Abstract
Considerable progress has recently been made in understanding the role of cardiolipin in mitochondria. In this brief review, we discuss new data that show how cardiolipin specifically contributes to the lateral organization of mitochondrial membranes. We argue that the function of cardiolipin has to be understood in the context of dynamic membrane assembly rather than static membrane structure, and we propose that remodeling of cardiolipin, i.e. the formation of uniformly substituted molecular species, may reduce the energy barrier of the assembly process.
The specific function of cardiolipin in mitochondria has been an unresolved question for many years. An answer seemed at reach when Miriam Greenberg’s laboratory succeeded to introduce a yeast strain lacking the enzyme cardiolipin synthase [1]. Initial work with that mutant has shown reduced activities in pathways, such as oxidative phosphorylation and mitochondrial protein import, but surprisingly, the fundamental functions of mitochondria remained intact even in the absence of cardiolipin [2]. Gradually, it has become clear that there is no essential requirement for cardiolipin; rather cardiolipin is an ingredient that makes mitochondria work better. It is fair to say however, that our insight into the role of cardiolipin has immensely benefited from studies of cardiolipin-deficient yeast as well as, more recently, from studies on Barth syndrome, a human disease in which the cardiolipin concentration is reduced and the cardiolipin composition is altered.
1. Protein interaction of cardiolipin: chaperone-like activity
Perhaps the most prominent physical property of cardiolipin is its ability to interact by non-covalent bonds with a number of unrelated proteins, which invokes analogy between cardiolipin and molecular chaperones. The list of proteins that bind cardiolipin with high affinity is long and includes, among others, the respiratory complexes I, III, IV, and V, the carrier family (ADP-ATP-carrier, phosphate carrier, uncoupling protein), and two peripheral membrane proteins (cytochrome c and creatine kinase) (for a review see ref. 3). Taken together they account for a large portion of the proteins of the inner mitochondrial membrane; thus most, if not all proteins of this membrane, are associated with at least some cardiolipin molecules.
To understand the nature of cardiolipin-protein interactions, several crystal structures have been analyzed. Indeed, two cardiolipin molecules have been resolved in each of the crystal structures of complex III [4, 5], complex IV [6], and the ADP-ATP carrier [7]. As expected, hydrogen bonds to the phosphate groups of cardiolipin and van-der–Waals bonds to the acyl chains of cardiolipin contributed to the forces that attract cardiolipin to the protein. The crystal structures also supported the notion that cardiolipin is well suited to act as a bridge between two interfacing proteins and may thus promote the formation of homodimers and other protein complexes.
Crystallographic data are typically interpreted in terms of specific and static interactions. However, cardiolipin is not the only phospholipid in protein crystals, and any notion of specificity is inconsistent with the promiscuity of cardiolipin-protein interactions. Indeed, cardiolipin must possess a great deal of flexibility to adapt to such different structures as complex III and the ADP-ATP carrier. At the same time, cardiolipin must exert strong binding forces because there is a surprising degree of immobilization of cardiolipin molecules at the protein surface, as revealed by 31P-NMR studies of the ADP-ATP carrier [8] and of complex V [9]. Broadening of the 31P resonance is specific for cardiolipin, i.e. it has not been observed for any other phospholipid. Cardiolipin competes effectively with sodium dodecylsulfate for binding sites at the ADP-ATP-carrier and only complete protein denaturation with heat and chaotropic agents can release cardiolipin from the carrier [8]. These experiments suggest that cardiolipin is an integral component of the protein, the presence of which is critical to folding. Chaperone-like activity of cardiolipin was also suggested by its ability to stabilize folding intermediates of the enzyme rhodanese [10]. Although the physiologic relevance of this observation remains unclear, it illustrates the remarkable versatility of cardiolipin to interact with various proteins. Unfolded rhodanese appeared to have more binding sites for cardiolipin than for phosphatidylserine [10], which may be the reason why cardiolipin is a more effective chaperone than other phospholipids.
2. Lateral segregation of cardiolipin: formation of membrane domains
Cluster formation is another important physical property of cardiolipin. Evidence of lateral segregation was first obtained in E. coli, where cardiolipin preferentially localizes to the polar and septal regions of the cytoplasmic membrane [11]. An elegant explanation for this phenomenon has been provided on the basis of the intrinsic curvature of the cardiolipin molecule [12]. According to this model, microphase separation occurs because the total energy of the membrane decreases if lipids with high intrinsic curvature, like cardiolipin, cluster at the bacterial poles. The underlying principle is that cardiolipin clusters of a certain size, but not individual cardiolipin molecules, stabilize the geometry of curved regions of the membrane. As a result, cardiolipin separates laterally from other phospholipids. It is not known whether curvature geometry drives cardiolipin clustering in mitochondria, but the abundance of high curvature zones in mitochondrial cristae, makes this idea attractive.
Besides intrinsic curvature, cardiolipin may encourage domain formation by interacting with proteins. In yeast mitochondria, there is now ample evidence that cardiolipin promotes the association of proteins involved in oxidative phosphorylation. Cardiolipin was found to be essential for the presence [13], or at least the stability [14] of respiratory supercomplexes III2IV and III2IV2, and for the association of respiratory supercomplexes with the ADP-ATP-carrier (AAC) [15]. The molecular mechanism of this phenomenon is not known and it has not been determined how many cardiolipin molecules are integrated into one respiratory supercomplex, i.e. it is unclear whether supercomplexes depend on cardiolipin clusters or just on isolated cardiolipin molecules. The proteins of oxidative phosphorylation have an innate tendency to form supramolecular associations, but the role of cardiolipin is to shift the equilibrium III2 + 2IV + AAC2↔III2IV2AAC2 towards the right side of the equation. Since these conclusions were drawn entirely from electrophoretic studies, it is important to note that a kinetic study of complex III/IV activity in isolated mitochondria also supported a role of cardiolipin in supercomplex formation [16]. The function of cardiolipin may not be limited to the aforementioned proteins but may encompass other complexes, including complex V/tafazzin assemblies [17] or caspase-8 oligomers [18]. Furthermore, proteins like octameric creatine kinase or cytochrome c, have been shown to induce cardiolipin clustering in artificial membranes [19].
3. Acid-base function of cardiolipin: shaping the proton gradient
The titration behavior of cardiolipin suggested the presence of a stable monovalent anion at neutral pH, implying that the second proton of cardiolipin dissociates only in a very alkaline milieu [20]. This second proton becomes part of an acid anion formed by hydrogen bonds between the two phosphates and the central hydroxyl group, which yields a stable symmetric structure. It has been suggested that cardiolipin acid anions trap protons at the outer face of the inner mitochondrial membrane and thus convert the proton gradient into a transmembrane electrical potential [21]. On the matrix side of the membrane, cardiolipin may also play a role in proton conduction. This idea was suggested by the specific localization of cardiolipin in the crystal structure of complex III, where cardiolipin was identified as part of the proton uptake pathway [4].
The proton buffering capacity of cardiolipin has been implicated in the formation of mitochondrial cristae. This idea sprung out of a study on giant unilamellar vesicles prepared from mitochondrial phospholipids [22]. When the vesicles were exposed to a brief pulse-injection of acid, they temporarily formed cristae-like invaginations, a process that specifically required cardiolipin but not any protein. Other acidic phospholipids were not able to reproduce the effect of cardiolipin in this experiment. The authors provided a plausible explanation of their observation by arguing that proton trapping by cardiolipin may lead to charge neutralization and consequently to area reduction in the outer leaflet. Since tubular invaginations will reduce the area of the outer leaflet relative to the area of the inner leaflet, they may simply represent the energetically most favorable geometry of the vesicle. Although the biological relevance of this experiment remains to be explored, it has demonstrated that sophisticated morphologies can arise in a minimalist system consisting of a cardiolipin/lipid vesicle and a local proton gradient.
4. Biosynthesis of cardiolipin: topological aspects
To understand the role of cardiolipin in mitochondria, it is necessary to know how it is formed and how it is integrated into mitochondrial membranes. Like all phospholipids, cardiolipin is synthesized from the universal precursor phosphatidic acid. However, the biosynthesis of phosphatidic acid occurs in the endoplasmic reticulum and in the outer mitochondrial membrane, while all subsequent steps of the de novo formation of cardiolipin are performed on the matix side of the inner mitochondrial membrane. This conclusion was drawn from a study in mitoplasts, where the first and the last enzymes of the de novo pathway, phosphatidate:cytidylyltransferase and cardiolipin synthase respectively, were inaccessible to exogenous proteases [23]. The topology of cardiolipin synthase was corroborated by the finding that the enzyme can be inhibited by blockage of the entry of the cofactor Mn2+ into the mitochondrial matrix [23]. In yeast, cardiolipin synthesis required Tam41, a protein that also binds to the matrix side of the inner membrane [24]. Although the function of this protein has not been identified, its effect on the intermediates of the cardiolipin pathway, suggested that Tam41 is involved in early steps of cardiolipin synthesis, i.e. steps that precede the formation of phosphatidylglycerol [24]. Subsequent to its de novo synthesis, cardiolipin “matures” by an acyl exchange mechanism that is catalyzed by tafazzin. Since tafazzin is localized in the mitochondrial periphery, that is the outer face of the inner membrane [25] and the inner face of the outer membrane [25, 26], cardiolipin has to be translocated across the inner membrane in order to be remodeled. Again, the translocation mechanism has remained obscure, although one potential translocator, phospholipid scramblase-3, has been identified in mitochondria [27, 28].
5. Molecular species of cardiolipin: thermodynamic theory of membrane assembly
Translocation and acyl remodeling are the last steps of cardiolipin biosynthesis and these steps must either immediately precede the incorporation of cardiolipin into the membrane or be an integral part of the assembly process. We have recently made surprising observations that have altered our idea of cardiolipin remodeling and that ultimately favor integration of remodeling and membrane assembly.
Conventional phospholipid remodeling occurs by repeated cycles of deacylation and reacylation, catalyzed by calcium-independent phospholipase A2 (iPLA2) and acyltransferase respectively. However, genetic inactivation of iPLA2 did not alter cardiolipin remodeling in Drosophila [29], and genetic ablation of iPLA2γ from mice led only to an about 30% decrease in remodeled cardiolipin [30], and inhibition of iPLA2 in insect cells failed to abolish cardiolipin remodeling [31]. These results do not support an essential requirement for iPLA2 in cardiolipin remodeling. On the other hand, iPLA2 seems to be involved in the hydrolysis of cardiolipin because deletion of iPLA2 prevented the accumulation of monolyso-cardiolipin in a Drosophila model of Barth syndrome [29]. If iPLA2 is involved in cardiolipin hydrolysis but not in cardiolipin remodeling, the question arises whether the two processes are necessarily linked. Of course, the current data do not exclude that other phospholipases participate in cardiolipin remodeling. While the involvement of phospholipases is still being debated, the enzyme tafazzin is most definitely involved in cardiolipin remodeling because tafazzin deficiency drastically alters cardiolipin composition in humans [32, 33], flies [34], and yeast [35]. Interestingly, we found that isolated tafazzin can exchange fatty acids between two phospholipids in vitro, inferring that a separate deacylation by iPLA2 may not be necessary for cardiolipin remodeling [31]. Although tafazzin is a phospholipid-lysophospholipid transacylase [36], it requires only trace amounts of lysophospholipids [31]. Tafazzin reacts with various phospholipid classes [31, 36] and with various acyl species both in sn-1 and sn-2 position [31]. Our current model of the transacylation mechanism of tafazzin is shown in Figure 1.
Fig. 1.
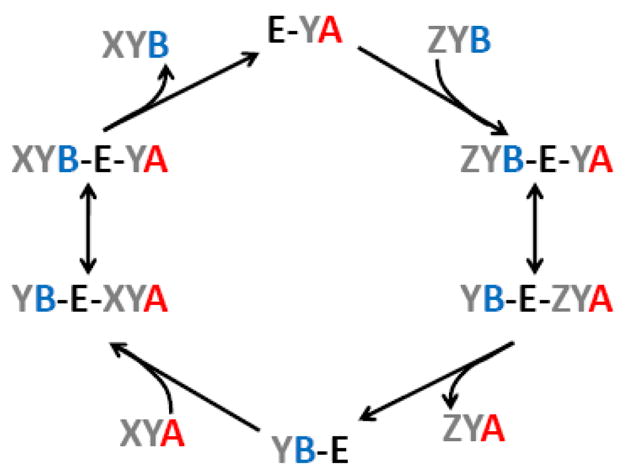
Proposed mechanism of transacylation by tafazzin. The enzyme (E) exchanges acyl groups (X, Y, Z) between phospholipid species with the head groups A and B. Two substrates, a phospholipid and a lysophospholipid, bind to the enzyme simultaneously. Acyl exchange between the substrates converts the phospholipid into a lysophospholipid and the lysophospholipid into a phospholipid. Only the phospholipid dissociates and is replaced by another phospholipid to begin another catalytic cycle. The scheme shows how tafazzin catalyzes the transacylation reaction XYA +ZYB → ZYA + XYB.
Since tafazzin exchanges acyl groups between phospholipids without much specificity for acyl species, head groups, or carbon positions, the question arises as to how the remarkable acyl specificity in cardiolipin is achieved. Throughout the eukaryotic kingdom, cardiolipin is dominated by molecular species that contain only one or two types of fatty acids, so structural uniformity and molecular symmetry prevail [37]. How can tafazzin select specific acyl groups for cardiolipin if it does indeed act as a facilitator of unrestricted acyl exchange between myriads of phospholipid species? A plausible if hypothetical answer has been given by a mathematical model that explores the consequences of the transacylation equilibrium [38]. Imagine a large set of molecular species (including different phospholipids) engages in free acyl exchange. The molecular composition of the entire ensemble may change in different directions and it is straightforward to define the boundaries within which the molecular composition can change for any given fatty acid composition. While tafazzin enables the ensemble of molecular species to adopt any composition within these boundaries, the ensemble must ultimately have an internal drive to move into the direction of the thermodynamic equilibrium. The question then becomes what is the molecular composition at equilibrium? If all acyl groups make equal contributions to the total free energy of the ensemble regardless of the phospholipids they are bound to, fatty acids would distribute randomly. If, on the other hand, some molecular species have a lower free energy than others, these species will accumulate in the transacylation equilibrium [38].
The model makes essentially two predictions. First, it suggests that acyl specificity may arise from the transacylation reaction in the absence of enzymatic specificity, merely due to specific interactions within the membrane. These interactions have not been defined, but they may include lipid-protein associations or lipid clusters as discussed in sections 1 and 2, or they may include other characteristics of mitochondrial membranes. It is intuitive that symmetric cardiolipin species are better suited to crosslink proteins and that uniformly substituted cardiolipin species have a greater tendency to cluster than heterogeneous cardiolipin molecules. Second, the model suggests that tafazzin confers compositional flexibility to phospholipids. Since tafazzin can “move” the molecular composition to any point within a mathematically defined spectrum of compositions, it can easily adapt the molecular composition to a changing microenvironment. A changing microenvironment is most likely encountered during the step-by-step assembly of mitochondrial membranes, in particular during the assembly of supramolecular structures such as respiratory supercomplexes. Thus we speculate that tafazzin lowers the energy barrier of the assembly process by constantly adapting the molecular composition of phospholipids.
In conclusion, tafazzin is a transacylase that redistributes acyl groups between phospholipids, which may generate specific patterns of molecular species in cardiolipin. The specific patterns are dependent on the type of mitochondria rather than the type of tafazzin. For instance, when human tafazzin was expressed in yeast [39], Drosophila (unpublished data), or Sf9 insect cells [31], the resulting changes in the cardiolipin pattern did not reflect the composition of human cardiolipin. In all three examples, human tafazzin was able to reshuffle the types of fatty acids that are characteristic of the host mitochondria. While it is clear from the above that transacylations are critical for the fatty acid composition of mitochondrial phospholipids, transacylations may also play a role in other aspects of phospholipid metabolism. For instance, transacylations are involved in the distribution of arachidonic acid among the molecular species of major glycerophospholipids [40].
Acknowledgments
Work in the author’s laboratories has been supported by the National Institutes of Health (HL078788), the Barth Syndrome Foundation, and the United Mitochondrial Disease Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jiang F, Rizavi HS, Greenberg ML. Cardiolipin is not essential for the growth of Saccharomyces cerevisiae on fermentable and non-fermentable carbon sources. Mol Microbiol. 1997;26:481–491. doi: 10.1046/j.1365-2958.1997.5841950.x. [DOI] [PubMed] [Google Scholar]
- 2.Jiang F, Ryan MT, Schlame M, Zhao M, Gu Z, Klingenberg M, Pfanner N, Greenberg ML. Absence of cardiolipin in crd1 null mutant results in decreased mitochondrial membrane potential and reduced mitochondrial function. J Biol Chem. 2000;275:22387–22394. doi: 10.1074/jbc.M909868199. [DOI] [PubMed] [Google Scholar]
- 3.Schlame M, Rua D, Greenberg ML. The biosynthesis and functional role of cardiolipin. Progr Lipid Res. 2000;39:257–288. doi: 10.1016/s0163-7827(00)00005-9. [DOI] [PubMed] [Google Scholar]
- 4.Lange C, Nett JH, Trumpower BL, Hunte C. Specific roles of protein-phospholipid interactions in the yeast cytochrome bc1 complex structure. EMBO J. 2001;20:6591–6600. doi: 10.1093/emboj/20.23.6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palsdottir H, Lojero CG, Trumpower BL, Hunte C. Structure of the yeast bc1 complex with a hydroxyquinone anion QO site inhibitor bound. J Biol Chem. 2003;278:31303–31311. doi: 10.1074/jbc.M302195200. [DOI] [PubMed] [Google Scholar]
- 6.Shinzawa-Itoh K, Aoyama H, Muramoto K, Terada H, Kurauchi T, Tadehara Y, Yamasaki A, Sugimura T, Kurono S, Tsujimoto K, Mizushima T, Yamashita E, Tsukihara T, Yoshikawa S. Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase. EMBO J. 2007;26:1713–1725. doi: 10.1038/sj.emboj.7601618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pebay-Peyroula E, Dahout-Gonzalez C, Kahn R, Trezeguet V, Lauquin GJM, Brandolin G. Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature. 2003;426:39–44. doi: 10.1038/nature02056. [DOI] [PubMed] [Google Scholar]
- 8.Beyer K, Klingenberg M. ADT/ATP carrier protein from beef heart mitochondria has high amounts of tightly bound cardiolipin, as revealed by 31P nuclear magnetic resonance. Biochemistry. 1985;24:3821–3826. doi: 10.1021/bi00336a001. [DOI] [PubMed] [Google Scholar]
- 9.Smith Eble K, Coleman WB, Hantgan RR, Cunningham CC. Tightly associated cardiolipin in the bovine heart mitochondrial ATP synthase as analyzed by 31P nuclear magnetic resonance spectroscopy. J Biol Chem. 1990;265:19434–19440. [PubMed] [Google Scholar]
- 10.Zardeneta G, Horowitz PM. Physical characterization of a reactivatable liposome-bound rhodanese folding intermediate. Biochemistry. 1993;32:13941–13948. doi: 10.1021/bi00213a025. [DOI] [PubMed] [Google Scholar]
- 11.Mileykovskaya E, Dowhan W. Visualization of phospholipids domains in Escherichia coli by using the cardiolipin-specific fluorescent dye 10-n-nonyl acridine orange. J Bacteriol. 2000;182:1172–1175. doi: 10.1128/jb.182.4.1172-1175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang KC, Mukhopadhyay R, Wingreen NS. A curvature-mediated mechanism for localization of lipids to bacterial poles. PLOS Comput Biol. 2006;2:e151. doi: 10.1371/journal.pcbi.0020151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang M, Mileykovskaya E, Dowhan W. Gluing the respiratory chain together: Cardiolipin is required for supercomplex formation in the inner mitochondrial membrane. J Biol Chem. 2002;277:43553–43556. doi: 10.1074/jbc.C200551200. [DOI] [PubMed] [Google Scholar]
- 14.Pfeiffer K, Gohil V, Stuart RA, Hunte C, Brandt U, Greenberg ML, Schagger H. Cardiolipin stabilizes respiratory chain supercomplexes. J Biol Chem. 2003;278:52873–52880. doi: 10.1074/jbc.M308366200. [DOI] [PubMed] [Google Scholar]
- 15.Claypool SM, Oktay Y, Boontheung P, Loo JA, Koehler CM. Cardiolipin defines the interactome of the major ADP/ATP carrier protein of the mitochondrial inner membrane. J Cell Biol. 2008;182:937–950. doi: 10.1083/jcb.200801152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang M, Mileykovskaya E, Dowhan W. Cardiolipin is essential for organization of complexes III and IV into a supercomplex in intact yeast mitochondria. J Biol Chem. 2005;280:29403–29408. doi: 10.1074/jbc.M504955200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Claypool SM, Boontheung P, McCaffery JM, Loo JA, Koehler CM. The cardiolipin transacylase, tafazzin, associates with two distinct respiratory components providing insight into Barth syndrome. Mol Biol Cell. 2008;19:5143–5155. doi: 10.1091/mbc.E08-09-0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonzalvez F, Schug ZT, Houtkooper RH, MacKenzie ED, Brooks DG, Wanders RJA, Petit PX, Vaz FM, Gottlieb E. Cardiolipin provides an essential activating platform for caspase-8 on mitochondria. J Cell Biol. 2008;183:681–696. doi: 10.1083/jcb.200803129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Epand RF, Tokarska-Schlattner M, Schlattner U, Wallimann T, Epand RM. Cardiolipin clusters and membrane domain formation induced by mitochondrial proteins. J Mol Biol. 2007;365:968–980. doi: 10.1016/j.jmb.2006.10.028. [DOI] [PubMed] [Google Scholar]
- 20.Kates M, Syz JY, Gosser D, Haines TH. pH-dissociation characteristics of cardiolipin and its 2′-deoxy analogue. Lipids. 1993;28:877–882. doi: 10.1007/BF02537494. [DOI] [PubMed] [Google Scholar]
- 21.Haines TH, Dencher NA. Cardiolipin: A proton trap for oxidative phosphorylation. FEBS Letters. 2002;528:35–39. doi: 10.1016/s0014-5793(02)03292-1. [DOI] [PubMed] [Google Scholar]
- 22.Khalifat N, Puff N, Bonneau S, Fournier JB, Angelova MI. Membrane deformation under local pH gradient: Mimicking mitochondrial cristae dynamics. Biophys J. 2008;95:4924–4933. doi: 10.1529/biophysj.108.136077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schlame M, Haldar D. Cardiolipin is synthesized on the matrix side of the inner membrane in rat liver mitochondria. J Biol Chem. 1993;268:74–79. [PubMed] [Google Scholar]
- 24.Kutik S, Rissler M, Guan XL, Guiard B, Shui G, Gebert N, Heacock PN, Rehling P, Dowhan W, Wenk MR, Pfanner N, Wiedemann N. The translocator maintenance protein Tam41 is required for mitochondrial cardiolipin biosynthesis. J Cell Biol. 2008;183:1213–1221. doi: 10.1083/jcb.200806048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Claypool SM, McCaffery JM, Koehler CM. Mitochondrial mislocalization and altered assembly of a cluster of Barth syndrome mutant tafazzins. J Cell Biol. 2006;174:379–390. doi: 10.1083/jcb.200605043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brandner K, Mick DU, Frazier AE, Taylor RD, Meisinger C, Rehling P. Taz1, an outer mitochondrial membrane protein, affects stability and assembly of inner membrane protein complexes: Implications for Barth syndrome. Mol Biol Cell. 2005;16:5202–5214. doi: 10.1091/mbc.E05-03-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu J, Dai Q, Chen J, Durrant D, Freeman A, Liu T, Grossman D, Lee RM. Phospholipd scramblase 3 controls mitochondrial structure, function, and apoptotic response. Mol Cancer Res. 2003;1:892–902. [PubMed] [Google Scholar]
- 28.Van Q, Liu J, Lu B, Feingold KR, Shi Y, Lee RM, Hatch GM. Phospholipid scramblase-3 regulates cardiolipin de novo biosynthesis and its resynthesis in growing HeLa cells. Biochem J. 2007;401:103–109. doi: 10.1042/BJ20060373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malhotra A, Edelman-Novemsky I, Xu Y, Plesken H, Ma J, Schlame M, Ren M. Role of calcium-independent phospholipase A2 in the pathogenesis of Barth syndrome. Proc Natl Acad Sci USA. 2009;106:2337–2341. doi: 10.1073/pnas.0811224106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mancuso DJ, Sims HF, Han X, Jenkins CM, Guan SP, Yang K, Moon SH, Pietka T, Abumrad NA, Schlesinger PH, Gross RW. Genetic ablation of calcium-independent phospholipase A2 leads to alterations in mitochondrial lipid metabolism and function in a deficient mitochondrial bioenergetic phenotype. J Biol Chem. 2007;282:34611–34622. doi: 10.1074/jbc.M707795200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malhotra A, Xu Y, Ren M, Schlame M. Formation of molecular species of mitochondrial cardiolipin: 1. A novel transacylation mechanism to shuttle fatty acids between sn-1 and sn-2 positions of multiple phospholipids species. Biochim Biophys Acta. 2009;1791:314–320. doi: 10.1016/j.bbalip.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vreken P, Valianpour F, Nijtmans LG, Grivell LA, Plecko B, Wanders RJA, Barth PG. Defective remodeling of cardiolipin and phosphatidylglycerol in Barth syndrome. Biochem Biophys Res Commun. 2000;279:378–382. doi: 10.1006/bbrc.2000.3952. [DOI] [PubMed] [Google Scholar]
- 33.Schlame M, Towbin JA, Heerdt PM, Jehle R, DiMauro S, Blanck TJJ. Deficiency of tetralinoleoyl-cardiolipin in Barth syndrome. Ann Neurol. 2002;51:634–637. doi: 10.1002/ana.10176. [DOI] [PubMed] [Google Scholar]
- 34.Xu Y, Condell M, Plesken H, Edelman-Novemsky I, Ma J, Ren M, Schlame M. A Drosophila model of Barth syndrome. Proc Natl Acad Sci USA. 2006;103:11584–11588. doi: 10.1073/pnas.0603242103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gu Z, Valianpour F, Chen S, Vaz FM, Hakkaart GA, Wanders RJA, Greenberg ML. Aberrant cardiolipin metabolism in the yeast taz1 mutant: A model for Barth syndrome. Mol Microbiol. 2004;51:149–158. doi: 10.1046/j.1365-2958.2003.03802.x. [DOI] [PubMed] [Google Scholar]
- 36.Xu Y, Malhotra A, Ren M, Schlame M. The enzymatic function of tafazzin. J Biol Chem. 2006;281:39217–39224. doi: 10.1074/jbc.M606100200. [DOI] [PubMed] [Google Scholar]
- 37.Schlame M, Ren M, Xu Y, Greenberg ML, Haller I. Molecular symmetry in mitochondrial cardiolipins. Chem Phys Lipids. 2005;138:38–49. doi: 10.1016/j.chemphyslip.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 38.Schlame M. Formation of molecular species of mitochondrial cardiolipin. 2. A mathematical model of pattern formation by phospholipid transacylation. Biochim Biophys Acta. 2009;1791:321–325. doi: 10.1016/j.bbalip.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vaz FM, Houtkooper RH, Valianpour F, Barth PG, Wanders RJA. Only one splice variant of the human TAZ gene encodes a functional protein with a role in cardiolipin metabolism. J Biol Chem. 2003;278:43089–43094. doi: 10.1074/jbc.M305956200. [DOI] [PubMed] [Google Scholar]
- 40.Leslie CC. Regulation of arachidonic acid availability for eicosanoid production. Biochem Cell Biol. 2004;82:1–17. doi: 10.1139/o03-080. [DOI] [PubMed] [Google Scholar]


