Summary
To ensure correct patterns of gene expression, eukaryotes use a variety of strategies to repress transcription. The transcriptional regulators mediating this repression can be broadly categorized as either passive or active repressors. While passive repressors rely on mechanisms such as steric hindrance of transcriptional activators to repress gene expression, active repressors display inherent repressive abilities commonly conferred by discrete repression domains. Recent studies have indicated that both categories of regulators function in a variety of plant processes, including hormone signal transduction, developmental pathways, and response to biotic and abiotic stresses.
Introduction
As sessile organisms, plants must perceive and respond to a wide range of biotic and abiotic signals in order to optimize their growth and development. Moreover, cells within a plant rely on positional information from their neighbors in order to adopt proper fates. A large part of these responses involves appropriate regulation of gene expression. To this end, eukaryotes employ a wide repertoire of transcriptional repression mechanisms. In general, such mechanisms can be separated into two main types: active and passive repression. Active repressors display an intrinsic repressive capacity conferred by defined repression domains [1,2]. For example, repression domains of sequence-specific transcription factors can be used to interact with non-DNA-binding proteins such as co-repressors. Co-repressors, in turn, recruit other regulators including chromatin remodeling factors that can promote the formation of a repressive chromatin state. Some of the best characterized of these factors are histone deacetylases (HDACs) which remove acetyl groups from lysine residues of histone amino terminal tails, generally resulting in a tightening of chromatin and gene silencing [3]. Contrasting active repression, regulatory proteins can employ steric hindrance mechanisms to counteract the function of transcriptional activators, such as preventing their binding to DNA. Such proteins that indirectly influence transcription by physically interfering with activators are termed passive repressors [1,2,4]. Interestingly, some transcription factors are able to repress gene expression both passively and actively. For instance, the mammalian retinoblastoma protein Rb passively interferes with E2F transcriptional activators by binding and “masking” their transactivation domain while recruiting histone modifiers such as HDACs to actively repress transcription [2,5]. In this review, we discuss various reports demonstrating that plants use a number of transcriptional repression methods to ensure correct gene expression. While we concentrate on mechanisms involving transcription factors, plants display numerous other strategies to silence genes [for reviews, see 6,7].
Transcriptional Repression in Hormone Signal Transduction
In recent years, a common theme has emerged regarding the induction of gene expression in response to a variety of plant hormones, including auxin, jasmonate (JA) and gibberellin (GA). In these signaling pathways, DNA-binding transcription factors are under the negative regulation of labile repressors. Upon exposure to the relevant hormone, the repressors are targeted for 26S proteosome-mediated degradation by Skp1-Cullin-F-box (SCF)-type E3 ubiquitin ligases. Following this degradation, transcriptional regulators are liberated to activate downstream target genes necessary for mediating the correct hormone response.
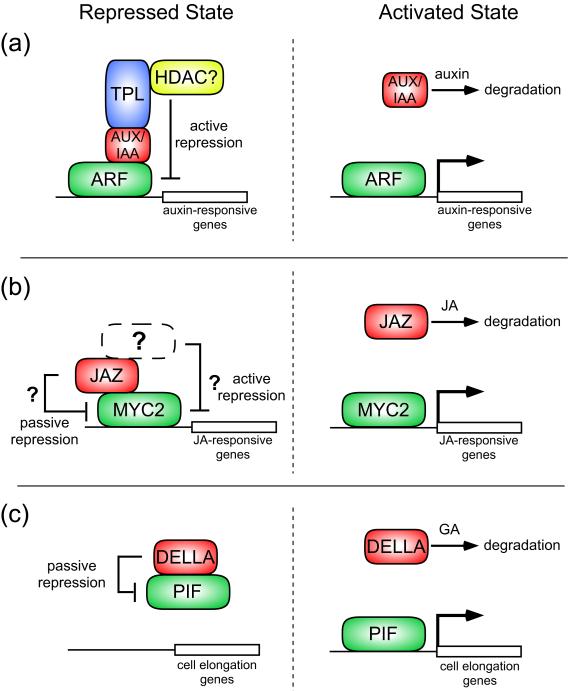
In the case of auxin signaling, AUX/IAA repressor proteins bind and negatively regulate AUXIN RESPONSE FACTORs (ARFs), a family of DNA-binding transcription factors involved in auxin-mediated developmental processes [8] (Figure 1a). Auxin relieves this repression by binding to its receptors, the F-box protein TRANSPORT INHIBITOR RESISTANT1 (TIR1) and its close homologs, resulting in increased affinity of SCFTIR1 for AUX/IAAs which are subsequently targeted for degradation via ubiquitination [9-12]. Repression by AUX/IAAs depends on a short sequence of amino acid residues (LxLxL), termed the ERF-associated amphiphilic repression (EAR) motif, located in their conserved domain I [13]. The motif is so named because it was originally identified as a strong transcriptional repression domain in members of the ethylene response factor (ERF) family [14]. However, the molecular mechanism behind EAR motif-conferred repression has remained unknown until recently. Insight was provided by a yeast 2-hybrid screen that identified IAA12/BODENLOS (BDL), an AUX/IAA which influences root and vascular pattern formation [15,16], as an interactor of the Groucho(Gro)/Tup1-like transcriptional co-repressor TOPLESS (TPL) [17*]. This interaction, which depends on the EAR motif of IAA12/BDL, supports a model whereby AUX/IAAs recruit TPL to actively repress ARF-mediated transcriptional regulation of target genes (Figure 1a).
Figure 1.
Transcriptional repression mechanisms in hormone signaling pathways.
(a) Active transcriptional repression of auxin-responsive genes.
(Left) In the absence of auxin, AUX/IAA repressor proteins bind directly to ARF transcriptional regulators through shared conserved carboxy-terminal domains [8]. AUX/IAAs recruit the transcriptional co-repressor TPL, an interaction that depends on the AUX/IAA EAR repression motif [17*]. As a Gro/Tup1-like co-repressor, TPL is predicted to recruit chromatin remodeling factors such as HDACs to negatively regulate target genes [88].
(Right) High auxin concentrations promote the interaction between AUX/IAAs and the auxin receptor TIR1, an F-box protein of an SCF-type E3 ligase [9-12]. This leads to degradation of AUX/IAAs in a 26S proteosome-dependent fashion and, because TPL is no longer recruited to the DNA, induction of auxin-responsive genes by activating ARFs.
(b) Transcriptional regulation of JA-inducible genes.
(Left) Through their conserved carboxy-terminal Jas domain, JAZ proteins physically interact with transcriptional regulators controlling JA-inducible gene expression such as MYC2 [19**-21]. Currently, the mode of JAZ-mediated repression is unknown. Possible passive mechanisms include JAZs interfering with the ability of MYC2 to bind DNA or to recruit factors involved in transcription initiation at target genes. JAZ proteins may act through an active repression mechanism analogous to AUX/IAA repressors by recruiting transcriptional co-repressors/chromatin remodeling factors to negatively regulate target gene expression.
(Right) Exposure to JA causes an increased association between JAZs and SCFCOI1, resulting in the ubiquitination and degradation of JAZs [19**,22**,23]. This liberates MYC2 to activate primary genes of the JA response.
(c) Passive repression of GA-mediated transcription.
(Left) DELLAs passively repress the transcriptional regulators PIF3 and PIF4 by directly associating with their bHLH domains and preventing their binding to DNA targets, including genes that promote hypocotyl elongation [33**,34**].
(Right) Binding of GA to its GID1 receptors increases their association with DELLAs, resulting in enhanced affinity of DELLAs for the SCFSLY1/GID2 complex and their 26S proteosome-mediated degradation [26-31]. Consequently, PIF transcription factors can activate the expression of genes responsible for hypocotyl growth.
Similar regulatory modes control the induction of genes by JA signaling, which functions in the defense response to various abiotic and biotic stresses [18]. Members of the JASMONATE ZIM-DOMAIN (JAZ) family of proteins bind and negatively regulate transcriptional regulators, such as MYC2, that confer JA responsive gene expression [19**-21] (Figure 1b). CORONATINE INSENSTIVE1 (COI1), an essential component of the JA receptor, is an F-box protein related to TIR1. In the presence of bioactive JA, COI1 displays an increased affinity for JAZ proteins and promotes their 26S proteosome-dependent degradation [19**,22**,23]. While identification and characterization of the JAZ repressors uncovered a key link between SCFCOI1 activity and JA-inducible gene expression, their mode of transcriptional repression remains to be determined. It has been shown that JAZ3 binds MYC2 at its amino-terminus, which harbors a putative transcriptional activation domain [19**,24]. This suggests JAZs may passively repress transcription by “masking” the ability of activators to recruit the transcriptional machinery (Figure 1b). However, if similarities to auxin signal transduction extend further, JAZ proteins may silence genes by recruiting transcriptional co-repressors. Such recruitment may occur through the conserved ZIM domain of JAZ proteins, as it was recently shown that this domain facilitates protein-protein interactions [21,25].
GA-mediated transcriptional regulation is subject to a repression mechanism involving DELLA domain proteins, a subfamily of the plant-specific GRAS transcriptional regulators. DELLA destabilization occurs upon GA binding to GIBBERELLIN INSENSITIVE DWARF1 (GID1) receptors, which complex with DELLAs and promote their association with the E3 ligase SCFSLEEPY(SLY1)/GID2 [26-31]. In Arabidopsis, there are five DELLAs, subsets of which have been implicated in a variety of GA-regulated processes [32].
Two recent reports have uncovered a role for DELLAs in the convergence of light and GA signaling and have described a mechanism of DELLA-mediated transcriptional repression [33**,34**]. In darkness, GA is required to maintain etiolated growth of seedlings, which includes hypocotyl elongation [35]. Phytochrome-interacting factors (PIFs) PIF3 and PIF4 are basic helix-loop-helix (bHLH) transcriptional regulators that also promote hypocotyl growth [36,37]. In response to light, however, PIF3 and PIF4 are degraded in a phytochrome-dependent fashion [33**,38-40]. Work by de Lucas et al. [33**] and Feng et al. [34**] has shown that the PIFs are also inactivated by DELLAs which directly bind the PIF bHLH DNA-recognition domain and prevent their binding to DNA targets (Figure 1c). Moreover, chromatin immunoprecipitation experiments were unable to detect association of affinity-tagged DELLAs with the promoters of GA-responsive genes [34**]. Collectively, these results suggest that DELLA-mediated repression occurs passively through the sequestration of transcription factors such as PIFs from DNA. Under conditions of increased GA levels, DELLAs are destabilized allowing PIF binding to target genes and the promotion of hypocotyl growth [33**,34**] (Figure 1c).
Transcriptional Repression in Developmental Responses
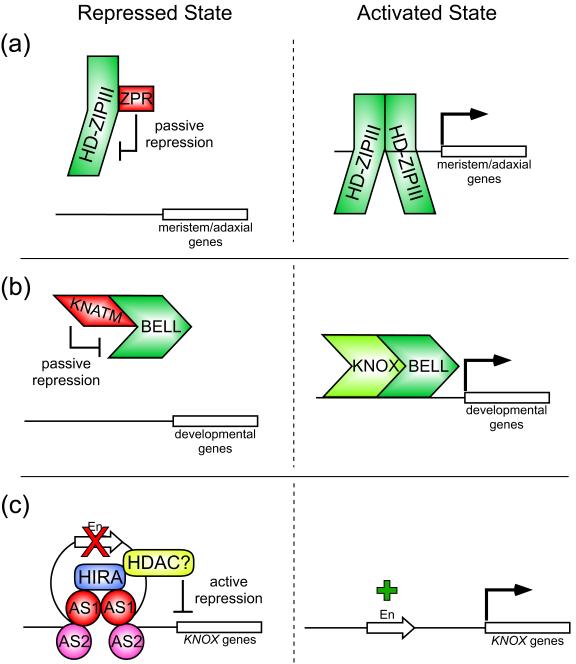
Similar to DELLA-dependent repression, a passive mechanism has been proposed for the regulation of class III homeodomain leucine zipper (HD-ZIPIII) proteins, which regulate shoot apical meristem maintenance and promote adaxial fate in lateral organs [41-43]. Arabidopsis HD-ZIPIII family members contain an HD immediately followed by a conserved leucine zipper domain that facilitates their dimerization, a requirement for DNA-binding of HD-ZIPs [44-47]. Interestingly, small negative regulators of HD-ZIPIIIs, termed LITTLE ZIPPERs (ZPRs), have been independently identified through analysis of genes induced by the HD-ZIPIII protein REVOLUTA (REV) and through a gain-of-function activation tagging screen [48**,49**]. These ZPRs, of which there are four in Arabidopsis (ZPR1-4), contain little more than a leucine zipper domain that facilitates their physical interaction with the structurally similar ZIP domain of HD-ZIPIIIs. Notably, ZPR association with HD-ZIPIIIs is believed to prevent HD-ZIPIII dimerization and disrupt DNA binding (Figure 2a). This hypothesis was strengthened by in vitro gel shift experiments where addition of ZPR3 abrogated the ability of REV to bind a probe containing its consensus recognition site [48**]. Since ZPR expression is positively regulated by HD-ZIPIIIs, the ZPRs appear to establish a negative feedback regulatory loop that dampens HD-ZIPIII activity [48**,49**]. It will be a future challenge to clarify if and how signals specifying cell fate (such as meristem and adaxial identity) influence the composition of HD-ZIPIII dimers, potentially promoting productive HD-ZIPIII/HD-ZIPIII or repressive HD-ZIPIII/ZPR interactions depending on the developmental context.
Figure 2.
Transcriptional repression mechanisms in developmental responses.
(a) Passive repression of HD-ZIPIII target genes.
(Left) ZPR proteins physically interact with HD-ZIPIII transcription factors through shared ZIP domains, preventing HD-ZIPIII/HD-ZIPIII dimerization [48**,49**]. Since HD-ZIPIII/HD-ZIPIII dimerization appears necessary for DNA binding [44-47], target genes are passively repressed.
(Right) In the absence of ZPR association with HD-ZIPIII transcription factors, the latter are able to dimerize, bind DNA, and activate targets, including genes involved in specifying meristem and adaxial identity [41-43].
(b) Passive repression of TALE homeodomain-mediated gene expression.
(Left) KNATM directly binds TALE homeodomain proteins, such as BELL transcription factors, and is proposed to render them inactive and/or sequester them in the cytoplasm [50**].
(Right) In the absence of repressive KNATM interactions, BELL proteins are able to enter the nucleus and/or bind gene targets. This may involve association with members of the KNOX family of transcriptional regulators, which have been shown to dimerize with BELL factors [89-91].
(c) Active repression of KNOX expression.
(Left) DNA recognition sites for AS1/AS2 heterodimers are present at two positions in the promoters of KNOX genes KNAT1/BP and KNAT2. Upon DNA-binding, heterodimers are proposed to associate with one another, likely due in part to the ability of AS1 to bind itself, resulting in a “looping” of the intervening promoter DNA [54,55**]. AS1 can also physically associate with the chromatin remodeling factor HIRA which plays a role in gene silencing [54,62-64], potentially due to interaction with HDACs [65]. This protein complex is predicted to produce a repressive chromatin state in this region of KNOX promoters, leading to the silencing of transcriptional enhancer (En) elements in the vicinity. These events are believed to effectively maintain KNOX gene silencing in domains of AS1/AS2 function, including leaf primoridia.
(Right) In the absence of AS1/AS2 activity, transcriptional enhancer elements in KNOX promoters are able to induce gene expression.
A newly characterized protein resembling KNOTTED1-LIKE HOMEOBOX (KNOX) transcription factors but lacking the conserved three amino acid loop extension (TALE) homeodomain may function similarly to ZPRs to passively repress transcription [50**]. This protein, KNATM, was originally identified from an in silico search for KNOX-related proteins in Arabidopsis [50**]. KNATM interacts with KNAT1/BREVIPEDICELLUS (BP) and BEL1-LIKE (BELL) homeodomain proteins through its amino-terminal acidic coiled-coil and conserved MEINOX (MEIS-KNOX) domains, respectively [50**,51]. Both the MEINOX domain and TALE homeodomain are shared between plant KNOX proteins and animal Myeloid ecotropic viral integration site (MEIS) proteins. Interestingly, isoforms of a mammalian MEIS homolog lacking a complete HD act as dominant-negative regulators of HD-containing variants [52]. KNATM, which is proposed to play a role in leaf proximal-distal patterning, may likewise act as a negative regulator of transcription factors by sequestering them in the cytoplasm and/or titrating them as inactive dimers [50**] (Figure 2b). In support of this hypothesis, bimolecular fluorescence complementation analysis showed that KNATM-BELL dimers preferentially accumulate in the cytoplasm of plant cells. Furthermore, combining overexpression lines of KNATM and the BELL gene SAWTOOTH1 [53] revealed an antagonistic relationship, as phenotypic abnormalities displayed by each individual transgenic line were mutually normalized, restoring a wild-type appearance. However, defining the precise role of KNATM in transcriptional regulation is complicated by the fact that it exhibits transcriptional activation activity [50**]. The isolation and analysis of a knatm loss-of-function allele should help clarify its function in the future.
A novel mechanism has been proposed for the transcriptional repression of the Arabidopsis KNOX meristem genes KNAT1/BP and KNAT2 in leaf primordia [54,55**]. In these developing organs, KNOX gene down-regulation corresponds with the expression of two transcriptional regulators, the MYB-domain factor ASYMMETRIC LEAVES 1 (AS1) and the LOB domain (LBD) protein AS2, which are necessary for maintaining repression and promoting determinate cell fate [56-59]. Chromatin immunoprecipitation experiments identified two distinct regions of both the KNAT1 and KNAT2 promoters bound by AS1, each comprised of a consensus MYB-binding site (motif I) followed by a previously uncharacterized motif (termed motif II) [55**]. Interestingly, in gel retardation experiments, AS1 only bound these regions when co-translated with AS2 in a cell-free expression system. Given that AS1 can physically interact with AS2 [54,60], cooperative association of AS1 and AS2 on motifs I and II is potentially required for DNA binding and repression of KNOX gene targets. Furthermore, since this binding module is repeated in a second position on both KNOX promoters, and since AS1 can homodimerize [54,61], the authors proposed a model in which two DNA-bound AS1/AS2 dimers associate with each other resulting in a looping-out of the intervening promoter region [55**] (Figure 2c). AS1 and its maize homolog ROUGH SHEATH2 (RS2) can physically interact with the chromatin remodeling factor HIRA [54], homologs of which in other eukaryotic systems associate with HDACs and function in gene silencing [62-65]. Since promoter regions in the vicinity of this proposed loop harbor enhancer elements necessary for ectopic KNOX expression, HIRA-mediated remodeling events are proposed to actively maintain KNOX silencing in developing lateral organs by negating enhancer activity. This activity closely resembles that of genetic insulators, which can form repressive chromatin loops that interfere with the ability of enhancer elements to communicate with promoters [66]. Lending support to this model, reduced levels of HIRA, like as1 and as2 mutants, result in ectopic KNOX expression in leaves [54].
HDAC Recruitment Confers Active Repression
HDACs are one of the best-studied classes of proteins recruited to facilitate active transcriptional repression. Histone acetylation is largely correlated with gene expression; therefore, removal of these modifications by HDACs generally leads to repression of transcription [3]. Mutation of the Arabidopsis Rpd3-like class I histone deacetylase HDA19, whose protein product exhibits HDAC activity in vitro [67], results in increased histone acetylation states in planta [68-73]. Furthermore, HDA19 functions cooperatively with co-repressors. For example, the Gro/Tup1-like transcriptional co-repressor LEUNIG (LUG), which shares structural homology with TPL, has been shown to physically interact with HDA19 in vitro [74**]. LUG plays a role in restricting the expression of the gene AGAMOUS (AG), which specifies the fate of the floral reproductive structures, to the inner two whorls of the flower [75,76]. DNA-binding transcription factors appear to recruit LUG to non-coding regulatory regions of AG through the intermediary adaptor protein SEUSS [77-79]. The direct and specific interaction between LUG and HDA19 in vitro implies that LUG negatively regulates genes such as AG by promoting the formation of a repressive chromatin structure. Interestingly, the observation that LUG can directly interact with Arabidopsis homologs of the Mediator complex indicates that LUG may also exert transcriptional repression by influencing RNA polymerase II activity [74**].
In other eukaryotes, class I Rpd3-like HDACs can function as part of multi-protein repressor complexes such as the Sin3 complex [80]. Arabidopsis homologs of some of these components have also been shown to associate with HDA19. For example, the putative transcriptional repressor ERF7 is proposed to function as a negative regulator of abscisic acid (ABA) and drought response by directly binding to ABA-inducible target genes and recruiting AtSIN3 and, in turn, HDA19 [81]. Similarly, the Arabidopsis homolog of Sin3-associated polypeptide of 18kDa (AtSAP18) interacts with the transcription factors ERF3 and AGAMOUS-LIKE 15 (AGL15) (which are expressed in response to salt stress and during embryogenesis, respectively) and is proposed to aid in the recruitment of HDA19 to repress target genes [82,83].
A variety of other reports have demonstrated the importance of HDA19 in regulating gene expression in response to environmental signals. For instance, HDA19 negatively regulates photomorphogenesis, and hda19 mutants exhibit increased levels of histone acetylation on a variety of light-responsive genes [72,73]. Conversely, HDA19 appears to be a positive regulator of plant defense by indirectly influencing the expression of PATHOGENESIS RELATED (PR) genes [71,84**]. For example, HDA19 is strongly induced by wounding, infection by Alternaria brassicicola (a pathogenic fungus), and the stress signals JA and ethylene [71]. Expression of PR genes co-regulated by JA and ethylene are increased in HDA19-overexpressing transgenic lines and decreased in lines with compromised HDA19 function, which show enhanced and weakened resistance to A. brassicicola, respectively [71]. Furthermore, in a recent study, HDA19 was identified as a physical interactor of the type III WRKY transcription factors WRKY38 and WRKY62 [84**]. These factors can activate transcription and are proposed to act on genes that, in turn, negatively regulate aspects of the plant defense response. Overexpression of HDA19, however, was shown to specifically reduce the ability of WRKY38 and WRKY62 to activate a reporter gene target in planta [84**]. Intriguingly, WRKY38 and WRKY62 are actually induced by the stress signal salicylic acid and infection by virulent Pseudomonas syringae strains. In this fashion, these WRKYs possibly prevent over-activation of the defense response at the onset of infection when pathogen levels are low. When a stronger effect becomes needed, HDA19, whose expression displays a delayed response to the stress signal, interacts with DNA-bound WRKYs to repress their target genes. This positively influences the plant defense response, including the induction of PR1 [84**].
Conclusions
It is becoming increasingly clear that plants rely heavily on transcriptional repression to control gene expression, which ensures proper development and responses to numerous environmental cues. Studies have shown that various repressive strategies are employed, including both passive and active mechanisms. It is only recently that active repression domains have been identified in plant proteins. This includes the EAR motif and two newly characterized repression domains [85*,86*]. The large number of transcriptional regulators containing one or more of these domains predicts a significant expansion of the plant repressor field in the near future. This will necessitate the identification and characterization of the co-repressors and/or chromatin remodeling factors that are recruited to confer repression. For example, while roles have been identified for HDA19, there are 17 other HDACs in Arabidopsis, most of which have not been functionally characterized [87]. Indeed, there will be numerous novel regulators that will remain silent no longer.
Acknowledgements
We thank R. Biddick, B. Chow, J. Meister and B. van Schooten for valuable comments on the manuscript. We apologize to colleagues whose work could not be included due to space constraints. Work on transcriptional repression in our laboratory is funded by National Institutes of Health Grant GM072764 to J.A.L. and a Natural Sciences and Engineering Research Council of Canada Postdoctoral Fellowship and a San Diego Foundation Blasker Science & Technology Grant to N.T.K.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cowell IG. Repression versus activation in the control of gene transcription. Trends Biochem Sci. 1994;19:38–42. doi: 10.1016/0968-0004(94)90172-4. [DOI] [PubMed] [Google Scholar]
- 2.Thiel G, Lietz M, Hohl M. How mammalian transcriptional repressors work. Eur J Biochem. 2004;271:2855–2862. doi: 10.1111/j.1432-1033.2004.04174.x. [DOI] [PubMed] [Google Scholar]
- 3.Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 4.Johnson AD. The price of repression. Cell. 1995;81:655–658. doi: 10.1016/0092-8674(95)90524-3. [DOI] [PubMed] [Google Scholar]
- 5.Gaston K, Jayaraman PS. Transcriptional repression in eukaryotes: repressors and repression mechanisms. Cell Mol Life Sci. 2003;60:721–741. doi: 10.1007/s00018-003-2260-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brodersen P, Voinnet O. The diversity of RNA silencing pathways in plants. Trends Genet. 2006;22:268–280. doi: 10.1016/j.tig.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 7.Henderson IR, Jacobsen SE. Epigenetic inheritance in plants. Nature. 2007;447:418–424. doi: 10.1038/nature05917. [DOI] [PubMed] [Google Scholar]
- 8.Guilfoyle TJ, Hagen G. Auxin response factors. Current Opin in Plant Biol. 2007;10:453–460. doi: 10.1016/j.pbi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 9.Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature. 2001;414:271–276. doi: 10.1038/35104500. [DOI] [PubMed] [Google Scholar]
- 10.Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- 11.Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435:446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- 12.Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jurgens G, Estelle M. Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell. 2005;9:109–119. doi: 10.1016/j.devcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 13.Tiwari SB, Hagen G, Guilfoyle TJ. Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell. 2004;16:533–543. doi: 10.1105/tpc.017384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M. Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell. 2001;13:1959–1968. doi: 10.1105/TPC.010127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamann T, Mayer U, Jurgens G. The auxin-insensitive bodenlos mutation affects primary root formation and apical-basal patterning in the Arabidopsis embryo. Development. 1999;126:1387–1395. doi: 10.1242/dev.126.7.1387. [DOI] [PubMed] [Google Scholar]
- 16.Hamann T, Benkova E, Baurle I, Kientz M, Jurgens G. The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev. 2002;16:1610–1615. doi: 10.1101/gad.229402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17*.Szemenyei H, Hannon M, Long JA. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science. 2008;319:1384–1386. doi: 10.1126/science.1151461.This paper provides insight into the mode of transcriptional repression by AUX/IAAs. The authors demonstrate that IAA12/BDL physically interacts with TPL through its domain I EAR motif. Genetic analyses show that the temperature-sensitive tpl-1 mutation, which under non-permissive conditions results in replacement of the shoot pole with a second root, suppresses the rootless defect of the dominant, protein-stabilizing bdl-1 mutation. Conversely, an iaa12 loss-of-function mutant enhances the severity of tpl-1 embryonic patterning defects. Furthermore, TPL is shown to be able to repress transcription in planta. Collectively, these results support a model whereby IAA12/BDL, and likely other AUX/IAAs, repress the activity of ARFs by recruiting TPL, resulting in the silencing of auxin responsive genes.
- 18.Chico JM, Chini A, Fonseca S, Solano R. JAZ repressors set the rhythm in jasmonate signaling. Curr Opin Plant Biol. 2008;11:486–494. doi: 10.1016/j.pbi.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 19**.Chini A, Fonseca S, Fernandez G, Adie B, Chico JM, Lorenzo O, Garcia-Casado G, Lopez-Vidriero I, Lozano FM, Ponce MR, et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448:666–671. doi: 10.1038/nature06006.Along with [22**], this work identifies members of the JAZ protein family as signaling components connecting SCFCOI1 function and JA-mediated gene regulation. Jasmonate induces the 26S proteosome-mediated degradation of JAZ3/JASMONATE INSENSITIVE3 (JAI3) in a COI1-dependent fashion. JAZ3/JAI3 is also shown to bind and negatively regulate MYC2, a transcriptional activator of JA-responsive gene expression. The protein product of jai3-1 lacks the Jas domain and fails to interact with MYC2, but interacts with COI1 in a hormone-independent fashion. Moreover, the jai3-1 mutation causes stabilization of other JAZs. Based on these observations, the authors propose a model whereby JAZ derivatives lacking the Jas domain bind and inactivate COI1, leading to the stabilization of other JAZs and, therefore, JA-insensitivity.
- 20.Melotto M, Mecey C, Niu Y, Chung HS, Katsir L, Yao J, Zeng W, Thines B, Staswick P, Browse J, et al. A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J. 2008;55:979–988. doi: 10.1111/j.1365-313X.2008.03566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chini A, Fonseca S, Chico JM, Fernandez-Calvo P, Solano R. The ZIM domain mediates homo- and heteromeric interactions between Arabidopsis JAZ proteins. Plant J. 2009 doi: 10.1111/j.1365-313X.2009.03852.x. DOI:10.1111/j.1365-313X.2009.03852.x. [DOI] [PubMed] [Google Scholar]
- 22**.Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J. JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature. 2007;448:661–665. doi: 10.1038/nature05960.Complementing [19**], the authors report the isolation of the JAZ family of repressors from analysis of JA-inducible transcripts. JAZ proteins are destabilized by JA treatment in a COI1- and 26S proteosome-dependent manner. The jasmonate-isoleucine conjugated form of the hormone specifically promotes JAZ1-COI1 interaction, while expression of JAZ1ΔJas confers JA-insensitivity. Therefore, this work identifies JAZ family members as the proposed negative regulators of JA signal transduction that are subject to SCFCOI1-mediated degradation.
- 23.Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc Natl Acad Sci U S A. 2008;105:7100–7105. doi: 10.1073/pnas.0802332105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lorenzo O, Chico JM, Sanchez-Serrano JJ, Solano R. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell. 2004;16:1938–1950. doi: 10.1105/tpc.022319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chung HS, Howe GA. A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell. 2009;21:131–145. doi: 10.1105/tpc.108.064097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, Chow TY, Hsing YI, Kitano H, Yamaguchi I, et al. GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature. 2005;437:693–698. doi: 10.1038/nature04028. [DOI] [PubMed] [Google Scholar]
- 27.Nakajima M, Shimada A, Takashi Y, Kim YC, Park SH, Ueguchi-Tanaka M, Suzuki H, Katoh E, Iuchi S, Kobayashi M, et al. Identification and characterization of Arabidopsis gibberellin receptors. Plant J. 2006;46:880–889. doi: 10.1111/j.1365-313X.2006.02748.x. [DOI] [PubMed] [Google Scholar]
- 28.Griffiths J, Murase K, Rieu I, Zentella R, Zhang Z-L, Powers SJ, Gong F, Phillips AL, Hedden P, Sun T-p, et al. Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell. 2006;18:3399–3414. doi: 10.1105/tpc.106.047415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ueguchi-Tanaka M, Nakajima M, Katoh E, Ohmiya H, Asano K, Saji S, Hongyu X, Ashikari M, Kitano H, Yamaguchi I, et al. Molecular interactions of a soluble gibberellin receptor, GID1, with a rice DELLA protein, SLR1, and gibberellin. Plant Cell. 2007;19:2140–2155. doi: 10.1105/tpc.106.043729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McGinnis KM, Thomas SG, Soule JD, Strader LC, Zale JM, Sun T-p, Steber CM. The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell. 2003;15:1120–1130. doi: 10.1105/tpc.010827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sasaki A, Itoh H, Gomi K, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Jeong D-H, An G, Kitano H, Ashikari M, et al. Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science. 2003;299:1896–1898. doi: 10.1126/science.1081077. [DOI] [PubMed] [Google Scholar]
- 32.Achard P, Genschik P. Releasing the brakes of plant growth: how GAs shutdown DELLA proteins. J Exp Bot. 2009;60:1085–1092. doi: 10.1093/jxb/ern301. [DOI] [PubMed] [Google Scholar]
- 33**.de Lucas M, Daviere J-M, Rodriguez-Falcon M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blazquez MA, Titarenko E, Prat S. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451:480–484. doi: 10.1038/nature06520.Complementing [34**], the authors show that DELLA proteins negatively regulate gene expression by binding the DNA-recognition domain of transcription factors such as PIF4. GA promotes DELLA destabilization, leading to PIF4 activation. PIF4, which promotes hypocotyl elongation, is negatively influenced by the photoreceptor phyB which promotes its destabilization. As such, this study identifies the transcriptional regulator PIF4 as a point of intersection between light and GA signaling pathways.
- 34**.Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S, et al. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature. 2008;451:475–479. doi: 10.1038/nature06448.This work, along with [33**], identifies a mechanism by which DELLA proteins confer transcriptional repression. DELLAs are shown to bind PIF3, a transcription factor involved in light signaling that positively regulates hypocotyl elongation, and prevent its association with target genes. GA treatment leads to increased affinity of GID1 receptors for the DELLA repressors, leading to their destabilization and the liberation of PIF3 activity. This work elucidates a mode by which plants integrate light and GA signal transduction in the control of hypocotyl growth.
- 35.Alabadi D, Gil J, Blazquez MA, Garcia-Martinez JL. Gibberellins repress photomorphogenesis in darkness. Plant Physiol. 2004;134:1050–1057. doi: 10.1104/pp.103.035451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huq E, Quail PH. PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 2002;21:2441–2450. doi: 10.1093/emboj/21.10.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim J, Yi H, Choi G, Shin B, Song P-S, Choi G. Functional characterization of Phytochrome Interacting Factor 3 in phytochrome-mediated light signal transduction. Plant Cell. 2003;15:2399–2407. doi: 10.1105/tpc.014498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bauer D, Viczian A, Kircher S, Nobis T, Nitschke R, Kunkel T, Panigrahi KC, Adam E, Fejes E, Schafer E, et al. Constitutive Photomorphogenesis 1 and multiple photoreceptors control degradation of Phytochrome Interacting Factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell. 2004;16:1433–1445. doi: 10.1105/tpc.021568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park E, Kim J, Lee Y, Shin J, Oh E, Chung W-I, Liu JR, Choi G. Degradation of Phytochrome Interacting Factor 3 in phytochrome-mediated light signaling. Plant Cell Physiol. 2004;45:968–975. doi: 10.1093/pcp/pch125. [DOI] [PubMed] [Google Scholar]
- 40.Al-Sady B, Ni W, Kircher S, Schafer E, Quail PH. Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol Cell. 2006;23:439–446. doi: 10.1016/j.molcel.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 41.Prigge MJ, Otsuga D, Alonso JM, Ecker JR, Drews GN, Clark SE. Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell. 2005;17:61–76. doi: 10.1105/tpc.104.026161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eshed Y, Baum SF, Perea JV, Bowman JL. Establishment of polarity in lateral organs of plants. Curr Biol. 2001;11:1251–1260. doi: 10.1016/s0960-9822(01)00392-x. [DOI] [PubMed] [Google Scholar]
- 43.McConnell JR, Emery J, Eshed Y, Bao N, Bowman J, Barton MK. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature. 2001;411:709–713. doi: 10.1038/35079635. [DOI] [PubMed] [Google Scholar]
- 44.Sessa G, Morelli G, Ruberti I. The Athb-1 and -2 HD-Zip domains homodimerize forming complexes of different DNA binding specificities. EMBO J. 1993;12:3507–3517. doi: 10.1002/j.1460-2075.1993.tb06025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sessa G, Steindler C, Morelli G, Ruberti I. The Arabidopsis Athb-8, -9 and -14 genes are members of a small gene family coding for highly related HD-ZIP proteins. Plant Mol Biol. 1998;38:609–622. doi: 10.1023/a:1006016319613. [DOI] [PubMed] [Google Scholar]
- 46.Tron AE, Bertoncini CW, Palena CM, Chan RL, Gonzalez DH. Combinatorial interactions of two amino acids with a single base pair define target site specificity in plant dimeric homeodomain proteins. Nucleic Acids Res. 2001;29:4866–4872. doi: 10.1093/nar/29.23.4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Palena CM, Gonzalez DH, Chan RL. A monomer-dimer equilibrium modulates the interaction of the sunflower homeodomain leucine-zipper protein Hahb-4 with DNA. Biochem J. 1999;341:81–87. [PMC free article] [PubMed] [Google Scholar]
- 48**.Wenkel S, Emery J, Hou BH, Evans MM, Barton MK. A feedback regulatory module formed by LITTLE ZIPPER and HD-ZIPIII genes. Plant Cell. 2007;19:3379–3390. doi: 10.1105/tpc.107.055772.A small family of leucine zipper-containing ZPR proteins, which physically interact with the HD-ZIPIII transcription factor REV, was identified by analyzing genes up-regulated by REV activation. In the presence of ZPR3, REV loses the ability to bind probes containing its consensus DNA-recognition sequence in vitro. Overexpression of ZPRs leads to phenotypic abnormalities consistent with reductions in HD-ZIPIII function. The authors present a model whereby ZPRs function in transcriptional repression by interacting with HD-ZIPIII family members to prevent HD-ZIPIII/HD-ZIPIII dimerization and DNA-binding.
- 49**.Kim YS, Kim SG, Lee M, Lee I, Park HY, Seo PJ, Jung JH, Kwon EJ, Suh SW, Paek KH, et al. HD-ZIP III activity is modulated by competitive inhibitors via a feedback loop in Arabidopsis shoot apical meristem development. Plant Cell. 2008;20:920–933. doi: 10.1105/tpc.107.057448.An activation tagging mutant resembling loss-of-function revoluta is shown to up-regulate the expression of ZPR3. ZPR3 can bind to HD-ZIPIII transcription factors through shared ZIP motifs, preventing both HD-ZIPIII/HD-ZIPIII interaction and HD-ZIPIII-mediated transcriptional activation in a heterologous yeast system. Furthermore, genetic analyses in planta indicate that ZPR3 and its homolog ZPR4 function antagonistically with HD-ZIPIII factors. Results from this study support the work described in [48**] and are consistent with the model that ZPRs directly bind to and negatively modulate HD-ZIPIII transcription factors by preventing their dimerization and association with target genes.
- 50**.Magnani E, Hake S. KNOX lost the OX: the Arabidopsis KNATM gene defines a novel class of KNOX transcriptional regulators missing the homeodomain. Plant Cell. 2008;20:875–887. doi: 10.1105/tpc.108.058495.An in silico search for KNOX-related proteins in Arabidopsis identifies KNATM, a novel KNOX-like protein that lacks a homeodomain but contains conserved coiled-coil and MEINOX domains. KNATM can interact with BELL homeodomain proteins through its MEINOX domain and with KNOX factors such as BP through its acidic coiled-coil domain (subsequently re-named the BP-interacting domain). The authors speculate that KNATM can function as a repressor by binding and sequestering transcription factors in a non-functional state. Notably, KNATM association with BP represents the first example of a homeodomain-independent KNOX-KNOX interaction.
- 51.Hake S, Smith HM, Holtan H, Magnani E, Mele G, Ramirez J. The role of KNOX genes in plant development. Annu Rev Cell Dev Biol. 2004;20:125–151. doi: 10.1146/annurev.cellbio.20.031803.093824. [DOI] [PubMed] [Google Scholar]
- 52.Yang Y, Hwang CK, D’Souza UM, Lee S-H, Junn E, Mouradian MM. Three-amino acid extension loop homeodomain proteins Meis2 and TGIF differentially regulate transcription. J Biol Chem. 2000;275:20734–20741. doi: 10.1074/jbc.M908382199. [DOI] [PubMed] [Google Scholar]
- 53.Kumar R, Kushalappa K, Godt D, Pidkowich MS, Pastorelli S, Hepworth SR, Haughn GW. The Arabidopsis BEL1-LIKE HOMEODOMAIN proteins SAW1 and SAW2 act redundantly to regulate KNOX expression spatially in leaf margins. Plant Cell. 2007;19:2719–2735. doi: 10.1105/tpc.106.048769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Phelps-Durr TL, Thomas J, Vahab P, Timmermans MCP. Maize rough sheath2 and its Arabidopsis orthologue ASYMMETRIC LEAVES1 interact with HIRA, a predicted histone chaperone, to maintain knox gene silencing and determinacy during organogenesis. Plant Cell. 2005;17:2886–2898. doi: 10.1105/tpc.105.035477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55**.Guo M, Thomas J, Collins G, Timmermans MCP. Direct repression of KNOX loci by the ASYMMETRIC LEAVES1 complex of Arabidopsis. Plant Cell. 2008;20:48–58. doi: 10.1105/tpc.107.056127.This work proposes a mechanism for the negative regulation of KNOX genes in leaf primordia by AS1 and AS2. The authors present evidence that AS1 functions as a transcriptional repressor in planta. In addition, cis-regulatory elements in KNOX gene promoters are identified that facilitate AS1/AS2 heterodimer association. A repressive chromatin arrangement is proposed to form due to the recruitment of the chromatin-remodeling factor HIRA, a WD40-repeat protein involved in gene silencing in other eukaryotic systems. These events are predicted to maintain KNOX gene silencing in lateral organ development.
- 56.Ori N, Eshed Y, Chuck G, Bowman JL, Hake S. Mechanisms that control knox gene expression in the Arabidopsis shoot. Development. 2000;127:5523–5532. doi: 10.1242/dev.127.24.5523. [DOI] [PubMed] [Google Scholar]
- 57.Byrne ME, Barley R, Curtis M, Arroyo JM, Dunham M, Hudson A, Martienssen RA. Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature. 2000;408:967–971. doi: 10.1038/35050091. [DOI] [PubMed] [Google Scholar]
- 58.Iwakawa H, Ueno Y, Semiarti E, Onouchi H, Kojima S, Tsukaya H, Hasebe M, Soma T, Ikezaki M, Machida C, et al. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 2002;43:467–478. doi: 10.1093/pcp/pcf077. [DOI] [PubMed] [Google Scholar]
- 59.Lin W-c, Shuai B, Springer PS. The Arabidopsis LATERAL ORGAN BOUNDARIES-domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial-abaxial patterning. Plant Cell. 2003;15:2241–2252. doi: 10.1105/tpc.014969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu L, Xu Y, Dong A, Sun Y, Pi L, Xu Y, Huang H. Novel as1 and as2 defects in leaf adaxial-abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying leaf adaxial identity. Development. 2003;130:4097–4107. doi: 10.1242/dev.00622. [DOI] [PubMed] [Google Scholar]
- 61.Theodoris G, Inada N, Freeling M. Conservation and molecular dissection of ROUGH SHEATH2 and ASYMMETRIC LEAVES1 function in leaf development. Proc Natl Acad Sci U S A. 2003;100:6837–6842. doi: 10.1073/pnas.1132113100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Spector MS, Raff A, DeSilva H, Lee K, Osley MA. Hir1p and Hir2p function as transcriptional corepressors to regulate histone gene transcription in the Saccharomyces cerevisiae cell cycle. Mol Cell Biol. 1997;17:545–552. doi: 10.1128/mcb.17.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kaufman PD, Cohen JL, Osley MA. Hir proteins are required for position-dependent gene silencing in Saccharomyces cerevisiae in the absence of chromatin assembly factor I. Mol Cell Biol. 1998;18:4793–4806. doi: 10.1128/mcb.18.8.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sharp JA, Fouts ET, Krawitz DC, Kaufman PD. Yeast histone deposition protein Asf1p requires Hir proteins and PCNA for heterochromatic silencing. Curr Biol. 2001;11:463–473. doi: 10.1016/s0960-9822(01)00140-3. [DOI] [PubMed] [Google Scholar]
- 65.Ahmad A, Takami Y, Nakayama T. WD dipeptide motifs and LXXLL motif of chicken HIRA are necessary for transcription repression and the latter motif is essential for interaction with histone deacetylase-2 in vivo. Biochem Biophys Res Commun. 2003;312:1266–1272. doi: 10.1016/j.bbrc.2003.11.073. [DOI] [PubMed] [Google Scholar]
- 66.Bushey AM, Dorman ER, Corces VG. Chromatin insulators: regulatory mechanisms and epigenetic inheritance. Mol Cell. 2008;32:1–9. doi: 10.1016/j.molcel.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fong PM, Tian L, Chen ZJ. Arabidopsis thaliana histone deacetylase 1 (AtHD1) is localized in euchromatic regions and demonstrates histone deacetylase activity in vitro. Cell Res. 2006;16:479–488. doi: 10.1038/sj.cr.7310059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tian L, Chen ZJ. Blocking histone deacetylation in Arabidopsis induces pleiotropic effects on plant gene regulation and development. Proc Natl Acad Sci U S A. 2001;98:200–205. doi: 10.1073/pnas.011347998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tian L, Wang J, Fong MP, Chen M, Cao H, Gelvin SB, Chen ZJ. Genetic control of developmental changes induced by disruption of Arabidopsis Histone Deacetylase 1 (AtHD1) expression. Genetics. 2003;165:399–409. doi: 10.1093/genetics/165.1.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tian L, Fong MP, Wang JJ, Wei NE, Jiang H, Doerge RW, Chen ZJ. Reversible histone acetylation and deacetylation mediate genome-wide, promoter-dependent and locus-specific changes in gene expression during plant development. Genetics. 2005;169:337–345. doi: 10.1534/genetics.104.033142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou C, Zhang L, Duan J, Miki B, Wu K. HISTONE DEACETYLASE19 is involved in jasmonic acid and ethylene signaling of pathogen response in Arabidopsis. Plant Cell. 2005;17:1196–1204. doi: 10.1105/tpc.104.028514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Benhamed M, Bertrand C, Servet C, Zhou DX. Arabidopsis GCN5, HD1, and TAF1/HAF2 interact to regulate histone acetylation required for light-responsive gene expression. Plant Cell. 2006;18:2893–2903. doi: 10.1105/tpc.106.043489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guo L, Zhou J, Elling AA, Charron JB, Deng XW. Histone modifications and expression of light-regulated genes in Arabidopsis are cooperatively influenced by changing light conditions. Plant Physiol. 2008;147:2070–2083. doi: 10.1104/pp.108.122929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74**.Gonzalez D, Bowen AJ, Carroll TS, Conlan RS. The transcription corepressor LEUNIG interacts with the histone deacetylase HDA19 and mediator components MED14 (SWP) and CDK8 (HEN3) to repress transcription. Mol Cell Biol. 2007;27:5306–5315. doi: 10.1128/MCB.01912-06.The authors use genome-wide transcript profiling to show that the co-repressor LUG regulates multiple genes in planta. The authors further demonstrate that LUG-mediated repression operates both in HDAC-independent and HDAC-dependent modes. Consistent with the latter, experiments in a heterologous yeast system show that the ability of LUG to repress transcription is abrogated in a mutant strain defective for the HDAC Rpd3. Moreover, LUG directly and specifically interacts with HDA19, a class I Rpd3-like HDAC. Finally, physical interactions between LUG and Arabidopsis Mediator components suggest a plausible HDAC-independent mechanism of LUG-mediated repression.
- 75.Liu Z, Meyerowitz EM. LEUNIG regulates AGAMOUS expression in Arabidopsis flowers. Development. 1995;121:975–991. doi: 10.1242/dev.121.4.975. [DOI] [PubMed] [Google Scholar]
- 76.Conner J, Liu Z. LEUNIG, a putative transcriptional corepressor that regulates AGAMOUS expression during flower development. Proc Natl Acad Sci U S A. 2000;97:12902–12907. doi: 10.1073/pnas.230352397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Franks RG, Wang C, Levin JZ, Liu Z. SEUSS, a member of a novel family of plant regulatory proteins, represses floral homeotic gene expression with LEUNIG. Development. 2002;129:253–263. doi: 10.1242/dev.129.1.253. [DOI] [PubMed] [Google Scholar]
- 78.Sridhar VV, Surendrarao A, Gonzalez D, Conlan RS, Liu Z. Transcriptional repression of target genes by LEUNIG and SEUSS, two interacting regulatory proteins for Arabidopsis flower development. Proc Natl Acad Sci U S A. 2004;101:11494–11499. doi: 10.1073/pnas.0403055101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sridhar VV, Surendrarao A, Liu Z. APETALA1 and SEPALLATA3 interact with SEUSS to mediate transcription repression during flower development. Development. 2006;133:3159–3166. doi: 10.1242/dev.02498. [DOI] [PubMed] [Google Scholar]
- 80.Cunliffe VT. Eloquent silence: developmental functions of Class I histone deacetylases. Curr Opin Genet Dev. 2008;18:404–410. doi: 10.1016/j.gde.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Song CP, Agarwal M, Ohta M, Guo Y, Halfter U, Wang P, Zhu JK. Role of an ArabidopsisAP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell. 2005;17:2384–2396. doi: 10.1105/tpc.105.033043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Song CP, Galbraith DW. AtSAP18, an orthologue of human SAP18, is involved in the regulation of salt stress and mediates transcriptional repression in Arabidopsis. Plant Mol Biol. 2006;60:241–257. doi: 10.1007/s11103-005-3880-9. [DOI] [PubMed] [Google Scholar]
- 83.Hill K, Wang H, Perry SE. A transcriptional repression motif in the MADS factor AGL15 is involved in recruitment of histone deacetylase complex components. Plant J. 2008;53:172–185. doi: 10.1111/j.1365-313X.2007.03336.x. [DOI] [PubMed] [Google Scholar]
- 84**.Kim KC, Lai Z, Fan B, Chen Z. Arabidopsis WRKY38 and WRKY62 transcription factors interact with Histone Deacetylase 19 in basal defense. Plant Cell. 2008;20:2357–2371. doi: 10.1105/tpc.107.055566.In this study, the authors identify HDA19 as a physical interactor of WRKY38 and WRKY62, both of which function as negative regulators of the Arabidopsis basal defense response. Specifically, wrky38 and wrky62 mutations enhance disease resistance and expression levels of the defense response gene PR1. Conversely, HDA19 appears to positively influence basal defense, as hda19 displays increased susceptibility to pathogen infection and decreased expression of PR1. Additionally, overexpression of HDA19 abrogates the ability of WRKY38 and WRKY62 to activate transcription in planta. Therefore, HDA19 appears to operate antagonistically to these WRKYs in basal defense by directly associating with them and repressing expression of their target genes.
- 85*.Matsui K, Umemura Y, Ohme-Takagi M. AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J. 2008;55:954–967. doi: 10.1111/j.1365-313X.2008.03565.x.A novel repression motif is identified in AtMYBL2, an R3-MYB domain transcriptional regulator that acts to negatively regulate anthocyanin production. AtMYBL2 is shown to function as a transcriptional repressor in planta, and this capacity is mediated by a short string of amino acids (TLLLFR) termed the “L2R” motif. The authors propose that AtMYBL2 negatively regulates gene expression by directly associating with the R-type bHLH protein TRANSPARENT TESTA8 (TT8), a transcription factor involved in the control of anthocyanin biosynthesis.
- 86*.Ikeda M, Ohme-Takagi M. A novel group of transcriptional repressors in Arabidopsis. Plant Cell Physiol. 2009;50:970–975. doi: 10.1093/pcp/pcp048.The authors identify a new domain, present in a number of B3 DNA-binding domain transcriptional regulators, that confers strong transcriptional repression. Fusion of this domain to previously characterized transcription factors appears to convert them into dominant repressors based on the phenotypic consequences of their expression in Arabidopsis. Deletion analysis identified an eight amino acid minimal repression motif within this domain containing a core consensus sequence of R/KLFGV. This motif is also present in other transcriptional repressors including members of the APETALA2 (AP2)/ERF and Heat shock transcription factor (Hsf) families.
- 87.Hollender C, Liu Z. Histone deacetylase genes in Arabidopsis development. J Integr Plant Biol. 2008;50:875–885. doi: 10.1111/j.1744-7909.2008.00704.x. [DOI] [PubMed] [Google Scholar]
- 88.Liu Z, Karmarkar V. Groucho/Tup1 family co-repressors in plant development. Trends Plant Sci. 2008;13:137–144. doi: 10.1016/j.tplants.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 89.Bellaoui M, Pidkowich MS, Samach A, Kushalappa K, Kohalmi SE, Modrusan Z, Crosby WL, Haughn GW. The Arabidopsis BELL1 and KNOX TALE homeodomain proteins interact through a domain conserved between plants and animals. Plant Cell. 2001;13:2455–2470. doi: 10.1105/tpc.010161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Muller J, Wang Y, Franzen R, Santi L, Salamini F, Rohde W. In vitro interactions between barley TALE homeodomain proteins suggest a role for protein-protein associations in the regulation of Knox gene function. Plant J. 2001;27:13–23. doi: 10.1046/j.1365-313x.2001.01064.x. [DOI] [PubMed] [Google Scholar]
- 91.Smith HMS, Boschke I, Hake S. Selective interaction of plant homeodomain proteins mediates high DNA-binding affinity. Proc Natl Acad Sci U S A. 2002;99:9579–9584. doi: 10.1073/pnas.092271599. [DOI] [PMC free article] [PubMed] [Google Scholar]