Abstract
Chronic Fatigue Syndrome (CFS) is characterized by debilitating fatigue, often accompanied by widespread muscle pain that meets criteria for Fibromyalgia Syndrome (FMS). Symptoms become markedly worse after exercise. Previous studies implicated dysregulation of the sympathetic nervous system (SNS), and immune system (IS) in CFS and FMS. We recently demonstrated that Acid Sensing Ion Channel (likely ASIC3), purinergic type 2X receptors (likely P2X4 and P2X5), and the transient receptor potential vanilloid type 1 (TRPV1) are molecular receptors in mouse sensory neurons detecting metabolites that cause acute muscle pain and possibly muscle fatigue. These molecular receptors are found on human leukocytes along with SNS and IS genes. Real-time, quantitative PCR showed that 19 CFS patients had lower expression of β-2 adrenergic receptors but otherwise did not differ from 16 controls before exercise. After a sustained moderate exercise test, CFS patients showed greater increases than controls in gene expression for metabolite detecting receptors ASIC3, P2X4 and P2X5, for SNS receptors α-2A, β-1, β-2 and COMT, and IS genes for IL10 and TLR4 lasting from 0.5–48 hours (P< .05). These increases were also seen in the CFS subgroup with comorbid FMS and were highly correlated with symptoms of physical fatigue, mental fatigue and pain. These new findings suggest dysregulation of metabolite detecting receptors as well as SNS and IS in CFS and CFS-FMS.
Perspective
Muscle fatigue and pain are major symptoms of CFS. Following moderate exercise, CFS and CFS-FMS patients show enhanced gene expression for receptors detecting muscle metabolites and for SNS and IS, which correlate with these symptoms. These findings suggest possible new causes, points for intervention and objective biomarkers for these disorders.
Keywords: fatigue, muscle pain, gene expression, ASIC, TRPV1, leukocytes
Introduction
Chronic Fatigue Syndrome (CFS) is defined by the CDC as “persistent or relapsing fatigue not relieved by rest that severely compromises activities of daily life for 6 months or longer, for which other possible medical causes have been ruled out.” In addition, the CDC diagnosis requires at least 4 of 8 additional symptoms, including muscle pain, joint pain, tender lymph nodes, sore throat, headaches of a new type, impairment of memory or concentration, unrefreshing sleep and postexertional malaise lasting longer than 24 hours 17. One of the most common conditions comorbid with CFS is Fibromyalgia (FMS, defined by chronic widespread pain and hyperalgesia at defined tender points), with up to 70% of patients with CFS also having FMS concurrently or in the past 1. All of the symptoms of CFS and FMS are subjective, making diagnosis and treatment difficult. Clearly, objective biomarkers for these syndromes are needed.
As with other diseases of unknown etiology, one strategy to find clues for its causes is to use gene expression microarrays to determine genes that are over or under expressed in CFS patients. A few attempts have been made to determine biomarkers utilizing gene expression with small populations of CFS patients, but results to date have been mixed 3; 15; 30; 61; 78. One explanation for this discordance may be that multiple types of CFS exist, with each type having a different gene expression profile. As many as 7 subtypes of CFS have been proposed based on gene expression 6; 30. Other investigators have used a more directed approach using quantitative mRNA measurements of genes related to immune function, again with little concordance between the genes investigated 3; 46; 65.
We have followed a different strategy in our attempts to use gene expression to determine useful biomarkers for CFS and to determine factors for initiation and maintenance of this syndrome. We focused on genes that might contribute to the primary symptom of CFS, fatigue, and on two of the most common additional symptoms, muscle pain and long-lasting post-exertional worsening of symptoms.
Fatigue has many definitions, from loss of voluntary muscle contraction to the perception of “feeling tired”. However, the symptoms described in CFS, and scored in fatigue inventories more closely approaches the latter phenomenon. This “fatigue” emanates from muscles and from a unique cognitive state in the brain. This sensation of fatigue from muscles (which can occur with or without muscle pain) is caused by metabolites produced during muscle contraction and is enhanced following exhausting exercise in normal subjects. However, moderate exercise causes little long-lasting post-exercise fatigue and usually no muscle pain in normal subjects, while these symptoms are often exacerbated following even mild exercise in CFS patients 76.
To learn more about sensory muscle fatigue and pain, we conducted mouse experiments determining the types of sensory neurons that encode metabolites produced by muscle contraction. We found at least two classes of sensory neurons 37. These two classes of sensory neurons likely represent 1) sensory neurons capable of sending signals interpreted as physical fatigue, and 2) sensory neurons capable of sending signals interpreted as muscle pain. Our analysis using antagonists and agonists suggested at least 4 molecular receptors acted synergistically to detect the metabolites produced by muscle contraction. These include an Acid Sensing Ion Channel (also called Amiloride Sensitive Ion Channel) or ASIC (most likely ASIC3), 2 Purinergic X type receptors (P2X5 and/or P2X4) that are activated by ATP, and Transient Receptor Potential Vanilloid Type 1 (TRPV1) that is activated by heat, acid or endocanabinoids. The difference in coding between the “fatigue” vs. the “nociceptive” sensing pathways appeared to be related to the P2X5 vs. P2X4 receptors with P2X5 conveying the increased sensitivity needed to detect low concentrations of metabolites associated with “fatigue”. The sharing of most (but not all) molecular receptors by “fatigue” and “nociceptive” muscle afferents predicts at least some overlap between fatigue symptoms in CFS and muscle pain symptoms. Other investigators have found that ASIC3 is greatly increased by muscle and joint inflammation, which induced hyperalgesia 22; 40; 41; 75. Thus, we suggest that the primary symptoms in CFS, fatigue and muscle pain, result from enhanced activation of “fatigue” and ”nociceptive” afferents supplying muscle. It follows that increased expression of the molecular receptors encoding metabolites could be a marker of enhanced fatigue and/or muscle pain.
The sympathetic nervous system (SNS) because of its major role in regulating regional blood flow in response to the metabolite buildup in working muscles, and the immune system (IS) because of its ability to alter the sensitivity of both peripheral and central sensory pathways could also contribute to symptoms of CFS and FMS. The Hypothalamic-Pituitary-Adrenal (HPA) axis has also been implicated in the induction and maintenance of CFS. These are the same systems in which dysregulated genes have been found using micro array analysis in CFS patients 3; 6; 30; 61. Specifically, 1) adrenergic receptors may be altered in CFS (see review by Johnson et al. 26), and 2) immune cell cytokines and receptors may be altered in CFS 46; 72, although many discrepancies exist (see review by Natelson et al. 44). 3) Finally, polymorphisms in HPA axis receptor genes, and dysregulated levels of HPA hormones and expression of HPA axis genes have also been implicated in CFS 24; 59; 73.
Because circulating immune cells respond to adrenergic agonists and metabolite increases in skeletal muscle 13; 14;33; 48; 56, and because the IS is implicated in CFS, we looked at alterations in mRNA from metabolite detecting, adrenergic, and immune function genes extracted from leukocytes of CFS patients and compared them with mRNA alterations in control subjects. Because fatigue and muscle pain in CFS and FMS are exacerbated by physical exercise to a much greater extent than in control subjects, we looked at gene expression before and after 25 minutes of mild exercise at times before, during and after we had found increased gene expression in these 3 systems in normal subjects exercising at maximal levels.
Initial experiments with normal subjects indicated that mRNA for Metabolite detecting (ASIC3, P2X4, P2X5, TRPV1), Adrenergic (α-2A, β-1, β-2, COMT), and Immune (IL6, IL10, TNFα, TLR4, CD14) was upregulated at 8 and 24 hours after strenuous exercise. The mRNA increases returned to near normal levels 48 hours after strenuous exercise 38. This time course of mRNA increases mimicked the reports of delayed onset muscle soreness (DOMS) and delayed muscle fatigue in these normal subjects. CFS patient report that even moderate exercise that does not cause DOMS or muscle fatigue in normal subjects causes physical fatigue and pain that lasts 48 hours or longer. Therefore, we tested the hypotheses that 25 minutes of moderate exercise would increase the gene expression of the aforementioned genes measured 8, 24, and 48 hours after exercise in CFS patients, but not in normal subjects. We further collected subjective measurements of fatigue and pain, and also measured some exercise physiological parameters to determine if the alterations in gene expression were related to fatigue and/or pain symptoms in CFS patients and controls.
Methods
Participants
All research reported here was approved by the University of Utah Institutional Review Board, and upon arrival to the laboratory informed consent was obtained from all subjects and patients. (These data were obtained in the same experiments reported in White A.T. et al. 2009 submitted). Due to some data loss during mRNA assays, the sample for the present report included 19 CFS patients (15 females) and 16 Control subjects (11 females). This ratio of 4 times as many females and males is typical in CFS research, and is also consistent with observations from large scale incidence and prevalence studies, including the Wichita sample 60. One female control subject developed Fibromyalgia within a year of being tested and was excluded from these analyses. All CFS patients met the CDC criteria for CFS (Fukuda et al., 1994). Prior screening had excluded all other known causes for persistent or relapsing fatigue in these CFS patients. Exclusion criteria included active upper respiratory infections, use of corticosteroids, SNS agonists, or prescription analgesics known to affect SNS, HPA or cytokine activity, and/or uncontrolled cardiovascular or pulmonary disease. All discontinued any such drugs for 3 days before and 3 days during the exercise protocol (verified at the time of testing). Due to the 4–6 week downward-titration required, use of antidepressants was not discontinued. Five of 15 Control subjects (33%) and 8 of 19 CFS patients (42%) had been previously diagnosed with clinical depression and were tested while continuing their physician prescribed antidepressants. In preliminary analyses, only one measure (baseline TLR4) showed a significant difference with lower levels for those on antidepressants (P < .05). All patients were also screened for FMS using the American College of Rheumatology (ACR) criteria, which includes presence of widespread pain for at least 6 months, and pain reported at 11 or more of 18 sites during tender point examination. Thirteen of the 19 CFS patients (68%) also met ACR criteria for FMS, similar to the high comorbidity of these disorders previously reported 2; 9; 27. For primary analyses, all 19 CFS patients were compared to the 15 Controls. In separate secondary analyses, the 13 patients meeting criteria for both CFS and FMS were compared to Controls. Because only 6 patients met criteria for CFS without FMS, our sample lacked sufficient statistical power to examine effects in this subgroup by itself.
Protocol Overview
All participants refrained from formal exercise other than the required exercise test for a period of 4 days beginning 48 hours preceding the exercise test until after the final (48 hr) blood sample was taken. Venous blood samples (from the arm) were obtained at baseline and at 0.5, 8, 24, and 48 hours post-exercise. To assess the severity of pre-existing and exercise-related fatigue and myalgia symptoms at the time of each blood draw and at the midpoint and immediately after completing the exercise task, the subject provided numerical ratings of mental fatigue, physical fatigue and overall body pain using a 0–100 scale where 100 was defined as the greatest level of fatigue or pain the subject could ever imagine experiencing. Immediately following the baseline blood draw, participants began the exercise session, as described below.
Exercise Protocol
A combined arm-leg cycle ergometer (Schwinn Air-Dyne) was used for the 25-min, whole-body exercise test. In the first five minutes of exercise, subjects were asked to increase pedaling rate until 70% age-predicted maximal heart rate was achieved. Thereafter, work rate was adjusted in order to maintain this target heart rate throughout the submaximal exercise protocol. Ratings of Perceived Exertion (RPE) were obtained on a scale of 1–10 every 5 minutes; heart rate was recorded each minute, and blood pressure was measured at baseline, every 10 minutes during exercise, and upon completion of the exercise. We elected to use a sustained moderate exercise rather than a maximal exercise test (which typically last only 5–9 min in CFS patients) because of closer similarity to the natural exercise experiences reported to exacerbate CFS symptoms in patients’ daily lives. Our 25 min submaximal exercise task did elicit consistent worsening of fatigue and pain symptoms from 8–48 hours post-exercise (see Results). In contrast, after a briefer maximal exercise task, reports of worsening CFS symptoms were inconsistent or absent until 5 days after the challenge 81, a pattern not typically observed in real life. Maximal exercise protocols have demonstrated few differences in cardiorespiratory and perceptual responses (RPE for example) between CFS patients and fitness matched controls 10. However, it is notable that responses to submaximal exercise including VO2 do predict peak exercise performance in CFS patients 52.
mRNA extraction and analysis
All blood processing and analyses were performed by personnel blinded to the subject’s group. At each of the 5 blood sampling times, blood was collected in EDTA tubes. Seven minutes after blood collection, the blood was centrifuged at 3200 rpm (1315 × g- Clay Adams Compact II Centrifuge) for 12 minutes, plasma removed, and the white layer carefully collected in RNase-Out (Invitrogen, Carlsbad, CA) and RNA-later (Qiagen, Valencia, CA), then quickly frozen using a methanol-dry ice slurry, and stored at −80°. RNA was extracted using RNeasy kits (Qiagen, Valencia, CA), according to manufacture’s directions, and treated with RNase-free DNase-I (Qiagen, Valencia, CA). Immediately following extraction, RNA was converted to a cDNA library using the ABI High Capacity cDNA Archive Kit (Applied Biosystems, Inc., Foster City, CA).
The cDNA samples were stored at −80°C until analysis by quantitative real-time PCR. The cDNA libraries were analyzed using the ABI quantitative, real-time PCR system on the ABI Prism 7900 Sequence Detection System (Applied Biosystems, Inc., Foster City, CA), using ABI TaqMan Master Mix (Applied Biosystems, Inc., Foster City, CA). Master Mix/primer probe solutions and template solutions were separately loaded onto 96 well preplates, with robot loading mixing these solutions when placed in the 384 well plates. Plates were centrifuged to remove any air bubbles in the wells. Each sample was run in duplicate with standards being run in quadruplicates, and with one “no template” control. Each 384 well plate contained samples from two subjects/patients, and all genes were analyzed on the same plate. Primer probes (all from TaqMan Gene Expression Assays, Applied Biosystems, Inc., Foster City, CA) were:
ASIC3 - Hs00245097_m1; P2X4 - Hs00175706_m1; P2X5 - Hs00175712_m1; TRPV1 - Hs00218912_m1; Adrenergic A2A - Hs00265081_s1; Adrenergic B-1 - Hs02330048_s1; Adrenergic B-2 - Hs00240532_s1; COMT - Hs00241349_m1; IL6 - Hs00174131_m1; IL10 - Hs00174086_m1; TNFα- Hs00236874_m1; TLR4 - Hs00152937_m1; CD14 - Hs00169122_g1. Control primer probes included TF2B - Hs00155321_m1; β-Actin - Hs99999903_m1; PSMB6 - Hs00382586_m1; 18S - 4333760T; and GAPDH - Hs99999905_m1.). All primer probes, except for the adrenergic receptors and CD14 (these genes do not have introns), recognize sequences that cross splice sites, and therefore, make detection of genomic DNA unlikely. In all cases we quenched the genomic DNA and ran no template control wells to ensure that contamination did not influence the final results. All of these primer probes were designed and tested to be used together, and have similar efficiencies to help eliminate inaccuracy. For the genes that have rarely been described in leukocytes (Adrenergic A2A, Adrenergic B-1, P2X5, TRPV1, ASIC3) we designed primers that contained 360 to 600 base-pairs, which included the mRNA regions ABI indicated the Primer probes listed above spanned. PCR product was generated from our cDNA libraries and sequenced. All of these sequences were >99–100% identical to the predicted mRNA sequences from transcribed these genes. Evaluation of Controls in this and previous experiments indicated that TF2B had less intrinsic variation, had a count range that was similar to the genes of interest, and did not increase or decrease due to the exercise protocol. Real-time PCR results were analyzed with SDS 2.1 (Applied Biosystems, Inc., Foster City, CA), inspected to determine artifacts, (loading errors, robot errors, thresholding errors, etc.). Count numbers were exported to an Excel spreadsheet, and analyzed according to the ddCT method described in ABI User Bulletin #2 (Applied Biosystems, Inc., Foster City, CA). Baseline levels for each gene were computed relative to TF2B, and these baselines were used as the comparator for all measures taken after the exercise period (see statistical methods below for further analysis details).
Complete Blood Cell Counts (CBC)
CBCs with differential subtyping were obtained by standard laboratory assays (ARUP, Salt Lake City, UT) from separate blood samples collected in EDTA tubes. Measures obtained included total red blood cells, hematocrit and hemoglobin, platelets, and total white blood cells plus subtypes including monocytes, eosinophils, lymphocytes and granulocytes. Group differences in these CBC measures have been reported separately (White A.T., 2009, submitted). These group differences were small and should not affect the mRNA results reported here (see Discussion).
Statistical Analysis
Because the ddCT method used for mRNA analysis necessarily creates a non-normal, rightward skewed distribution, data were log transformed to yield distributions that could be appropriately analyzed with parametric statistics. Baseline differences between Controls and CFS patients (and secondarily between CFS-FMS patients and Controls) were determined with one-way ANOVA. Similar approaches were used to compare cardiovascular, work rate and RPE measures obtained during the exercise task.
Post-exercise values for each gene expression measure were first normalized relative to the same subject’s baseline levels (1.00 = baseline). To minimize the increase in the likelihood of obtaining false positive results when examining multiple outcomes, we took two conservative steps. First, instead of examining group differences separately at each of the 4 post-exercise time points, we combined them into a single measure labeled Area Under the Curve (AUC). The post-exercise AUC for each gene expression measure was computed by summing the 0.5, 8, 24, and 48 hour values, after which these AUC measures were log transformed. Second, before examining univariate ANOVA results, we first grouped our AUC measures into 3 categories (metabolite detecting markers, adrenergic markers and immune markers), and then employed multivariate analyses of variance (MANOVA) to examine whether there were reliable group differences in each of these 3 outcome categories. These three MANOVAs each yielded significant overall effects of Group in the direction of greater post-exercise mRNA increases among CFS patients vs. Controls (F 4,23 = 2.21, P < .05, F 4,23 = 3.29, P < .015 and F5,23 = 3.29, P < .005 respectively one-tailed), which allowed us then to examine univariate effects of Group on each specific AUC measure using one-way ANOVAs.
To examine whether Group differences were related to differences in age, gender, or body mass index (BMI), the MANOVAs and univariate ANOVAs were repeated with each of these included in the model as covariates. In no instance was age or gender a significant covariate, while BMI was significant for two measures: β-1 and β-2 AUC; thus, results for these measures are reported both before and after adjusting for BMI. Based on a priori hypotheses that CFS patients would show greater post-exercise AUC increases than Controls in gene expression for metabolite detecting measures ASIC3, P2X4 and P2X5, these three group comparisons employed one-tailed tests, while all other analyses employed two-tailed tests. In addition to group comparisons, Pearson r correlations were used to examine relationships between exercise variables, pain and fatigue ratings, and gene expression measures. All data are presented as means and standard errors, with significance set at P < 0.05.
Results
Primary Analyses
Table 1 gives age and BMI as well as the cardiovascular, work performance and subjective exertion data for all CFS patients vs. Controls. Groups did not differ in age, blood pressure, or heart rate levels at baseline or during the exercise task. Both groups maintained the task objective of 70% of maximum age-predicted HR during exercise. The CFS patients did have significantly higher BMI than Controls, and they reported significantly higher mean ratings of perceived exertion (RPE) even though they exercised at significantly lower work rates than Controls.
Table 1.
Subject Characteristics for Controls and CFS Patients
| Controls (n=15; 11F) | CFS Patients (n=19; 15F) | df | F | P | |
|---|---|---|---|---|---|
| Age (years) | 35.6 ± 3.0 | 42.2 ± 2.7 | 1,32 | 2.69 | NS |
| Body Mass Index (kg/m2) | 22.9 ± 0.73 | 27.0 ±1.18* | 1,32 | 7.69 | .009 |
| Resting SBP (mmHg) | 126 ± 3.1 | 122 ± 3.4 | 1,32 | 0.59 | NS |
| Resting DBP (mmHg) | 77 ± 2.5 | 79 ± 2.2 | 1,32 | 0.13 | NS |
| Exercise SBP (mmHg) | 159 ± 8.9 | 146 ± 5.5 | 1,32 | 0.60 | NS |
| Exercise DBP (mmHg) | 89 ± 3.4 | 88 ± 2.1 | 1,32 | 0.19 | NS |
| Exercise HR (BPM) | 130 ±1.6 | 123 ± 3.0 | 1,32 | 3.44 | NS |
| Exercise HR (%PMHR) | 70.6 ± 0.78 | 69.3 ± 1.18 | 1,32 | 0.82 | NS |
| Exercise WR (Kcal/kg/min) | 7.5 ± 0.42 | 4.3 ± 0.36* | 1,32 | 34.85 | .0001 |
| Exercise RPE | 3.0 ± 0.21 | 4.8 ± 0.34* | 1,32 | 17.60 | .0001 |
BMI = body mass index; SBP = systolic blood pressure; DBP = diastolic blood pressure; %PMHR = percent of age-predicted maximal heart rate; WR = work rate; RPE = rating of perceived exertion; NS = non-significant;
and bold indicates significant difference between CFS and Controls, P<.05.
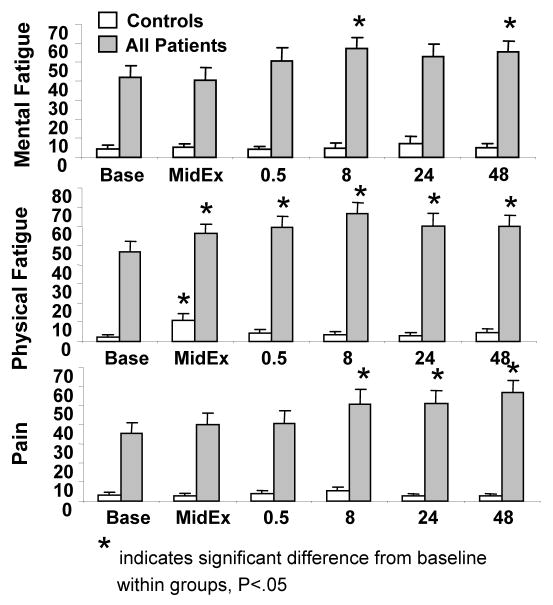
Figure 1 depicts fatigue and pain ratings before, during and for 48 hours after the exercise task. CFS patients had significantly higher ratings than Controls for all three symptoms even at baseline, and these differences continued at all other times of measurement (Group Effect F 1,32 = 30.88, 52.33, and 25.60, P < .0001 for mental fatigue, physical fatigue and pain, respectively). There were also increases from baseline levels in these 3 symptoms during and after exercise (Time Effects F 4,128 = 2.62, 2.66 and 3.07, P < .04). At mid-exercise, both Controls and CFS patients showed increases over baseline levels in physical fatigue, but only CFS patients continued to show increased physical fatigue at 0.5, 8, 24 and 48 hours post-exercise. Also, only CFS patients showed significantly increased levels of pain at 8, 24 and 48 hours and increased mental fatigue at 8 and 48 hours after exercise.
Figure 1.
Ratings of Mental Fatigue, Physical Fatigue, and Pain at the times indicated. Ratings were based on a 0-100 scale from none to the worst the patient could even imagine experiencing. Base = before exercise; MidEx = at midpoint of 25 minute exercise (min 13); 0.5 = one half hour after end of exercise; 8 = 8 hours after exercise; 24 = 24 hours after exercise; 48 = 48 hours after exercise.
Gene expression measures
Baseline levels of gene expression markers are provided in Table 2. β-2 adrenergic receptor expression was significantly lower in CFS patients vs. Controls, while α-2A adrenergic receptor expression was marginally higher (P < .09). None of the other gene expression measures differed at baseline between Groups.
Table 2.
Baseline means + SEs and ANOVA results for all mRNAs relative to β-Actin, and ANOVA results for post-exercise Area Under Curve (AUC) mRNA changes from baseline in Controls and CFS Patients.
| Metabolite Detecting | ||||||||
|---|---|---|---|---|---|---|---|---|
| Controls | Patients | df | F | P | AUC df | AUC F | AUC P | |
| Baseline | Baseline | Baseline | Baseline | Baseline | ||||
| ASIC3 | 8.72E-03 ± 1.05E-03 | 1.01E-02 ± 1.19E-03 | 1,26 | 0.73 | NS | 1,29 | 5.55 | 0.013 |
| P2X4 | 1.29E-01 ± 1.86E-02 | 1.45E-01 ± 2.64E-02 | 1,27 | 0.21 | NS | 1,32 | 8.37 | 0.004 |
| P2X5 | 2.37E-01 ± 3.68E-02 | 2.47E-01 ± 4.69E-02 | 1,26 | 0.03 | NS | 1,30 | 3.78 | 0.031 |
| TRPV1 | 9.82E-03 ± 1.33E-03 | 1.13E-02 ±1.37E-03 | 1,23 | 0.57 | NS | 1,26 | 1.50 | NS |
|
| ||||||||
| Adrenergic | ||||||||
|
| ||||||||
| Controls | Patients | |||||||
|
| ||||||||
| α2-a | 7.37E-03 ± 1.59E-03 | 1.53E-02τ ± 3.56E-03 | 1,25 | 3.11 | .09 | 1,28 | 6.68 | 0.015 |
| β-1 | 4.73E-03 ± 4.73E-03 | 1.61E-01 ± 1.49E-01 | 1,27 | 0.89 | NS | 1,30 | 10.06 | 0.003 |
| β-2 | 1.47E+00 ± 2.75E-01 | 9.06E-01* ± 1.09-01 | 1,27 | 4.26 | 0.049 | 1,30 | 12.80 | 0.001 |
| COMT | 2.07E-01 ± 2.80E-02 | 2.28E-01 ± 3.96E-02 | 1,25 | 0.17 | NS | 1,29 | 5.61 | 0.025 |
|
| ||||||||
| Immune | ||||||||
|
| ||||||||
| Controls | Patients | |||||||
|
| ||||||||
| IL6 | 6.83E-03 ± 4.33E-03 | 3.71E-03 ± 8.77E-04 | 1,27 | 0.60 | NS | 1,28 | 1.37 | 0.119 |
| IL10 | 3.93E-03 ± 6.60E-04 | 4.81E-02 ± 4.47E-02 | 1,25 | 0.90 | NS | 1,28 | 13.30 | 0.001 |
| TNFα | 4.70E-02 ± 7.84E-03 | 7.35E-02 ± 2.24E-02 | 1,27 | 1.05 | NS | 1,30 | 1.21 | NS |
| TLR4 | 7.38E-01 ± 1.93E-01 | 5.82E-01 ± 8.35E-02 | 1,27 | 0.63 | NS | 1,30 | 4.33 | 0.046 |
| CD14 | 1.81E+00 ± 2.32E-01 | 2.00E+00 ± 3.89E-01 | 1,25 | 0.17 | NS | 1,28 | 2.95 | NS |
indicates significant difference from Controls, P<.05,
indicates borderline difference from Controls, P=.09
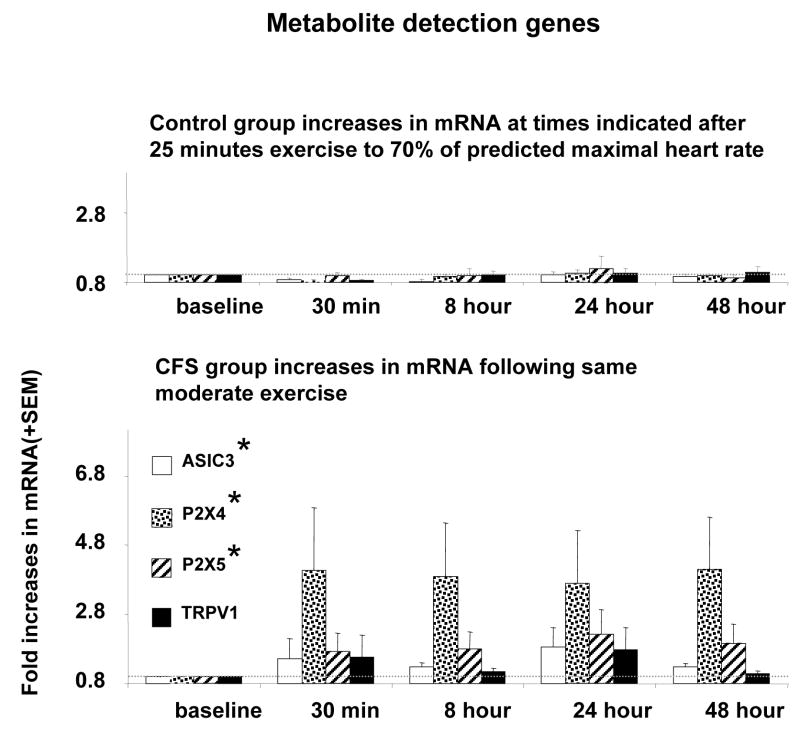
As previously described for post-exercise AUC measures, initial MANOVAs yielded significant effects of Group for metabolite detecting genes, adrenergic genes, and immune genes. Subsequent univariate ANOVAs for metabolite sensing markers (Table 2) yielded significant Group differences in post-exercise AUC between CFS patients and Controls for ASIC3, P2X4 and P2X5, but not TRPV1. Figure 2 depicts these data as fold increases relative to baseline levels (where 1.0 fold = baseline). CFS patients showed consistently greater mean increases in ASIC3, P2X4 and P2X5 gene expression than Controls across all post-exercise time points. TRPV1 increases also demonstrated a consistent although non-significant trend to be higher in the CFS group.
Figure 2.
Amount of mRNA for ASIC3, P2X4, P2X5, and TRPV1 expressed as fold increases relative to baseline levels at each of the times indicated before (baseline) and after the end of 25 minutes of moderate exercise. * Indicates P< 0.05 compared to control subjects for the area under curve (AUC) of mRNA across all time points after exercise. Faint dotted line indicates the baseline levels.
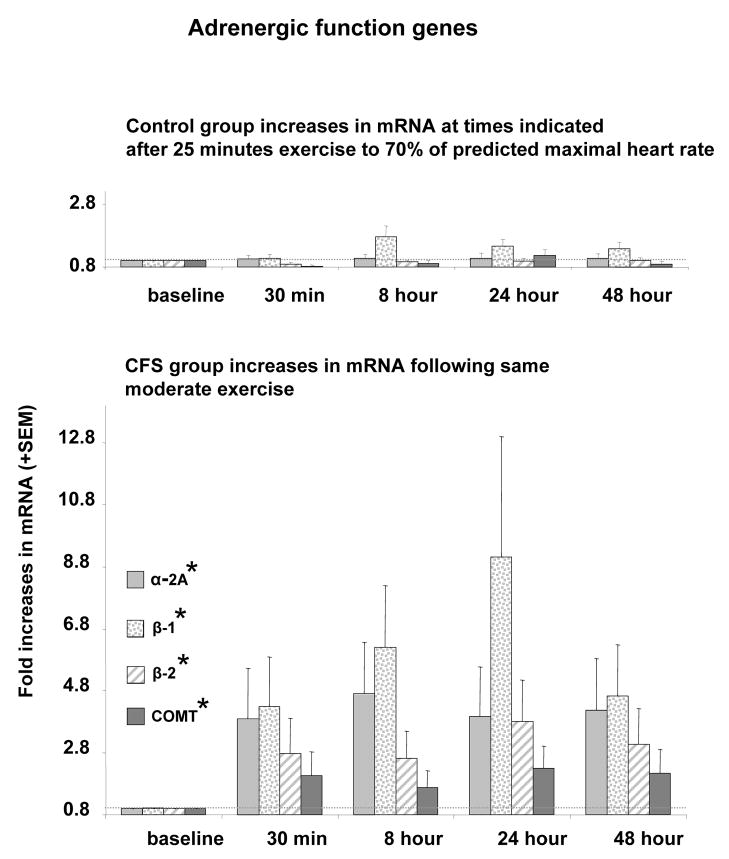
Significant effects of Group were also obtained in the univariate ANOVAs for all of the post-exercise adrenergic AUC markers (Table 2), due to consistently greater mean increases in α-2a, β-1 and β-2 adrenergic receptor expression, and greater COMT gene expression in the CFS patients vs. Controls, as shown in Figure 3.
Figure 3.
Amount of mRNA for α-2A, β-1, β-2 adrenergic receptors and catechol-o-methyl-transferase (COMT) relative to baseline levels. * Indicates P< 0.05 compared to control subjects for the area under curve (AUC) of mRNA across all time points after exercise. Faint dotted line indicates the baseline levels.
For β-1 and β-2 where BMI was observed to be a significant covariate, ANOVAs were repeated with BMI included in the model. Group differences in post-exercise AUC β-2 receptor expression were still significant. Surprisingly, for the post-exercise AUC increase in β-1 receptor expression, where mean group differences had been greater than any other gene expression marker, inclusion of BMI in the model caused the effect of Group to become non-significant (P = 0.19).
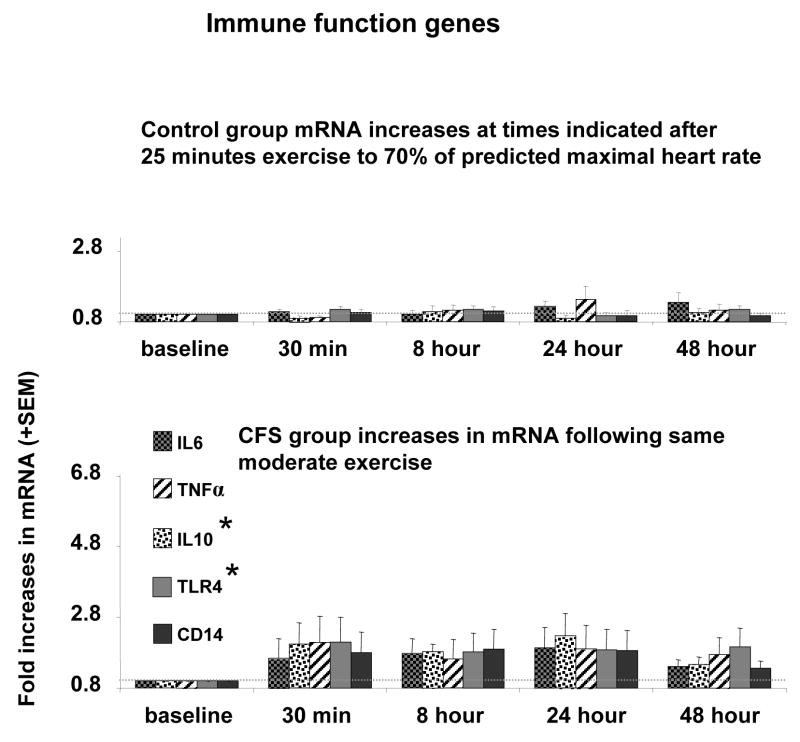
For post-exercise immune markers, univariate ANOVAs (Table 2) showed significant group differences in TLR4 and IL-10 AUC expression, but not in CD14, IL-6, or TNF-α AUC expression. As depicted in Figure 4, CFS patients showed greater post-exercise increases in TLR4 and IL-10 than Controls.
Figure 4.
Amount of mRNA for IL6, TNFα, IL10, TLR4, CD14 relative to baseline levels. * Indicates P< 0.05 compared to control subjects for the area under curve (AUC) of mRNA across all time points after exercise. Faint dotted line indicates the baseline levels.
Correlations between post-exercise fatigue and pain and the post-exercise AUC gene expression measures are depicted in Table 3, along with the intercorrelations among the gene expression measures. As indicated by these relationships, post-exercise increases in metabolite detecting receptors, adrenergic receptors, and certain immune markers occurred in parallel, and greater increases in all of these receptors were associated with greater fatigue. Post-exercise pain symptoms were not associated with increases in P2X4, P2X5, β-1 or TLR4 receptor expression, but were linked to ASIC3, TRPV1, α2-a, β-2 and IL-10 increases. Interestingly, when examined among the CFS patients by themselves, exercise work rate was not correlated with post-exercise fatigue, pain, or with an increase in any metabolite detecting or immune marker, and it was inversely related to increases in β-1 and β-2 AUC receptor expression (r = −.71 and −.60, P < .05).
Table 3.
Correlations of Post-exercise AUC Gene Expression Measures with Post-exercise AUC Fatigue and Pain and with Each Other
| ASIC3 | P2X4 | P2X5 | TRPV1 | α-2a | β-1 | β-2 | COMT | IL10 | TLR4 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Mental fatigue | +.53 | +.62 | +.38 | +.43 | +.63 | +.41 | +.58 | +.51 | +.64 | +.48 |
| Physical fatigue | +.53 | +.58 | +.37 | +.41 | +.60 | +.43 | +.58 | +.48 | +.63 | +.45 |
| Pain | +.54 | NS | NS | +.38 | +.43 | NS | +.50 | NS | +.47 | NS |
| ASIC3 | +.60 | +.76 | +.83 | +.51 | NS | +.62 | +.77 | +.53 | +.55 | |
| P2X4 | +.58 | +.63 | +.70 | +.51 | +.65 | +.72 | +.80 | +.64 | ||
| P2X5 | +.72 | +.38 | NS | +.51 | +.78 | NS | +.39 | |||
| TRPV1 | +.39 | NS | +.49 | +.79 | +.59 | +.61 | ||||
| α-2a | +.77 | +.86 | +.35 | +.63 | +.48 | |||||
| β-1 | +.89 | NS | +.63 | +.50 | ||||||
| β-2 | +.42 | +.70 | +.59 | |||||||
| COMT | +.43 | +.61 | ||||||||
| IL10 | +.67 |
NS = nonsignificant. For all listed correlations P < .05. For those correlations > +.44, P < .01.
Secondary Analyses
Although heart rate level and percent of age predicted maximum heart rate were quite closely matched between Groups during exercise, Controls did exercise at a higher work rate than CFS patients. However, when we repeated our univariate ANOVAs for post-exercise AUC gene expression comparing Groups with exercise work rate as a covariate in the model, we found that it was not a significant covariate for any measure except β-1 receptor expression (P < .02). As with inclusion of BMI as a covariate, covarying for work rate did cause the Group effect for post-exercise β-1 AUC receptor expression to become non-significant. This pattern indicates that high BMI, low exercise work rate, and post-exercise increases in β-1 receptor expression are all interlinked in CFS.
Univariate ANOVAs were also repeated after dropping the 6 CFS patients who did not also meet ACR criteria for FMS. The findings were essentially unchanged from those reported above except for the immune receptor, TLR4; differences between CFS-FMS patients and Controls were then only marginally significant (P =.07) for this gene. For all other measures where significant Group differences were obtained in the primary analyses, the same measures showed significant differences between the CFS-FMS patients and Controls in the secondary analyses. Although the group size (n=6) of patients with CFS-only was insufficient to examine separately in formal analyses, mean post-exercise AUC measures for this subgroup were similar in magnitude and variance to those of the CFS-FMS subgroup in most cases, with lesser ASIC3 and TRPV1 increases being exceptions.
To address the issue of fitness differences, ANOVAs comparing post-exercise AUC measures were also repeated after dropping the most fit Controls and least fit CFS patients. The remaining 11 Controls (9 females) and 10 Patients (8 females) were closely matched for actual work-rates required to achieve 70% of maximum predicted HR levels. These fitness-matched subgroups did not differ in age, BMI, work-rate during exercise, or any BP or HR measure before or during exercise. Nevertheless, the fitness-matched CFS patients still showed greater or marginally greater increases than Controls in post-exercise ASIC3 (P < .036), P2X4 (P < .07), P2X5 (P < .028), β-1 (P < .045) β-2 (P < .002) COMT (P < .085), and IL-10 (P < .006), and only the TLR4 differences were abolished. Despite exercising at the same workrate, the patients also still reported greater perceived exertion during the task (RPE = 4.65 vs. 2.95, P < .001).
To address whether sampling time was an important factor, additional repeated measures ANOVAs were performed using Group and Time (post-exercise 0.5, 8, 24, and 48 hours) as factors and also examining Group x Time interactions. The same main effects of Group reported above were replicated with this approach, and no effects involving Times were significant, suggesting that CFS patients differed from Controls similarly at all sampling times.
Discussion
Main findings
Baseline mRNA levels were not different between Controls and CFS patients for any of the metabolite detecting or immune genes. Among the adrenergic measures, baseline β-2 was lower and baseline α-2A tended to be higher in the CFS patients. Although these baseline differences were modest in magnitude, decreased β-2 and increased α-2A receptors in the vasculature would lead to enhanced total vascular resistance as was previously reported by Light et al 39 for patients with FMS. No prior study has examined metabolite detecting gene expression, and prior studies comparing immune function gene expression in CFS patients vs. controls at rest have likewise reported few consistent increases in any of these mRNAs with the exception of TNFα. 58.
In contrast to the very few group differences at baseline, CFS patients showed greater mRNA increases than healthy Controls in the majority of the metabolite sensing, adrenergic, and immune function genes examined here following 25 minutes of moderate, whole-body exercise (when summed across 30 minutes, 8, 24 and 48 hours following exercise). With this moderate exercise task, healthy Controls exhibited no significant increases from baseline in expression of any of the genes, while CFS patients demonstrated increases in expression of ASIC3, P2X4, P2X5, α-2A, β-1, β-2, COMT, IL10, and TLR4 that reliably exceeded responses of Controls. TRPV1 expression increased significantly above baseline levels only in CFS patients, although the CFS-Control group differences remained a non-significant trend for this measure.
Groups also did not differ in increased expression of IL-6 or TNFα. While gene expression of IL6 and TNFα was increased above baseline after exercise in the CFS group, controls showed similar post-exercise increases. These CFS patients and controls showed similar post-exercise increases in circulating serum levels of IL6 and TNFα (White A.T. et al. 2009, submitted) that has been shown in other reports as well 49. Other studies have indicated that while serum levels of IL6 and TNFα were increased after exercise, mRNA expression in leukocytes was not increased for either of these two cytokines 43; 46; 51. Cytokine mRNA taken from exercised muscle (as opposed to leukocytes), however, did show increased TNFα mRNA 50. Our study (White A.T. et al. 2009, submitted) also observed that serum IL10 levels actually decreased after exercise in Controls (who showed no reliable changes in IL10 mRNA) but did not decrease in CFS patients (whose IL10 mRNA increased).
Surprisingly, we observed a trend of mRNA increases in patients as early as 30 minutes following exercise, several hours sooner than we had observed increases in control subjects after higher intensity exercise 38. Increases in mRNA, with the exception of β-1, were not influenced by BMI or work rate, and were present in both CFS patients with, and without co-morbid CFS. Underscoring the relationship between the primary symptoms defining CFS, strong correlations were found between both summed post exercise physical and mental fatigue and the summed increases in mRNA of the genes listed above (see Table 3). Strong correlations were similarly found between summed post exercise pain and ASIC3, TRPV1, α-2A, β-2, and IL10 increases. ASIC3, TRPV1 and β-2 have all recently been shown to play a role in muscle pain 37; 63.
The fact that 68% of patients in our sample met criteria for both CFS and FMS is somewhat problematic because we cannot yet discern whether the observed responses are typical of all CFS patients or only of the subgroup that has both disorders. The present data indicated that the group of patients that did not meet criteria for FMS (CFS-only group) had AUC means and variances similar to the CFS-FMS group for most of the 13 genes examined here. Notable exceptions were ASIC3 and TRPV1 which tended to show less increases in the CFS-only group. Thus, while most of the gene alterations apply to both groups, there may be gene alterations that discriminate these two groups. Our current research is intended to include an expanded sub-sample of patients with CFS alone in order to clarify this important issue.
It is worth noting that complete blood counts (CBC) were taken from all patients and Controls at each time point. At pre-exercise baseline (as detailed in White et al 2009, submitted) total white blood cell counts from CFS patients were slightly higher than Controls, and specifically, monocyte, eosinophil, and granulocyte counts were higher in the patient group. Counts in both patients and Controls were significantly increased from baseline at 8 hours after exercise but returned to baseline levels at 24 and 48 hours. However, hemoglobin levels were lower in patients than controls, and decreased at 8 hours. These directionally opposite effects in white vs. red cell counts indicate that hydration differences were not responsible. These CBC differences were not clinically significant, and similar differences have been reported by others (see review 44).
mRNAs as biomarkers for fatigue, muscle pain, CFS and FMS
The marked alterations in gene expression from circulating leukocytes of CFS patients after exercise suggest that such alterations could be used as objective markers for CFS. Kerr recently reviewed potential biomarkers for CFS including several microarray experiments looking at gene expression in large numbers of genes from leukocytes 29. Sakai et al 61 also recently determined that mRNA alterations in 9 leukocyte genes (culled from 1400 genes related to stress) were potential biomarkers for CFS. None of the genes identified as specific for one or more subtype of CFS in these previous publications were those examined in the present report. However, the functional groups of genes found in the microarray experiments were similar to those measured here. Immunity and defense, energy metabolism, signal transduction, and ion-channel genes have all been identified in these previous experiments. All of the genes included in the present report can be included in one or more of these groups.
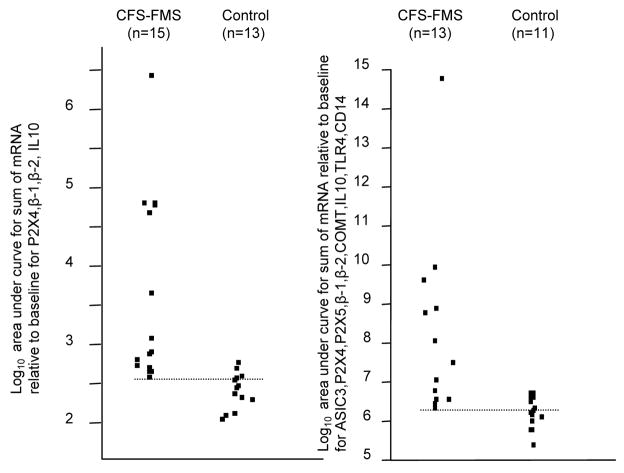
In the present sample, ~90% of the CFS patients could be distinguished from Controls using 4 of the genes measured (P2X4, β-1, β-2, IL10; see Figure 5). If all of the 9 genes that increased in CFS-FMS patients relative to Controls were used, the results are similar (Figure 5). However, we cannot yet conclude that gene expression changes in these measures after mild or moderate exercise are specific biomarkers for CFS.
Figure 5.
Point plots of AUC for sum of 4 genes (P2X4, β-1, β-2, and IL10) (left) and 9 genes (ASIC3, P2X4, β-1, β-2, COMT, IL10, TLR4, and CD14) (right). All data converted to Log base 10 so the points could be plotted together. Dotted lines indicate the lowest values for CFS-FMS patients, indicating little overlap with Control Subjects.
Potentially, some of the genes used here may discriminate other causes of exaggerated post-exercise fatigue, and/or other genes could be used to discriminate known causes of fatigue such as viral infections, abnormalities in metabolism, etc. A simple blood-based test that provides an objective measure for excessive and debilitating fatigue would be diagnostically valuable to physicians and patients with CFS (or with chronic fatigue from other causes). In addition, tests of this type could help distinguish subtypes of CFS, and the different contributing dysregulation involved in each of these subtypes. Finally, these tests could be used to objectively evaluate the success of different treatments for Chronic Fatigue and Fibromyalgia Syndromes.
Function of genes whose expression was altered in CFS patients compared to Controls
We measured the mRNA of several receptors that have rarely been described on white blood cells, particularly those that are essential for detecting exercise produced metabolites by sensory neurons 37. ASIC3, P2X4, P2X5, TRPV1 (both mRNA and protein) have been found on monocytes 12; 23;12; 35; 62; 64; 77. The function of ASIC3, P2X4 and TRPV1 receptors on leukocytes is not known, but could be related to the recruitment of monocytes and lymphocytes that occurs following exercise 33; 34. P2X5 protein in most humans lacks an essential portion of the pore forming part of the channel, rendering it incapable of gating ions, and possibly of being inserted in the plasma membrane 53. Whether the truncated form of P2X5 found in most humans can modify responses of ASIC receptors is unknown. In humans The P2X5 gene codes for a minor histocompatibility protein, and its expression has been shown to be related to some tumor treatments 54. Perhaps, in humans, the ability to detect small increases in metabolites is a function of other P2X heteromers rather than P2X5 function.
β-adrenergic receptors are normally associated with cardiovascular function. Activation of β-1 receptors is known to enhance heart rate and contractility, and activation of β2 receptors allows dilation of arteries and arterioles that feed skeletal muscles. These receptors play a major role in maintaining sufficient blood flow to skeletal muscles during exercise preventing excessive accumulation of metabolites. β2 adrenergic receptors may also mediate SNS effects on the IS 13; 14. Less certain are alpha adrenergic receptor effects on circulating immune cells. Our results show that leukocytes have substantial levels of adrenergic α2-A, β-1, and β-2 receptor mRNAs, as well as high levels of COMT mRNA - a major enzyme involved in inactivating epinephrine and norepinephrine. Polymorphisms in COMT have been implicated in depression and a number of pain disorders 16. Exercise increased the mRNA of all these adrenergic function genes in CFS patients much more than in Controls. This greatly enhanced upregulation suggests powerful upstream signaling to the IS in CFS.
IL10 mRNA was upregulated after exercise in CFS patients compared with Controls. IL10 is an anti-inflammatory cytokine, inhibiting the production of pro-inflammatory cytokines such as TNFα. The increase in IL10 mRNA observed here is consistent with a recent report suggesting that FMS patients express an anti-inflammatory profile that could be related to some of their symptoms 72. In our patients, serum levels of anti-inflammatory IL10 and IL13 increased at 8 hours post-exercise only in those CFS patients reporting greater and more prolonged increases in fatigue and pain. However, these patients also showed increases in pro-inflammatory IL1β, IL8 and IL12 as well as IL6, suggesting overall immune activation. (for details, see White A.T. et al., 2009, submitted). The mRNA for TLR4 was also increased by exercise in CFS patients but not in Controls, although in the subgroups matched for fitness this difference was absent suggesting that it was due to lesser fitness in the CFS group. TLR4 is an immune function receptor that transduces bacterial invasion by detecting the lipopolysaccaride coat on bacteria. It is of obvious importance in preventing infection.
Implications of dysregulated mRNAs for CFS
Increases in mRNA can be caused either by increases in transcription, and/or increased stability of mRNA (decreases in degradation) 42. Both transcription and stability can be altered by environmental factors. For most of the genes investigated here, transcriptional alterations via known transcription factors have been documented. Regulation of these genes by RNA modulatory factors is also likely. The correlations between alterations in many of the genes assayed suggest that common upstream transcription factors may be activated in CFS patients. These findings also support previous studies suggesting interactive alterations between SNS, IS and sensory systems in CFS.
Evidence supporting metabolite detecting and adrenergic involvement in enhanced fatigue in CFS
The rapid and sustained increases in mRNA from sensory genes (ASIC3, P2X4, P2X5) and adrenergic β-1, and β-2 receptors as well as strong correlations between these receptors in CFS patients following moderate exercise suggest a possible mechanism for the hallmark symptom of CFS, sensory fatigue, and its enhancement following exercise. We and others recently discovered that ASIC3 and P2X5, and possibly P2X4 and TRPV1 on muscle sensory neurons synergize to detect the metabolites produced by muscle contraction that may lead to the signaling of muscle fatigue and pain 37; 47; 67. If the number of these receptors were greatly increased in these sensory neurons, resting levels of metabolites could activate sensory fatigue afferents, sending a continuous signal of muscle sensory fatigue to the central SNS causing dysregulation of SNS reflexes, and to the central nervous system, producing the cognitive recognition of enhanced fatigue.
Other recent work in our laboratory suggests that βadrenergic receptors on sensory neurons may play a role in both muscle sensory fatigue and muscle pain. First, muscle afferent neurons from mice that have inflamed muscles respond to much lower concentrations of metabolites if β agonists are co-applied with the metabolites (Light, A.R., unpublished observations. Secondly, adrenergic β-1, and β-2 receptor mRNAs were upregulated in male mice dorsal root ganglia 24 hours to 8 days following carrageenan inflammation of a hindlimb skeletal muscle 36. Thirdly, clinical pain was quickly reduced in patients with FMS or temporomandibular disorder (TMD) when the non-specific β antagonist propranolol was administered in low doses 39. Finally, Khasar et al 32 suggest a mechanism involving the SNS and hypothalamic-pituitary-adrenal axis (HPA) by which stress could contribute to hyperalgesia mediated by adrenergic and hormone receptors on sensory neurons. All of these data suggest that βadrenergic receptors on muscle sensory afferents can enhance metabolite signals, particularly in patients with FMS 39. Thus, if the upregulation of mRNA for sensory and adrenergic receptors observed in leukocytes of CFS patients also occurs in their muscle sensory afferents (sensory fatigue afferents), CFS patients may have an enhanced sensory signal for fatigue that is further increased following exercise. The similarity in transcriptional control of all these molecular receptors may also explain why a large percentage of CFS patients also have FMS. However, because the sensory afferent neurons detecting muscle sensory fatigue vs. muscle pain must use somewhat different molecular receptor combinations that may be differentially regulated, patients may experience both muscle sensory fatigue and muscle pain, or each symptom independently.
Another possibility is that leukocytes use the same sensory receptors to detect metabolites produced by muscle contraction. If these receptors are increased in CFS patients, it is possible that low levels of metabolites can activate leukocytes, and moderate exercise could increase the signal, thereby increasing cytokine levels that sensitize muscle sensory fatigue signaling afferents. Considerable evidence for cytokine sensitization of sensory afferents exists 4; 8; 25; 31; 66. This and the previous mechanism could act together.
Supporting these hypotheses, enhancement of peripheral sensory signals has been demonstrated in patients with FMS and CFS 69; 74. Since the muscle sensory fatigue signal is likely to activate SNS reflexes that normally maintain adequate blood flow to the brain and skeletal muscles, a tonic signal from fatigue afferents may lead to SNS dysregulation because vascular smooth muscle adrenergic receptors desensitize to the constant release of catecholamines (see review by 28). This dysregulation could lead to muscle ischemia and bouts of increased metabolites that would further activate sensory receptors. It could also lead to orthostatic intolerance often associated with CFS 20; 45; 80. Interestingly, ASIC3, P2X and TRPV1 receptors have been implicated in enhanced signals from the gut in animal models of irritable bowel syndrome 5; 70; 71, and TRPV1 is associated with multiple chemical sensitivity 55. ASICs have also been associated with alterations in hearing21; 57, and hyperacusis has also been associated with CFS and FMS 18. Perhaps common transcriptional regulation of these receptors can occur in a variety of tissues, resulting in some of the co-morbidities commonly associated with CFS.
Finally, long-term sensory receptor activation can lead to sensitization of spinal cord and brain systems that transmit fatigue signals, causing long-term fatigue enhancement within the CNS. Such alterations in brain transmission has been demonstrated in Chronic Pain 7; 68 and in CFS and FMS 11; 19; 62; 79.
Conclusions
The experiments reported here show that 25 minutes of moderate exercise generates large and rapid increases in gene expression in leukocytes of CFS patients but not in control subjects. Increases in mRNA were found for genes that can detect increases in muscle produced metabolites (ASIC3, P2X4, P2X5), genes that are essential for SNS processes (adrenergic α-2A, β-1, and β-2, as well as COMT), and immune function genes (IL10, and TLR4). These findings confirm previous hypotheses suggesting that alterations in all parts of the HPA axis may mediate and sustain the symptoms of CFS and FMS. These gene alterations suggest a potential role for alterations of peripheral sensory signaling in the symptoms of CFS, as has been proposed for FMS 69; 74. They also suggest that a blood test could be devised as an objective biomarker for sensory muscle fatigue and muscle pain in CFS.
Acknowledgments
The research presented here was primarily supported by NIH grant R21 NS057821 from NINDS and NIAMS, with additional ancillary support from NIH R21 AT0002209 from NCAM, a Catalyst grant from the University of Utah Health Sciences Center, and the Dept. of Anesthesiology, University of Utah. We also wish to acknowledge Dr. Lucinda Bateman for referral of patients and clinical advice.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Aaron LA, Buchwald D. A review of the evidence for overlap among unexplained clinical conditions. Ann Intern Med. 2001;134:868–881. doi: 10.7326/0003-4819-134-9_part_2-200105011-00011. [DOI] [PubMed] [Google Scholar]
- 2.Aaron LA, Herrell R, Ashton S, Belcourt M, Schmaling K, Goldberg J, Buchwald D. Comorbid clinical conditions in chronic fatigue: a co-twin control study. J Gen Intern Med. 2001;16:24–31. doi: 10.1111/j.1525-1497.2001.03419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aspler AL, Bolshin C, Vernon SD, Broderick G. Evidence of inflammatory immune signaling in chronic fatigue syndrome: A pilot study of gene expression in peripheral blood. Behav Brain Funct. 2008;4:44. doi: 10.1186/1744-9081-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brenn D, Richter F, Schaible HG. Sensitization of unmyelinated sensory fibers of the joint nerve to mechanical stimuli by interleukin-6 in the rat: an inflammatory mechanism of joint pain. Arthritis Rheum. 2007;56:351–359. doi: 10.1002/art.22282. [DOI] [PubMed] [Google Scholar]
- 5.Brierley SM, Carter R, Jones W, III, Xu L, Robinson DR, Hicks GA, Gebhart GF, Blackshaw LA. Differential chemosensory function and receptor expression of splanchnic and pelvic colonic afferents in mice. J Physiol. 2005;567:267–281. doi: 10.1113/jphysiol.2005.089714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carmel L, Efroni S, White PD, Aslakson E, Vollmer-Conna U, Rajeevan MS. Gene expression profile of empirically delineated classes of unexplained chronic fatigue. Pharmacogenomics. 2006;7:375–386. doi: 10.2217/14622416.7.3.375. [DOI] [PubMed] [Google Scholar]
- 7.Chapman CR, Tuckett RP, Song CW. Pain and stress in a systems perspective: reciprocal neural, endocrine, and immune interactions. J Pain. 2008;9:122–145. doi: 10.1016/j.jpain.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng JK, Ji RR. Intracellular signaling in primary sensory neurons and persistent pain. Neurochem Res. 2008;33:1970–1978. doi: 10.1007/s11064-008-9711-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciccone DS, Natelson BH. Comorbid illness in women with chronic fatigue syndrome: a test of the single syndrome hypothesis. Psychosom Med. 2003;65:268–275. doi: 10.1097/01.psy.0000033125.08272.a9. [DOI] [PubMed] [Google Scholar]
- 10.Cook DB, Nagelkirk PR, Poluri A, Mores J, Natelson BH. The influence of aerobic fitness and fibromyalgia on cardiorespiratory and perceptual responses to exercise in patients with chronic fatigue syndrome. Arthritis Rheum. 2006;54:3351–3362. doi: 10.1002/art.22124. [DOI] [PubMed] [Google Scholar]
- 11.Cook DB, O’Connor PJ, Lange G, Steffener J. Functional neuroimaging correlates of mental fatigue induced by cognition among chronic fatigue syndrome patients and controls. Neuroimage. 2007;36:108–122. doi: 10.1016/j.neuroimage.2007.02.033. [DOI] [PubMed] [Google Scholar]
- 12.de Rijke B, Horssen-Zoetbrood A, Beekman JM, Otterud B, Maas F, Woestenenk R, Kester M, Leppert M, Schattenberg AV, de Witte T, van de Wiel-van Kemenade, Dolstra H. A frameshift polymorphism in P2X5 elicits an allogeneic cytotoxic T lymphocyte response associated with remission of chronic myeloid leukemia. J Clin Invest. 2005;115:3506–3516. doi: 10.1172/JCI24832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elenkov IJ. Neurohormonal-cytokine interactions: implications for inflammation, common human diseases and well-being. Neurochem Int. 2008;52:40–51. doi: 10.1016/j.neuint.2007.06.037. [DOI] [PubMed] [Google Scholar]
- 14.Elenkov IJ, Wilder RL, Chrousos GP, Vizi ES. The sympathetic nerve--an integrative interface between two supersystems: the brain and the immune system. Pharmacol Rev. 2000;52:595–638. [PubMed] [Google Scholar]
- 15.Fang H, Xie Q, Boneva R, Fostel J, Perkins R, Tong W. Gene expression profile exploration of a large dataset on chronic fatigue syndrome. Pharmacogenomics. 2006;7:429–440. doi: 10.2217/14622416.7.3.429. [DOI] [PubMed] [Google Scholar]
- 16.Fillingim RB, Wallace MR, Herbstman DM, Ribeiro-Dasilva M, Staud R. Genetic contributions to pain: a review of findings in humans. Oral Dis. 2008;14:673–682. doi: 10.1111/j.1601-0825.2008.01458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fukuda K, Straus SE, Hickie I, Sharpe MC, Dobbins JG, Komaroff A. The chronic fatigue syndrome: a comprehensive approach to its definition and study. International Chronic Fatigue Syndrome Study Group. Ann Intern Med. 1994;121:953–959. doi: 10.7326/0003-4819-121-12-199412150-00009. [DOI] [PubMed] [Google Scholar]
- 18.Geisser ME, Glass JM, Rajcevska LD, Clauw DJ, Williams DA, Kileny PR, Gracely RH. A psychophysical study of auditory and pressure sensitivity in patients with fibromyalgia and healthy controls. J Pain. 2008;9:417–422. doi: 10.1016/j.jpain.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 19.Geisser ME, Strader DC, Petzke F, Gracely RH, Clauw DJ, Williams DA. Comorbid somatic symptoms and functional status in patients with fibromyalgia and chronic fatigue syndrome: sensory amplification as a common mechanism. Psychosomatics. 2008;49:235–242. doi: 10.1176/appi.psy.49.3.235. [DOI] [PubMed] [Google Scholar]
- 20.Goldstein DS, Robertson D, Esler M, Straus SE, Eisenhofer G. Dysautonomias: clinical disorders of the autonomic nervous system. Ann Intern Med. 2002;137:753–763. doi: 10.7326/0003-4819-137-9-200211050-00011. [DOI] [PubMed] [Google Scholar]
- 21.Hildebrand MS, de Silva MG, Klockars T, Rose E, Price M, Smith RJ, McGuirt WT, Christopoulos H, Petit C, Dahl HH. Characterisation of DRASIC in the mouse inner ear. Hear Res. 2004;190:149–160. doi: 10.1016/S0378-5955(04)00015-2. [DOI] [PubMed] [Google Scholar]
- 22.Ikeuchi M, Kolker SJ, Burnes LA, Walder RY, Sluka KA. Role of ASIC3 in the primary and secondary hyperalgesia produced by joint inflammation in mice. Pain. 2008;137:662–669. doi: 10.1016/j.pain.2008.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jahr H, van Driel M, van Osch GJ, Weinans H, van Leeuwen JP. Identification of acid-sensing ion channels in bone. Biochem Biophys Res Commun. 2005;337:349–354. doi: 10.1016/j.bbrc.2005.09.054. [DOI] [PubMed] [Google Scholar]
- 24.Jerjes WK, Taylor NF, Wood PJ, Cleare AJ. Enhanced feedback sensitivity to prednisolone in chronic fatigue syndrome. Psychoneuroendocrinology. 2007;32:192–198. doi: 10.1016/j.psyneuen.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 25.Jin X, Gereau RW. Acute p38-mediated modulation of tetrodotoxin-resistant sodium channels in mouse sensory neurons by tumor necrosis factor-alpha. J Neurosci. 2006;26:246–255. doi: 10.1523/JNEUROSCI.3858-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson SK, Deluca J, Natelson BH. Chronic fatigue syndrome: reviewing the research findings. Ann Behav Med. 1999;21:258–271. doi: 10.1007/BF02884843. [DOI] [PubMed] [Google Scholar]
- 27.Kato K, Sullivan PF, Evengard B, Pedersen NL. Chronic widespread pain and its comorbidities: a population-based study. Arch Intern Med. 2006;166:1649–1654. doi: 10.1001/archinte.166.15.1649. [DOI] [PubMed] [Google Scholar]
- 28.Kaufman MP, Hayes SG. The exercise pressor reflex. Clin Auton Res. 2002;12:429–439. doi: 10.1007/s10286-002-0059-1. [DOI] [PubMed] [Google Scholar]
- 29.Kerr JR, Christian P, Hodgetts A, Langford PR, Devanur LD, Petty R, Burke B, Sinclair LI, Richards SC, Montgomery J, McDermott CR, Harrison TJ, Kellam P, Nutt DJ, Holgate ST. Current research priorities in chronic fatigue syndrome/myalgic encephalomyelitis: disease mechanisms, a diagnostic test and specific treatments. J Clin Pathol. 2007;60:113–116. doi: 10.1136/jcp.2006.042374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kerr JR, Petty R, Burke B, Gough J, Fear D, Sinclair LI, Mattey DL, Richards SC, Montgomery J, Baldwin DA, Kellam P, Harrison TJ, Griffin GE, Main J, Enlander D, Nutt DJ, Holgate ST. Gene expression subtypes in patients with chronic fatigue syndrome/myalgic encephalomyelitis. J Infect Dis. 2008;197:1171–1184. doi: 10.1086/533453. [DOI] [PubMed] [Google Scholar]
- 31.Khan AA, Diogenes A, Jeske NA, Henry MA, Akopian A, Hargreaves KM. Tumor necrosis factor alpha enhances the sensitivity of rat trigeminal neurons to capsaicin. Neuroscience. 2008;155:503–509. doi: 10.1016/j.neuroscience.2008.05.036. [DOI] [PubMed] [Google Scholar]
- 32.Khasar SG, Burkham J, Dina OA, Brown AS, Bogen O, Alessandri-Haber N, Green PG, Reichling DB, Levine JD. Stress induces a switch of intracellular signaling in sensory neurons in a model of generalized pain. J Neurosci. 2008;28:5721–5730. doi: 10.1523/JNEUROSCI.0256-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kruger K, Lechtermann A, Fobker M, Volker K, Mooren FC. Exercise-induced redistribution of T lymphocytes is regulated by adrenergic mechanisms. Brain Behav Immun. 2008;22:324–338. doi: 10.1016/j.bbi.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 34.Kruger K, Mooren FC. T cell homing and exercise. Exerc Immunol Rev. 2007;13:37–54. [PubMed] [Google Scholar]
- 35.Le KT, Paquet M, Nouel D, Babinski K, Seguela P. Primary structure and expression of a naturally truncated human P2X ATP receptor subunit from brain and immune system. Febs Lett. 1997;418:195–199. doi: 10.1016/s0014-5793(97)01380-x. [DOI] [PubMed] [Google Scholar]
- 36.Light AR, Hughen RW, Zhang J. Increases in receptor mRNA in mouse dorsal root ganglion (DRG) neurons following carrageenan induced inflammation of mouse hindlimb muscle. Society for Neuroscience. 2008;#174(16) [Google Scholar]
- 37.Light AR, Hughen RW, Zhang J, Rainier J, Liu Z, Lee J. Dorsal root ganglion neurons innervating skeletal muscle respond to physiological combinations of protons, ATP, and lactate mediated by ASIC, P2X, and TRPV1. J Neurophysiol. 2008;100:1184–1201. doi: 10.1152/jn.01344.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Light AR, Hughen RW, Zhang J, White A, Light KC, Jensen BT, Fitschen KL. Molecular receptors for pH found on sensory neurons are also found on mouse and human leukocytes and increase 8–48 hours post-exercise in both control subjects and fibromyalgia and chronic fatigue patients. Society for Neuroscience. 2007;#510(3) [Google Scholar]
- 39.Light KC, Bragdon EE, Grewen KM, Brownley KA, Girdler SS, Maixner W. Adrenergic Dysregulation and Pain With and Without Acute Beta-blockade in Women with Fibromyalgia and Temporomandibular Disorder. J Pain. 2009;10:542–552. doi: 10.1016/j.jpain.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mamet J, Baron A, Lazdunski M, Voilley N. Proinflammatory mediators, stimulators of sensory neuron excitability via the expression of acid-sensing ion channels. J Neurosci. 2002;22:10662–10670. doi: 10.1523/JNEUROSCI.22-24-10662.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mamet J, Lazdunski M, Voilley N. How nerve growth factor drives physiological and inflammatory expressions of acid-sensing ion channel 3 in sensory neurons. J Biol Chem. 2003;278:48907–48913. doi: 10.1074/jbc.M309468200. [DOI] [PubMed] [Google Scholar]
- 42.McKee AE, Silver PA. Systems perspectives on mRNA processing. Cell Res. 2007;17:581–590. doi: 10.1038/cr.2007.54. [DOI] [PubMed] [Google Scholar]
- 43.Moldoveanu AI, Shephard RJ, Shek PN. Exercise elevates plasma levels but not gene expression of IL-1beta, IL-6, and TNF-alpha in blood mononuclear cells. J Appl Physiol. 2000;89:1499–1504. doi: 10.1152/jappl.2000.89.4.1499. [DOI] [PubMed] [Google Scholar]
- 44.Natelson BH, Haghighi MH, Ponzio NM. Evidence for the presence of immune dysfunction in chronic fatigue syndrome. Clin Diagn Lab Immunol. 2002;9:747–752. doi: 10.1128/CDLI.9.4.747-752.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Natelson BH, Intriligator R, Cherniack NS, Chandler HK, Stewart JM. Hypocapnia is a biological marker for orthostatic intolerance in some patients with chronic fatigue syndrome. Dyn Med. 2007;6:2. doi: 10.1186/1476-5918-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Natelson BH, Zhou X, Ottenweller JE, Bergen MT, Sisto SA, Drastal S, Tapp WN, Gause WL. Effect of acute exhausting exercise on cytokine gene expression in men. Int J Sports Med. 1996;17:299–302. doi: 10.1055/s-2007-972850. [DOI] [PubMed] [Google Scholar]
- 47.Naves lA, McCleskey EW. An acid-sensing ion channel that detects ischemic pain. Braz J Med Biol Res. 2005;38:1561–1569. doi: 10.1590/s0100-879x2005001100001. [DOI] [PubMed] [Google Scholar]
- 48.Nehlsen-Cannarella SL, Nieman DC, Jessen J, Chang L, Gusewitch G, Blix GG, Ashley E. The effects of acute moderate exercise on lymphocyte function and serum immunoglobulin levels. Int J Sports Med. 1991;12:391–398. doi: 10.1055/s-2007-1024700. [DOI] [PubMed] [Google Scholar]
- 49.Nieman DC, Davis JM, Henson DA, Gross SJ, Dumke CL, Utter AC, Vinci DM, Carson JA, Brown A, McAnulty SR, McAnulty LS, Triplett NT. Muscle cytokine mRNA changes after 2.5 h of cycling: influence of carbohydrate. Med Sci Sports Exerc. 2005;37:1283–1290. doi: 10.1249/01.mss.0000175054.99588.b1. [DOI] [PubMed] [Google Scholar]
- 50.Nieman DC, Davis JM, Henson DA, Walberg-Rankin J, Shute M, Dumke CL, Utter AC, Vinci DM, Carson JA, Brown A, Lee WJ, McAnulty SR, McAnulty LS. Carbohydrate ingestion influences skeletal muscle cytokine mRNA and plasma cytokine levels after a 3-h run. J Appl Physiol. 2003;94:1917–1925. doi: 10.1152/japplphysiol.01130.2002. [DOI] [PubMed] [Google Scholar]
- 51.Nieman DC, Henson DA, Davis JM, Dumke CL, Utter AC, Murphy EA, Pearce S, Gojanovich G, McAnulty SR, McAnulty LS. Blood leukocyte mRNA expression for IL-10, IL-1Ra, and IL-8, but not IL-6, increases after exercise. J Interferon Cytokine Res. 2006;26:668–674. doi: 10.1089/jir.2006.26.668. [DOI] [PubMed] [Google Scholar]
- 52.Nijs J, Demol S, Wallman K. Can submaximal exercise variables predict peak exercise performance in women with chronic fatigue syndrome? Arch Med Res. 2007;38:350–353. doi: 10.1016/j.arcmed.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 53.North RA. Molecular physiology of P2X receptors. Physiol Rev. 2002;82:1013–1067. doi: 10.1152/physrev.00015.2002. [DOI] [PubMed] [Google Scholar]
- 54.Overes IM, de Rijke B, Horssen-Zoetbrood A, Fredrix H, de Graaf AO, Jansen JH, van Krieken JH, Raymakers RA, van d V, de Witte TM, Dolstra H. Expression of P2X5 in lymphoid malignancies results in LRH-1-specific cytotoxic T-cell-mediated lysis. Br J Haematol. 2008;141:799–807. doi: 10.1111/j.1365-2141.2008.07125.x. [DOI] [PubMed] [Google Scholar]
- 55.Pall ML, Anderson JH. The vanilloid receptor as a putative target of diverse chemicals in multiple chemical sensitivity. Arch Environ Health. 2004;59:363–375. doi: 10.3200/AEOH.59.7.363-375. [DOI] [PubMed] [Google Scholar]
- 56.Peake J, Peiffer JJ, Abbiss CR, Nosaka K, Okutsu M, Laursen PB, Suzuki K. Body temperature and its effect on leukocyte mobilization, cytokines and markers of neutrophil activation during and after exercise. Eur J Appl Physiol. 2008;102:391–401. doi: 10.1007/s00421-007-0598-1. [DOI] [PubMed] [Google Scholar]
- 57.Peng BG, Ahmad S, Chen S, Chen P, Price MP, Lin X. Acid-sensing ion channel 2 contributes a major component to acid-evoked excitatory responses in spiral ganglion neurons and plays a role in noise susceptibility of mice. J Neurosci. 2004;24:10167–10175. doi: 10.1523/JNEUROSCI.3196-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Powell R, Ren J, Lewith G, Barclay W, Holgate S, Almond J. Identification of novel expressed sequences, up-regulated in the leucocytes of chronic fatigue syndrome patients. Clin Exp Allergy. 2003;33:1450–1456. doi: 10.1046/j.1365-2222.2003.01745.x. [DOI] [PubMed] [Google Scholar]
- 59.Rajeevan MS, Smith AK, Dimulescu I, Unger ER, Vernon SD, Heim C, Reeves WC. Glucocorticoid receptor polymorphisms and haplotypes associated with chronic fatigue syndrome. Genes Brain Behav. 2007;6:167–176. doi: 10.1111/j.1601-183X.2006.00244.x. [DOI] [PubMed] [Google Scholar]
- 60.Reyes M, Nisenbaum R, Hoaglin DC, Unger ER, Emmons C, Randall B, Stewart JA, Abbey S, Jones JF, Gantz N, Minden S, Reeves WC. Prevalence and incidence of chronic fatigue syndrome in Wichita, Kansas. Arch Intern Med. 2003;163:1530–1536. doi: 10.1001/archinte.163.13.1530. [DOI] [PubMed] [Google Scholar]
- 61.Saiki T, Kawai T, Morita K, Ohta M, Saito T, Rokutan K, Ban N. Identification of marker genes for differential diagnosis of chronic fatigue syndrome. Mol Med. 2008;14:599–607. doi: 10.2119/2007-00059.Saiki. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schweinhardt P, Sauro KM, Bushnell MC. Fibromyalgia: a disorder of the brain? Neuroscientist. 2008;14:415–421. doi: 10.1177/1073858407312521. [DOI] [PubMed] [Google Scholar]
- 63.Sluka KA, Radhakrishnan R, Benson CJ, Eshcol JO, Price MP, Babinski K, Audette KM, Yeomans DC, Wilson SP. ASIC3 in muscle mediates mechanical, but not heat, hyperalgesia associated with muscle inflammation. Pain. 2006 doi: 10.1016/j.pain.2006.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sluyter R, Barden JA, Wiley JS. Detection of P2X purinergic receptors on human B lymphocytes. Cell Tissue Res. 2001;304:231–236. doi: 10.1007/s004410100372. [DOI] [PubMed] [Google Scholar]
- 65.Sorensen B, Jones JF, Vernon SD, Rajeevan MS. Transcriptional control of complement activation in an exercise model of chronic fatigue syndrome. Mol Med. 2009;15:34–42. doi: 10.2119/molmed.2008.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sorkin LS, Xiao WH, Wagner R, Myers RR. Tumour necrosis factor-alpha induces ectopic activity in nociceptive primary afferent fibres. Neuroscience. 1997;81:255–262. doi: 10.1016/s0306-4522(97)00147-4. [DOI] [PubMed] [Google Scholar]
- 67.Spelta V, Fierro L, Naves lA, McCleskey EW. Abstract Viewer/Itinerary Planner . Washington, DC: Society for Neuroscience; 2004. Extracellular ATP ehances ASIC3-Like Current in Ischemia Sensing Neurons Through an Electrically Quiet Ion Channel. Online Program No. 859.4: 2004. [Google Scholar]
- 68.Staud R, Robinson ME, Price DD. Temporal summation of second pain and its maintenance are useful for characterizing widespread central sensitization of fibromyalgia patients. J Pain. 2007;8:893–901. doi: 10.1016/j.jpain.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Staud R, Vierck CJ, Robinson ME, Price DD. Overall fibromyalgia pain is predicted by ratings of local pain and pain-related negative affect--possible role of peripheral tissues. Rheumatology (Oxford) 2006;45:1409–1415. doi: 10.1093/rheumatology/kel121. [DOI] [PubMed] [Google Scholar]
- 70.Sugiura T, Bielefeldt K, Gebhart GF. Mouse colon sensory neurons detect extracellular acidosis via TRPV1. Am J Physiol Cell Physiol. 2007;292:C1768–C1774. doi: 10.1152/ajpcell.00440.2006. [DOI] [PubMed] [Google Scholar]
- 71.Sugiura T, Dang K, Lamb K, Bielefeldt K, Gebhart GF. Acid-sensing properties in rat gastric sensory neurons from normal and ulcerated stomach. J Neurosci. 2005;25:2617–2627. doi: 10.1523/JNEUROSCI.2894-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Togo F, Natelson BH, Adler GH, Ottenweller JE, Goldenberg DL, Struzik ZR, Yamamoto Y. Plasma Cytokine Fluctuations Over Time in Healthy Controls and Patients with Fibromyalgia. Exp Biol Med (Maywood) 2008 doi: 10.3181/0808-RM-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Van Den EF, Moorkens G, Van Houdenhove B, Cosyns P, Claes SJ. Hypothalamic-pituitary-adrenal axis function in chronic fatigue syndrome. Neuropsychobiology. 2007;55:112–120. doi: 10.1159/000104468. [DOI] [PubMed] [Google Scholar]
- 74.Vierck CJ., Jr Mechanisms underlying development of spatially distributed chronic pain (fibromyalgia) Pain. 2006;124:242–263. doi: 10.1016/j.pain.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 75.Voilley N, de Weille J, Mamet J, Lazdunski M. Nonsteroid anti-inflammatory drugs inhibit both the activity and the inflammation-induced expression of acid-sensing ion channels in nociceptors. J Neurosci. 2001;21:8026–8033. doi: 10.1523/JNEUROSCI.21-20-08026.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wagner D, Nisenbaum R, Heim C, Jones JF, Unger ER, Reeves WC. Psychometric properties of the CDC Symptom Inventory for assessment of chronic fatigue syndrome. Popul Health Metr. 2005;3:8. doi: 10.1186/1478-7954-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang L, Jacobsen SE, Bengtsson A, Erlinge D. P2 receptor mRNA expression profiles in human lymphocytes, monocytes and CD34+ stem and progenitor cells. BMC Immunol. 2004;5:16. doi: 10.1186/1471-2172-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Whistler T, Unger ER, Nisenbaum R, Vernon SD. Integration of gene expression, clinical, and epidemiologic data to characterize Chronic Fatigue Syndrome. J Transl Med. 2003;1:10. doi: 10.1186/1479-5876-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Williams DA, Gracely RH. Biology and therapy of fibromyalgia. Functional magnetic resonance imaging findings in fibromyalgia. Arthritis Res Ther. 2006;8:224. doi: 10.1186/ar2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wyller VB, Saul JP, Walloe L, Thaulow E. Sympathetic cardiovascular control during orthostatic stress and isometric exercise in adolescent chronic fatigue syndrome. Eur J Appl Physiol. 2008;102:623–632. doi: 10.1007/s00421-007-0634-1. [DOI] [PubMed] [Google Scholar]
- 81.Yoshiuchi K, Cook DB, Ohashi K, Kumano H, Kuboki T, Yamamoto Y, Natelson BH. A real-time assessment of the effect of exercise in chronic fatigue syndrome. Physiol Behav. 2007;92:963–968. doi: 10.1016/j.physbeh.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]