Abstract
Natural products display impressive activities against a wide range of targets, including viruses, microbes and tumors. However, their clinical use is hampered frequently by their scarcity and undesirable toxicity. Not only can engineering Escherichia coli for plasmid-based pharmacophore biosynthesis offer alternative means of simple and easily-scalable production of valuable yet hard-to-obtain compounds, but also carries a potential for providing a straightforward and efficient means of preparing natural product analogs. The quinomycin family of nonribosomal peptides, including echinomycin, trtiostin A and SW-163s, are important secondary metabolites imparting antibiotic antitumor activity via DNA bisintercalation. Previously we have shown the production of echinomycin and trtiostin A in E. coli using our convenient and modular plasmid system to introduce these heterologous biosynthetic pathways into E. coli. However, we have yet to develop a novel biosynthetic pathway capable of producing bioactive unnatural natural products in E. coli. Here we report an identification of a new gene cluster responsible for the biosynthesis of SW-163s that involves previously unknown biosynthesis of (+)-(1S, 2S)-norcoronamic acid and generation of aliphatic side chains of various sizes via iterative methylation of an unactivated carbon center. Substituting an echinomycin biosynthetic gene with a gene from the newly identified SW-163 biosynthetic gene cluster, we were able to rationally re-engineer the plasmid-based echinomycin biosynthetic pathway for the production of a novel bioactive compound in E. coli.
Keywords: depsipeptide, hybrid molecule, nonribosomal peptide synthetase, engineered biosynthesis, E. coli
Introduction
With the growing global threat of multi-drug-resistant microbes and increasing economic and environmental concerns on synthetic production of natural products and their analogs, there is an increasing interest in developing alternate platforms for the discovery, production and development of pharmaceutically valuable compounds. To date, many biosynthetic gene clusters encoding polyketide synthases (PKSs), nonribosomal peptide (NRP) synthetases (NRPSs), mixed PKS—NRPSs and associated auxiliary enzymes have been found in various different organisms through recent genome and metagenome sequencing efforts1. However, many organisms are difficult to culture in large scale2, and some are impractical to handle due to pathogenicity. Others produce natural products at very low level or maintain their gene clusters silent under conventional culture conditions3. One way to exploit the potential of these organisms and the genomic information for drug discovery and development is to establish a heterologous production system using a convenient host organism capable of expressing the exogenous genes and biosynthesizing the desired compounds. Several organisms, including certain Streptomyces strains and yeast, have been used as such hosts4, but Escherichia coli carries certain advantages over others. First, a wealth of tools are available for incorporation, controlling expression, and modification of foreign genes in E. coli, such as plasmids with various useful properties, including plasmid compatibility and selectability. Additionally, methods of manipulating E. coli and its vectors are well-established and straightforward to perform. Furthermore, E. coli is forgiving in expressing exogenous genes and readily grows with a very short doubling time in a simple, inexpensive growth medium without requiring particularly difficult-to-obtain culture conditions. Although E. coli is not known for its ability to produce diverse secondary metabolites, the fact that E. coli is naturally capable of producing siderophore NRPs5 and a PK—NRP hybrid molecule colibactin6 suggests that its metabolic system is not entirely foreign to natural product biosynthesis. To introduce a heterologous biosynthetic pathway into E. coli, we chose a plasmid-based system. In this approach, plasmids are used to assemble and deliver genes necessary to reconstitute the target biosynthetic pathway into the host. Although this approach demands initial efforts of constructing plasmids bearing multiple genes of the target biosynthetic pathway, we have lowered the hurdle significantly by developing a cloning method that exploits iterative ligation of DNA fragments bearing compatible cohesive-ends. This led to the first successful production of a bioactive NRP natural product echinomycin 1 and triostin A 2 in E. coli (Figure 1)7.
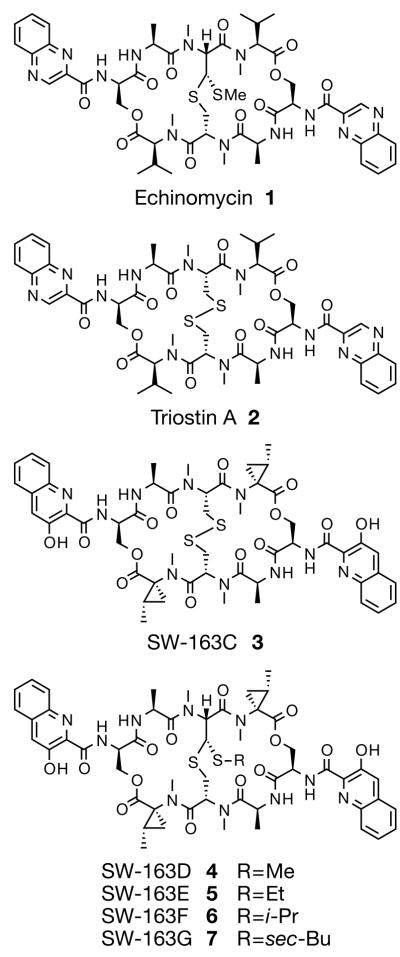
Figure 1.
Chemical structures of quinomycin antibiotics.
In parallel with the development of the E. coli-based system for heterologous production of secondary metabolites, we are also trying to identify new biosynthetic pathways that give rise to unique chemical structures found among natural products. Such knowledge is crucial in broadening the scope of combinatorial biosynthesis of various natural products and their analogs. Discovery of new enzymes that are capable of performing different transformations permits expansion of the toolbox with which we can devise novel biosynthetic pathways for production of interesting new compounds. Combined with the advantages of using E. coli as a heterologous production host described earlier, E. coli-based plasmid-borne heterologous secondary metabolite biosynthetic system can not only facilitate the reconstitution of various biosynthetic gene clusters but also provide a flexible platform suitable for fast and simple rational re-designing of biosynthetic pathways through modular reassembling of the biosynthetic genes from various biosynthetic systems. Here, to put this concept to work, we have identified a new biosynthetic gene cluster responsible for the biosynthesis of the SW-163 series of NRPs involving several previously uncharacterized enzymatic transformations. Then we took this new knowledge and applied it to our previously established E. coli-based plasmid-borne NRP biosynthetic system to achieve a rationally engineered biosynthesis of unnatural bioactive NRP we named ecolimycin C, a hybrid compound of 2 and SW-163 C 3.
Results
Isolation of SW-163 Biosynthetic Gene Cluster
Previously, we reported the total biosynthesis of two antitumor NRPs, 1 and its intermediate 2 in E. coli7. Originally identified and isolated from Streptomyces strains, both compounds were categorized as quinomycin antibiotics because of their quinoxaline or quinoline chromophores attached to the C2-symmetric bicyclic depsipeptide core8. This family of antibiotics is well-characterized, and exhibits potent antibacterial, anticancer and antiviral activities9. While both compounds bis-intercalate into GC-rich DNA sequences, 1 and 2 exhibit distinct sequence preference. Now we have isolated an additional family of quinomycin antibiotics, SW-163C 3, D 4, E 5, F 6 and G 710,11 from Streptomyces sp. SNA15896 (Figure 1). All of them carry a pair of 3-hydroxyquinaldic acid (HQA, 8) chromophores along with a non-proteinogenic amino acid, norcoronamic acid (NCA), whose biosynthetic mechanism has not been described to date. The absolute configuration of 4, and hence the SW-163s, was determined by establishing the absolute configuration of the NCA residue to be (+)-(1S, 2S)-norcoronamic acid [(+)-NCA, 9]11. Also, like in 1, compounds 4, 5, 6 and 7 all possess a central thioacetal cross-bridge formed by the chemical rearrangement of a disulfide bridge catalyzed by a post-NRPS modification enzyme, methyltransferase7. Most interestingly, however, the thioacetal bond is appended by a distinct alkyl side chain in these compounds, each varying by a single carbon unit.
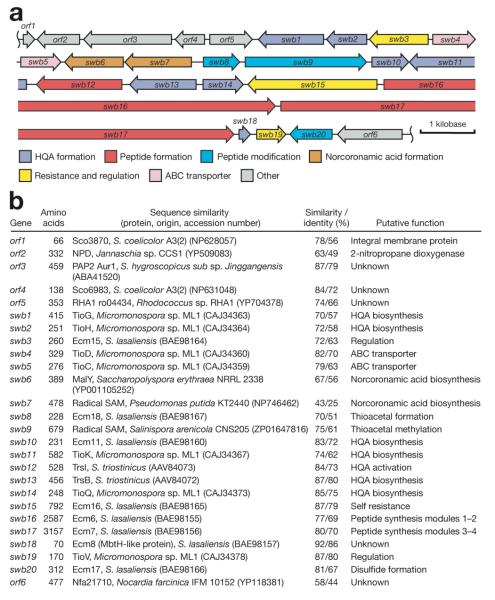
Closely resembling the echinomycin biosynthetic gene cluster, the SW-163 biosynthetic gene cluster (Figure 2) was identified and isolated from the total DNA of Streptomyces sp. SNA15896 (Supporting Information Experimental Procedures). DNA sequence analysis of this 38 kilobase-long cluster revealed the presence of fifteen genes considered to be involved directly in the biosynthesis of SW-163s: seven genes (swb1, swb2, swb10, swb11, swb13, swb14 and swb18) likely forming the proposed SW-163 primer unit 8 (Figure 3a), three genes (swb6, swb7, swb9) assembling 9 (Figure 3b), and five genes (swb8, swb12, swb16, swb17, swb20) constructing and modifying the peptide backbone (Figure 3c). The cluster also contains a presumed resistance gene (swb15) and other genes, including swb3, swb4, swb5 and swb19, presumably involved in the regulation of biosynthesis or export of the products.
Figure 2.
Biosynthetic cluster of SW-163 from Streptomyces sp. SNA15896. (a) Organization of the SW-163 biosynthetic gene cluster. (b) Deduced functions of the ORFs identified within the SW-163 biosynthetic gene cluster based on the percentage sequence similarity/identity to known proteins determined using BLAST.
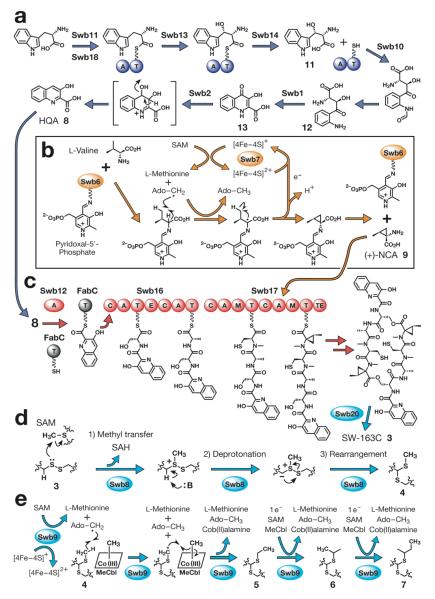
Figure 3.
Proposed mechanism for the SW-163 biosynthesis. (a) Proposed pathway for the biosynthesis of HQA 8. (b) Proposed mechanism for the formation of (+)-(1S, 2S)-norcoronamic acid [9 (+)-NCA]. (c) Proposed pathway for the biosynthesis of the octadepsipeptide core. (d) Proposed mechanism for the formation of a thioacetal bridge. (e) Proposed mechanism for the formation of a series of alkyl substituents differing by one carbon unit. NRPS domain name abbreviations are: A, adenylation; C, condensation; E, epimerization; M, N-methyltransferase ; M*, inactive methylation; T, thiolation; and TE, thioesterase. Other abbreviations are: Ado-CH3, 5′-deoxyadenosine; AMP adenosine 5′-monophosphate ; HQA, 3-hydroxyquinaldic acid; MeCbl, methylcobalamine; (+)-NCA, (+)-(1S, 2S)-norcoronamic acid; SAM, S-adenosyl-L-methionine; and SAH, S-adenosyl-L-homocysteine.
Based on the results from our feeding experiments using isotope-labeled substrates14 in conjunction with the previous BLAST12 analyses of the genes from the SW-163, thiocoraline13 and echinomycin7 biosynthetic clusters, biosynthesis of 8 is thought to proceed similarly as previously proposed for the biosynthesis of quinoxaline-2-carboxylic acid (QXC 10, see Figure 4b). That is, the biosynthesis proceeds via the initial β-hydroxylation of L-tryptophan to form (2S, 3S)-β-hydroxytryptophan 11, followed by the formation of β-hydroxykynurenine 12. Transamination of 12 by Swb1 can generate 13, and oxidation and dehydration of 13 by Swb2 can afford 8 (Figure 3a).
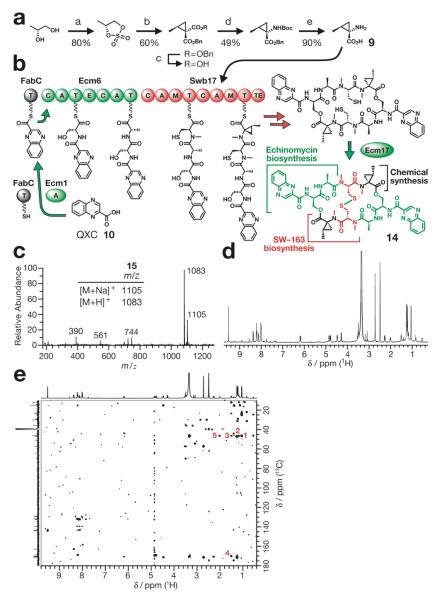
Figure 4.
Biosynthesis of a novel antibiotic ecolimycin C 14 in E. coli. (a) Chemical synthesis of the extender unit 9 fed to E. coli for the biosynthesis of 14. a, i) SOCl2, ClCH2CH2Cl, ii) cat. RuCl3, NaIO4, CH3CN—H2O; b, dibenzyl malonate, NaH, 1,2-dimethoxyethane; c, 1 M NaOH, MeOH; d, diphenylphosphoryl azide, Et3N, t-BuOH; e, i) H2, 10% Pd/C, EtOAc, ii) 6 M HCl, MeOH. Product yield of each chemical reaction step is reported in percentages. (b) Schematics of the pathway for the E. coli biosynthesis of 14. Abbreviation: QXC, quinoxaline-2-carboxylic acid. (c) LC—MS data, (d) 1H NMR spectrum and (e) HMBC spectrum of 14. Important correlations (in red) are 1: δ 1.05 (NCA—CH3) and 46.4 (NCA—Cα); 2: δ 1.18 (NCA—CH2) and 46.4 (NCA—Cα); 3: δ 1.51 (NCA—CH2) and 46.4 (NCA—Cα); 4: δ 1.51 (NCA—CH2) and 170.1 (NCA—CO); 5: δ 2.01 (NCA—CH) and 46.4 (NCA—Cα).
Despite the structural similarity of 9 to coronamic acid, whose biosynthesis involves a cryptic chlorination step15, no homologs of the coronamic acid biosynthetic genes were identified in the SW-163 biosynthetic gene cluster. Although the mechanism of biosynthesis of 9 remains unclear, based on previous reports of a radical SAM dehydrogenase and lysine 2, 3-aminomutase16,17, we hypothesize that predicted radical SAM protein Swb7 and pyridoxal-5′-phosphate (PLP)-dependent aminotransferase Swb6 can work together closely to form 9 from L-Val via radical cyclopropanation (Figure 3b).
Assembling of the SW-163 peptide scaffold from L-Ser, L-Ala, L-Cys and 9 appears to be accomplished by two bimodular NRPSs, Swb16 and Swb17. The terminal TE domain in Swb17 likely homodimerizes and cycloreleases the peptide chain (Figure 3c)18. Following cyclization of the linear peptide, an oxidoreductase Swb20 presumably catalyzes an oxidation reaction to generate the disulfide bond in 3. Subsequently, the disulfide bridge in 3 can be converted to a thioacetal bridge to form 4 (Figure 3d) by a predicted methyltransferase Swb8, homolog of the SAM-dependent methyltransferase Ecm18 that catalyzes the equivalent step in the echinomycin biosynthetic pathway7.
Lastly but most notably, we propose that Swb9, a predicted radical SAM protein, performs the further iterative methylation of the thioacetal bridge. With the finding of Swb9, we are able to come up with a more plausible mechanism for the iterative methylation of the thioacetal bridge (Figure 3e) than our previously proposed mechanism7. Using the well-established method19 for isotopically enriching the SAM-derived methyl groups with a labeled L-methionine, we were able to observe that, in addition to the four N-methyl groups of the cyclic depsipeptide core, every proton in the S-alkyl chains of 4, 5, 6 and 7 became deuterated upon feeding of [methyl-D3]-L-methionine to the SW-163-producing host (Supporting Information Figure 1). These results were consistent with the S-alkyl side chains formed by step-wise transfer of a methyl group derived from SAM by a methyltransferase. As proposed for the methylation during the fosfomycin biosynthesis20, a radical SAM protein can perform a methyl transfer reaction on a non-activated carbon at the cost of one molecule of SAM and methylcobalamine (MeCbl). We hypothesize that this unique radical SAM methyltransferase can iteratively methylate 4 to generate ethyl- 5, i-propyl- 6, and sec-butyl 7 derivatives (Figure 3e). Further studies to support this idea are currently underway.
E. coli Biosynthesis of Novel NRP with Antibiotic Activity
To further challenge the versatility of our system, we aimed to biosynthesize in vivo in E. coli a new, bioactive compound. For this purpose, as an initial attempt we chose to replace L-Val in the depsipeptide scaffold of 2 with 9 to yield a hybrid molecule 14 we termed ecolimycin C. The crystal structure of the complex between 2 and its DNA substrate showed L-Val minimally participating in the DNA recognition21. Also, the fact that the SW-163 antibiotics carry this moiety indicated that the new compound would most likely retain antibiotic activity.
To convert the triostin A biosynthetic pathway into a biosynthetic pathway for 14, Ecm7 was replaced with Swb17, a bimodular NRPS showing high production level in E. coli (Supporting Information Figure 2a) that contains the adenylation domain capable of accepting 9 as an extender unit and the thioesterase domain capable of homodimerizing and cycloreleasing the peptide chain containing 9. To minimize plasmid instability, gene assemblies were kept to a minimal length while maintaining as many genes within a contiguous metabolic pathway on a single plasmid as possible; the genes for the biosynthesis of 10 were put together on one plasmid (pKW532), two megasynthetases Ecm6 and Swb17 on another (pKW755, Supporting Information Figure 3a), and the rest of the genes on the third plasmid (pKW539, Supporting Information Figure 3b). Also, to prevent potential premature termination and mRNA degradation22 while transcribing an extremely long polycistronic gene assembly, all genes were assembled in a multi-monocistronic fashion.
Because 14 may possess antibiotic activity, the growth of E. coli can be hampered by its accumulation in the cytosol. Thus, a self-resistance mechanism was included to circumvent this potential problem. Previously, the echinomycin resistance-conferring gene ecm16, a close homolog of the daunorubicin resistance-conferring gene drrC that provides non-destructive resistance against daunorubicin in S. lividans and E. coli23, was used successfully for our E. coli in vivo production of 1 and 27. Considering this previous report, along with the observation that the expression level of swb15 in E. coli (Supporting Information Figure 2b) was comparable to that of ecm16, we reasoned that either Ecm16 or Swb15, UvrA-like protein with high (90%) sequence homology to Ecm16 (Figure 2b), can provide E. coli resistance against 14. To ascertain the effectiveness of the two genes on the culture viability and titer of 14, two separate constructs were prepared to include either ecm16 (pKW539) or swb15 (pKW756) in the expression system (Supporting Information Figure 3b). However, since there was almost no difference in the titer between the two systems, Ecm16 was used throughout the studies.
BL21 (DE3) was transformed with three plasmids pKW532, pKW755 and pKW539, and subjected to shake-flask cultivation for seven days in M9 minimal medium supplemented with chemically synthesized 9 (Figure 4a). After subjecting the E. coli culture extract to a series of chromatographic steps to obtain purity necessary for collecting well-resolved 2D NMR spectra, the final yield of 14 was 0.6 mg per liter of culture. The expected mass of [M+H]+ ion at m/z = 1083.3849 was obtained on the purified 14 by FAB—HR—MS (Figure 4c and Supporting Information Table 3). The compound was characterized further by 1H, 13C and 2D NMR (Figure 4d, 4e and Supporting Information Table 3, Figures 4 and 5) to reveal a characteristic resonance at δ 9.50 (s, 1 H, QXC H-3′). Ultimately, total correlated spectroscopy (TOCSY) (Supporting Information Figure 5b) confirmed unambiguously that our engineered bacterium was capable of producing 14. Furthermore, the antibiotic activity of this new hybrid NRP compound was established to be as effective if not more potent than 3 as determined by halo formation assay against Bacillus subtilis (Figure 5).
Figure 5.
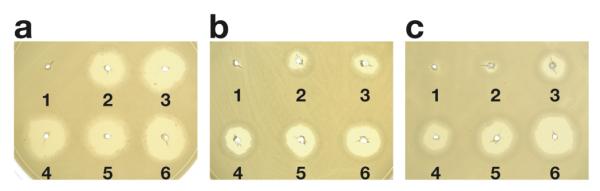
Halo-formation assay for assessing the antibiotic activity of 2, 3 and 14. A tryptic soy agar plate plated with B. subtilis was spotted with (a) 2, (b) 3 and (c) 14. Each well contained the corresponding compound at an incremental amount of 2: 1.0 ng; 3: 2.0 ng; 4: 4.0 ng; 5: 8.0 ng and 6: 16 ng. Well 1 is a negative control.
Discussion
The E. coli-based biosynthetic system clearly exhibits its ease of manipulation when one attempts to introduce planned modifications into the target biosynthetic pathway. Taking full advantage of these features and challenging our plasmid-borne NRP biosynthetic system further, we decided to combine two NRP biosynthetic pathways from two different sources to biosynthesize a novel hybrid NRP which we named ecolimycin C 14. To prepare this hybrid molecule, we applied part of the biosynthetic gene cluster for the quinomycin antibiotics SW163C—G that we have newly discovered to our plasmid-borne triostin A biosynthetic pathway. Apart from the very interesting thioacetal bridge alkyl side chain formation that appears to be accomplished by unprecedented iterative methylation of non-activated carbon centers by a radical SAM methyltransferase Swb9 (Figure 3e), the SW-163 biosynthetic pathway involves the production of non-proteinogenic amino acid 9 (Figure 3b) and its incorporation into the homodimeric cyclic depsipeptide core by the NRPS Swb17 (Figure 3c). By replacing Ecm7 in the triostin A biosynthetic pathway with Swb17, we envisioned the production of 14, which carries the chromophores of 2 while having a sterically more constrained, α-branched amino acid residue of 3 in its backbone (Figure 3b). The HQA-to-QXC and L-Val-to-(+)-NCA switch seemed modest enough to preserve the antibiotic activity of the parent compounds 2 and 3, yet different enough to see possible alterations in the substrate DNA affinity and/or sequence selectivity24.
Although it was necessary to feed E. coli chemically synthesized 9 to accomplish the biosynthesis of 14, the observed titer of 14 was significantly higher than that of 1 or 2 without requiring feeding of 10. This unexpected high yield of an unnatural NRP could be due to higher expression level of swb17 (Supporting Information Figure 2a). Also, lower metabolic turnover of the building block 9 may have helped boost the yield further. Nevertheless, this result indicates that, in addition to Ecm7, another NRPS Swb17 can also accept, elongate, homodimerize and cyclorelease unnatural peptide chain. Furthermore, Ecm17 exhibited relaxed substrate specificity by accepting the unnatural intermediate to 14. Most importantly, however, our result demonstrates that two NRPSs from differing origin were able to establish productive in vivo intermodular interactions and produce 14 in E. coli. The fact that 14 was obtained at a comparable or better yield than the parent compounds in E. coli by a simple substitution of an enzyme in the corresponding heterologous biosynthetic pathway without any extensive modifications of the pathway nor its constituents is especially encouraging for the future prospect of our system. In addition, the efficiency and versatility of the E. coli platform can be improved further by incorporating various technologies currently being investigated, including the development of efficient fed-batch fermentation protocols25, use of quantitative metabolic flux analyses for identifying the possible routes to improving the system26, application of directed evolution for improving the protein-protein interactions among foreign enzymes and their domains27, and manipulation of the transcriptional machineries for tailoring the global cellular phenotype for improved heterologous secondary metabolite production28. Taken together, we believe that our results have laid down the foundation for the future development and refinement of E. coli as a flexible, robust platform for fast and simple large-scale production of pharmaceutically useful molecules and their analogs in an environmentally friendly fashion from simple carbon and nitrogen sources.
Supplementary Material
Acknowledgment
This paper is dedicated to the memory of Professor Akitami Ichihara (1934–2009), for his pioneering studies in bioorganic chemistry, including total synthesis of tautomycin. We thank DAIICHI SANKYO Co., Ltd. for providing a sample of Streptomyces sp. SNA15896. This study was supported by Special Coordination Funds for Promoting Sciences and Technology of the Ministry of Education, Sport, Culture, Science and Technology of Japan (K.W.), by National Institute of General Medical Sciences grant GM 075857-01 (C.C.C.W. and K.W.), by American Cancer Society grant RSG-06-010-01-CDD (C.C.C.W. and K.W.), by Novozymes Japan Research Foundation (K.W.) and by Northern Advancement Center for Science and Technology Foundation (K.W.).
Footnotes
Supporting Information Available: Experimental procedures, two tables of DNA sequence information for the oligonucleotides, a figure for mass spectra of 3, 4, 5, 6, 7 isolated from labeled methionine-feeding experiment, and a table and two figures of spectrometric data on the E. coli-produced 14.
REFERENCES
- (1).Bode HB, Muller R. Angew. Chem. Int. Ed. Engl. 2005;44:6828–46. doi: 10.1002/anie.200501080. [DOI] [PubMed] [Google Scholar]
- (2).Riesenfeld CS, Schloss PD, Handelsman J. Annu. Rev. Genet. 2004;38:525–52. doi: 10.1146/annurev.genet.38.072902.091216. [DOI] [PubMed] [Google Scholar]
- (3).Wenzel SC, Müller R. Curr. Opin. Biotechnol. 2005;16:594–606. doi: 10.1016/j.copbio.2005.10.001. [DOI] [PubMed] [Google Scholar]
- (4).Zhang H, Wang Y, Pfeifer BA. Mol. Pharm. 2008;5:212–25. doi: 10.1021/mp7001329. [DOI] [PubMed] [Google Scholar]
- (5).Valdebenito M, Crumbliss AL, Winkelmann G, Hantke K. Int. J. Med. Microbiol. 2006;296:513–20. doi: 10.1016/j.ijmm.2006.06.003. [DOI] [PubMed] [Google Scholar]
- (6).Nougayrède JP, Homburg S, Taieb F, Boury M, Brzuszkiewicz E, Gottschalk G, Buchrieser C, Hacker J, Dobrindt U, Oswald E. Science. 2006;313:848–51. doi: 10.1126/science.1127059. [DOI] [PubMed] [Google Scholar]
- (7).Watanabe K, Hotta K, Praseuth AP, Koketsu K, Migita A, Boddy CN, Wang CC, Oguri H, Oikawa H. Nat. Chem. Biol. 2006;2:423–8. doi: 10.1038/nchembio803. [DOI] [PubMed] [Google Scholar]
- (8).Martin DG, Mizsak SA, Biles C, Stewart JC, Bacynsky L, Meulman PA. J. Antibiot. (Tokyo) 1975;28:332–6. doi: 10.7164/antibiotics.28.332. [DOI] [PubMed] [Google Scholar]
- (9).Sieber SA, Marahiel MA. Chem. Rev. 2005;105:715–38. doi: 10.1021/cr0301191. [DOI] [PubMed] [Google Scholar]
- (10).Kurosawa K, Takahashi K, Tsuda E. J. Antibiot. (Tokyo) 2001;54:615–21. doi: 10.7164/antibiotics.54.615. [DOI] [PubMed] [Google Scholar]
- (11).Nakaya M, Oguri H, Takahashi K, Fukushi E, Watanabe K, Oikawa H, Koketsu K. Biosci. Biotechnol. Biochem. 2007;71:2969–76. doi: 10.1271/bbb.70371. [DOI] [PubMed] [Google Scholar]
- (12).McGinnis S, Madden TL. Nucleic Acids Res. 2004;32:W20–5. doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Lombo F, Velasco A, Castro A, de la Calle F, Brana AF, Sanchez-Puelles JM, Mendez C, Salas JA. Chembiochem. 2006;7:366–76. doi: 10.1002/cbic.200500325. [DOI] [PubMed] [Google Scholar]
- (14).Koketsu K, Oguri H, Watanabe K, Oikawa H. Org. Lett. 2006;8:4719–22. doi: 10.1021/ol061738+. [DOI] [PubMed] [Google Scholar]
- (15).Vaillancourt FH, Yeh E, Vosburg DA, O’Connor SE, Walsh CT. Nature. 2005;436:1191–4. doi: 10.1038/nature03797. [DOI] [PubMed] [Google Scholar]
- (16).Frey PA, Magnusson OT. Chem. Rev. 2003;103:2129–48. doi: 10.1021/cr020422m. [DOI] [PubMed] [Google Scholar]
- (17).Yokoyama K, Numakura M, Kudo F, Ohmori D, Eguchi T. J. Am. Chem. Soc. 2007;129:15147–55. doi: 10.1021/ja072481t. [DOI] [PubMed] [Google Scholar]
- (18).Trauger JW, Kohli RM, Mootz HD, Marahiel MA, Walsh CT. Nature. 2000;407:215–8. doi: 10.1038/35025116. [DOI] [PubMed] [Google Scholar]
- (19).Rawlings BJ. Nat. Prod. Rep. 1997;14:523–56. doi: 10.1039/np9971400523. [DOI] [PubMed] [Google Scholar]
- (20).Woodyer RD, Li G, Zhao H, van der Donk WA. Chem. Commun. 2007:359–61. doi: 10.1039/b614678c. [DOI] [PubMed] [Google Scholar]
- (21).Wang AH, Ughetto G, Quigley GJ, Hakoshima T, van der Marel GA, van Boom JH, Rich A. Science. 1984;225:1115–21. doi: 10.1126/science.6474168. [DOI] [PubMed] [Google Scholar]
- (22).Sørensen HP, Mortensen KK. J. Biotechnol. 2005;115:113–28. doi: 10.1016/j.jbiotec.2004.08.004. [DOI] [PubMed] [Google Scholar]
- (23).Lomovskaya N, Hong SK, Kim SU, Fonstein L, Furuya K, Hutchinson RC. J. Bacteriol. 1996;178:3238–45. doi: 10.1128/jb.178.11.3238-3245.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Fox KR, Gauvreau D, Goodwin DC, Waring MJ. Biochem. J. 1980;191:729–42. doi: 10.1042/bj1910729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Lau J, Tran C, Licari P, Galazzo J. J. Biotechnol. 2004;110:95–103. doi: 10.1016/j.jbiotec.2004.02.001. [DOI] [PubMed] [Google Scholar]
- (26).Gonzalez-Lergier J, Broadbelt LJ, Hatzimanikatis V. Biotechnol. Bioeng. 2006;95:638–44. doi: 10.1002/bit.20925. [DOI] [PubMed] [Google Scholar]
- (27).Fischbach MA, Lai JR, Roche ED, Walsh CT, Liu DR. Proc. Natl. Acad. Sci. USA. 2007;104:11951–6. doi: 10.1073/pnas.0705348104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Alper H, Stephanopoulos G. Metab. Eng. 2007;9:258–67. doi: 10.1016/j.ymben.2006.12.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.