Summary
Cardiolipin (CL) plays a key role in dynamic organization of bacterial and mitochondrial membranes. CL forms membrane domains in bacterial cells, and these domains appear to participate in binding and functional regulation of multi-protein complexes involved in diverse cellular functions including cell division, energy metabolism, and membrane transport. Visualization of CL domains in bacterial cells by the fluorescent dye 10-N-nonyl acridine orange is critically reviewed. Possible mechanisms proposed for CL dynamic localization in bacterial cells are discussed. In the mitochondrial membrane CL is involved in organization of multi-subunit oxidative phosphorylation complexes and in their association into higher order supercomplexes. Evidence suggesting a possible role for CL in concert with ATP synthase oligomers in establishing mitochondrial cristae morphology is presented. Hypotheses on CL-dependent dynamic reorganization of the respiratory chain in response to changes in metabolic states and CL dynamic re-localization in mitochondria during the apoptotic response are briefly addressed.
Keywords: Cardiolipin, membrane domains, nonyl acridine orange, supercomplexes, cristae membrane
Introduction
The ability of lipids to promote formation of membrane sub-domains with unique protein and lipid composition provides a mechanism to compartmentalize and regulate biological processes. Cardiolipin (CL), also called diphosphatidylglycerol, with its unique dimeric molecular structure in which two phosphatidyl moieties are linked by a glycerol, is a major participant in the formation of membrane domains in bacteria and mitochondria [1, 2]. Although CL has two phosphate groups, it is not fully ionized at physiological pH due to intra-molecular hydrogen bonding between the free hydroxyl of the central glycerol and a protonated phosphate [3]. Due to the four acyl chains and a small headgroup, CL can organize into domains, which have the potential to serve as a proton sink in the membranes particularly when in close proximity to oxidative phosphorylation (OXPHOS) complexes that generate and consume ΔμH+ [4]. Being a dimeric lipid it can serve as a “flexible linker”, which fills cavities at protein interfaces and thus stabilizes interactions between individual subunits of oligomeric proteins as well as between multi-subunit individual complexes organized into higher order supra-molecular structures or supercomplexes (for review and references see [5-8]). Due to its propensity to form non-bilayer structures dependent on pH or divalent counter cations, CL has the potential to participate in formation of dynamic protein-lipid membrane domains of higher curvature, for example at bacterial division sites [9] or in the cristae of mitochondrial membranes ([1] and references within). In this review we will discuss roles of CL in dynamic organization and functional regulation of supra-molecular membrane structures, such as lipid domains and protein supercomplexes in bacterial and mitochondrial membranes.
Detection of CL in Whole Cells
Due to the specific interaction of the hydrophobic fluorescent dye 10-N-nonyl acridine orange (NAO) with CL, it was initially used to visualize mitochondria of eukaryotic cells [10-12]. Subsequently, NAO was used in the first visualization of CL-enriched membrane domains located at cell poles and near potential division sites of Escherichia coli [13]. The dye has a higher affinity for CL than for other anionic phospholipids [11, 12], and only association with CL but not with other phospholipids induces a green to red shift in its fluorescence emission maximum due to π–π bond stacking (see below). Let's first discuss use of NAO as specific reagent for CL detection
NAO as specific reagent for CL detection
According to [10] who introduced NAO for fluorescence microscopy, it stains specifically the mitochondria of living HeLa-cells. Sub μmolar concentrations of NAO were sufficient to specifically stain mitochondria generating a bright green fluorescence. Importantly, ionophores, uncoupling agents, or ATPase inhibitors did not affect dye accumulation suggesting that in contrast to the flourescent dye Rhodamine 123 the transmembrane potential is not the driving force for NAO mitochondrial localization, and NAO is bound to the mitochondrial membrane through hydrophobic interactions. These low levels of NAO did not affect respiratory activity after short incubation times. However, increasing inhibition of respiration with increasing NAO concentration and incubation time was observed. Similar to the respiratory activity there was no difference in the mitochondrial ultrastructure at low concentrations and short incubation times, but ultrastructure changed rapidly with increasing NAO concentration and incubation time eventuating in cristae transformation into multi–lamellar stacked membranes and final collapse of the mitochondria. These experiments showed that NAO is specifically accumulated at the inner mitochondrial membrane and the cristae.
Mitochondrial CL was identified as the NAO target, and binding was associated with appearance of red emission maximum in the fluorescence spectrum of the dye. The green to red shift was explained by π–π bond stacking of two positively charged NAO molecules bound to the two ionized phosphate groups of CL [11, 12], which differentiates CL from other anionic phospholipids containing a single phosphate group. An alternative model suggests that binding of NAO to CL microdomains results in the red shift [14]. This model takes into account the central hydroxyl of the glycerol connecting the two phosphate groups, which results in one acidic pKa and one basic pKa > 8.5 for CL [3]. Therefore, at physiological pH CL is not fully ionized. High affinity association between NAO and CL and the red shift were attributed to the ability of CL molecules with four chains and a small headgroup to make arrays (microdomains in the membrane), which provide enough space for the NAO molecules also to form arrays of parallel stacks between CL arrays [14]. The fluorescence spectrum of NAO at high concentrations in solution mimics the spectrum of NAO bound to CL in membranes, which is consistent with concentration and self-association of NAO in CL-containing membranes. This phenomenon is similar to the interaction of acridine orange (AO) with nucleic acids. The staining pattern of AO (green fluorescence for DNA, but red fluorescence for single-stranded RNA) is a function of the dye concentration due to interaction with DNA in a monomeric form and RNA as a complex of dye polymers and the nucleic acid (for references see http://www.micro-scope.de/fluoro.html).
Comparison of NAO with hydrophilic AO [15] and NAO analogues with different alkyl chain length [16] in their interaction with CL in model systems demonstrated that the hydrophobic interaction between NAO and CL plays a more important role than electrostatic interaction. On the other hand results from spectroscopy studies of the NAO binding to archaebacterial cardiolipin analogues were similar to those occurring with other diacidic phospholipids and sulfoglycolipids independent of the number of phytanyl chains (2 or 4) suggesting that two acidic residues or an acidic sulfocarbohydrate residue are determinants for NAO binding to archaebacterial lipids [17].
Although in many experiments the changes in NAO fluorescence upon binding to mitochondria were independent of the proton electrochemical gradient across the mitochondrial membrane, a number of experiments reported that NAO fluorescence in living eukaryotic cells was affected by addition of uncouples and inhibitors of the respiratory chain [18]. Depolarization of mitochondria resulted in changes in the NAO signal and redistribution of the dye inside the cells [19]. Indeed, NAO belongs to a group of membrane permeable lipophilic cations, which has ability to sense membrane potential (negative inside). However, due to the high affinity of NAO for CL, the dye demonstrates high passive (membrane potential independent) binding to the mitochondrial membrane. Thus, it was concluded that dependence of NAO binding on membrane potential might be a function of experimental conditions and the type of eukaryotic cell used. In addition depolarization of the mitochondrial membrane appears to affect spatial arrangement of CL in the mitochondrial membrane (see below), making interpretation of the experiments with uncouplers even more difficult.
Finally, yeast mutants lacking CL analyzed by flow cytometry emitted the same level of red fluorescence as wild type cells. Therefore, other components of eukaryotes appear to bind and concentrate NAO [20]. However, the ratios of red to green fluorescence for wild type and mutant cannot be determined in flow cytometry experiments. This parameter is necessary to distinguish the tailing of high green fluorescence into the red region from a true maximum of red fluorescence of CL. But in any case, flow cytometry is not suitable for a quantitative assessment of CL levels in living eukaryotic cells [20].
Nevertheless, staining of astrocyte cells fixed by glutaraldehyde revealed specific membrane potential-independent binding of NAO to mitochondria consistent with selective binding of NAO to CL in the mitochondrial membrane [19]. The data obtained on purified mitochondria, as well as inverted sub-mitochondrial particles and model systems such as liposomes, were highly consistent with NAO binding to CL. Capillary electrophoresis with post-column laser-induced fluorescence detection was successfully used to measure the CL content in individual NAO pre-stained mitochondria [21]. NAO also can be added to the running electrolyte buffer for visible spectrophotometric detection of CL by capillary electrophoresis; the technique was used for estimation of CL levels in purified inner and outer membranes of mitochondria before and after UV-induced apoptosis events [22]. A method was developed for fluorescent determination of CL in methanol/water solutions by NAO based on the decrease in fluorescence emission corresponded to the disappearance of monomeric form of NAO and demonstrated higher specificity for CL compared to other anionic phospholipids and lack of reaction with zwitterionic phospholipids [15]. Importantly, the fluorescence parameters of NAO bound to CL reflected also the state of CL organization in the mitochondrial membrane. Fluorescence resulting from CL interaction with NAO changed with the spatial arrangement of CL molecules within artificial membranes [23]. In isolated mitochondria fixed by glutaraldehyde at different respiratory states (3 and 4), the NAO fluorescence parameters (ratio between intensities of red and green fluorescence) are directly related to the mitochondrial energetic state. This was interpreted as reorganization of CL in the cristae membrane upon mitochondrial transition from one energetic state to another [23]. Consistent with this observation, experiments with phosphatidylcholine-CL containing liposomes demonstrated that the binding of NAO to the same amount of CL either organized in domains or dispersed as individual molecules in a phosphatidylcholine matrix resulted in a higher red/green fluorescence ratio for the former compared to the latter (Piccotti, L., Mileykovskaya, E. and Dowhan, W., unpublished data).
CL Domains in Bacteria
Visualization of CL domains in bacteria
As is the case for purified mitochondria, staining of different bacterial species with NAO was highly dependent on CL content. Addition of NAO at 100 to 200 nM to the growth medium resulted in staining of E. coli cells without any influence on their growth rate [13, 24]. Three-dimensional image reconstruction obtained by optical sectioning and a deconvolution algorithm revealed NAO-binding domains in the plane of the cell membrane. The fluorescent domains are observed mostly in the septal region and on the poles (for example see Fig.1 A). Substantial red fluorescence emission of bound NAO supported labeling of CL-containing domains while no red fluorescence was detected in mutants deficient in CL biosynthesis [13, 24]. A critical concentration of CL in the membrane was found at which domains with red emission maximum were observed [24].
Fig. 1.
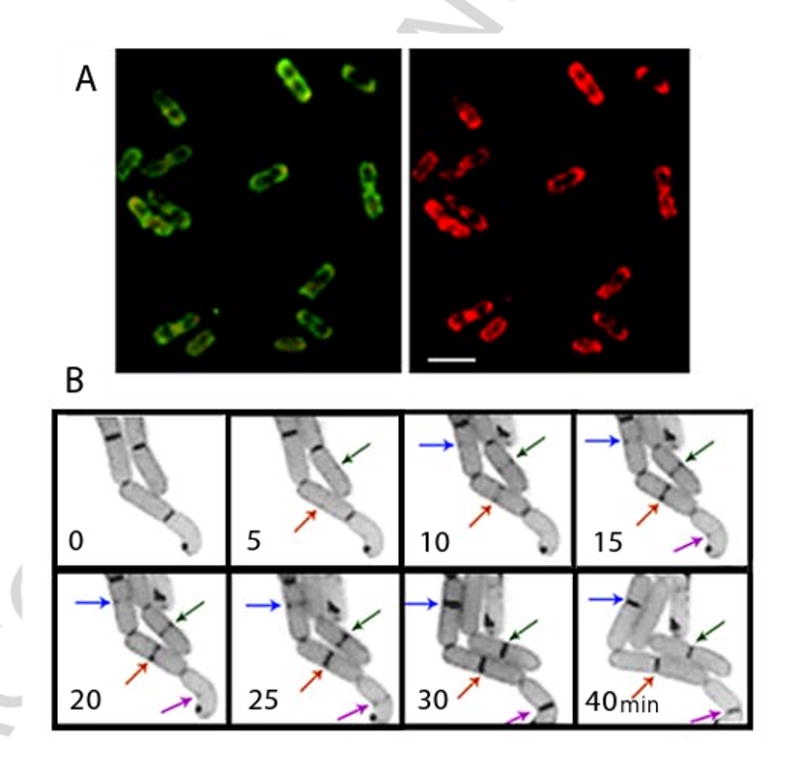
Staining of living wild type E. coli cells with fluorescent dye NAO revealed dynamic localization of CL enriched membrane domains. (A) Deconvoluted images of an optical section of cells grown in LB media supplemented with 200 nM NAO. Excitation was at 490 nm and emission was at either 528 (left) or 617 (right) nm; bar, 3 μm. (B) E. coli cells stained with NAO growing on a thin layer of LB agar on a microscopy glass slide. Images were taken with time intervals as indicated. Arrows point to CL domains as they appeared in the mid-cell areas of individual cells and further evolved into complete septa structures enriched in CL. (Fluorescent microscopy was performed by Lu Yang and Eugenia Mileykovskaya)
CL domains were also visualized with NAO in Bacillus subtilis [25, 26] and Pseudomonas putida [27]. Septal and polar localization of fluorescent domains was observed in exponentially growing B. subtilis cells. Importantly, the domains were not observed in the clsA null mutants lacking CL synthase and, consequently were due to the presence of CL. In addition fixation of B. subtilis cells by paraformaldehyde as well as depolarization with uncoupling agents did not alter the membrane distribution pattern of NAO fluorescence. All these data together establish that NAO fluorescence reflects a spatially heterogeneous distribution of CL in B. subtilis membranes. Cell pole enrichment in CL in E. coli was supported by an independent approach employing mass spectrometric lipid analysis of minicells that spontaneously bud off from the cell poles of the minCDE null mutant [28]. CL was approximately four-fold enriched at the expense of phosphatidylglycerol (PG) in the minicells over whole cells.
Mechanism of CL localization
Studies of the subcellular localization of green fluorescent protein fusions to the following enzymes involved in CL biosynthesis in B. subtilis showed septal localization [29]: phosphatidylglycerophosphate synthase (catalyses the committed step for the synthesis of PG and CL) and CL synthase. Experiments with FtsZ-depleted cells demonstrate that the localization of these synthases was Z-ring (forms at the septum to organize the cell division machinery) dependent. This is in a good agreement with the requirement to synthesize phospholipids in concert with the synthesis of peptidoglycan at the leading edge of the invaginating septum. However, the mechanism that prevents CL diffusion into the lateral part of the membrane was not addressed [29].
An equilibrium mechanism based on lipid microphase separation was proposed to explain the observed polar and division site localization of CL in rod-shaped bacteria [30, 31]. CL is classified as “high curvature lipid” due to the smaller cross-sectional area of its head group relative to its hydrophobic domain that induces high negative curvature strain in the lipid bilayer. A thermodynamic model of multicomponent inner membrane organization in bacterial cells indicates that in the presence of thermal fluctuations aggregates of high-curvature lipids may localize to the cell poles by sensing differences in cell curvature that are not sensed by individual lipid molecules. Such aggregation of CL is driven, in part, by short-range interactions between lipid molecules. It was assumed that two components contribute to the elastic energy of the membrane, the bending energy and a pinning potential. The balance between the osmotic-pressure difference across the membrane and the inward pressure of the cell wall determines the strength of the membrane pinning. Pinning of the cytoplasmic membrane by the cell wall results in the formation of stable finite-sized clusters of high-intrinsic-curvature lipids (e.g. CL), and subsequent targeting to the cell poles. At areas where the osmotic-pressure difference is reduced, the membrane is less constrained by the shape of the cell wall, and hence it is more likely to contain larger clusters of high-intrinsic-curvature lipids. Since the new septum during cell division separates two cell compartments with similar membrane pinning osmolyte concentrations, the osmotic-pressure difference across the septum should be much smaller than that across the rest of the cell membrane and the reduction in membrane pinning can lead to localization of the larger CL clusters at the division site, even in the absence of differences in cell wall curvature. The model also predicts a critical cellular CL concentration of 1% of total phospholipid for the formation of CL clusters large enough for polar targeting. The calculated numbers are in quantitative agreement with the observed critical concentration of CL required for visualization by NAO [24]. Thus the areas illuminated by NAO are not proposed to be a continuous CL-enriched large-scale domain resulting from phase segregation of lipids but rather a lattice of CL clusters.
Studies with polycationic antimicrobial compounds demonstrated that such compounds sequester anionic lipids from zwitterionic lipids also resulting in anionic lipid clusters, which in this case lead to phase boundary defects and increase in the membrane permeability [32]. The formation of these clusters of anionic lipids was suggested to alter the stability or composition of existing membrane domains and affect bacterial function.
CL domains and multi-protein complexes
The self-organizing nature of the lipid domains suggests that they might recruit proteins to the poles and division site in the living cell. A large number of proteins are localized to the polar regions in bacterial cells [24]. The co-localization with CL domains suggests CL involvement in the polar (and also septal) positioning of at least some of these proteins [9, 26]. The direct dependence of polar/septal localization of a membrane protein on formation of CL domains recorded by staining with NAO in E. coli was recently found for the osmosensory transporter ProP [24, 33]. The Eps system, involved in the export of cholera toxin in Vibrio cholerae, is localized to the poles and shows an in vitro requirement for CL, which strongly stimulates ATPase activity of the complex between cytoplasmic EpsE and membrane associated EpsL. Thus CL might promote interaction of EpsE with the C-terminal region of EpsL and its oligomerization on the membrane [34].
We suggested that formation of CL domains at cell pole/division sites plays an important role in selection and recognition of the division site by amphitropic cell cycle and cell division proteins, such as DnaA (initiation of DNA replication at oriC), MinD (a part of MinCDE system preventing positioning of the divisome at cell poles) and FtsA (bacterial actin which is a linker protein for cytoskeletal protein FtsZ (bacterial tubulin) responsible for targeting the Z-ring to the mid-cell membrane domain). These proteins interact directly with membrane phospholipids through specific amphipathic motifs enriched in basic amino acids, which confer the preference for anionic lipids ([9, 35] and references within). A mutant lacking PE and containing highly elevated levels of PG and CL showed a strong inhibition of cell division and an aggregation of FtsZ/FtsA and MinD proteins at domains enriched in CL [36, 37]. In vitro studies also demonstrate high affinity of these proteins for anionic phospholipids, specifically for CL [37, 38]. MinD preference for anionic phospholipids might play a role in the nucleation of the MinD oligomers at the cell poles [39]. The assumption of a nucleation site for MinD polymerization has been formulated [40] and was suggested in the “Min proteins polymerization-depolymerization model” [41]. The multi-stranded MinDE polymerization model [42] also supports this assumption (however, see [43]).
If activation of DnaA depends on an interaction with CL domains, then formation of CL domains at the cell center should precede or be concurrent with initiation of chromosome replication at oriC at the mid-cell membrane domain by DnaA. In a round of DNA replication the origin moves towards the cell poles leaving the CL domain in the center available for interaction with FtsZ/FtsA [9]. However, a recent report on B. subtilis demonstrated by using two membrane binding fluorescent dyes, FM4-64 and NAO, that in addition to cell pole/division site CL domains, a lipid spiral formed by PG extended along the long axis of the rod-shaped cell [44]. FRET was observed between green fluorescent protein-MinD and FM 4-64 labeled lipid spirals in wild type cells but not in a mutant lacking anionic lipids, thus supporting a direct interaction of MinD with anionic lipid spirals. Correct placement of the Z-ring in the cell might be a result of the dynamic interaction of MinD and cell division proteins FtsA/FtsZ with anionic lipid spirals [44]. MinD/FtsA/FtsZ-anionic lipid dynamic spiral structure might be classified as the division site placement hyperstructure in the bacterial cell [45]. In vitro experiments demonstrate that MinD binding to model membranes in which anionic and zwitterionic phospholipids are either uniformly mixed or segregated into domains is enhanced by segregation of anionic phospholipids to fluid domains in a gel-phase environment. Moreover, MinD stabilizes such domains [46, 47]. Taking into account the permanent nature of PG-spirals in the cell, it is not necessary in this case to postulate appearance of mid-cell anionic lipid domains immediately before initiation of DNA replication. In this scenario CL targeting to mid-cell would occur at the time of septum formation, in good agreement with the minimum osmotic-pressure difference across the membrane at the septum [30] (see above). An enrichment of the central domain with non-bilayer forming CL fulfills requirements for the remodeling of the membrane bilayer at the mid-cell domain into the cell poles resulting in membrane invagination and cell constriction. Our initial experiments clearly show positioning of CL domains at the future division sites and their gradual development with time into septa enriched in CL (Fig.1 B). Further studies of CL-domain dynamics are required to determine the precise step of the bacterial cell cycle at which the mid-cell domain positioning takes place.
CL in Higher Order Organization of Oxidative Phosphorylation Complexes
Multimeric protein complexes
CL is specifically required for organization and optimal activity of oxidative phosphorylation and photophosphorylation complexes in bacteria and mitochondria. It is preferentially bound at subunit interfaces in multi-subunit proteins as well as at the monomer interfaces of oligomeric proteins and acts as a flexible amphipathic linkage. CL at such positions is observed in the crystal structure of reaction centers [48], E. coli succinate dehydrogenase [49] and formate dehydrogenase-N [50]. CL plays both a structural and functional role in yeast bc1 complex (Complex III) [51], mammalian cytochrome c oxidase (Complex IV) [52, 53] and ATP/ADP carrier (AAC) [54]. In bc1 complex one CL molecule resides at the Qi site suggesting direct involvement of CL in proton uptake at the ubiquinone reduction site. This CL lies at the interface with cytochrome c oxidase in the supercomplex formed with complex IV [55] (see below). A second CL was found in the hydrophobic cleft close to the bc1 homodimer interface [5, 56]. Two CLs were resolved in Complex IV [53]. One CL together with two phosphatidylethanolamines and one PG stabilize the bovine Complex IV dimmer; however, the contacts made by phosphatidylethanolamine and PG molecules between monomers are much weaker than those made by CL. Interaction of this CL molecule with protein monomers demonstrate how CL stabilizes contacts between proteins. The four acyl chains of CL interact through van der Waals contacts with hydrophobic amino acid residues belonging to both monomers, and the two phosphate groups interact with both monomers via hydrogen bonds. A third CL, found by photolabeling with arylazido-containing CL analogues [52], is located near the entrance to the putative proton pumping channel and thus might facilitate proton entry into the channel. Another type of interaction mediated by CL tightly bound to a protein is observed in the crystal structure of AAC. Two CL molecules are sandwiched between the monomers of AAC, suggesting that this is the putative interface for protein dimerization.
Supercomplex formation
Individual Complexes I-IV (NADH:ubiquinone oxidoreductase, sucinate:ubiquinone oxidoreductase, bc1 complex or ubiqunol:cytochrome c oxidoreductase, and cytochrome c oxidase, respectively) of the mammalian mitochondrial respiratory chain are in equilibrium with supercomplexes or ‘respirasomes’ composed of all the above individual complexes [57]. Metabolic conditions could shift this equilibrium and consequently the mode of electron transfer within the respiratory chain between substrate channeling of small red-ox carriers (ubiquinone, between I (or II) and III or cytochrome c between III and IV) within the supercomplexes and random diffusion of these carriers among individual complexes [58, 59] (see below). This equilibrium in S. cerevisiae mitochondria appears to favor supercomplex organization of Complexes III and IV and possibly Complex II and two peripheral NADH dehydrogenases [60-62].
The 3-D structure for a respiratory supercomplex I1III2IV1 from bovine heart mitochondria [63] and a 2-D model of S. cerevisiae supercomplex IV1III2IV1 [64] have been recently proposed based on single particle analysis of negatively stained purified supercomplexes visualized by electron microscopy. The S. cerevisiae supercomplex (IV1III2IV1) is organized as a dimer of Complex III flanked by two Complex IV monomers while in the bovine supercomplex (I1III2IV1) the Complex III dimer interacts with a Complex IV monomer and both are attached to Complex I [65, 66]. Positions of ubiquinone- and cytochrome c binding sites in both supercomplexes support a substrate channeling mechanism.
Single particle analysis of respiratory supercomplexes from potato demonstrated four different types of supercomplexes, including I1III2, III2IV1, I1III2IV1 and I2III2 [67]. Based on 2-D projection maps of the potato I1III2IV1 supercomplex the authors suggest several differences in comparison with the 3-D structure of the bovine respirasome [63]. Specifically, complex III2 associates with complex I from the site that is opposite to the binding site in bovine supercomplex and complex III2 interacts with complex IV in a manner rather similar to the yeast supercomplex. Observation of a I2III2 structure allowed the authors to suggest a megacomplex organization of the respiratory complexes, in which complex III2 binds two copies of complex I (I1III2I1). Complex IV dimers then may connect I2III2 supercomplexes to form a “string” structure in the mitochondrial membrane.
Association of these individual complexes into a functional higher order “respirasome” is dependent on CL levels. In wild type S. cerevisiae Complexes III and IV remain tightly associated after extraction by mild detergents. However, in S. cerevisiae mutants lacking CL, which is replaced by elevated levels of PG, there is no stable supercomplex in the detergent extracts as revealed by native gel electrophoresis [55, 68]. Functional analysis of the organization of the respiratory chain of S. cerevisiae [60] demonstrated that in the CL-lacking mutant, in contrast to wild type cells, cytochrome c exhibited pool behavior consistent with the absence of respirasome organization [61].
A similar requirement for CL in respirosome organization in mammalian cells is supported by studies of mitochondria from patients with Barth syndrome, an X-linked genetic disorder characterized by cardiomyopathy, skeletal myopathy, neutropenia, and growth retardation (see [69, 70] for reviews). Mitochondria from Barth syndrome patients display a lower CL content and a polydispersity in acyl chain composition of CL, which departs significantly from normal due to mutations in the TAZ gene product, which is responsible for remodeling CL fatty acid composition. Analysis of the I1III2IV1 supercomplex in lymphoblasts from Barth syndrome patients after mild detergent extraction revealed an increase in level of complex IV monomer and a decrease in the level of the I1III2 supercomplex [71]. Mutants in the S. cerevisiae TAZ1 gene display reduced Complex IV levels and reduced incorporation of Complex IV into the supercomplex [72, 73].
CL-dependent association of respiratory supercomplexes with the ATP/ADP carrier into an “interactome” has also been demonstrated in yeast mitochondria. This association would place the carrier in an environment that promotes its optimal activity and explains the more efficient ATP production in S. cerevisiae wild type mitochondria compared with mitochondria from mutants lacking CL [74].
CL in Dynamic Organization of the Mitochondrial Membrane
CL and self-organization of cristae membranes
Two morphologically distinct domains, the inner boundary membrane and the cristae membrane, are observed in the inner mitochondrial membrane. The narrow tubular cristae junctions connect these domains [75, 76]. Respiratory chain supercomplexes and Complex V (ATP synthase) are significantly enriched in the cristae over the boundary membranes. The high molecular weight of these supercomplexes may prevent their diffusion through the cristae junctions, which act as a filter to regulate distribution of proteins between the two domains of the inner membrane [77].
Complex V dimers are also organized into oligomeric supercomplexes, which are required for cristae formation [65, 76, 78-82]. Electron cryo-tomography of mammalian mitochondria demonstrated long rows of dimeric ATP synthase located at the apex of cristae membranes. Based on the angle between the monomers of the dimer structure, the organization of ATPase into oligomeric ribbons should enforce a strong local curvature for the cristae membrane. Based on calculations of the electrostatic field strength, regions of high membrane curvature display a significant increase in charge density resulting in a local pH gradient of about 0.5 units. Thus, the mitochondrial cristae might act as proton traps, and the organization of ATP synthase may optimize its own performance [80]. CL segregation to membrane compartments of high curvature may further contribute to the proton sink and enhance ATP synthase function ([7] and references within). Studies in model systems composed of CL-containing giant unilamellar vesicles demonstrated that creation of a localized pH gradient induced cristae–like morphology in the vesicles. This phenomenon was not observed for lipid vesicles without CL. The results support the idea that self-organization of ATP synthase is coupled in the cristae membrane with self-organization of CL to produce maximum curvature of the membrane [83]. The authors also came to the conclusion that once established, the cristae morphology becomes self-maintaining as it limits the lateral diffusion of the oxidative phosphorylation protein complexes as well as protons in the vicinity of the complexes. The proton trapping potential of CL would increase protonic coupling between respiratory supercomplexes and Complex V in cristae membranes and prevent delocalization of the proton electrochemical gradient [4].
Association of ATP synthase with Taz1p into an interactome was recently demonstrate for S. cerevisiae mitochondria [84]. Interestingly, electron microscopy of the Δtaz1 mutant mitochondria demonstrated alterations in their morphology, such as elongated cristae and onion-like structures, which have been observed in ATP synthase oligomer mutants. However, ATP synthase oligomerization was normal in the taz1 mutant suggesting that oligomerization is required but not sufficient for normal cristae morphology. Reduced levels of CL and alterations in its composition in the mutant might be the second factor resulting in the aberrant morphology of the mitochondrial cristae membrane. Electron microscopy of mitochondria purified from a Δcrd1 S. cerevisiae mutant lacking CL supports this suggestion (Fig. 2, Mullapudi, S., Mileykovskaya. E., and Dowhan, W.; unpublished result).
Fig. 2.
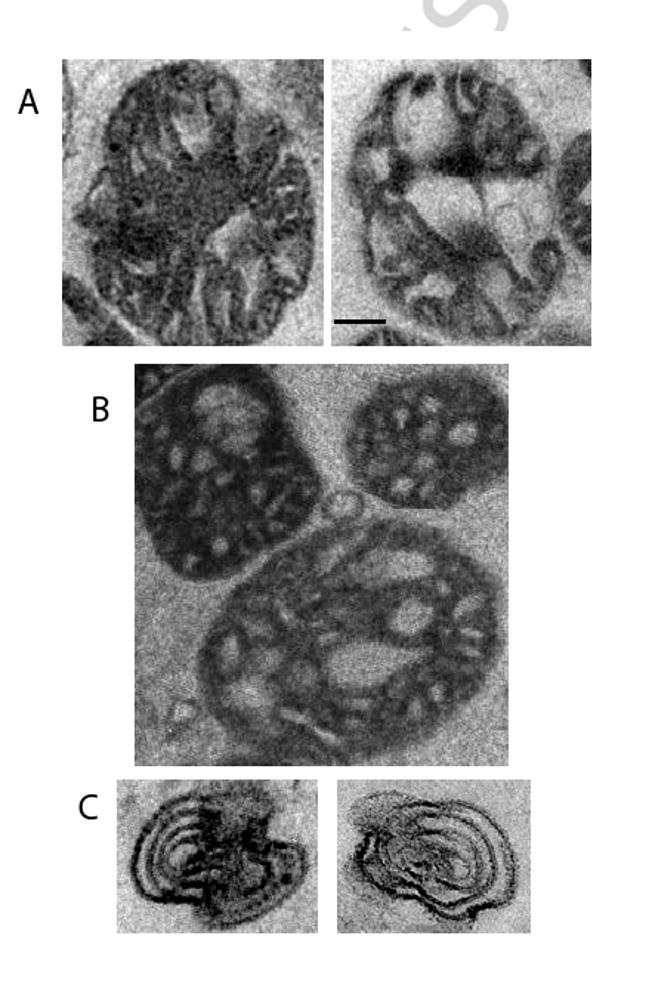
Altered cristae morphology in mitochondria from S. cerevisiae mutant lacking CL. Purified mitochondria were fixed for electron microscopy by glutaraldehyde followed by further fixation with osmium tetroxide and stained with uranyl acetate. (A) Wild type mitochondria; (B) Δcrd1 mitochondria; (C) onion-like structures in crd1Δ mitochondrial preparation. Bar, 25 nm. (Electron microscopy was performed by Srinivas Mullapudi).
The structural reorganization of the OXPHOS system
Metabolic flux control analysis was used to evaluate the impact of ΔμH+ on the control of cell respiration by cytochrome c oxidase [59]. Conditions mimicking switching between mitochondrial State 3 and 4 of respiration (respiration coupled with ATP production and uncoupled respiration, respectively), changed the apparent control efficiency of cytochrome c oxidase. The results were interpreted as changes in the assembly state of redox supercomplexes (respirasomes) depending on “energy demand” since the ‘respirasomes’ should gain a higher catalytic efficiency because of the channeling of mobile carries, ubiquinon and cytochrome c. Thus the model implies that a change in ΔμH+ triggers the structural reorganization of the OXPHOS system, which led to the hypothesis that CL might be the molecular sensor of the energy state of the mitochondrial membrane as well as an active player enabling the complexes of the OXPHOS system to change their aggregation state in response to the changes in ΔμH+ [59]. The hypothesis is consistent with a number of experimental facts, such as CL requirement for supercomplex formation (see above) and re-localization of CL in the cristae membrane upon transition of mitochondrion from one energetic state to another [23]. The level of CL biosynthesis by CL synthase in mitochondria depends on the ΔpH component of the proton-motive force generated by the electron transport chain [85]. All these data together suggest that changes in the respiratory state and ΔμH+ might induce re-localization and/or changes in CL levels in mitochondrial membrane, and thus might trigger changes in distribution between free individual complexes and complexes organized into respirasomes. Since morphology of the inner mitochondrial membrane affects function, changes in cristae membrane organization may be a means of regulating mitochondrial metabolism [75].
CL dynamics in apoptosis
Re-localization of CL among inner membrane domains and between inner and outer membranes may play a direct role in the regulation of early steps of apoptosis (see [86] for review and references). Movement of CL from the inner membrane to the outer membrane may be due to increased presence of CL in the outer leaflet of the inner membrane due to transmembrane movement from the matrix side. Alternatively, CL may redistribute from the cristae membrane to the inner boundary membrane (see [7] for review). In either case more CL would be available for movement to the outer membrane possibly via membrane contact sites. The CL decrease in the cristae membrane might induce dissociation of respiratory supercomplexes [7] as well as release of ATP/ADP carrier from interactomes (see above). Since cytochrome c has high affinity for CL, cytochrome c would also redistribute along with CL reducing its availability as an electron carrier (see [87, 88] for reviews). Moreover, interaction with CL causes structural changes in cytochrome c converting it into a peroxidase, which catalyzes CL peroxidation, that in turn would promote release of cytochrome c to the cytoplasm thus enhancing induction of apoptosis [89]. Translocation of Bid protein from the cytosol to mitochondria and formation of the active truncated form, tBid, seem also to be associated with the apoptotic migration of CL [87]. In vitro experiments demonstrated that mitochondrial creatine kinase, which in vivo is localized at contact sites between the inner and outer mitochondrial membranes, in its octameric form, promotes lipid segregation and clustering of CL in liposomes [90] and CL-containing monolayers [91] suggesting that a similar phenomenon might take place in vivo in the mitochondrial contact sites. The exact mechanisms driving apoptotic redistribution of CL are still poorly understood. However, it is clear that CL dynamics is directly involved in the apoptotic response.
Future Directions
The importance of CL in normal cell function and the possibility that aberrant CL levels are the molecular basis for several diseases have received recent attention. In this review we focused specifically on the role of CL in bacterial and mitochondrial membrane dynamics as it is related to the regulation of cellular function. The themes reviewed correlated CL dynamics to cellular processes but much remains to be done in order to elucidate molecular mechanisms. What is a molecular mechanism underlying CL localization in bacterial cells? The mathematical models built on experimental observations await experimental testing. Why and how do membrane proteins co-localized with CL-enriched domains and what are the exact molecular details of the resulting lipid-protein interactions? Similar questions can be asked about the mitochondrial membrane. During last decade it was demonstrated that CL is involved in association of OXPHOS complexes into higher ordered supercomplexes. Experimental data and hypothesis were summarized that support a role for CL dynamics in mitochondrial structural reorganization, which affects function and regulation, and, importantly, switching between different modes of operation of this organelle. However, details of mechanism are lacking. Future progress in this area will be dependent on more reliable means of detecting and quantifying CL and following CL movement in whole cells.
Acknowledgments
We thank Dr Kerwyn Casey Huang for reading the manuscript and helpful suggestions, Dr Srinivas Mullapudi for providing electron micrographs, and Lu Yang for performing experiments with NAO. The work of the authors cited in this review was supported in part by grants GM020478 and GM056289 to W.D. from the National Institutes of Health, U.S.A.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dowhan W, Bogdanov M, Mileykovskaya E. Functional roles of lipids in membranes. In: Vance DE, Vance JE, editors. Biochemistry of Lipids Lipoproteins and Membranes. Elsevier Press; Amsterdam: 2008. pp. 1–37. [Google Scholar]
- 2.Schlame M. Cardiolipin synthesis for the assembly of bacterial and mitochondrial membranes. J Lipid Res. 2008;49:1607–1620. doi: 10.1194/jlr.R700018-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kates M, Syz JY, Gosser D, Haines TH. pH-dissociation characteristics of cardiolipin and its 2′-deoxy analogue. Lipids. 1993;28:877–882. doi: 10.1007/BF02537494. [DOI] [PubMed] [Google Scholar]
- 4.Haines TH, Dencher NA. Cardiolipin: a proton trap for oxidative phosphorylation. FEBS Lett. 2002;528:35–39. doi: 10.1016/s0014-5793(02)03292-1. [DOI] [PubMed] [Google Scholar]
- 5.Hunte C. Specific protein-lipid interactions in membrane proteins. Biochem Soc Trans. 2005;33:938–942. doi: 10.1042/BST20050938. [DOI] [PubMed] [Google Scholar]
- 6.Mileykovskaya E, Zhang M, Dowhan W. Cardiolipin in energy transducing membranes. Biochemistry (Mosc) 2005;70:154–158. doi: 10.1007/s10541-005-0095-2. [DOI] [PubMed] [Google Scholar]
- 7.Bogdanov M, Mileykovskaya E, Dowhan W. Lipids in the Assembly of Membrane Proteins and Organization of Protein Supercomplexes: Implications for Lipid-linked Disorders. Subcell Biochem. 2008;49:197–239. doi: 10.1007/978-1-4020-8831-5_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joshi AS, Zhou J, Gohil VM, Chen S, Greenberg ML. Cellular functions of cardiolipin in yeast. Biochim Biophys Acta. 2008;1793:212–218. doi: 10.1016/j.bbamcr.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mileykovskaya E, Dowhan W. Role of membrane lipids in bacterial division-site selection. Curr Opin Microbiol. 2005;8:135–142. doi: 10.1016/j.mib.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 10.Septinus M, Berthold T, Naujok A, Zimmermann HW. Hydrophobic acridine dyes for fluorescent staining of mitochondria in living cells. 3. Specific accumulation of the fluorescent dye NAO on the mitochondrial membranes in HeLa cells by hydrophobic interaction. Depression of respiratory activity, changes in the ultrastructure of mitochondria due to NAO. Increase of fluorescence in vital stained mitochondria in situ by irradiation. Histochemistry. 1985;82:51–66. doi: 10.1007/BF00502091. [DOI] [PubMed] [Google Scholar]
- 11.Petit JM, Maftah A, Ratinaud MH, Julien R. 10-N-nonyl acridine orange interacts with cardiolipin and allows the quantification of this phospholipid in isolated mitochondria. Eur J Biochem. 1992;209:267–273. doi: 10.1111/j.1432-1033.1992.tb17285.x. [DOI] [PubMed] [Google Scholar]
- 12.Petit JM, Huet O, Gallet PF, Maftah A, Ratinaud MH, Julien R. Direct analysis and significance of cardiolipin transverse distribution in mitochondrial inner membranes. Eur J Biochem. 1994;220:871–879. doi: 10.1111/j.1432-1033.1994.tb18690.x. [DOI] [PubMed] [Google Scholar]
- 13.Mileykovskaya E, Dowhan W. Visualization of phospholipid domains in Escherichia coli by using the cardiolipin-specific fluorescent dye 10-N-nonyl acridine orange. J Bacteriol. 2000;182:1172–1175. doi: 10.1128/jb.182.4.1172-1175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mileykovskaya E, Dowhan W, Birke RL, Zheng D, Lutterodt L, Haines TH. Cardiolipin binds nonyl acridine orange by aggregating the dye at exposed hydrophobic domains on bilayer surfaces. FEBS Lett. 2001;507:187–190. doi: 10.1016/s0014-5793(01)02948-9. [DOI] [PubMed] [Google Scholar]
- 15.Kaewsuya P, Danielson ND, Ekhterae D. Fluorescent determination of cardiolipin using 10-N-nonyl acridine orange. Anal Bioanal Chem. 2007;387:2775–2782. doi: 10.1007/s00216-007-1135-0. [DOI] [PubMed] [Google Scholar]
- 16.Rodriguez ME, Azizuddin K, Zhang P, Chiu SM, Lam M, Kenney ME, Burda C, Oleinick NL. Targeting of mitochondria by 10-N-alkyl acridine orange analogues: role of alkyl chain length in determining cellular uptake and localization. Mitochondrion. 2008;8:237–246. doi: 10.1016/j.mito.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lobasso S, Saponetti MS, Polidoro F, Lopalco P, Urbanija J, Kralj-Iglic V, Corcelli A. Archaebacterial lipid membranes as models to study the interaction of 10-N-nonyl acridine orange with phospholipids. Chem Phys Lipids. 2009;157:12–20. doi: 10.1016/j.chemphyslip.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 18.Keij JF, Bell-Prince C, Steinkamp JA. Staining of mitochondrial membranes with 10-nonyl acridine orange, MitoFluor Green, and MitoTracker Green is affected by mitochondrial membrane potential altering drugs. Cytometry. 2000;39:203–210. doi: 10.1002/(sici)1097-0320(20000301)39:3<203::aid-cyto5>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 19.Jacobson J, Duchen MR, Heales SJ. Intracellular distribution of the fluorescent dye nonyl acridine orange responds to the mitochondrial membrane potential: implications for assays of cardiolipin and mitochondrial mass. J Neurochem. 2002;82:224–233. doi: 10.1046/j.1471-4159.2002.00945.x. [DOI] [PubMed] [Google Scholar]
- 20.Gohil VM, Gvozdenovic-Jeremic J, Schlame M, Greenberg ML. Binding of 10-N-nonyl acridine orange to cardiolipin-deficient yeast cells: implications for assay of cardiolipin. Anal Biochem. 2005;343:350–352. doi: 10.1016/j.ab.2005.04.039. [DOI] [PubMed] [Google Scholar]
- 21.Fuller KM, Duffy CF, Arriaga EA. Determination of the cardiolipin content of individual mitochondria by capillary electrophoresis with laser-induced fluorescence detection. Electrophoresis. 2002;23:1571–1576. doi: 10.1002/1522-2683(200206)23:11<1571::AID-ELPS1571>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 22.Qi L, Danielson ND, Dai Q, Lee RM. Capillary electrophoresis of cardiolipin with on-line dye interaction and spectrophotometric detection. Electrophoresis. 2003;24:1680–1686. doi: 10.1002/elps.200305448. [DOI] [PubMed] [Google Scholar]
- 23.Garcia Fernandez MI, Ceccarelli D, Muscatello U. Use of the fluorescent dye 10-N-nonyl acridine orange in quantitative and location assays of cardiolipin: a study on different experimental models. Anal Biochem. 2004;328:174–180. doi: 10.1016/j.ab.2004.01.020. [DOI] [PubMed] [Google Scholar]
- 24.Romantsov T, Helbig S, Culham DE, Gill C, Stalker L, Wood JM. Cardiolipin promotes polar localization of osmosensory transporter ProP in Escherichia coli. Mol Microbiol. 2007;64:1455–1465. doi: 10.1111/j.1365-2958.2007.05727.x. [DOI] [PubMed] [Google Scholar]
- 25.Kawai F, Shoda M, Harashima R, Sadaie Y, Hara H, Matsumoto K. Cardiolipin domains in Bacillus subtilis marburg membranes. J Bacteriol. 2004;186:1475–1483. doi: 10.1128/JB.186.5.1475-1483.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsumoto K, Kusaka J, Nishibori A, Hara H. Lipid domains in bacterial membranes. Mol Microbiol. 2006;61:1110–1117. doi: 10.1111/j.1365-2958.2006.05317.x. [DOI] [PubMed] [Google Scholar]
- 27.Bernal P, Munoz-Rojas J, Hurtado A, Ramos JL, Segura A. A Pseudomonas putida cardiolipin synthesis mutant exhibits increased sensitivity to drugs related to transport functionality. Environ Microbiol. 2007;9:1135–1145. doi: 10.1111/j.1462-2920.2006.01236.x. [DOI] [PubMed] [Google Scholar]
- 28.Koppelman CM, Aarsman ME, Postmus J, Pas E, Muijsers AO, Scheffers DJ, Nanninga N, den Blaauwen T. R174 of Escherichia coli FtsZ is involved in membrane interaction and protofilament bundling, and is essential for cell division. Mol Microbiol. 2004;51:645–657. doi: 10.1046/j.1365-2958.2003.03876.x. [DOI] [PubMed] [Google Scholar]
- 29.Nishibori A, Kusaka J, Hara H, Umeda M, Matsumoto K. Phosphatidylethanolamine domains and localization of phospholipid synthases in Bacillus subtilis membranes. J Bacteriol. 2005;187:2163–2174. doi: 10.1128/JB.187.6.2163-2174.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang KC, Mukhopadhyay R, Wingreen NS. A curvature-mediated mechanism for localization of lipids to bacterial poles. PLoS Comput Biol. 2006;2:e151. doi: 10.1371/journal.pcbi.0020151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mukhopadhyay R, Huang KC, Wingreen NS. Lipid localization in bacterial cells through curvature-mediated microphase separation. Biophys J. 2008;95:1034–1049. doi: 10.1529/biophysj.107.126920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Epand RM, Epand RF. Lipid domains in bacterial membranes and the action of antimicrobial agents. Biochim Biophys Acta. 2009;1788:289–294. doi: 10.1016/j.bbamem.2008.08.023. [DOI] [PubMed] [Google Scholar]
- 33.Mileykovskaya E. Subcellular localization of Escherichia coli osmosensory transporter ProP: focus on cardiolipin membrane domains. Mol Microbiol. 2007;64:1419–1422. doi: 10.1111/j.1365-2958.2007.05766.x. [DOI] [PubMed] [Google Scholar]
- 34.Camberg JL, Johnson TL, Patrick M, Abendroth J, Hol WG, Sandkvist M. Synergistic stimulation of EpsE ATP hydrolysis by EpsL and acidic phospholipids. Embo J. 2007;26:19–27. doi: 10.1038/sj.emboj.7601481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dowhan W, Mileykovskaya E, Bogdanov M. Diversity and versatility of lipid-protein interactions revealed by molecular genetic approaches. Biochim Biophys Acta. 2004;1666:19–39. doi: 10.1016/j.bbamem.2004.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mileykovskaya E, Sun Q, Margolin W, Dowhan W. Localization and function of early cell division proteins in filamentous Escherichia coli cells lacking phosphatidylethanolamine. J Bacteriol. 1998;180:4252–4257. doi: 10.1128/jb.180.16.4252-4257.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mileykovskaya E, Fishov I, Fu X, Corbin BD, Margolin W, Dowhan W. Effects of phospholipid composition on MinD-membrane interactions in vitro and in vivo. J Biol Chem. 2003;278:22193–22198. doi: 10.1074/jbc.M302603200. [DOI] [PubMed] [Google Scholar]
- 38.Sekimizu K, Kornberg A. Cardiolipin activation of dnaA protein, the initiation protein of replication in Escherichia coli. J Biol Chem. 1988;263:7131–7135. [PubMed] [Google Scholar]
- 39.Shih YL, Le T, Rothfield L. Division site selection in Escherichia coli involves dynamic redistribution of Min proteins within coiled structures that extend between the two cell poles. Proc Natl Acad Sci U S A. 2003;100:7865–7870. doi: 10.1073/pnas.1232225100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Norris V, Woldringh C, Mileykovskaya E. A hypothesis to explain division site selection in Escherichia coli by combining nucleoid occlusion and Min. FEBS Lett. 2004;561:3–10. doi: 10.1016/S0014-5793(04)00135-8. [DOI] [PubMed] [Google Scholar]
- 41.Drew DA, Osborn MJ, Rothfield LI. A polymerization-depolymerization model that accurately generates the self-sustained oscillatory system involved in bacterial division site placement. Proc Natl Acad Sci U S A. 2005;102:6114–6118. doi: 10.1073/pnas.0502037102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cytrynbaum EN, Marshall BD. A multistranded polymer model explains MinDE dynamics in E. coli cell division. Biophys J. 2007;93:1134–1150. doi: 10.1529/biophysj.106.097162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sengupta S, Rutenberg A. Modeling partitioning of Min proteins between daughter cells after septation in Escherichia coli. Phys Biol. 2007;4:145–153. doi: 10.1088/1478-3975/4/3/001. [DOI] [PubMed] [Google Scholar]
- 44.Barak I, Muchova K, Wilkinson AJ, O'Toole PJ, Pavlendova N. Lipid spirals in Bacillus subtilis and their role in cell division. Mol Microbiol. 2008;68:1315–1327. doi: 10.1111/j.1365-2958.2008.06236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Norris V, den Blaauwen T, Cabin-Flaman A, Doi RH, Harshey R, Janniere L, Jimenez-Sanchez A, Jin DJ, Levin PA, Mileykovskaya E, Minsky A, Saier M, Jr, Skarstad K. Functional taxonomy of bacterial hyperstructures. Microbiol Mol Biol Rev. 2007;71:230–253. doi: 10.1128/MMBR.00035-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mazor S, Regev T, Mileykovskaya E, Margolin W, Dowhan W, Fishov I. Mutual effects of MinD-membrane interaction: II. Domain structure of the membrane enhances MinD binding. Biochim Biophys Acta. 2008;1778:2505–2511. doi: 10.1016/j.bbamem.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mazor S, Regev T, Mileykovskaya E, Margolin W, Dowhan W, Fishov I. Mutual effects of MinD-membrane interaction: I. Changes in the membrane properties induced by MinD binding. Biochim Biophys Acta. 2008;1778:2496–2504. doi: 10.1016/j.bbamem.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fyfe PK, McAuley KE, Roszak AW, Isaacs NW, Cogdell RJ, Jones MR. Probing the interface between membrane proteins and membrane lipids by X-ray crystallography. Trends Biochem Sci. 2001;26:106–112. doi: 10.1016/s0968-0004(00)01746-1. [DOI] [PubMed] [Google Scholar]
- 49.Yankovskaya V, Horsefield R, Tornroth S, Luna-Chavez C, Miyoshi H, Leger C, Byrne B, Cecchini G, Iwata S. Architecture of succinate dehydrogenase and reactive oxygen species generation. Science. 2003;299:700–704. doi: 10.1126/science.1079605. [DOI] [PubMed] [Google Scholar]
- 50.Jormakka M, Tornroth S, Byrne B, Iwata S. Molecular basis of proton motive force generation: structure of formate dehydrogenase-N. Science. 2002;295:1863–1868. doi: 10.1126/science.1068186. [DOI] [PubMed] [Google Scholar]
- 51.Lange C, Nett JH, Trumpower BL, Hunte C. Specific roles of protein-phospholipid interactions in the yeast cytochrome bc1 complex structure. Embo J. 2001;20:6591–6600. doi: 10.1093/emboj/20.23.6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sedlak E, Panda M, Dale MP, Weintraub ST, Robinson NC. Photolabeling of cardiolipin binding subunits within bovine heart cytochrome c oxidase. Biochemistry. 2006;45:746–754. doi: 10.1021/bi050870z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shinzawa-Itoh K, Aoyama H, Muramoto K, Terada H, Kurauchi T, Tadehara Y, Yamasaki A, Sugimura T, Kurono S, Tsujimoto K, Mizushima T, Yamashita E, Tsukihara T, Yoshikawa S. Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase. Embo J. 2007;26:1713–1725. doi: 10.1038/sj.emboj.7601618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nury H, Dahout-Gonzalez C, Trezeguet V, Lauquin G, Brandolin G, Pebay-Peyroula E. Structural basis for lipid-mediated interactions between mitochondrial ADP/ATP carrier monomers. FEBS Lett. 2005;579:6031–6036. doi: 10.1016/j.febslet.2005.09.061. [DOI] [PubMed] [Google Scholar]
- 55.Pfeiffer K, Gohil V, Stuart RA, Hunte C, Brandt U, Greenberg ML, Schagger H. Cardiolipin stabilizes respiratory chain supercomplexes. J Biol Chem. 2003;278:52873–52880. doi: 10.1074/jbc.M308366200. [DOI] [PubMed] [Google Scholar]
- 56.Palsdottir H, Hunte C. Lipids in membrane protein structures. Biochim Biophys Acta. 2004;1666:2–18. doi: 10.1016/j.bbamem.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 57.Schagger H, Pfeiffer K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. Embo J. 2000;19:1777–1783. doi: 10.1093/emboj/19.8.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Genova ML, Baracca A, Biondi A, Casalena G, Faccioli M, Falasca AI, Formiggini G, Sgarbi G, Solaini G, Lenaz G. Is supercomplex organization of the respiratory chain required for optimal electron transfer activity? Biochim Biophys Acta. 2008;1777:740–746. doi: 10.1016/j.bbabio.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 59.Piccoli C, Scrima R, Boffoli D, Capitanio N. Control by cytochrome c oxidase of the cellular oxidative phosphorylation system depends on the mitochondrial energy state. Biochem J. 2006;396:573–583. doi: 10.1042/BJ20060077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boumans H, Grivell LA, Berden JA. The respiratory chain in yeast behaves as a single functional unit. J Biol Chem. 1998;273:4872–4877. doi: 10.1074/jbc.273.9.4872. [DOI] [PubMed] [Google Scholar]
- 61.Zhang M, Mileykovskaya E, Dowhan W. Cardiolipin is essential for organization of complexes III and IV into a supercomplex in intact yeast mitochondria. J Biol Chem. 2005;280:29403–29408. doi: 10.1074/jbc.M504955200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bornhovd C, Vogel F, Neupert W, Reichert AS. Mitochondrial membrane potential is dependent on the oligomeric state of F1F0-ATP synthase supracomplexes. J Biol Chem. 2006;281:13990–13998. doi: 10.1074/jbc.M512334200. [DOI] [PubMed] [Google Scholar]
- 63.Schafer E, Dencher NA, Vonck J, Parcej DN. Three-dimensional structure of the respiratory chain supercomplex I1III2IV1 from bovine heart mitochondria. Biochemistry. 2007;46:12579–12585. doi: 10.1021/bi700983h. [DOI] [PubMed] [Google Scholar]
- 64.Heinemeyer J, Braun HP, Boekema EJ, Kouril R. A structural model of the cytochrome c reductase/oxidase supercomplex from yeast mitochondria. J Biol Chem. 2007;282:12240–12248. doi: 10.1074/jbc.M610545200. [DOI] [PubMed] [Google Scholar]
- 65.Vonck J, Schafer E. Supramolecular organization of protein complexes in the mitochondrial inner membrane. Biochim Biophys Acta. 2008 doi: 10.1016/j.bbamcr.2008.05.019. [DOI] [PubMed] [Google Scholar]
- 66.Dudkina NV, Sunderhaus S, Boekema EJ, Braun HP. The higher level of organization of the oxidative phosphorylation system: mitochondrial supercomplexes. J Bioenerg Biomembr. 2008 doi: 10.1007/s10863-008-9167-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bultema JB, Braun HP, Boekema EJ, Kouril R. Megacomplex organization of the oxidative phosphorylation system by structural analysis of respiratory supercomplexes from potato. Biochim Biophys Acta. 2009;1787:60–67. doi: 10.1016/j.bbabio.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 68.Zhang M, Mileykovskaya E, Dowhan W. Gluing the respiratory chain together. Cardiolipin is required for supercomplex formation in the inner mitochondrial membrane. J Biol Chem. 2002;277:43553–43556. doi: 10.1074/jbc.C200551200. [DOI] [PubMed] [Google Scholar]
- 69.Schlame M, Ren M. Barth syndrome, a human disorder of cardiolipin metabolism. FEBS Lett. 2006;580:5450–5455. doi: 10.1016/j.febslet.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 70.Hauff KD, Hatch GM. Cardiolipin metabolism and Barth Syndrome. Prog Lipid Res. 2006;45:91–101. doi: 10.1016/j.plipres.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 71.McKenzie M, Lazarou M, Thorburn DR, Ryan MT. Mitochondrial respiratory chain supercomplexes are destabilized in Barth Syndrome patients. J Mol Biol. 2006;361:462–469. doi: 10.1016/j.jmb.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 72.Brandner K, Mick DU, Frazier AE, Taylor RD, Meisinger C, Rehling P. Taz1, an outer mitochondrial membrane protein, affects stability and assembly of inner membrane protein complexes: implications for Barth Syndrome. Mol Biol Cell. 2005;16:5202–5214. doi: 10.1091/mbc.E05-03-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li G, Chen S, Thompson MN, Greenberg ML. New insights into the regulation of cardiolipin biosynthesis in yeast: implications for Barth syndrome. Biochim Biophys Acta. 2007;1771:432–441. doi: 10.1016/j.bbalip.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 74.Claypool SM, Oktay Y, Boontheung P, Loo JA, Koehler CM. Cardiolipin defines the interactome of the major ADP/ATP carrier protein of the mitochondrial inner membrane. J Cell Biol. 2008;182:937–950. doi: 10.1083/jcb.200801152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mannella CA. Structure and dynamics of the mitochondrial inner membrane cristae. Biochim Biophys Acta. 2006;1763:542–548. doi: 10.1016/j.bbamcr.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 76.Zick M, Rabl R, Reichert AS. Cristae formation-linking ultrastructure and function of mitochondria. Biochim Biophys Acta. 2008 doi: 10.1016/j.bbamcr.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 77.Vogel F, Bornhovd C, Neupert W, Reichert AS. Dynamic subcompartmentalization of the mitochondrial inner membrane. J Cell Biol. 2006;175:237–247. doi: 10.1083/jcb.200605138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Giraud MF, Paumard P, Soubannier V, Vaillier J, Arselin G, Salin B, Schaeffer J, Brethes D, di Rago JP, Velours J. Is there a relationship between the supramolecular organization of the mitochondrial ATP synthase and the formation of cristae? Biochim Biophys Acta. 2002;1555:174–180. doi: 10.1016/s0005-2728(02)00274-8. [DOI] [PubMed] [Google Scholar]
- 79.Dudkina NV, Sunderhaus S, Braun HP, Boekema EJ. Characterization of dimeric ATP synthase and cristae membrane ultrastructure from Saccharomyces and Polytomella mitochondria. FEBS Lett. 2006;580:3427–3432. doi: 10.1016/j.febslet.2006.04.097. [DOI] [PubMed] [Google Scholar]
- 80.Strauss M, Hofhaus G, Schroder RR, Kuhlbrandt W. Dimer ribbons of ATP synthase shape the inner mitochondrial membrane. Embo J. 2008;27:1154–1160. doi: 10.1038/emboj.2008.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wittig I, Schagger H. Structural organization of mitochondrial ATP synthase. Biochim Biophys Acta. 2008;1777:592–598. doi: 10.1016/j.bbabio.2008.04.027. [DOI] [PubMed] [Google Scholar]
- 82.Devenish RJ, Prescott M, Rodgers AJ. The structure and function of mitochondrial F1F0-ATP synthases. Int Rev Cell Mol Biol. 2008;267:1–58. doi: 10.1016/S1937-6448(08)00601-1. [DOI] [PubMed] [Google Scholar]
- 83.Khalifat N, Puff N, Bonneau S, Fournier JB, Angelova MI. Membrane deformation under local pH gradient: mimicking mitochondrial cristae dynamics. Biophys J. 2008;95:4924–4933. doi: 10.1529/biophysj.108.136077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Claypool SM, Boontheung P, McCaffery JM, Loo JA, Koehler CM. The cardiolipin transacylase, tafazzin, associates with two distinct respiratory components providing insight into Barth Syndrome. Mol Biol Cell. 2008;19:5143–5155. doi: 10.1091/mbc.E08-09-0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gohil VM, Hayes P, Matsuyama S, Schagger H, Schlame M, Greenberg ML. Cardiolipin biosynthesis and mitochondrial respiratory chain function are interdependent. J Biol Chem. 2004;279:42612–42618. doi: 10.1074/jbc.M402545200. [DOI] [PubMed] [Google Scholar]
- 86.Gonzalvez F, Gottlieb E. Cardiolipin: setting the beat of apoptosis. Apoptosis. 2007;12:877–885. doi: 10.1007/s10495-007-0718-8. [DOI] [PubMed] [Google Scholar]
- 87.Lucken-Ardjomande S, Martinou JC. Newcomers in the process of mitochondrial permeabilization. J Cell Sci. 2005;118:473–483. doi: 10.1242/jcs.01654. [DOI] [PubMed] [Google Scholar]
- 88.Ott M, Zhivotovsky B, Orrenius S. Role of cardiolipin in cytochrome c release from mitochondria. Cell Death Differ. 2007;14:1243–1247. doi: 10.1038/sj.cdd.4402135. [DOI] [PubMed] [Google Scholar]
- 89.Basova LV, Kurnikov IV, Wang L, Ritov VB, Belikova NA, Vlasova II, Pacheco AA, Winnica DE, Peterson J, Bayir H, Waldeck DH, Kagan VE. Cardiolipin switch in mitochondria: shutting off the reduction of cytochrome c and turning on the peroxidase activity. Biochemistry. 2007;46:3423–3434. doi: 10.1021/bi061854k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Epand RF, Tokarska-Schlattner M, Schlattner U, Wallimann T, Epand RM. Cardiolipin clusters and membrane domain formation induced by mitochondrial proteins. J Mol Biol. 2007;365:968–980. doi: 10.1016/j.jmb.2006.10.028. [DOI] [PubMed] [Google Scholar]
- 91.Maniti O, Lecompte MF, Marcillat O, Desbat B, Buchet R, Vial C, Granjon T. Mitochondrial creatine kinase binding to phospholipid monolayers induces cardiolipin segregation. Biophys J. 2009;96:2428–2438. doi: 10.1016/j.bpj.2008.12.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]


