Abstract
The Bacillus subtilis Spx protein is a global transcription factor that interacts with the C-terminal domain of the RNA polymerase α subunit (αCTD) and regulates transcription of genes involved in thiol-oxidative stress, sporulation, competence, and organosulfur metabolism. Here we determined the X-ray crystal structure of the Spx/αCTD complex from an entirely new crystal form than previously reported (Newberry et al. 2005. Proc. Natl. Acad. Sci. USA. 102, 15839–15844). Comparison of the previously reported sulfate-bound complex and our sulfate-free complex reveals subtle conformational changes that may be important for the role of Spx in regulating organosulfur metabolism.
Keywords: Bacillus subtillus, Spx, RNA polymerase, transcription
In bacteria, transcription is ensured by a single multisubunit DNA-dependent RNA polymerase (RNAP), with a molecular mass of ~400 kDa and a subunit composition of α2ββ′ω. The α subunits play a key role in transcription initiation and activation (Gourse et al., 2000). The N-terminal domain of α (αNTD) is essential for α dimerization (Zhang et al., 1998) and RNAP assembly (Igarashi et al., 1991). The C-terminal domain (αCTD) binds to A/T rich regions (UP elements) at many promoters (Ross et al., 1993; Estrem et al., 1998) and contains several determinants for interacting with transcription factors (Savery et al., 1998; Busby et al., 1999; Lee et al., 2000; Savery et al., 2002; Nakano et al., 2003; Ross et al., 2003; Shah et al., 2004).
The Bacillus subtilis (Bsu) Spx protein, conserved among low GC-Gram-positive bacteria, is a thiol-based transcriptional regulator that registers thiol-oxidative stress via an N-terminal C-X-X-C motif (Zuber et al., 2004; Nakano et al., 2005). During thiol-oxidative stress, oxidised Spx binds to the αCTD of RNAP and appropriates RNAP to promoters upstream of genes whose products combat thiol-oxidative stress to induce their expression (Nakano et al., 2003b). In addition to its stimulatory role, Spx exerts negative control over a number of cellular processes; such as sporulation, genetic competence (Nakano et al., 2000; Nakano et al., 2001; Nakano et al., 2003) and organosulfur metabolism (Erwin et al., 2005; Choi et al., 2006).
The crystal structure of the in vitro-assembled Bsu Spx/αCTD complex, crystallized in space group R3, has been reported (Newberry et al., 2005). Spx adopts a thioredoxin fold (Martin, 1995), has substantial secondary structure homology to the Escherichia coli (Eco) ArsC protein (Martin et al., 2001; Newberry et al., 2005) and has an internal disulphide bond formed between residues C10 and C13, on the face of Spx opposite to the αCTD-binding determinants. It is apparent from the structure that Spx modulates the activity of αCTD by interacting with αCTD determinants that are essential for DNA-, transcription factor- and σ factor-binding. In both the Spx and ArsC crystal structures, a sulfate ion, derived from the crystallization solution, is bound to an arginine residue (R92 in Spx) that is part of an invariant RPI motif conserved in all Spx homologs and ArsC (Zuber, 2004; Newberry et al., 2005). Newberry and co-workers speculated that R92 could bind the sulfate ion in vivo, resulting in the coordinated regulation of genes involved in organosulfur metabolism (Zuber, 2004; Newberry et al., 2005).
In this work, we describe the crystal structure of the in vivo-assembled Spx/αCTD complex crystallized in space group P212121. Although the two structures were determined from crystals with very different crystal packing environments, the overall structures are very similar. In our structure, no sulfate ion can be observed near Spx residue Arg92, and the conformation of the residues surrounding the disulfide bond formed between Cys10 and Cys13 is altered. Thus, the structure of Spx in the in vivo assembled complex reveals a ”sulfate-free” conformation that Spx may adopt when regulating the expression of genes other than those required for organosulfur metabolism.
The Bsu spx gene was amplified from Bsu strain 168 genomic DNA by the polymerase chain reaction (PCR) using primers Bsu NdeI spx F (sense; 5′-gggattcccatatggttacactatacacatcaccaagc-3′; the NdeI site is underlined) and Bsu BamHI spx R (antisense; 5′-gttaggatccttagtttgccaaacgctgtgcttc-3; the BamHI site is underlined); the resulting DNA fragment was digested with NdeI and BamHI and cloned between the NdeI and BamHI sites of pET21a (Novagen) to create pET21aBsuSpx. The DNA encoding the Bsu αCTD (amino acid residues 240–314) was amplified by PCR from Bsu strain 168 genomic DNA using primers Bsu NdeI rpoA (240) F (sense; 5′-gggattcccatatgaaagaagaagatcaaaaagagaaagttcttg-3′; the NdeI site is underlined) and Bsu HindIII rpoA R (antisense; 5′-gttaaagctttcaatcgtctttgcgaagtccgag-3′; the HindIII site is underlined); the resulting PCR product was digested with NdeI and HindIII and cloned between the NdeI and HindIII sites of a pET28a-derived plasmid, creating pET28aBsu(His)6αCTD. The bicistronic Spx/αCTD co-expression system was created by amplifying the DNA encoding Bsu αCTD amino acid residues 240 to 314, using pET28aBsu(His)6αCTD as a template and primers Liz 13 (sense; 5′-gagcggatccaattcccctc-3′; the BamHI site is underlined), which anneals upstream of the plasmid-encoded translational enhancer, ribosome binding site, and (His)6 tag; and the T7 terminator primer (antisense; 5′-gctagttattgctcagcgg-3′). The DNA fragment was digested with BamHI and HindIII and cloned between the BamHI and HindIII sites of pET21aBsuSpx creating pET21aBsuSpx/(His)6αCTD. All cloning was confirmed to be correct by DNA sequencing.
The plasmid pET21aBsuSpx/(His)6αCTD was transformed into Eco Rosetta (DE3) cells (Novagen) and transformants were grown at 310 K in Luria-Bertani media supplemented with ampicillin (200 µg mL−1) and chloramphenicol (25 µg mL−1) to an A650 nm between 0.6 and 0.8. Subsequently, ampicillin (100 µg mL−1) and isopropyl-β,D-thiogalactopyranoside (1 mM) were added to the culture. After incubation at 303 K for 3 hours the cells were harvested by centrifugation, resuspended in 20 mM Tris-HCl (pH 8.0 at 277 K), 200 mM NaCl, 5% (v/v) glycerol, 5 mM imidazole, 1 mM phenylmethylsulphonylfluoride; lysed using a continuous-flow homogenizer (Avestin), and then centrifuged to remove insoluble debris. The clarified cell lysate was applied to a Ni2+-charged HiTrap column (GE Healthcare) equilibrated in buffer A (20 mM Tris-HCl, [pH 8.0 at 277 K], 200 mM NaCl, 5% (v/v) glycerol, 0.5 mM β-mercaptoethanol) + 5 mM imidazole. The column was washed with 5 column volumes (cv) of buffer A + 20 mM imidazole, 5 cv of buffer A + 40 mM imidazole and 5 cv of buffer A + 60 mM imidazole. Proteins bound to the column were eluted with buffer A + 250 mM imidazole. After overnight cleavage with PreScission protease (GE Healthcare) to remove the (His)6 tag and dialysis against buffer A + 20 mM imidazole, a subtractive Ni2+-chelating chromatographic step removed uncleaved (His)6αCTD and the cleaved (His)6 tag, and a GST HiTrap column (GE Healthcare) removed the protease. The sample was precipitated with ammonium sulphate (90% saturation) and the pellet resuspended in buffer B (10 mM Tris-HCl [pH 8.0 at 277 K], 50 mM NaCl, 5 mM DTT) to give a final protein concentration of 10 mg mL−1. The sample was applied to a Superdex 200 gel filtration column (GE Healthcare). Peak fractions eluted from the gel filtration column were applied to a Q-Sepharose HiTrap column (GE Healthcare) equilibrated in buffer B, and the column was developed with a linear gradient from 50 mM to 500 mM NaCl. Finally, the purified sample was exchanged into storage buffer (10 mM Tris-HCl [pH 8.0 at 277 K], 50 mM NaCl, 5 mM DTT). The purity of the complex was judged to be >95% as analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis and Coomassie blue staining (data not shown).
Using hanging-drop vapor diffusion, initial crystals grew at 295 K from 8% (w/v) PEG 4000, 100 mM sodium acetate (pH 4.6). Crystals for structure determination were obtained at 295 K using hanging-drop vapor diffusion against 6% (w/v) PEG 6000, 100 mM sodium acetate (pH 5.0), 10 µM CuCl2, with a protein concentration of 11.5 mg mL−1 and a protein to crystallant ratio of 1:1.
The crystals were soaked in a solution composed of the mother liquor supplemented with 20% (v/v) glycerol prior to freezing in liquid ethane. A full dataset was collected on a single crystal to 2.2 Å at X25, National Synchrotron Light Source (NSLS), Brookhaven National Laboratory (BNL). No anomalous signal due to copper ions was observed.
Using the structure of the in vitro-assembled Spx/αCTD complex (PDB ID code 1Z3E; Newberry et al., 2005) as a search model, a solution for the in vivo-assembled complex was obtained using MOLREP (Vagin et al., 1997). An initial rigid body between 20 to 3 Å corrected a slight conformational change in the Spx molecule. Flexible loop regions and the N-terminus of αCTD; which differed from the search model, were built manually in O (Jones et al., 1991). After iterative rounds of building and minimization to 2.3 Å, the final model was refined to R = 22.8%, Rfree = 28.25% using CNS (Brünger et al., 1998). The final model contained 259 water molecules and an additional nine amino acid residues (four residues are vector-derived) appended to the N-terminus of αCTD compared to the in vitro-assembled complex.
The crystal packing environment observed in both the in vivo- and in vitro-assembled structures is substantially different and is probably due to the additional nine amino acid residues appended to the N-terminus of αCTD in the in vivo-assembled complex. In the in vivo-assembled complex, which crystallized in space group P212121, the N-terminus of αCTD docks into a crevice between the two domains of an Spx molecule in a symmetry related complex (Figure 1A). Whereas in the in vitro-assembled complex, which crystallized in space group R3, the N-terminus of αCTD contacts a symmetry related αCTD molecule and both αCTD/αCTD and Spx/Spx contacts are required to maintain the crystal packing (Figure 1B).
Figure 1.
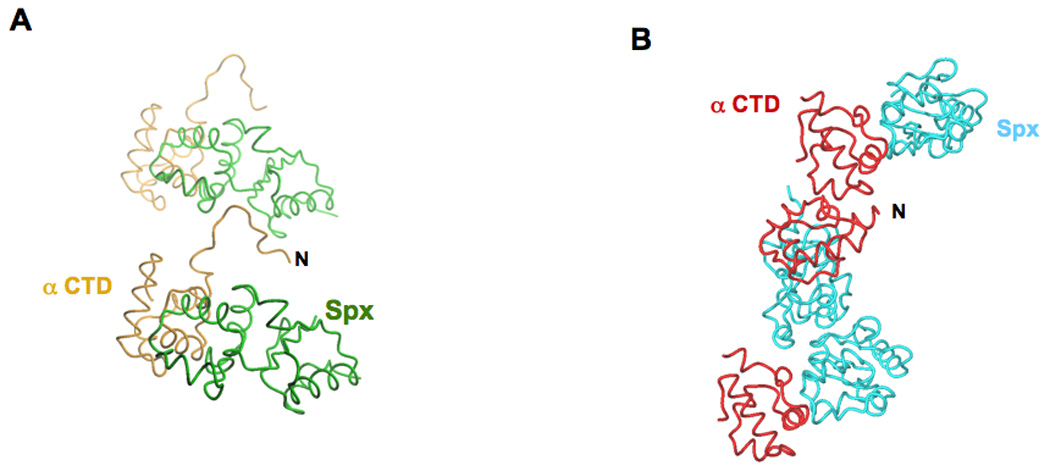
Crystal packing interactions. A. Crystal packing of the in vivo-assembled Spx/αCTD complex: the N-terminus of αCTD (labeled N) docks into a crevice formed between the two domains of a symmetry related Spx molecule. B. Crystal packing of the in vitro-assembled Spx/αCTD complex: crystal packing is maintained by both αCTD/αCTD interactions and Spx/Spx interactions. The in vivo-assembled complex αCTD is colored orange and its cognate Spx molecule is colored green. The in vitro-assembled complex αCTD is colored red and its cognate Spx molecule is colored cyan.
Overall, the structure of the in vivo-assembled Spx/αCTD complex is very similar to the structure of the in vitro-assembled complex (Figure 2; Newberry et al., 2005). The αCTD in the two different space groups superimpose with a RMSD of 0.504 Å over 62 α carbon positions. Spx is composed of two domains; the first domain (D1) is not contiguous in the primary sequence and encompasses amino acid residues 1 to 31 and 91 to 118, while the second domain (D2) interacts with αCTD and comprises amino acid residues 32 to 90. Superimposition of the Spx molecules from the two structures yields a RMSD of 0.732 Å over 118 α carbon positions; providing further confirmation that the in vivo- and in vitro-assembled complexes are very similar.
Figure 2.
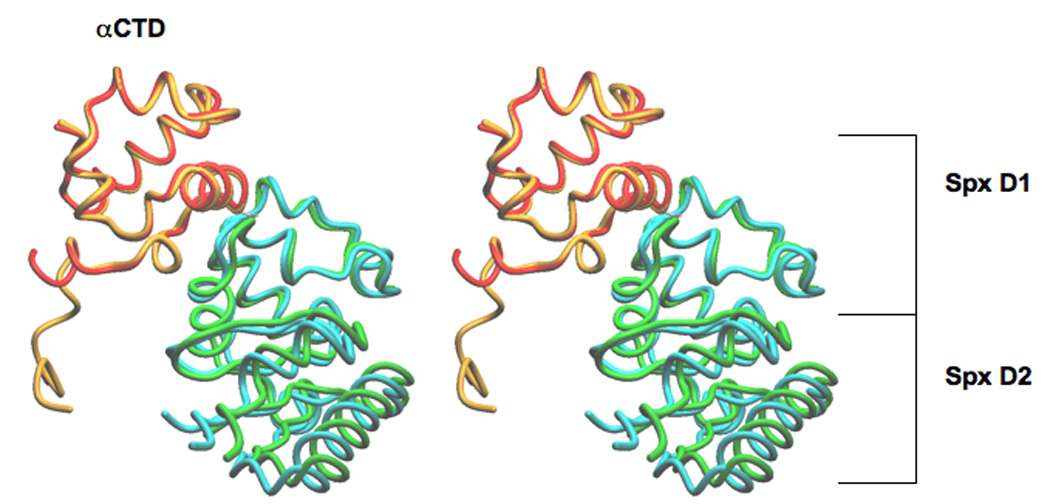
Stereoview of the superimposition of the in vivo- and in vitro-assembled complexes, crystallized in space groups P212121 and R3, respectively. The superimposition using LSQ was over the αCTD domain only (lsq_exp command, http://xray.bmc.uu.se/~alwyn/Essential_O/lsq_frameset.html). The N-terminus of αCTD in the in vivo-assembled complex is nine residues longer than in the in vitro-assembled complex. The αCTD molecules superimpose with a RMSD of 0.504 Å over 62 α carbon positions and the Spx molecules superimpose with a RMSD of 0.732 Å over 118 α carbon positions. In the in vivo-assembled complex αCTD is colored orange and Spx is colored green; in the in vitro-assembled complex αCTD is colored red and the Spx is colored cyan.
Although the crystal packing in both the in vivo- and the in vitro-assembled complexes is different; the molecular details of many of the interactions between Spx and αCTD are similar despite small movements of the Spx/αCTD interface. In both structures, three major interactions define the interface between αCTD and Spx. The side chain of α residue Asn264 interacts with the side chain of Spx Thr53 and the side chain of α Arg268 interacts with the side chains of both Spx Asp51 and Asp54 (Figure 3A). Finally, the side chain of α Val260 projects into a hydrophobic pocket composed of Spx residues Ile46, Gly52, Val71, Met74, Ile76 and Ile79 (Figure 3B).
Figure 3.
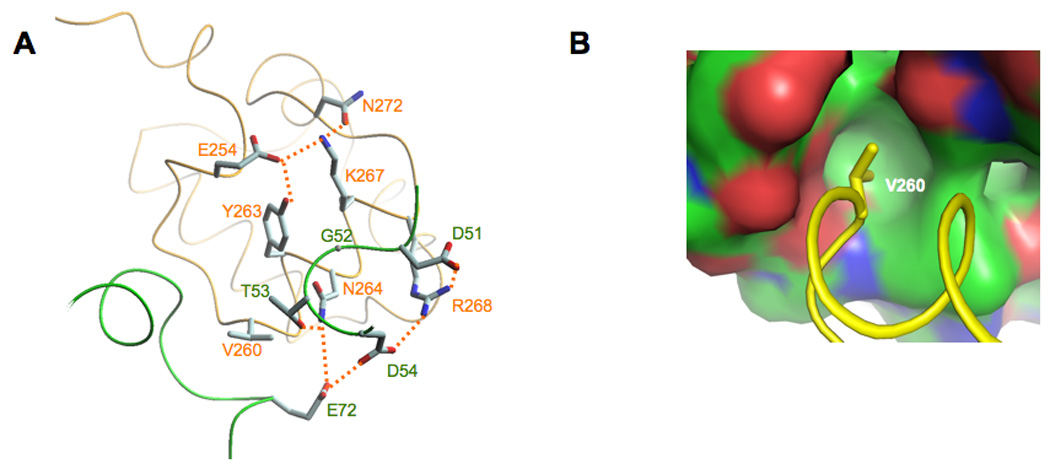
Molecular details of the Spx/αCTD interface. A. In both the in vivo- and the in vitro-assembled structures, αCTD residue Asn264 and Arg268 interact with Spx residues Asp51, Thr53 and Asp54. The residues that contribute to the overall Spx/αCTD interface are highlighted (hydrogen bonds are shown as dashed lines). B. αCTD residue Val260 projects into a hydrophobic pocket in Spx (the surface is colored in green for carbon atoms, red for oxygens and blue for nitrogens).
Accumulated genetic and biochemical data suggest that Spx G52 and α Y263 are important for Spx/αCTD complex formation (Nakano et al., 2000; Nakano et al., 2001; Nakano et al., 2003). Supporting these findings, the structure of the in vitro-assembled Spx/αCTD complex revealed that Spx G52 and α Y263 are in van der Waals contact. Y263 is part of a hydrogen bond network involving α residues E254, K267 and N272, all highly conserved in the RNAP α subunits encoded by Gram-positive bacteria, but not in the α subunits encoded by Gram-negative bacteria (data not shown). We hypothesize that one role of this hydrogen bond network is to maintain α residues N264 and R268 in the correct position for interacting with Spx.
In the in vitro-assembled complex, a sulfate ion forms a hydrogen bond with the hydroxyl group of S12 - which is in close proximity to the disulfide bond formed between residues C10 and C13 - and two hydrogen bonds with the side chain of R92 (Figure 4A; Newberry et al., 2005). In our structure of the in vivo-assembled complex, both C10 and C13 form a disulfide bond, but rather than a sulfate ion, the hydroxyl group of S12 is stabilized by a water molecule network and the side chain of R92 is stabilized by hydrogen bonds formed between a water molecule, the hydroxyl group of S7, the carbonyl group of G88 and the carbonyl group of L90 (Figure 3B). Additionally, the side chain of residue S7 is rotated almost 180° relative to that observed in the in vitro-assembled complex.
Figure 4.
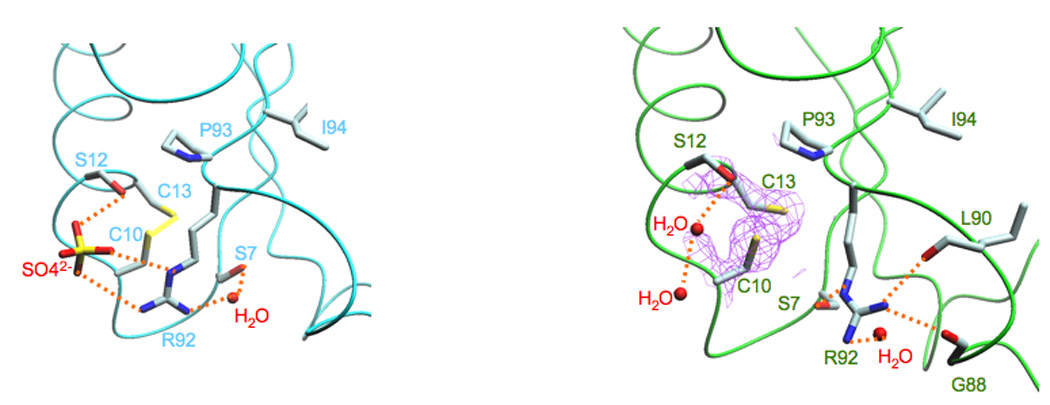
The “sulfate-free” conformation of Spx. A. In the structure of the in vitro-assembled complex (Newberry et al., 2005), a sulfate ion binds to Spx residues Ser12, which is adjacent to the disulfide bond formed between residues Cys10 and Cys13, and Arg92, which is part of the conserved Arg-Pro-Ile motif (Arg92, Pro93, Ile94). B. In the structure of the in vivo-assembled complex, no sulfate ion is present. The amine group of Arg92 is oriented “away” from the disulfide bond and forms an interaction with Ser7 and the carbonyl groups of residues Gly88 and Leu90. It is clear that residues Cys10 and Cys13 are oxidized, as revealed by the 2fo-fc electronic density (colored purple and contoured at 1.3σ).
Conclusions
We have solved the structure of the in vivo-assembled Spx/αCTD complex crystallized in space group P212121. Interestingly, in our structure, rather than interacting directly with Spx G52, as observed in the in vitro-assembled Spx/αCTD complex, α Y263 participates in a hydrogen bond network that could function to maintain α N264 and R268 in the correct orientation for direct binding to Spx. In both the in vitro-assembled Spx/αCTD complex and Eco ArsC structures, a sulfate ion interacts with an arginine residue (R92 in Spx) that is part of an RPI motif conserved in all Spx homologs and ArsC (Martin et al., 2001; Newberry et al., 2005). Newberry and co-workers hypothesized that the binding of sulfate to Spx R92 could function to modulate the activity of Spx during organosulfur metabolism. The structure of Spx in the in vivo-assembled complex is devoid of sulfate and presumably reveals a “sulfate-free” conformation that Spx may adopt when regulating the expression of genes other than those required for organosulfur metabolism.
Despite these differences, many of the interactions between Spx and αCTD are conserved, as is the oxidized state of Spx, suggesting that the movement of R92 or the presence or absence of sulfate, does not modulate either the binding interface greatly or disulphide bond formation.
Table 1.
Crystallographic Analysis
| Diffraction data | |||||
|---|---|---|---|---|---|
| Data set | Resolution (Å) | Number of reflections (Total/Unique) | Completeness | I/σ | Rsyma(%) |
| Native | 50-2.2 (2.28-2.20) | 82278/11313 | 99.4 (75.3) | 32.9 (13.8) | 5.1 (11.5) |
| Crystal space group | P212121 | ||||
| Unit cell | a = 41.8 Å, b = 44.3 Å, c = 117.2 Å | ||||
| Solvent content | 58 % (1 complex in the asymmetric unit) | ||||
| Refinementb | |||||
| Resolution | 15.0 - 2.3Å | ||||
| Rcryst/Rfree c(%) | 22.8 / 28.2 | ||||
| 259 water molecules | |||||
| 94.7% (180/190) of all residues were in favored regions and 98.4% (187/190) of all residues were in allowed regions d | |||||
Rsym = Σ|I-<I>|/ΣI, where I is observed intensity and <I> is average intensity obtained from multiple observations of symmetry-related reflections. Data reduction and scaling were performed using HKL2000 (http://www.hkl-xray.com/hkl_web1/hkl/HKL_2000.html).
The refinement was performed using CNS (http://cns-online.org/v1.2/).
Rcryst = Σ‖Fobserved|−|Fcalculated‖/Σ|Fobserved|, Rfree = Rcryst calculated using 5.2 % random data omitted from the refinement.
calculated from Molprobity (http://molprobity.biochem.duke.edu)
Acknowledgments
The authors wish to thank all the members of the Darst laboratory, Peter Zuber, Kate Newberry and Dick Brennan for their advice and enthusiasm for this project. We are grateful to both Andrea Feucht and Jeff Errington (Newcastle University, UK) for their generous gift of purified Bsu strain 168 genomic DNA. We are indebted to the staff at the National Synchrotron Light Source beamline X25 for support during data collection.
All figures, except for figure 3B, were generated using the program DINO (http://www.dino3d.org). Figure 3B was generated using the program PyMOL (http://www.pymol.org). V. L. was supported by a Women & Science Postdoctoral Fellowship from the Rockefeller University. This work was supported by NIH GM063759 to S. A. D.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data deposition: The structure coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3GFK)
References
- Benoff B, Yang H, Lawson CL, Parkinson G, Liu J, Blatter E, Ebright YW, Berman HM, Ebright RH. Structural basis of transcription activation: The CAP-αCTD-DNA complex. Science. 2002;297:1562–1566. doi: 10.1126/science.1076376. [DOI] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D. Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Busby S, Ebright RH. Transcription activation by catabolite activator protein (CAP) J. Mol. Biol. 1999;293:199–213. doi: 10.1006/jmbi.1999.3161. [DOI] [PubMed] [Google Scholar]
- Choi S-Y, Reyes D, Leelakriangsak M, Zuber P. The global regulator Spx functions in the control of organosulfur metabolism in Bacillus subtilis. J. Bacteriol. 2006;188:5741–5751. doi: 10.1128/JB.00443-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin KN, Nakano S, Zuber P. Sulfate-dependent repression of genes that function in organosulfur metabolism in Bacillus subtilis requires Spx. J. Bacteriol. 2005;187:4042–4049. doi: 10.1128/JB.187.12.4042-4049.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrem ST, Gaal T, Ross W, Gourse RL. Identification of an UP element consensus sequence for bacterial promoters. Proc. Natl. Acad. Sci. USA. 1998;95:9761–9766. doi: 10.1073/pnas.95.17.9761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourse RL, Ross W, Gaal T. UPs and downs in bacterial transcription initiation: The role of the alpha subunit of RNA polymerase in promoter recognition. Mol. Microbiol. 2000;37:687–695. doi: 10.1046/j.1365-2958.2000.01972.x. [DOI] [PubMed] [Google Scholar]
- Igarashi K, Fujita N, Ishihama A. Identification of a subunit assembly domain in the alpha subunit of Escherichia coli RNA polymerase. J. Mol. Biol. 1991;218:1–6. [PubMed] [Google Scholar]
- Jones TA, Zou J-Y, Cowan S, Kjeldgaard M. Improved methods for building protein models in electron denstiy maps and the location of errors in these models. Acta crystallographica. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Lee DJ, Wing HJ, Savery NJ, Busby SJW. Analysis of interactions between activating region 1 of Escherichia coli FNR protein and the C-terminal domain of the RNA polymerase α subunit: use of alanine scanning and suppression genetics. Mol. Microbiol. 2000;37:1032–1040. doi: 10.1046/j.1365-2958.2000.02086.x. [DOI] [PubMed] [Google Scholar]
- Martin JL. Thioredoxin - a fold for all reasons. Structure. 1995;3:245–250. doi: 10.1016/s0969-2126(01)00154-x. [DOI] [PubMed] [Google Scholar]
- Martin P, DeMel S, Shi J, Gladysheva T, Gatti DL, Rosen BP, Edwards BF. Insights into the structure, solvation, and mechanism of ArsC arsenate reductase, a novel arsenic detoxification enzyme. Structure. 2001;9:1071–1081. doi: 10.1016/s0969-2126(01)00672-4. [DOI] [PubMed] [Google Scholar]
- Nakano MM, Zhu Y, Liu J, Reyes DY, Yoshikawa H, Zuber P. Mutations conferring amino acid residue substitutions in the carboxy-terminal domain of RNA polymerase alpha can suppress clpX and clpP with respect to developmentally regulated transcription in Bacillus subtilis. Mol. Microbiol. 2000;37:869–884. doi: 10.1046/j.1365-2958.2000.02052.x. [DOI] [PubMed] [Google Scholar]
- Nakano MM, Hajarizadeh F, Zhu Y, Zuber P. Loss-of-function mutations in yjbD result in ClpX- and ClpP-independent competence development of Bacillus subtilis. Mol. Microbiol. 2001;42:383–394. doi: 10.1046/j.1365-2958.2001.02639.x. [DOI] [PubMed] [Google Scholar]
- Nakano S, Nakano MM, Zhang Y, Leelakriangsak M, Zuber P. A regulatory protein that interferes wtih activator-stimulated transcription in bacteria. Proc. Natl. Acad. Sci. USA. 2003;100:4233–4238. doi: 10.1073/pnas.0637648100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano S, Küster-Schöck E, Grossman AD, Zuber P. Spx-dependent global transcriptional control is induced by thiol-specific oxidative stress in Bacillus subtilis. Proc. Natl. Acad. Sci. USA. 2003b;100:13603–13608. doi: 10.1073/pnas.2235180100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano S, Erwin KN, Ralle M, Zuber P. Redox-sensitive transcriptional control by a thiol/disulphide switch in the global regulator. Spx. Mol. Microbiol. 2005;55:498–510. doi: 10.1111/j.1365-2958.2004.04395.x. [DOI] [PubMed] [Google Scholar]
- Newberry KJ, Nakano S, Zuber P, Brennan RG. Crystal structure of the Bacillus subtilis anti-alpha, global transcriptional regulator, Spx, in complex with the alpha C-terminal domain of RNA polymerase. Proc. Natl. Acad. Sci. USA. 2005;102:15839–15844. doi: 10.1073/pnas.0506592102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross W, Gosink K, Salomon J, Igarashi K, Zou C, Ishihama A, Severinov K, Gourse RL. A third recognition element in bacterial promoters: DNA binding by the alpha subunit of RNA polymerase. Science. 1993;262:1407–1413. doi: 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- Ross W, Schneider DA, Paul BJ, Mertens A, Gourse RL. An intersubunit contact stimulating transcription initiation by E. coli RNA polymerase: interaction of the alpha C-terminal domain and sigma region 4. Genes Dev. 2003;17:1293–1307. doi: 10.1101/gad.1079403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savery NJ, Lloyd GS, Kainz M, Gaal T, Ross W, Ebright RH, Gourse RL, Busby SJW. Transcription activation at class II CRP-dependent promoters: Identification of determinants in the C-terminal domain of the RNA polymerase alpha subunit. EMBO J. 1998;17:3439–3447. doi: 10.1093/emboj/17.12.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savery NJ, Lloyd GS, Busby SJW, Thomas MS, Ebright RH, Gourse RL. Determinants of the C-terminal domain of the Escherichia coli RNA polymerase α subunit important for transcription at class I cyclic AMP receptor protein-dependent promoters. J. Bacteriol. 2002;184:2273–2280. doi: 10.1128/JB.184.8.2273-2280.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah IM, Wolf RE., Jr Novel protein-protein interaction between Escherichia coli SoxS and the DNA binding determinant of the RNA polymerase a subunit: SoxS functions as a co-sigma factor and redeploys RNA polymerase from UP-element-containing promoters to SoxS-dependent promoters during oxidative stress. J. Mol. Biol. 2004;343:513–532. doi: 10.1016/j.jmb.2004.08.057. [DOI] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A. MOLREP: An automated program for molecular replacement. J. Appl. Crystallogr. 1997;30:1022–1025. [Google Scholar]
- Zhang G, Darst SA. Structure of the Escherichia coli RNA polymerase α subunit amino-terminal domain. Science. 1998;281:262–266. doi: 10.1126/science.281.5374.262. [DOI] [PubMed] [Google Scholar]
- Zuber P. Spx-RNA polymerase interaction and global transcriptional control during oxidative stress. J. Bacteriol. 2004;186:1911–1918. doi: 10.1128/JB.186.7.1911-1918.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]


