Summary
We have previously shown that the PEGylated LPD (liposome-polycation-DNA) nanoparticles were highly efficient in delivering siRNA to the tumor with low liver uptake. Its mechanism of evading the reticuloendothelial system (RES) is reported here. In LPD, nucleic acids were condensed with protamine into a compact core, which was then coated by two cationic lipid bilayers with the inner bilayer stabilized by charge-charge interaction (also called the supported bilayer). Finally, a detergent-like molecule, polyethylene glycol (PEG)-phospholipid is post-inserted into the lipid bilayer to modify the surface of LPD. The dynamic light scattering (DLS) data showed that LPD had improved stability compared to cationic liposomes after incubation with a high concentration of DSPE-PEG2000, which is known to disrupt the bilayer. LPD prepared with a multivalent cationic lipid, DSGLA, had enhanced stability compared to those containing DOTAP, a monovalent cationic lipid, suggesting that stronger charge-charge interaction in the supported bilayer contributed to a higher stability. Distinct nanoparticle structure was found in the PEGylated LPD by transmission electron microscopy, while the cationic liposomes were transformed into tubular micelles. Size exclusion chromatography data showed that approximately 60% of the total cationic lipids, which were located in the outer bilayer of LPD, were stripped off during the PEGylation; and about 20% of the input DSPE-PEG2000 was incorporated into the inner bilayer with about 10.6 mol% of DSPE-PEG2000 presented on the particle surface. This led to complete charge shielding, low liver sinusoidal uptake, and 32.5% injected dose delivered to the NCI-H460 tumor in a xenograft model.
Keywords: LPD nanoparticles, PEGylation, supported bilayer, pharmacokinetics, reticuloendothelial system
Introduction
Prolongation of the circulation half-life by surface incorporation of polyethylene glycol (PEG) in liposomes was demonstrated by us [1] and others [2] in early 1990s. Macrophages in the reticuloendothelial system (RES) located in the liver and the spleen vividly take up particles bound with serum proteins; and surface modification by PEG reduces the opsonization of liposomes and minimizes the clearance by the RES, leading to improved pharmacokinetic properties. This approach has been used in a variety of nanoparticle systems to improve the circulation half-life and enhance the drug delivery. There are two methods for surface PEGylation of liposomes. The first is to hydrate a lipid film consisting of PEG-phospholipids and other lipid components, and this self-assembling process allows PEG to extend from the lipid bilayer to both the inner aqueous core and the outer aqueous phase. The second method, post-insertion, is also commonly used in which PEG-phospholipids are incubated, usually at an elevated temperature, with pre-formed liposomes [3]. The PEG conjugates are inserted into the liposome bilayer due to hydrophobic interaction of the acyl chains of the conjugates with the liposomal lipids. This method permits that only the outer surface of liposomes is modified with PEG. However, due to the detergent-like activity of the PEG-phospholipids, the degree of surface PEGylation is usually less than 5 mol % if the liposome integrity is to be preserved [3, 4].
LPD (liposome-polycation-DNA) nanoparticle was developed earlier in our lab [5] and has been used for the delivery of peptide [6] and nucleic acid, including plasmid DNA [5], oligonucleotide and siRNA [7–10]. LPD was prepared by mixing cationic liposomes, protamine and nucleic acids at a fixed ratio. The self-assembled nanoparticles were 100–150 nm in diameter. Recently, we have modified LPD nanoparticles with a high level, i.e., 10 mol %, of PEG-phospholipid conjugate and showed that the i.v. injected nanoparticles accumulated in the NCI-H460 human lung tumor at approximately ∼60 % injected dose (ID)/g tissue in a xenograft model [7]. More importantly, liver and spleen uptakes of the nanoparticles were unusually low. Such RES evasion property of the nanoparticles is obviously interesting and important for drug and gene delivery. We have decided to study the mechanism in some detail.
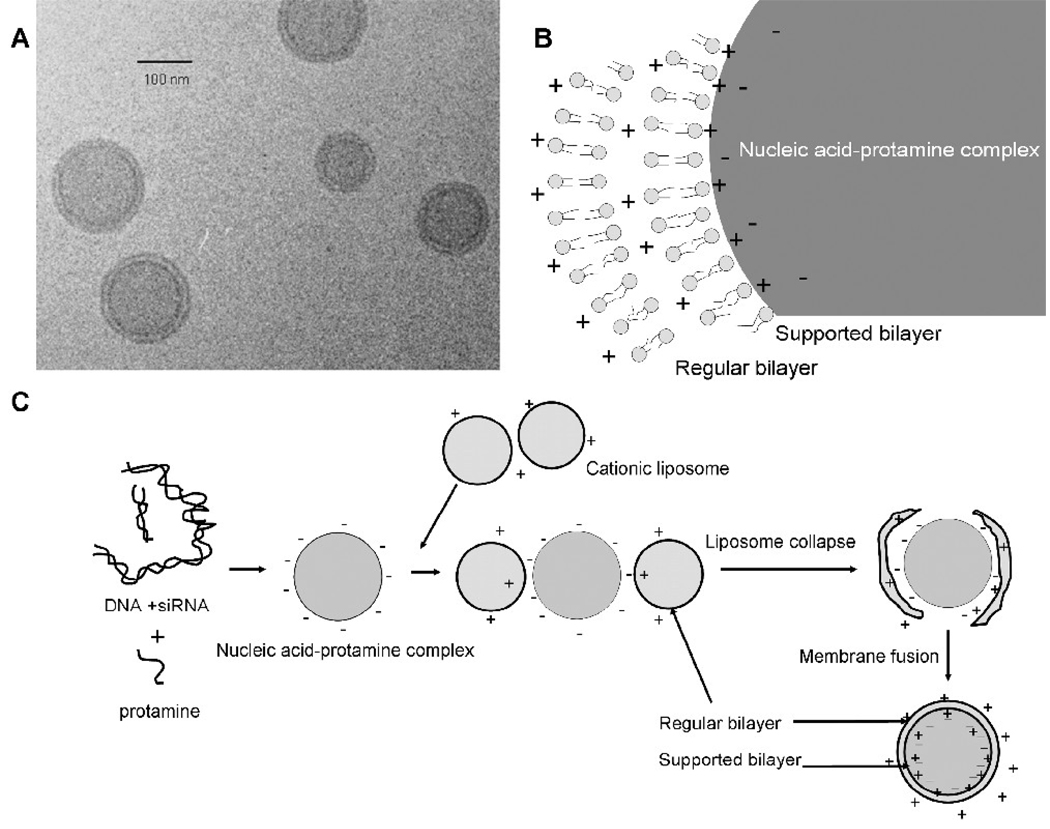
The detailed structure of the LPD was demonstrated by the cryo-TEM (transmission electron microscopy) (figure 1A) [11], showing that the nucleic acid was complexed by protamine to form a compact core, which was coated with two cationic lipid bilayers (figure 1B). The formation mechanism of the LPD has been proposed (figure 1C) [11]. First, the polycation interacts with the nucleic acid to form a negatively charged compact core. The subsequently added cationic liposomes collapse onto the core by charge-charge interaction. Finally, two separate lipid bilayer membranes appear on the surface of the LPD nanoparticles as the result of bilayer fusion and re-organization. In this model, the inner bilayer is directly in contact with the core and is supported and stabilized by the charge-charge interaction of the cationic lipids with the negatively charged core. We hypothesized that a supported bilayer can tolerate a high level of DSPE-PEG2000, which is a detergent-like surfactant, better than a regular bilayer. This unique feature of LPD may provide an opportunity to modify the formulation with a high amount of DSPE-PEG2000 to achieve an enhanced surface shielding and thus improve the pharmacokinetic properties of the nanoparticle formulation.
Figure 1.
Illustration of the structure and formation of LPD. Cryo-TEM photograph (A), the illustration of the double lipid bilayer structure (B) and proposed mechanism for the formation of the LPD nanoparticles (C). Figure 1A was reproduced from Tan et al. with permission [11].
In this study, we evaluated the tolerance of liposomes and LPD to DSPE-PEG2000 micelles by using dynamic light scattering (DLS) and TEM. We also titrated the input amount of DSPE-PEG2000 micelles for the post-insertion and monitored the stability of the resulting nanoparticles. Size exclusion chromatography was performed to purify and characterize the PEGylated LPD. The surface shielding effect of the PEGylated LPD was assessed by measuring the zeta potential and the non-specific uptake by the liver sinusoidal cells in the isolated liver perfusion model. Finally, the tissue distribution of the PEGylated LPD was studied in a tumor-bearing mouse model.
Materials and Methods
Materials
DOTAP, NBD-DOTAP, NBD-PE, cholesterol, DSPE-PEG2000, and DSPE-PEG2000-CF were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL). Protamine sulfate (fraction X from salmon sperm), calf thymus DNA (for hybridization, phenol-chloroform extracted and ethanol precipitated), and Sepharose CL 2B were from Sigma-Aldrich (St. Louis, MO). DSGLA was synthesized in our lab as previously reported [12].
Anti-luciferase siRNA (GL3) (target sequence 5’- CTT ACG CTG AGT ACT TCG A −3’) was purchased from Dharmacon (Lafayette, CO) in deprotected, desalted, annealed form. FAM labeled siRNA (3’ end of the sense strand) was used to evaluate the incorporation efficiency for siRNA in the LPD. Cy3 labeled siRNA was used for the isolated liver perfusion study.
NCI-H460 and CT26 cells were purchased from the American Type Culture Collection, and TC-1 cells were obtained from Dr. T.C. Wu (Johns Hopkins University, Baltimore, MD). Cells were cultured in PRMI-1640 medium supplemented with 10% fetal bovine serum and antibiotics.
Experimental animals
Female C57BL/6 mice of age 6–8 week (16–18 g) and female athymic nude mice of age 6–8 week were purchased from Charles River Laboratories (Wilmington, MA). All work performed with animals was in accordance with and approved by the IACUC committee at the University of North Carolina at Chapel Hill (UNC).
Preparation of siRNA containing LPD nanoparticles
LPD was prepared as previously described [8]. Briefly, unmodified LPD were obtained by quickly mixing suspension A (8.3 mM liposomes (DOTAP: cholesterol = 1: 1, molar ratio) and 0.2 mg/ml protamine in 150 µl nuclease free water) with solution B (0.16 mg/ml siRNA and 0.16 mg/ml calf thymus DNA in 150 µl nuclease free water) followed by incubation at room temperature for 10 min. PEGylated LPD was prepared by incubating the LPD suspension (300 µl) with 37.8 µl micelles solution of DSPE-PEG2000 (10 mg/ml) at 50°C for 10 min. PEGylated LPD was allowed to stand at room temperature for 10 min. The charge ratio of the formulation was about 1:5 (−:+). The particle size was measured using a submicron particle sizer (NICOMP particle sizing systems, AutodilutePAT Model 370, Santa Barbra, CA) in the NICOMP mode. The zeta potential of various LPD formulations diluted in 1 mM KCl was determined by using a Zeta Plus zeta potential analyzer (Brookhaven Instruments Corporation, Holtsville, NY). PEGylated LPD was freshly prepared and used within 20 min for the following experiments. For size exclusion chromatography, either 10 mol% NBD-DOTAP labeled liposomes, 10 mol% DSPE-PEG2000-CF labeled DSPE-PEG2000 or FAM-siRNA was used for the preparation of the PEGylated LPD. For the liver perfusion study, cy3-siRNA was used.
Negative-stain transmission electron microscopy
TEM images were acquired using a Phillips CM12 (FEI, Hillsboro, OR). Briefly, freshly prepared formulations (5 µl) were dropped onto 300 mesh carbon-coated copper grids (Ted Pella, Inc., Redding, CA) and allowed a short incubation (5 min) at room temperature. Grids were then stained with 1% uranyl acetate (40 µl) and wicked dry. All images were acquired at an accelerating voltage of 100 kV. Gatan Digital Micrograph software was used to analyze the images.
Size exclusion chromatography
Ten µl of the samples was loaded onto a PBS pre-equilibrated Sepharose CL 2B column (1 × 10 cm). Column was eluted with PBS at a flow rate around 0.5 ml/min. Elute fractions (200–300 µl) were collected, diluted 1:1 in ethanol, and analyzed for fluorescence intensity by using a plate reader (λex: 485 nm, λem: 535 nm) (PLATE CHAMELEON Multilabel Detection Platform, Bioscan Inc., Washington, DC).
Isolated liver perfusion study
C57BL/6 mice were sacrificed and the interior vena cava was incised to allow the blood flush out when 3 ml warm PBS was infused into the mouse liver through the portal vein. cy3-siRNA containing LPD formulations (300 µl) were incubated with 50 µl mouse serum at 37°C for 10 min, and then diluted with PBS ( final volume = 1 ml). The complex was infused into the isolated liver via the portal vein. Finally, the liver was perfused with 3 ml warm PBS, excised, fixed in 3.6% paraformaldehyde in PBS for overnight, and frozen sectioned (5 µm in thickness). Sections were washed with PBS, permeabilized with 0.1% Triton X-100 in PBS, stained with Alexa Fluor® 488 Phalloidin (Invitrogen, Eugene, OR), mounted with the DAPI containing medium (Vectashield®, Vector Laboratories Inc., Burlingame, CA) and imaged using a Leica SP2 confocal microscope.
Tissue distribution study
Tissue distribution of the FAM-labeled siRNA formulation in the PEGylated LPD was analyzed 4 h after i.v. injection as previously described [7].
Statistical analysis
Data are presented as the mean ± SD. The statistical significance was determined by using the analysis of variance (ANOVA, one way) or the two-sided student t-test. P values of <0.05 were considered to be significant.
Results and Discussion
Characterization of the nanoparticles after PEGylation
PEG-phospholipids have been used widely in lipid based nanoparticle formulations, such as liposomes, to avoid the non-specific uptake by RES and increase the circulation half-life [13]. It is also known that incorporation of too much PEG-phospholipids will disrupt the integrity of the lipid membrane due to its detergent-like properties [14], which increases membrane permeability and premature drug release. Since the unmodified LPD contains two lipid bilayer membranes, incubation with PEG-phospholipids may strip off the membranes from the nanoparticles and form micelles of smaller particle size. Here, we examined the stability of nanoparticle formulations after the addition of different amounts of DSPE-PEG2000 by measuring their particle size distribution. Two cationic lipids, a monovalent lipid DOTAP and a trivalent lipid DSGLA (figure 2A), were used in the study. By using DLS, only one narrow size distribution around 100 nm was revealed for all four formulations (polydispersity index <0.1), i.e., liposomes and LPD composed of either DOTAP or DSGLA, before the addition of DSPE-PEG2000 (figure 2B). However, after the addition of DSPE-PEG2000, a population of smaller particles appeared in a dose dependent manner for both DOTAP and DSGLA liposomes. It is noted that pure DSPE-PEG2000 micelles was undetectable at the concentrations used in this experiment. Thus, the smaller size particles must come off from the nanoparticles upon the introduction of DSPE-PEG2000. The LPD formulations showed significantly higher stability compared to the liposomes. When 10 mol% of DSPE-PEG2000 was added, the LPD formulations remained relatively stable, whereas the liposome formulations showed a significant increase in the population of small particles (∼5%). The DLS data (figure 2B) is consistent with the observation by TEM (figure 3), in which a mixture of disrupted particles and tubular micelles was found in the DOTAP liposomes after PEGylation. On the other hand, intact nanoparticles around 100 nm in diameter were still found in the PEGylated LPD formulation containing DOTAP (figure 3). It is also noticed that DSGLA (containing 3 positive charges)-LPD showed an improved stability compared to DOTAP (containing 1 positive charge)-LPD (figure 2B), suggesting that LPD was stabilized by charge-charge interaction between the cationic lipid and the negatively charged core. DLS was determined to be a convenient method to assess the relative stability of the particles. For example, smaller size particles were found in the TEM photographs of the PEGylated LPD (figure 3, arrow heads); while DLS data showed no presence of smaller particles (figure 2B). It is known that particles of larger sizes showed significant higher light scattering compared to smaller size particles at the same concentration. Nevertheless, the dynamic light scattering data provided a quantitative comparison of the relative stability of different nanoparticle formulations.
Figure 2.
Stability of the nanoparticle formulations upon different degree of PEGylation. (A) Chemical structures of DOTAP and DSGLA. (B) Size distribution of different PEGylated formulations. Data = mean ± SD, n = 4–6
Figure 3.
TEM photographs of liposomes/PEGylated liposomes and LPD/PEGylated LPD. Arrows indicate the “sprouts” of the particles and arrow heads indicate the small particles. Bar = 100 nm.
As can be seen in figure 3, after 10 mol% PEGylation, DOTAP liposomes were greatly disrupted and most of them were transformed into tubular micellar structures, indicating the instability of the formulation. In the PEGylated LPD, a mixture of particles around 10 nm (figure 3, arrow head), “sprouts” on the 100 nm-particles (figure 3, arrows), and 100 nm-particles was found, suggesting that the lipids on the surface of the LPD (∼100 nm) were being stripped off (forming sprouts) and eventually became smaller DSPE-PEG2000 containing particles. The possibility of presence of pure DSPE-PEG micelles in the PEGylated LPD sample cannot be ruled out, since the DSPE-PEG2000 micelles showed similar size (TEM data not shown) as the smaller particles (figure 3, arrow head).
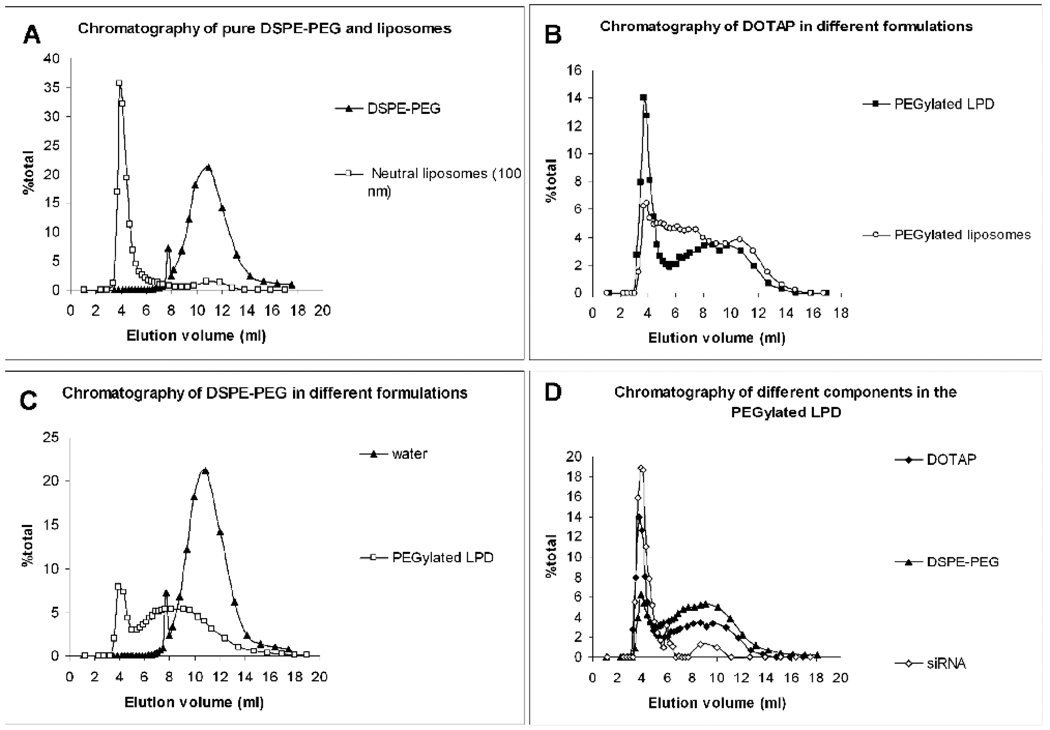
To further characterize the LPD nanoparticles, we studied in more details the nanoparticle formulation containing DOTAP after incubating with 10 mol % DSPE-PEG2000. We have used either 10 mol% NBD-DOTAP labeled liposomes, 10 mol% DSPE-PEG2000-carboxyfluorescein (DSPE-PEG2000-CF) labeled DSPE-PEG2000 or FAM-siRNA to prepare the PEGylated LPD. A Sepharose CL 2B column was used to separate particles. In this study, a neutral liposome formulation (DOPC/Cholesterol/NBD-PE = 49/49/2, molar ratio, mean particle size around 100 nm) was used to calibrate the column. Cationic liposomes and unmodified LPD containing excess positive surface charges formed aggregates in the elution medium (phosphate-buffered saline, PBS) and thus, could not be studied by the chromatography. As shown in figure 4A, pure DSPE-PEG2000 could be clearly separated from the 100 nm-nanoparticles by the size exclusion column chromatography. At least two particle populations were observed in the NBD-DOTAP labeled PEGylated LPD (figure 4B). The first major peak coincided with that of the 100 nm-nanoparticles, and the second peak appeared between the peaks of micelles and 100 nm-nanoparticles. The PEGylated liposomes (NBD-DOTAP labeled) showed less significant first peak but a smear of particle size distribution, suggesting the lipid membrane was disrupted in this formulation. The observation is consistent with DLS (figure 2B) and TEM data (figure 3). Moreover, the area under the curve (AUC) of the first peak of the PEGylated LPD (NBD-DOTAP labeled) was about 37.2% (37.2 ± 4.6%, n = 3). The data suggest that approximately 37.2% of the total lipids were still associated with the nanoparticles, while the rest of the lipids were stripped off by the DSPE-PEG2000 and formed smaller particles (< 100 nm). From the cryoTEM data (figure 2a), it can be calculated that approximately 36.4% (36.4 ± 3.2%, n = 5) of the total lipids should be located in the inner lipid bilayer of the LPD, suggesting that only the inner lipid bilayer remained with the nanoparticles. The inner cationic lipid bilayer was in direct contact with the negatively charged surface of the nucleic acid/protamine core and therefore, could accommodate an increased amount of DSPE-PEG2000 compared to the outer bilayer. It is noted that further increase of the input DSPE-PEG2000 to 20 mol% caused damage of the supported bilayer. Only 24.2% of the total cationic lipids remained associated with the nanoparticles (data not shown), indicating that substantial amount of the lipids in the supported bilayer was removed in this high concentration of DSPE-PEG2000. Figure 4C shows that around 20.6% (20.6 ± 3.4%, n = 3) of the input DSPE-PEG2000 (DSPE-PEG2000-CF labeled) was incorporated with the nanoparticles (first peak) after incubation. Figure 4D indicates that the nanoparticles eluted in the first peak contained 90% of siRNA, 37.2% of the total lipids and 20.2% of the input DSPE-PEG2000. These nanoparticles should be the major contributor for siRNA delivery for the PEGylated LPD formulation.
Figure 4.
Size exclusion chromatography of different samples. (A) chromatography of neutral liposomes and pure DSPE-PEG2000 ; (B) chromatography of DOTAP in different formulations; (C) chromatography of DSPE-PEG2000 in different formulations; (D) chromatography of different components in the PEGylated LPD. Data are representative chromatography from 2–3 batches of formulations.
Ten µl of the PEGylated LPD (loading amount for column) contained 0.71 µg siRNA, 36.71 nmol lipids and 3.99 nmol DSPE-PEG2000. The nanoparticles eluted in the first peak contained 0.64 µg siRNA, 13.66 nmol lipids and 0.81 nmol DSPE-PEG2000. Assuming that DSPE-PEG2000 was inserted only into the outer leaflet of the supported bilayer and the lipid content in the outer and inner leaflet of the bilayer were the same, we calculated that 10.6 mol% of the outer leaflet was modified with DSPE-PEG2000. This led to a complete charge shielding. The zeta potential of the unmodified LPD was about 40 mV, but the purified PEGylated LPD was −5.6 ± 4.5 mV. Approximately 90% siRNA was encapsulated in the purified PEGylated LPD, which was a neutral delivery vehicle that is desirable for in vivo drug delivery because of its improved pharmacokinetics and reduced non-specific interaction with cells or serum protein.
Depending on the surface density and molecular weight of the PEG grafted to the lipid bilayer, three PEG conformations can be identified [15]. Factors controlling the PEG conformation include the distance between the PEG chains in the lipid bilayer (D) and the Flory dimension, RF, which is defined as aN3/5 (a is the persistence length of the monomer, N is the number of monomer units in the PEG) [16]. Three regimes can be defined: (a) when D>2 RF (interdigitated mushrooms); (b) when D<2 RF (mushrooms); and (c) when D< RF (brushes) [16]. For a 100 nm-liposomal particle grafted with DSPE-PEG2000, PEG chains should be arranged in the mushroom mode in the presence of <4 mol% DSPE-PEG2000; in the transition mode with a 4–8 mol% modification; and in the brush mode with >8 mol% PEGylation [17]. The brush configuration ensures that the entire surface of nanoparticles is covered [18] and provides the nanoparticles with full protection from opsonization. It is difficult to prepare stable PEGylated liposomes with the brush conformation of PEG while maintaining the integrity of lipid membrane. When PEG is arranged in the brush mode, the repulsion force among PEGs may cause disruption of the lipid bilayer, as what have been shown in this study. On the other hand, the LPD nanoparticles contained two lipid bilayers; the regular outer layer was stripped off by the DSPE-PEG2000, while the supported bilayer that was stabilized by charge-charge interaction remained intact after accommodated 10.6 mol% DSPE-PEG2000 with the PEG arranged in the brush conformation.
Applying another force to antagonize the repulsion among the PEGs; i.e., charge-charge interaction from the supported bilayer of LPD, is the key to maintain a high mol% of PEG arranged in the brush mode. Hu and Wu reported another method to prepare highly PEGylated particles with PEG in the brush configuration [19, 20]. They synthesized a thermally sensitive polymer (PNIPAM (poly[N-isopropylacrylamide]) grafted with linear PEG chains) that self-assembled into microgel at 25ºC, followed by increasing the temperature to ∼35ºC, which shrank the particle size by 3-fold and reduced the surface area by 10-fold and thus the PEGs were forced to settle in the brush mode. In this case, the force from the hydrophobic interaction between the polymers stabilized the PEG conformation [20].
Uptake of the LPD nanoparticles in the isolated liver and the tissue distribution after i.v. administration
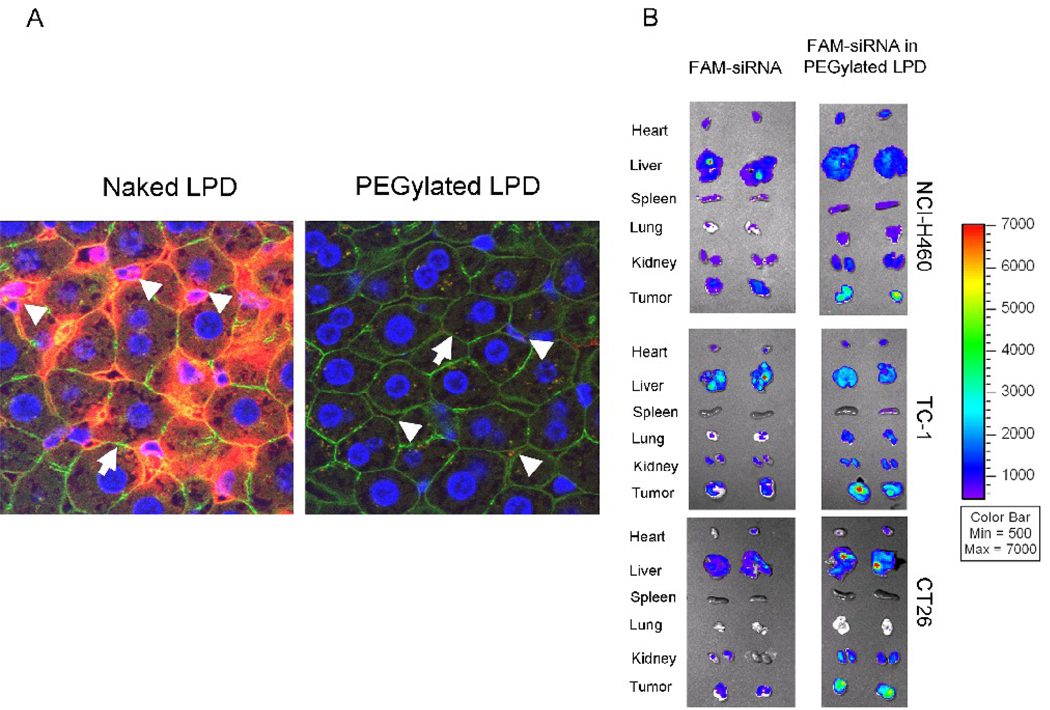
To further investigate if the PEGylated LPD showed low RES uptake in the mouse liver, we performed a liver perfusion assay. Cy3 labeled siRNA was formulated in the PEGylated LPD containing DOTAP and mixed with the mouse serum for 10 min before infused into the liver. After the infusion, the liver was perfused with warm PBS and excised, fixed, sectioned, and examined by confocal microscopy. As shown in figure 5A, the nuclei of the liver tissue were stained as blue, the F-actin outlining the cell morphology was stained with Alexa Fluor® 488 Phalloidin as green, and the siRNA was labeled with cy3 and shown as red. Thus, the sinus where the Kupffer cells reside and hepatocytes can be distinguished by their distinct morphologies as indicated by the arrow (hepatocyte) and arrow head (sinus). The PEGylated LPD showed little sinusoidal uptake in the liver, while the positive control, unmodified LPD that have been demonstrated to accumulate in the RES [5], displayed intense liver sinusoidal uptake. The data indicate that the PEG on the LPD surface arranged in the brush mode prevented serum opsonization and abolished the non-specific RES uptake in the isolated liver. Since the RES uptake in the liver and spleen often contributes to the major loss of injected nanoparticles, the data also suggest that the fully protected LPD of low RES uptake might have improved chance to reach the tumor via the enhanced permeability and retention (EPR) effect [21].
Figure 5.
Tissue distribution analysis. (A) Liver sinusoidal uptake of cy3-siRNA in naked LPD (positive control) and PEGylated LPD. Nuclei (blue), F-actin (green), cy3-siRNA (red). Magnification = 1,600 x. Data are representative pictures from 3 mice in each group. (B) Fluorescence signal of FAM-labeled siRNA in different tissues detected by the Xenogen IVIS imaging system. Data are from two representative animals in each group. Data of the NCI-H460 model are reproduced from a previously published article with permission [7].
Then, we examined the tissue distribution of FAM-labeled siRNA formulated in PBS or in the PEGylated LPD containing DOTAP in 3 different tumor models (NCI-H460 in nude mice, TC-1 in C57BL/6 mice, and CT26 in Balb/c mice). Four h after the i.v. injection, we euthanized the mice, collected tissues and used the Xenogen IVIS imaging system to examine the FAM-siRNA signals. As can be seen in figure 5B, free FAM-siRNA exhibited little bioavailability for the major tissues from all 3 models except the liver of the C57BL/6 mice. PEGylated LPD, however, showed very high delivery efficiency for FAM-siRNA to the tumors, while leaving other normal tissues with minimal to moderate uptake, particularly in the NCI-H460 model. We have also extracted the accumulated FAM-siRNA from the tissues in the NCI-H460 model and measured the amount of the fluorescent siRNA. The accumulated doses of free FAM-siRNA were 5.0 ± 2.2% injected dose (ID) in the tumor, 0.8 ± 1.4% in the heart, 1.5 ± 0.5% in the lung, 0.8 ± 1.6% in the kidney and 21.8 ± 5.1% in the liver, respectively (data = mean ± SD, n = 3–4). PEGylated LPD delivered 32.5 ± 11.3% ID to the tumor, 3.9 ± 2.1% to the lung and 12.0 ± 5.7% to the liver, respectively, while the levels of FAM-siRNA were under the detection limit in the rest of the tissues (data = mean ± SD, n = 3–4). The data showed that the PEGylated LPD displayed efficient delivery of siRNA to the tumor with low uptake by the RES in the liver and the spleen. The tumor uptake, which greatly exceeded the liver uptake, had become the major clearance mechanism of the nanoparticles from the circulation [7]. Tumor uptake occurred very early after i.v. administration. In fact, when the tissues were examined 0.5 h after injection of the PEGylated LPD, 13.8 ± 2.3% ID was already accumulated in the tumor, leaving the liver with only 4.0 ± 2.3% and the spleen with 1.3 ± 0.3% (data = mean ± SD, n = 4). The initial low uptake by the RES was likely due to the low level of opsonization of the PEGylated LPD resulting from the PEG brush on the particle surface. It should be noted that the neovasculature of the tumor is significantly leakier than that of the liver sinusoidal membrane [22, 23] which provides a significant opportunity for the nanoparticles to extravasate into the tumor. In the tumor free animals, the nanoparticles circulated for a longer period of time, but were still eventually taken up by the liver just as all “stealth liposomes” [7].
Proposed model for the formation of PEGylated LPD nanoparticles and the mechanism of avoiding RES uptake
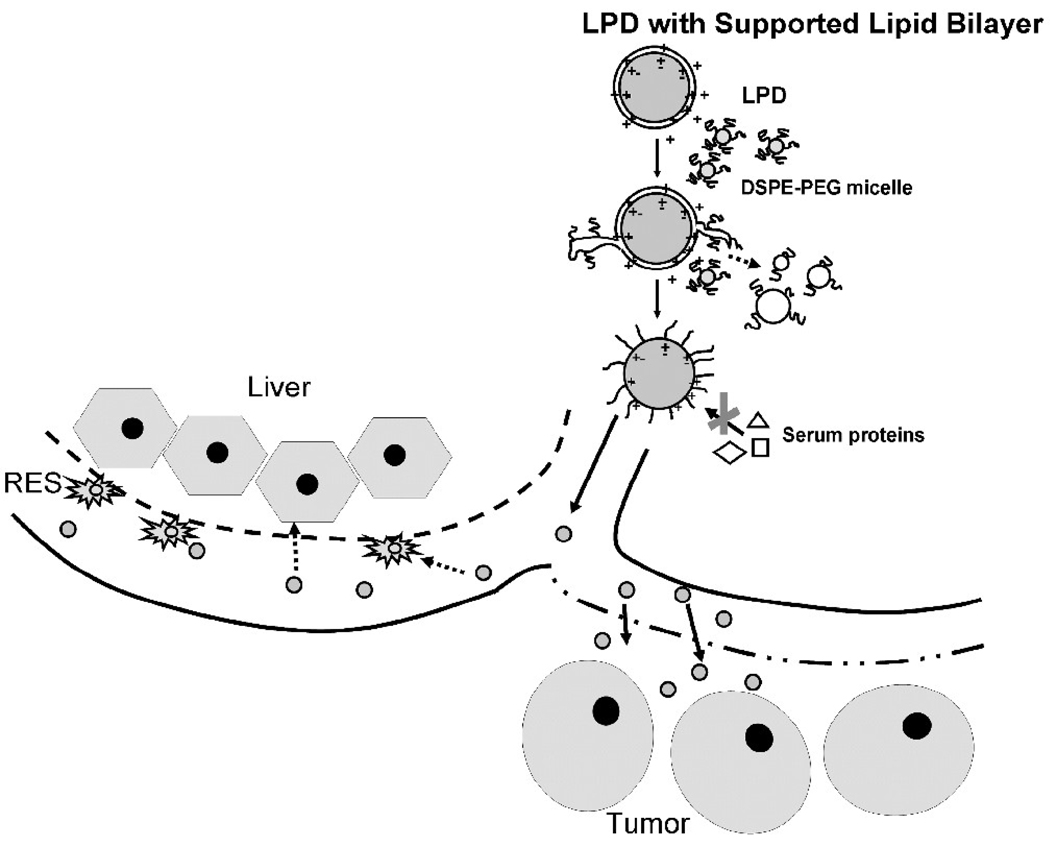
After the addition of DSPE-PEG2000 micelles at 50°C, the micelles act like detergents and strip off the outer lipid bilayer of the LPD (figure 6). In the intermediate phase, “sprouts” (outer bilayer being stripped off from the LPD) are formed and eventually break down into smaller PEGylated lipid particles. The inner lipid bilayer is stabilized by charge-charge interaction and therefore, remains intact. A brush layer of PEG is present on the surface of the nanoparticles due to the insertion of a large amount of DSPE-PEG2000, which completely shields the surface charge of the unmodified LPD. The well protected nanoparticles with presumably reduced opsonization can thus avoid the initial clearance by the RES and are favored for penetration into the tumor through its leaky neovasculature. We have thus demonstrated the role of the supported bilayer in improving the in vivo stability and tumor delivery of the PEGylated LPD.
Figure 6.
Proposed model for the formation of PEGylated LPD and the mechanism of favored tumor uptake.
Conclusion
Using a variety of analytical techniques, we have demonstrated that the LPD nanoparticles contained two lipid bilayers with the inner bilayer stabilized by charge-charge interaction. During the process of post-insertion of PEGylated lipids, the regular outer bilayer was stripped off from the particles due to the detergent-like effect and the PEGylated lipids were incorporated into the outer layer of the supported bilayer with increased stability, which accommodated up to 10.6 mol% of DSPE-PEG. The highly PEGylated nanoparticles with the PEG arranged in the brush mode displayed neutral surface charge and minimal protein binding when mixed with serum, resulting in little RES uptake; and thus showed high tumor accumulation. Here, we have shown the importance of incorporating a stabilizing mechanism (i.e. supported bilayer) in a nanoparticle formulation that allows an increased level of PEGylation to further decrease the opsonization effect and RES uptake, leading to much improved tumor delivery.
Acknowledgement
We thank Dr. Reddy Bathula (UNC) for synthesizing the DSGLA lipid and Drs. M. Joseph Costello (UNC), Christine C Conwell (UNC) and Sumio Chono (UNC) for their assistance on the TEM. Drs. Russell Mumper (UNC) and Moo J. Cho (UNC) are greatly appreciated for their valuable comments on this project. We also thank the Department of Histopathology (UNC) and the Michael Hooker Microscopy Center (UNC) for the preparation of tissue sections and the assistance on the microscopy imaging. Qi Angela Yang and Yun-Ching Chen are acknowledged for their help in preparing the manuscript.
Abbreviations
- LPD
liposome-polycation-DNA
- DLS
dynamic light scattering
- DSPE-PEG2000
1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[( polyethylene glycol)-2000]
- DSGLA
N,N-Distearyl-N-methyl-N-2[N’-(N2-guanidino-L-lysinyl)] aminoethyl ammonium chloride
- DOTAP
1,2 -dioleoyl-3-trimethylammonium-propane
- PEG
polyethylene glycol
- RES
reticuloendothelial system
- TEM
transmission electron microscopy
- NBD-DOTAP
1-oleoyl-2-[6-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino] hexanoyl]-3-trimethylammonium propane
- DSPE-PEG2000-CF
1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[poly(ethylene glycol)2000-N'-carboxyfluorescein]
- FAM
fluorescein
- DOPC
1, 2-dioleoyl-sn-Glycero-3-Phosphocholine
- NBD-PE
1-Oleoyl-2-[12-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]dodecanoyl]-sn-Glycero-3-Phosphoethanolamine
- PBS
phosphate-buffered saline
- AUC
area under the curve
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Klibanov AL, Maruyama K, Torchilin VP, Huang L. Amphipathic polyethyleneglycols effectively prolong the circulation time of liposomes. FEBS Lett. 1990;268:235–237. doi: 10.1016/0014-5793(90)81016-h. [DOI] [PubMed] [Google Scholar]
- 2.Woodle MC, Lasic DD. Sterically stabilized liposomes. Biochim Biophys Acta. 1992;1113:171–199. doi: 10.1016/0304-4157(92)90038-c. [DOI] [PubMed] [Google Scholar]
- 3.Uster PS, Allen TM, Daniel BE, Mendez CJ, Newman MS, Zhu GZ. Insertion of poly(ethylene glycol) derivatized phospholipid into pre-formed liposomes results in prolonged in vivo circulation time. FEBS Lett. 1996;386:243–246. doi: 10.1016/0014-5793(96)00452-8. [DOI] [PubMed] [Google Scholar]
- 4.Silvander M, Johnsson M, Edwards K. Effects of PEG-lipids on permeability of phosphatidylcholine/cholesterol liposomes in buffer and in human serum. Chem Phys Lipids. 1998;97:15–26. doi: 10.1016/s0009-3084(98)00088-7. [DOI] [PubMed] [Google Scholar]
- 5.Li S, Huang L. In vivo gene transfer via intravenous administration of cationic lipid-protamine-DNA (LPD) complexes. Gene Ther. 1997;4:891–900. doi: 10.1038/sj.gt.3300482. [DOI] [PubMed] [Google Scholar]
- 6.Vangasseri DP, Han SJ, Huang L. Lipid-protamine-DNA-mediated antigen delivery. Curr Drug Deliv. 2005;2:401–406. doi: 10.2174/156720105774370168. [DOI] [PubMed] [Google Scholar]
- 7.Li SD, Chen YC, Hackett MJ, Huang L. Tumor-targeted Delivery of siRNA by Self-assembled Nanoparticles. Mol Ther. 2008;20:163–169. doi: 10.1038/sj.mt.6300323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li SD, Chono S, Huang L. Efficient gene silencing in metastatic tumor by siRNA formulated in surface-modified nanoparticles. J Control Release. 2008;126:77–84. doi: 10.1016/j.jconrel.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li SD, Chono S, Huang L. Efficient oncogene silencing and metastasis inhibition via systemic delivery of siRNA. Mol Ther. 2008;16:942–946. doi: 10.1038/mt.2008.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li SD, Huang L. Targeted delivery of antisense oligodeoxynucleotide and small interference RNA into lung cancer cells. Mol Pharm. 2006;3:579–588. doi: 10.1021/mp060039w. [DOI] [PubMed] [Google Scholar]
- 11.Tan Y, Whitmore M, Li S, Frederik P, Huang L. LPD nanoparticles--novel nonviral vector for efficient gene delivery. Methods Mol Med. 2002;69:73–81. doi: 10.1385/1-59259-141-8:073. [DOI] [PubMed] [Google Scholar]
- 12.Chen Y, Sen J, Bathula SR, Yang Q, Fittipaldi R, Huang L. Novel Cationic Lipid That Delivers siRNA and Enhances Therapeutic Effect in Lung Cancer Cells. Mol Pharm. 2009 doi: 10.1021/mp800136v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan X, Scherphof GL, Kamps JA. Liposome opsonization. J Liposome Res. 2005;15:109–139. doi: 10.1081/lpr-64971. [DOI] [PubMed] [Google Scholar]
- 14.Dos Santos N, Allen C, Doppen AM, Anantha M, Cox KA, Gallagher RC, Karlsson G, Edwards K, Kenner G, Samuels L, Webb MS, Bally MB. Influence of poly(ethylene glycol) grafting density and polymer length on liposomes: relating plasma circulation lifetimes to protein binding. Biochim Biophys Acta. 2007;1768:1367–1377. doi: 10.1016/j.bbamem.2006.12.013. [DOI] [PubMed] [Google Scholar]
- 15.Kenworthy AK, Hristova K, Needham D, McIntosh TJ. Range and magnitude of the steric pressure between bilayers containing phospholipids with covalently attached poly(ethylene glycol) Biophys J. 1995;68:1921–1936. doi: 10.1016/S0006-3495(95)80369-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nicholas AR, Scott MJ, Kennedy NI, Jones MN. Effect of grafted polyethylene glycol (PEG) on the size, encapsulation efficiency and permeability of vesicles. Biochim Biophys Acta. 2000;1463:167–178. doi: 10.1016/s0005-2736(99)00192-3. [DOI] [PubMed] [Google Scholar]
- 17.Garbuzenko O, Barenholz Y, Priev A. Effect of grafted PEG on liposome size and on compressibility and packing of lipid bilayer. Chem Phys Lipids. 2005;135:117–129. doi: 10.1016/j.chemphyslip.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 18.Owens DE, 3rd, Peppas NA. Opsonization, biodistribution, and pharmacokinetics of polymeric nanoparticles. Int J Pharm. 2006;307:93–102. doi: 10.1016/j.ijpharm.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 19.Hu T, Wu C. Clustering induced collapse of a polymer brush. Physical Review Letters. 1999;83:4105–4107. [Google Scholar]
- 20.Hu T, Wu C. Grafting density induced stretching and collapse of tethered poly(ethylene oxide) chains on a thermally sensitive microgel Macromolecules. 2001;34:6802–6805. [Google Scholar]
- 21.Brannon-Peppas L, Blanchette JO. Nanoparticle and targeted systems for cancer therapy. Adv Drug Deliv Rev. 2004;56:1649–1659. doi: 10.1016/j.addr.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 22.Braet F, Wisse E. Structural and functional aspects of liver sinusoidal endothelial cell fenestrae: a review. Comp Hepatol. 2002;1:1. doi: 10.1186/1476-5926-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monsky WL, Fukumura D, Gohongi T, Ancukiewcz M, Weich HA, Torchilin VP, Yuan F, Jain RK. Augmentation of transvascular transport of macromolecules and nanoparticles in tumors using vascular endothelial growth factor. Cancer Res. 1999;59:4129–4135. [PubMed] [Google Scholar]