI] [PubMed] [Google Scholar]
- John M, Constien R, Akinc A, Goldberg M, Moon YA, Spranger M, Hadwiger P, Soutschek J, Vornlocher HP, Manoharan M, Stoffel M, Langer R, Anderson DG, Horton JD, Koteliansky V, Bumcrot D. Effective RNAi-mediated gene silencing without interruption of the endogenous microRNA pathway. Nature. 2007 Oct 11;449(7163):745–7. doi: 10.1038/nature06179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapsimali M, Kloosterman WP, de Bruijn E, Rosa F, Plasterk RH, Wilson SW. MicroRNAs show a wide diversity of expression profiles in the developing and mature central nervous system. Genome Biol. 2007;8(8):R173. doi: 10.1186/gb-2007-8-8-r173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller JN, Hanni KB, Markesbery WR. Impaired proteasome function in Alzheimer's disease. J Neurochem. 2000;75:436–439. doi: 10.1046/j.1471-4159.2000.0750436.x. [DOI] [PubMed] [Google Scholar]
- Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, Hannon G, Abeliovich A. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007 Aug 31;317(5842):1220–4. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky AM, Sonntag KC, Isacson O, Kosik KS. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells. 2006;24:857–864. doi: 10.1634/stemcells.2005-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005 Dec 1;438(7068):685–9. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- Kumar P, Wu H, McBride JL, Jung KE, Kim MH, Davidson BL, Lee SK, Shankar P, Manjunath N. Transvascular delivery of small interfering RNA to the central nervous system. Nature. 2007 Jul 5;448(7149):39–43. doi: 10.1038/nature05901. [DOI] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001 Oct 26;294(5543):858–62. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005 Feb 17;433(7027):769–73. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lukiw WJ, Zhao Y, Cui JG. An NF-kappaB-sensitive micro RNA-146a-mediated inflammatory circuit in Alzheimer disease and in stressed human brain cells. J Biol Chem. 2008 Nov 14;283(46):31315–22. doi: 10.1074/jbc.M805371200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev, et al. The microRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP. Pathways towards and away from Alzheimer's disease. Nature. 2004a Aug 5;430(7000):631–9. doi: 10.1038/nature02621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP, Maudsley S, Martin B. BDNF and 5-HT: a dynamic duo in age-related neuronal plasticity and neurodegenerative disorders. Trends Neurosci. 2004b Oct;27(10):589–94. doi: 10.1016/j.tins.2004.08.001. [DOI] [PubMed] [Google Scholar]
- McBride JL, Boudreau RL, Harper SQ, Staber PD, Monteys AM, Martins I, Gilmore BL, Burstein H, Peluso RW, Polisky B, Carter BJ, Davidson BL. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc Natl Acad Sci U S A. 2008 Apr 15;105(15):5868–73. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellios N, Huang HS, Grigorenko A, Rogaev E, Akbarian S. A set of differentially expressed miRNAs, including miR-30a-5p, act as post-transcriptional inhibitors of BDNF in prefrontal cortex. Hum Mol Genet. 2008 Oct 1;17(19):3030–42. doi: 10.1093/hmg/ddn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington's disease. J Neurosci. 2008 Dec 31;28(53):14341–6. doi: 10.1523/JNEUROSCI.2390-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins DO, Jeffries CD, Jarskog LF, Thomson JM, Woods K, Newman MA, Parker JS, Jin J, Hammond SM. microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biol. 2007;8(2):R27. doi: 10.1186/gb-2007-8-2-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redell JB, Liu Y, Dash PK. Traumatic brain injury alters expression of hippocampal microRNAs: potential regulators of multiple pathophysiological processes. J Neurosci Res. 2009 May 1;87(6):1435–48. doi: 10.1002/jnr.21945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G, Li T, Lan JQ, Wilz A, Simon RP, Boison D. Lentiviral RNAi-induced downregulation of adenosine kinase in human mesenchymal stem cell grafts: a novel perspective for seizure control. Exp Neurol. 2007;208(1):26–37. doi: 10.1016/j.expneurol.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saba R, Goodman CD, Huzarewich RL, Robertson C, Booth SA. A miRNA signature of prion induced neurodegeneration. PLoS ONE. 2008;3(11):e3652. doi: 10.1371/journal.pone.0003652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehi A, Delcroix JD, Belichenko PV, Zhan K, Wu C, Valletta JS, Takimoto-Kimura R, Kleschevnikov AM, Sambamurti K, Chung PP, Xia W, Villar A, Campbell WA, Kulnane LS, Nixon RA, Lamb BT, Epstein CJ, Stokin GB, Goldstein LS, Mobley WC. Increased App expression in a mouse model of Down's syndrome disrupts NGF transport and causes cholinergic neuron degeneration. Neuron. 2006;51:29–42. doi: 10.1016/j.neuron.2006.05.022. [DOI] [PubMed] [Google Scholar]
- Sapru MK, Yates JW, Hogan S, Jiang L, Halter J, Bohn MC. Silencing of human alpha-synuclein in vitro and in rat brain using lentiviral-mediated RNAi. Exp Neurol. 2006;198:382–390. doi: 10.1016/j.expneurol.2005.12.024. [DOI] [PubMed] [Google Scholar]
- Schwamborn JC, Berezikov E, Knoblich JA. The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell. 2009;136(5):913–25. doi: 10.1016/j.cell.2008.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008 Sep 4;455(7209):58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Sheedy FJ, O'Neill LA. Adding fuel to fire: microRNAs as a new class of mediators of inflammation. Ann Rheum Dis. 2008 Dec;67 3:iii50–5. doi: 10.1136/ard.2008.100289. [DOI] [PubMed] [Google Scholar]
- Silahtaroglu AN, Nolting D, Dyrskjøt L, Berezikov E, Møller M, Tommerup N, Kauppinen S. Detection of microRNAs in frozen tissue sections by fluorescence in situ hybridization using locked nucleic acid probes and tyramide signal amplification. Nat Protoc. 2007;2(10):2520–8. doi: 10.1038/nprot.2007.313. [DOI] [PubMed] [Google Scholar]
- Singer O, Marr RA, Rockenstein E, Crews L, Coufal NG, Gage FH, Verma IM, Masliah E. Targeting BACE1 with siRNAs ameliorates Alzheimer disease neuropathology in a transgenic model. Nat Neurosci. 2005 Oct;8(10):1343–9. doi: 10.1038/nn1531. [DOI] [PubMed] [Google Scholar]
- Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J, Hulihan M, Peuralinna T, Dutra A, Nussbaum R, Lincoln S, Crawley A, Hanson M, Maraganore D, Adler C, Cookson MR, Muenter M, Baptista M, Miller D, Blancato J, Hardy J, Gwinn-Hardy K. alpha-Synuclein locus triplication causes Parkinson's disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- Singleton A, Myers A, Hardy J. The law of mass action applied to neurodegenerative disease: a hypothesis concerning the etiology and pathogenesis of complex diseases. Hum Mol Genet. 2004;13(Spec No 1):R123–126. doi: 10.1093/hmg/ddh093. [DOI] [PubMed] [Google Scholar]
- Smirnova L, Gräfe A, Seiler A, Schumacher S, Nitsch R, Wulczyn FG. Regulation of miRNA expression during neural cell specification. Eur J Neurosci. 2005 Mar;21(6):1469–77. doi: 10.1111/j.1460-9568.2005.03978.x. [DOI] [PubMed] [Google Scholar]
- Tsang J, Zhu J, van Oudenaarden A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell. 2007 Jun 8;26(5):753–67. doi: 10.1016/j.molcel.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbich C, Kuehbacher A, Dimmeler S. Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc Res. 2008 Sep 1;79(4):581–8. doi: 10.1093/cvr/cvn156. Epub 2008 Jun 11. [DOI] [PubMed] [Google Scholar]
- van Solingen C, Seghers L, Bijkerk R, Duijs JM, Roeten MK, van Oeveren-Rietdijk AM, Baelde HJ, Monge M, Vos JB, de Boer HC, Quax PH, Rabelink TJ, van Zonneveld AJ. Antagomir-Mediated Silencing of Endothelial Cell Specific MicroRNA-126 Impairs Ischemia-Induced Angiogenesis. J Cell Mol Med. 2008 Dec 16; doi: 10.1111/j.1582-4934.2008.00613.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Várallyay E, Burgyán J, Havelda Z. MicroRNA detection by northern blotting using locked nucleic acid probes. Nat Protoc. 2008;3(2):190–6. doi: 10.1038/nprot.2007.528. [DOI] [PubMed] [Google Scholar]
- Wang WX, Rajeev BW, Stromberg AJ, Ren N, Tang G, Huang Q, Rigoutsos I, Nelson PT. The expression of microRNA miR-107 decreases early in Alzheimer's disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J Neurosci. 2008a;28:1213–1223. doi: 10.1523/JNEUROSCI.5065-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, van der Walt JM, Mayhew G, Li YJ, Züchner S, Scott WK, Martin ER, Vance JM. Variation in the miRNA-433 binding site of FGF20 confers risk for Parkinson disease by overexpression of alpha-synuclein. Am J Hum Genet. 2008b;82:283–289. doi: 10.1016/j.ajhg.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Deng W, Gage FH. Mechanisms and functional implications of adult neurogenesis. Cell. 2008 Feb 22;132(4):645–60. doi: 10.1016/j.cell.2008.01.033. [DOI] [PubMed] [Google Scholar]
- Zhao C, Sun G, Li S, Shi Y. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat Struct Mol Biol. 2009;16(4):365–371. doi: 10.1038/nsmb.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Yuan P, Wang Y, Hunsberger JG, Elkahloun A, Wei Y, Damschroder-Williams P, Du J, Chen G, Manji HK. Evidence for Selective microRNAs and their Effectors as Common Long-Term Targets for the Actions of Mood Stabilizers. europsychopharmacology. 2008 Aug 13; doi: 10.1038/npp.2008.131. [DOI] [PMC free article] [PubMed] [Google Scholar]


