Summary
The bacterial transcription-repair coupling factor (TRCF) is a large, multi-domain, SF2 ATPase that is widely conserved. It couples nucleotide excision repair with transcription by dislodging inactive RNA polymerase molecules stalled at template DNA lesions, and by increasing the rate at which the Uvr(A)BC excinuclease acts at these sites. The recent elucidation of Escherichia coli TRCF X-ray crystal structures reveals its architectural details and will allow the design of more incisive experiments addressing how TRCF translocates on double-stranded DNA, destabilizes the RNAP ternary elongation complex, and recruits the Uvr(A)BC system.
Introduction
DNA repair asymmetry in genomes has long been linked to transcription [1]. Following its initial discovery by Hanawalt and co-workers in eukaryotes and later in Escherichia.coli [2,3], the preferential repair of DNA damage in the transcribed strand of genes, termed transcription-coupled repair (TCR), has emerged as a subpathway of nucleotide excision repair (NER) that is elicited by a stalled RNA polymerase (RNAP).
While eukaryotic TCR has only very recently been reconstituted in vitro [4], bacterial TCR has been relatively well studied in E. coli, where the product of the mfd gene was identified by in vitro complementation with purified protein to be the (only) Transcription-Repair Coupling Factor (TRCF) [5]. Work from the Sancar laboratory began to dissect the structure-function relationship of TRCF, and established that TRCF, a large monomeric SF2 ATPase, binds UvrA, RNAP and DNA, and has a dual function: it dislodges RNAP ternary elongation complexes (TECs) using energy of ATP hydrolysis, and it recruits the Uvr(A)BC excinuclease to the sites of damage by binding to the UvrA subunit [6–8].
So how exactly does TRCF perform these multiple functions? TRCF-mediated UvrA recruitment is poorly characterized mechanistically, mainly because it has not been possible to isolate coupling intermediates, such as a TRCF/RNAP/UvrA ternary complex. In contrast, understanding of TRCF function in RNAP release has been facilitated by the realization that TRCF employs a motor module homologous to that of RecG, a Holliday branch migration protein involved in replication fork damage bypass [6]. This sequence homology provided considerable insight into how TRCF may act on TECs. Structural and biochemical analysis of the RecG motor revealed a molecular architecture composed of a RecA-type dsDNA translocase motor and a separate strand-separating module consisting of a junction-DNA binding "wedge" domain across which DNA is dragged by the action of the motor [9]. Instead of possessing a DNA strand-separating module, TRCF was shown by two-hybrid methods to be equipped with an ancillary domain termed the "RID" (RNAP Interacting Domain), which was hypothesized to serve as an anchorage point onto the RNAP within the TEC [10,11], explaining why TRCF is devoid of helicase activity on a variety of substrates, including transcription bubble mimics [7]. The experimental finding that provided the most insight into TRCF function came from biochemical experiments demonstrating that TRCF can rescue backtracked TECs by binding to dsDNA upstream of the transcription bubble and, using the energy of ATP hydrolysis, cause forward translocation of the RNAP [11]. Taken together, these data suggested an "induced forward translocation" model, whereby TRCF uses its RecG-like SF2 ATPase module to track on upstream dsDNA and push RNAP downstream, causing the RNAP to dissociate if forward translocation is impeded by protein roadblocks or DNA damage that prevents translesional bypass. In the absence of obstacles to RNAP forward translocation, TRCF action causes elongation to be resumed by repositioning the 3'-OH of the nascent RNA in the catalytic center [11]. TEC release competes ineffectively with elongation at cellular concentrations of NTPs [11], which reflects a kinetic competition, at least in vitro, that probably ensures that only TECs stalled at sites of DNA damage or by roadblocks are targeted for TRCF-mediated release.
TRCF function is not restricted to TCR, but extends to cellular processes that are either regulated by, or are interfered with by, roadblocks to RNAP elongation [12–15]. In these cases, it is likely that only the RNAP elongation complex release function of TRCF is needed.
TRCF - a molecular motor with many parts
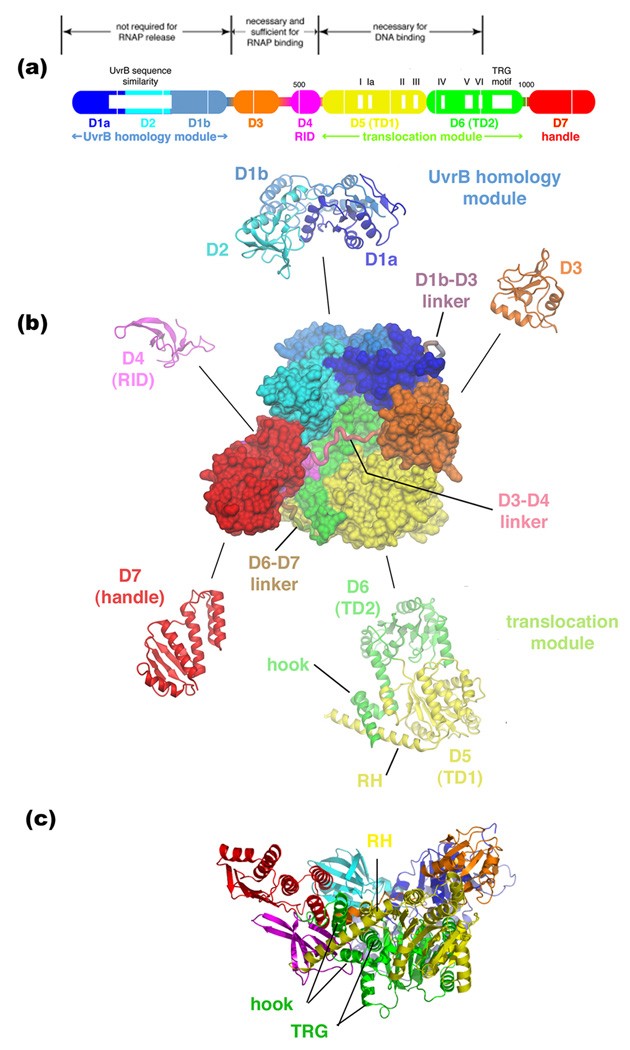
The recent elucidation of the 3.2 Å crystal structure of E. coli TRCF represents a key contribution to our understanding of bacterial TCR because it paves the way to more targeted, structure-guided experiments to dissect the details of the TRCF functional cycle [16,17]. The structure revealed a compact protein organized into five modules (corresponding to eight structural domains) connected by flexible linkers (Figure 1). The five modules correspond to two protein/protein interaction modules (the N-terminal UvrB homology module and the RID), the ATP-dependent translocase (D5/D6, or TD1/TD2, for Translocation Domain), a species-specific module of unknown function (D3), and a C-terminal, novel α/β fold with a proposed UvrA-recruitment regulatory function (D7). Such an architectural design serves to maintain a tight coupling of functional activities [17].
Figure 1. Overall architecture of E. coli TRCF.
(a) Schematic represenation of TRCF domain organization, with labeled translocase motifs and sequence characteristics. Thin white vertical lines are 100-residue marks. Structural domains are represented as thick bars, thin bars represent linkers connecting the domains. The domains are color-coded as follows: D1a, dark blue; D2, cyan; D1b, light blue; D3, orange; D4, magenta; D5, yellow; D6, green; D7, red. D1a/D2/D1b contain a region of sequence similarity with UvrB (white box) [6], and make up the UvrB homology module. D4 is an RNAP Interacting Domain (RID) [17]. D5/D6 (also called TD1/TD2 for Translocation Domains) contain the seven SF2 ATPase motifs (denoted by white boxes and labeled, share the TRG motif with RecG [23, 24], and make up the translocation module.
(b) In the middle is shown the top view of E. coli TRCF, color-coded as in (a). Structural domains are shown as molecular surfaces, with the linkers connecting the domains shown as bacbone worms. Ribbon representations of the individual modules are shown around the outside.
(c) Side view of TRCF, shown as a ribbon representation and color-coded as in (a). Adapted from [17].
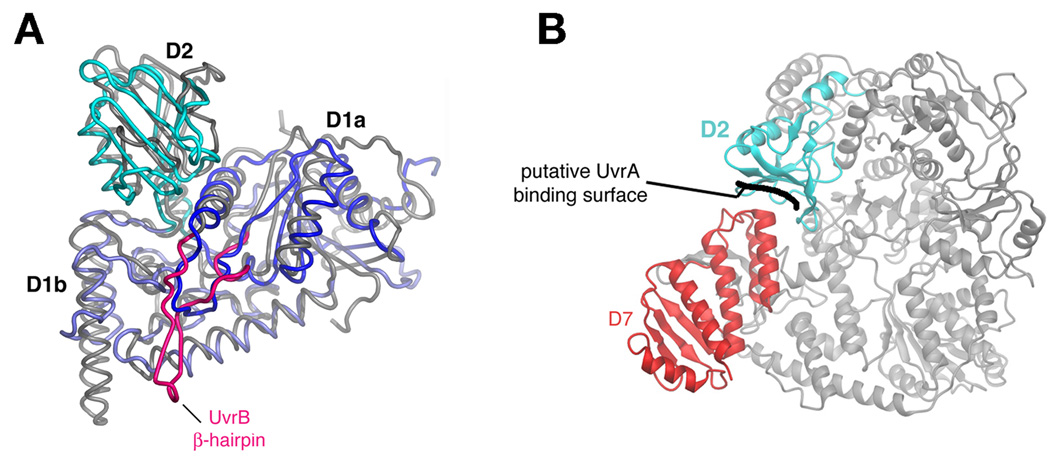
An N-terminal region of TRCF (approximately residues 82–219) shows weak sequence similarity with the NER component UvrB [6]. The 2.1 Å-resolution crystal structure of the N-terminal 333 residues of E. coli TRCF revealed that, unexpectedly, the close structural similarity with UvrB extends over the entire fragment, although TRCF lacks key elements of UvrB, such as an active ATPase function and the UvrB β-hairpin involved in DNA damage recognition (Figure 2a) [18]. Included within this region of structural similarity between TRCF and UvrB is the interface thought to be critical for the interaction between UvrB and UvrA [17–19] located in D2 (Figure 2), leading to the proposal that the TRCF/UvrA interaction structurally mimics the UvrB/UvrA interaction [17,19]. Remarkably, in the structure of intact TRCF, the putative UvrA binding surface is occluded due to inter-domain packing with D7 (Figure 2b), and residues in both D2 and D7 participating in this interaction are highly conserved across the TRCF family [17]. The hypothesis of Deaconescu et al. [17], consistent with both in vitro and in vivo data on the effects of C-terminally truncated TRCF on TCR and global NER [8], is that during its functional cycle, TRCF undergoes conformational changes that expose the buried surface and allow for recruitment of UvrAB. TRCF may actually aid formation of the UvrB/DNA preincision complex by promoting dissociation of UvrA from the UvrAB complex, as suggested by the fact that immobilized TRCF only retains UvrA upon incubation with UvrAB [8]. It is interesting to note that C-terminally truncated TRCF, unlike full-length TRCF, can translocate on naked DNA in triplex destabilizing assays (N.S., unpublished). This also supports the notion that repositioning of D7 is a prerequisite for initiation of the coupling pathway. Although this model is consistent with all available data, a caveat is that D7 is extensively involved in crystal packing. Thus the conformation observed in the crystal structure might not be representative of the conformation in solution.
Figure 2. UvrB homology module.
(A) Backbone worms of D1a/D2/D1b of B. caldotenax UvrB (gray worm, except the β-hairpin is highlighted in magenta) [19] and TRCF (D1a, dark blue; D2, cyan; D1b, light blue) superimposed on D1a and D1b.
(B) Ribbon representation showing the top view of TRCF, with D2 (cyan) and D7 (red) highlighted. The putative UvrA binding surface on D2 (blocked in an interface with D7) is indicated. Adapted from [17].
The RID is an all β, Tudor-like domain. Surprisingly, the closest structural neighbor of the RID is the KOW domain of the NusG antiterminator, itself an RNAP binding protein [20,21]. This is an intriguing observation as NusG, like TRCF, enhances escape from class II pause sites, but not class I sites, and both proteins relieve backtracking [22]. There are, however, no striking similarities in sequence or surface properties between the NusG-KOW domain and the TRCF-RID.
The translocase and the TRG motif
The translocase module (TD1/TD2 in Figure 1), directly connected to the RID via the "relay helix" (RH), represents the ATP-hydrolyzing, dsDNA-tracking motor. DNA binding is most likely mediated by two DNA binding sites, which may alternate in cycles of DNA binding and release during translocation, as in other DNA-tracking enzymes such as Swi/Snf2 [23]. While the structure of TRCF allowed identification of a putative DNA binding site on TD1 [17], details of the secondary binding site remain obscure. ATP binding is required for TRCF to bind DNA, and hydrolysis of the ATP γ-phosphate results in TRCF dissociation from the DNA. Stable binding is only achieved with nonhydrolyzable ATP analogs such as ATP-γ-S [8]. The TRCF crystal structure represents the apo-form (no nucleotide). Thus the translocase assumes an open conformation that does not stably bind DNA [17].
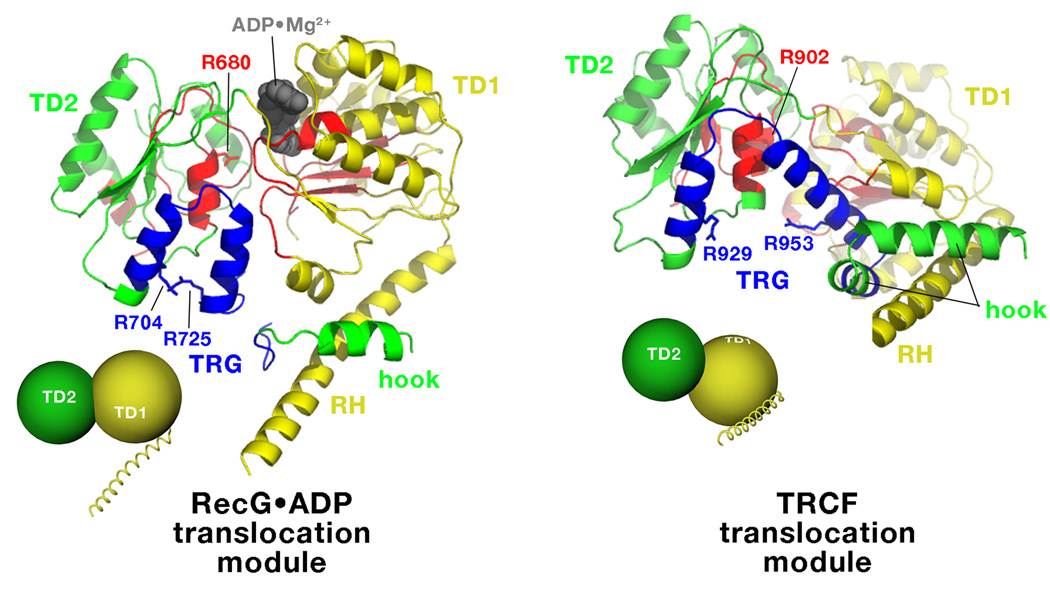
The TRG motif is a key element in the transduction of the energy of ATP hydrolysis into mechanical work [24,25]. Mechanistically, the role of the TRG motif was suggested from comparison of the RecG•ADP and TRCF structures (Figure 3). The helical TRG hairpin may sense the γ-phosphate indirectly, via a conserved arginine finger in ATPase motif VI (TRCF R902/RecG R680; Figure 3), and appears to undergo an opening motion upon nucleotide dissociation triggered by two invariant arginines (TRCF R929 and R953/RecG R704 and R725; Figure 3). This motion engages the hook and the RH, which undergoes a straight/bent transition that appears to be regulated by the hook though evolutionarily conserved interactions. The RH, hook, and the TRG are all mechanically coupled because the hook is located immediately C-terminal to the TRG, while the RH is surrounded by the hook.
Figure 3. TRCF and RecG translocation modules.
Side-by-side comparisons of the translocation modules of RecG•ADP (left) and TRCF (right), illustrating the overall domain reorientation between TD1 (yellow) and TD2 (green) and the associated conformational change in the TRG motif (blue). The SF2 ATPase motifs (Figure 1a) are colored red. Individual amino acid side chains discussed in the text are shown and labeled. The small insets schematically illustrate the reorientation of TD1/TD2 between the ADP-bound state (RecG•ADP) and the nucleotide-free state (TRCF), which involves a 97° rigid body rotation of TD1 with respect to TD2 [17].
The TRCF/TEC model and TEC destabilization
Destabilization and release of the highly stable TEC is in itself quite a feat, and only one other protein factor in bacteria, Rho, is able to terminate transcription. The third termination mechanism, the intrinsic terminator, is entirely nucleic-acid based, but interestingly, like Rho and TRCF, it appears to lead to forward translocation of the RNAP on the template along the pathway of RNA release [26]. In fact, forward translocation of RNAP is a common theme underlying all transcription termination mechanisms, but the manner in which this is achieved is unique for each termination mechanism. For example, Rho mechanically peels the RNA due to its 5'-3' translocase activity [27], the RNA terminator hairpin destabilizes the DNA/RNA hybrid during its folding and interaction with the RNAP [28], while TRCF likely imposes torque upsteam of RNAP due to helical tracking on DNA. Recent experiments have demonstrated that in the case of TRCF, transcription complex release is preceded by the reannealing of basepairs at the upstream edge of the transcription bubble. Indeed, TECs reconstituted on DNA heteroduplexes that inhibit rewinding of the upstream duplex are less susceptible to both TRCF attack, and Rho-induced RNA release [29].
In addition to the potential for topological stress generated by the TRCF helical tracking motor, specific protein/protein interactions between TRCF and RNAP are required for RNAP release. This is supported by findings that trailing, elongating RNAPs cannot dissociate a leading, stalled (downstream) RNAP [15], despite RNAP being a powerful molecular motor [30,31]. In this regard, the interaction between the RID and the RNAP β-subunit, particularly with the RNAP-β "IKE" motif (a motif corresponding to E. coli β residues 117–119; [32]), appears to play a critical role, as substitution of TRCF-L499 (surface exposed and positioned close to the "IKE" motif in the TEC/TRCF model [17]) abolishes TEC displacement as well as RID/RNAP β-subunit interactions detected by a bacterial two-hybrid assay [17].
Conclusions and Perspectives
The bacterial TRCF is a complex, macromolecular machine. Understanding the details of its functional mechanism is important from two perspectives. From a biological perspective, TRCF is the link connecting the central cellular processes of transcription and NER. In addition, the TRCF uses its multiple domains and functions to manipulate the RNAP as well as the NER machinery in interesting ways – understanding the details of these manipulations will shed light on the functional mechanism of these macromolecular machines as well.
While the manner in which TRCF interfaces with the NER machinery remains murky, the last few years have seen significant advances in our understanding of TRCF/RNAP interactions. Biochemical and biophysical experiments have revealed how the TRCF dsDNA-tracking motor induces forward translocation of the RNAP, fleshed out the requirements for the TRCF interaction with the TEC [11], and shed light on the transcription termination mechanism [29]. Mutational studies and biochemical analyses have begun to provide details of TRCF/RNAP protein/protein interactions [17,32]. The new structural results outlined in this review [17,19] now provide a structural framework with which to design and interpret more incisive experiments addressing the TRCF structure/function relationship.
The model for the TRCF/TEC assembly put forth by Deaconescu et al. [17] clearly has limitations. Most importantly, the apo-TRCF structure used for modeling does not represent the DNA-bound state being modeled, which would require the binding of ATP. Furthermore, the model is a hybrid, based on combined atomic coordinates of Thermus aquaticus RNAP and E. coli TRCF. Much work remains to uncover the details of protein/protein interactions between TRCF/RNAP and TRCF/UvrA.
Of utmost importance in understanding TCR will be to elucidate the nature and timing of the conformational changes that TRCF undergoes during its functional cycle [17]. Structural studies of nucleotide-bound TRCF by X-ray crystallography, as well as RNAP/TEC complexes by cryo-electron microscopy combined with alternative solution-based techniques such as small angle X-ray scattering and TRCF inter-domain crosslinking via engineered disulfide cross-links will be highly informative.
Acknowledgements
Figures were prepared using Dino (http://cobra.mih.unibas.ch/dino/) and Pymol (http://pymol.sourceforge.net/). The authors work was supported by Biotechnology and Biological Sciences Research Council research grant BB/C507053/1 to N.J.S., and National Institutes of Health grant GM073829 to S.A.D.
References and recommending reading
Papers of particular interest, published within the annual period of the review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Bohr VA, Smith CA, Okumoto DS, Hanawalt PC. DNA repair in an active gene: Removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- 2.Mellon I, Hanawalt PC. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature. 1989;342:95–98. doi: 10.1038/342095a0. [DOI] [PubMed] [Google Scholar]
- 3.Mellon I, Spivak G, Hanawalt PC. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- *4. Laine JP, Egly JM. Initiation of DNA repair mediated by a stalled RNA polymerase IIO. EMBO J. 2006;25:387–397. doi: 10.1038/sj.emboj.7600933.. Reports the first in vitro reconstitution of eukaryotic TCR.
- 5.Selby CP, Witkin EM, Sancar A. Escherichia coli mfd mutant deficient in "mutation frequency decline" lacks strand-specific repair: in vitro complementation with purified coupling factor. Proc Natl Acad Sci U S A. 1991;88:11574–11578. doi: 10.1073/pnas.88.24.11574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Selby CP, Sancar A. Molecular mechanism of transcription-repair coupling. Science. 1993;260:53–58. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]
- 7.Selby CP, Sancar A. Structure and function of transcription-repair coupling factor. II. Catalytic properties. J Biol Chem. 1995;270:4890–4895. doi: 10.1074/jbc.270.9.4890. [DOI] [PubMed] [Google Scholar]
- 8.Selby CP, Sancar A. Structure and function of transcription-repair coupling factor. I. Structural domains and binding properties. J Biol Chem. 1995;270:4882–4889. doi: 10.1074/jbc.270.9.4882. [DOI] [PubMed] [Google Scholar]
- 9.Singleton MR, Scaife S, Wigley DB. Structural analysis of DNA replication fork reversal by RecG. Cell. 2001;107:79–89. doi: 10.1016/s0092-8674(01)00501-3. [DOI] [PubMed] [Google Scholar]
- 10.Rain JC, Selig L, De Reuse H, Battaglia V, Reverdy C, Simon S, Lenzen G, Petel F, Wojcik J, Schachter V, et al. The protein-protein interaction map of Helicobacter pylori. Nature. 2001;409:211–215. doi: 10.1038/35051615. [DOI] [PubMed] [Google Scholar]
- 11.Park J-S, Marr MT, Roberts JW. E. coli transcription repair coupling factor (Mfd protein) rescues arrested complexes by promoting forward translocation. Cell. 2002;109:757–767. doi: 10.1016/s0092-8674(02)00769-9. [DOI] [PubMed] [Google Scholar]
- 12.Zalieckas JM, Wray LVJ, Ferson AE, Fisher SH. Transcription-repair coupling factor is involved in carbon catabolite repression of the Bacillus subtilis hut and gnt operons. Mol. Microbiol. 1998;27:1031–1038. doi: 10.1046/j.1365-2958.1998.00751.x. [DOI] [PubMed] [Google Scholar]
- 13.Zeng X, Galinier A, Saxild HH. Catabolite repression of dra-nupC-pdp operon expression in Bacillus subtilis. Microbiol. 2000;146:2901–2908. doi: 10.1099/00221287-146-11-2901. [DOI] [PubMed] [Google Scholar]
- 14.Washburn RS, Wang Y, Gottesman ME. Role of E. coli transcription-repair coupling factor Mfd in Nun-mediated transcription termination. J. Mol. Biol. 2003;329:655–662. doi: 10.1016/s0022-2836(03)00465-0. [DOI] [PubMed] [Google Scholar]
- 15.Trautinger BW, Jaktaji RP, Rusakova E, Lloyd RG. RNA polymerase modulators and DNA repair activities resolve conflicts between DNA replication and transcription. Mol. Cell. 2005;19:247–258. doi: 10.1016/j.molcel.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 16.Deaconescu AM, Darst SA. Crystallization and preliminary structure determination of Escherichia coli Mfd, the transcription-repair coupling factor. Acta Crystallogr. F. 2006;61:1062–1064. doi: 10.1107/S1744309105035876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **17. Deaconescu AM, Chambers AL, Smith AJ, Nickels BE, Hochschild A, Savery NJ, Darst SA. Structural basis for bacterial transcription-coupled DNA repair. Cell. 2006;124:507–520. doi: 10.1016/j.cell.2005.11.045.. Reports the 3.2 Å-resolution X-ray crystal structure of full-length E. coli TRCF, along with a model for a TRCF/TEC complex based on in vivo and in vitro data on a TRCF mutant defective in binding RNAP.
- **18. Truglio JJ, Croteau DL, Skorvaga M, DellaVecchia MJ, Theis K, Mandavilli BS, Van Houten B, Kisker C. Interactions between UvrA and UvrB: The role of UvrB's domain 2 in nucleotide excision repair. EMBO J. 2004;23:2498–2509. doi: 10.1038/sj.emboj.7600263.. Reports the 2.1 Å-resolution X-ray crsytal strcuture of the N-terminus (residues 1–333) of E. coli TRCF, revealing structural similarity with the N-terminus of UvrB.
- 19.Assenmacher N, Wenig K, Lammens A, Hopfner KP. Structural basis for transcription-coupled repair: The N-terminus of Mfd resembles UvrB with degenerate ATPase motifs. J. Mol. Biol. 2006;355:675–683. doi: 10.1016/j.jmb.2005.10.033. [DOI] [PubMed] [Google Scholar]
- 20.Butland G, Peregrin-Alvarez JM, Li J, Yang W, Yang X, Canadien V, Starostine A, Richards D, Beattie B, Krogan N, et al. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 2005;433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- 21.Li J, Horwitz R, McCracken S, Greenblatt J. NusG, a new Escherichia coli elongation factor involved in transcriptional antitermination by the N protein of phage lambda. J. Biol. Chem. 1992;267:6012–6019. [PubMed] [Google Scholar]
- 22.Pasman Z, von Hippel PH. Regulation of rho-dependent transcription termination by NusG is specific to the Escherichia coli elongation complex. Biochemistry. 2000;39:5573–5585. doi: 10.1021/bi992658z. [DOI] [PubMed] [Google Scholar]
- 23.Durr H, Korner C, Muller M, Hickmann V, Hopfner KP. X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA. Cell. 2005;121:363–373. doi: 10.1016/j.cell.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 24.Chambers AL, Smith AJ, Savery NJ. A DNA translocation motif in the bacterial transcription-repair coupling factor, Mfd. Nucleic Acids Res. 2003;31:6409–6418. doi: 10.1093/nar/gkg868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahdi AA, Briggs GS, Sharples GJ, Wen Q, Lloyd RG. A model for dsDNA translocation revealed by a structural motif common to RecG and Mfd proteins. EMBO J. 2003;22:724–734. doi: 10.1093/emboj/cdg043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santangelo TJ, Roberts JW. Forward translocation is the natural pathway of RNA release at an intrinsic terminator. Mol. Cell. 2004;14:117–126. doi: 10.1016/s1097-2765(04)00154-6. [DOI] [PubMed] [Google Scholar]
- 27.Skordalakes E, Berger JM. Structure of the Rho transcription terminator: Mechanism of mRNA recognition and helicase loading. Cell. 2003;114:135–146. doi: 10.1016/s0092-8674(03)00512-9. [DOI] [PubMed] [Google Scholar]
- 28.Toulokhonov I, Landick R. The flap domain is required for pause RNA hairpin inhibition of catalysis by RNA polymerase and can modulate intrinsic termination. Mol. Cell. 2003;12:1125–1136. doi: 10.1016/s1097-2765(03)00439-8. [DOI] [PubMed] [Google Scholar]
- *29. Park JS, Roberts JW. Role of DNA bubble rewinding in enzymatic transcription termination. Proc. Natl. Acad. Sci. USA. 2006;103:4870–4875. doi: 10.1073/pnas.0600145103.. Biochemical analyses indicating that DNA bubble rewinding is a prerequisite for Rho-and TRCF-mediated transcription termination, and that forward translocation of RNAP, either by Rho or TRCF, represents an intermediate in RNA release.
- 30.Gelles J, Landick R. RNA polymerase as a molecular motor. Cell. 1998;93:13–16. doi: 10.1016/s0092-8674(00)81140-x. [DOI] [PubMed] [Google Scholar]
- 31.Wang MD, Schnitzer MJ, Yin H, Landick R, Gelles J, Block SM. Force and velocity measured for single molecules of RNA polymerase. Science. 1998;282:920–907. doi: 10.1126/science.282.5390.902. [DOI] [PubMed] [Google Scholar]
- *32. Smith AJ, Savery NJ. RNA polymerase mutants defective in the initiation of transcription-coupled DNA repair. Nucleic Acids Res. 2005;33:755–764. doi: 10.1093/nar/gki225.. Reports the identification of mutations within the N-terminal domain of the RNAP β-subunit that render RNAP immune to TRCF-mediated release and reactivation of backtracked TEC's. The authors proposed that these substitutions affect the initial steps of TCR, namely recruitment of TRCF to the TEC.