Abstract
When monoclonal ANAs and non-ANAs generated from a genetically simplified mouse model of lupus, B6.Sle1, were recently compared, the ANAs exhibited 3 sequence motifs in their immunoglobulin heavy chains, including increased cationicity in CDR3 (“motif A”), reduced anionicity in CDR2 (“motif B”) and increased aspartate at H50 (“motif C”). The present study was designed to elucidate the extent to which these ANA-associated sequence motifs might be hard-wired into the primary B-cell repertoire in lupus. The immunoglobulin heavy chain sequence of total splenic B cells, follicular B-cells and marginal zone B-cells from B6.Sle1 congenic mice and C57BL/6 controls were amplified by single cell PCR and compared. Analysis of the primary immunoglobulin heavy chain repertoire indicated that the first 2 sequence motifs “A” and “B” were already encoded in the naïve repertoire of B6.Sle1z mice, whereas the third motif “C” was introduced in part by somatic mutation. Site-directed mutagenesis confirmed that non-anionic CDR2 and cationic CDR3 residues in the immunoglobulin heavy chain facilitated nuclear antigen binding in concert, whereas aspartate at H50 strongly vetoed DNA-binding, while preserving nucleosome reactivity. Hence, antinuclear antibodies appear to arise as a consequence of 2 distinct processes–genetically programmed selection of specific CDR charge motifs into the primary immunoglobulin repertoire, with secondary contribution from somatic mutation. Polymorphisms in the lupus susceptibility gene Ly108 that impair central B-cell tolerance may be mechanistically responsible for these early repertoire differences in lupus.
Keywords: Autoimmunity, B-cells, autoantibodies, genetics, immunoglobulin repertoire
Introduction
Antinuclear antibodies (ANAs) constitute an important hallmark of systemic lupus erythematosus, as extensively reviewed (1-3), though autoantibody-independent mechanisms leading to lupus nephritis have also been described (4-8). Sequence comparisons of ANAs against non-nuclear antigen reactive Abs have highlighted several interesting molecular features, particularly in the immunoglobulin (Ig) heavy chains (HC), including the prominence of “R” residues in the CDR3 regions (8-12). The importance of these residues in facilitating DNA-reactivity has been unequivocally demonstrated through site-directed mutagenesis (13-16). The evidence for distinct molecular signatures that distinguish the CDR2 regions of ANA HCs from those of non-ANAs has been less convincing. Nevertheless, sequence comparison studies and site-directed mutagenesis has helped demonstrate the potential importance of polarity at selected CDR2 positions in conferring or enhancing DNA-reactivity (12, 14, 16, 17). In contrast to the HC, the light chains (LCs) of ANAs possess few molecular “signatures” that consistently light up across different data sets (9, 18), though LCs have proven to be important in defining specificity in BCR HC Tg model (19-22).
The above findings were reproduced and extended in a genetically simplified mouse model of lupus-B6 mice rendered congenic for the NZM2410-derived lupus susceptibility interval, Sle1z (23, 24). Whereas B6 mice do not exhibit anti-nuclear autoantibodies, B6.Sle1z congenics (which are on the same B6 genetic background) exhibit high titers of anti-nuclear autoantibodies, with preferential binding to nucleosomes and DNA/histone complexes (23, 24). When the Ig HC sequences of ANAs and non-ANA mAbs derived from this strain were compared, 3 distinct sequence motifs emerged, including increased cationic residues in CDR3 (termed ‘motif A’), reduced anionic residues in CDR2 H52-H56 (termed ‘motif B’) and increased ‘D’ residues at H50 in CDR2 (termed ‘motif C’), as recently reported (25). Importantly, the presences of all 3 motifs within the same HC sequence increased the likelihood of the Ab being nuclear-antigen reactive by ~4 fold, with an odds ratio of 5 (25). In contrast, no significant differences were noted in the Ig LC repertoire between ANAs and non-ANAs drawn from these mice (25).
It was of particular interest to note that in the above hybridoma study, several of the ANA-associated HC sequence motifs were already germline-encoded by a subset of HC genes (as opposed to being introduced though somatic mutation). This observation suggested that the ANA-associated sequence motifs may already be imprinted in the primary B-cell repertoire in lupus, presumably as a result of early tolerance deficits. To test this hypothesis, in the present report, the primary Ig repertoire of B6.Sle1z congenics and B6 healthy controls were elucidated using single cell PCR amplification, using well documented approaches, as described (26-30). Through this approach, we trace the origins of the 3 key sequence motifs that characterize anti-nuclear antibodies. Viewed in the context of our previous mechanistic studies (31), it appears that culprit genes within the Sle1z lupus susceptibility interval, notably Ly108z, may be infringing early checkpoints in B-cell tolerance, resulting in a primary Ig repertoire that is already nuclear antigen skewed to begin with. Taken together with Ig repertoire monitoring studies in human SLE (32, 33), these findings underscore the importance of early tolerance deficits in shaping the anti-nuclear autoantibody repertoire in lupus.
Materials and Methods
Mice
B6.Sle1z are C57BL/6 (B6) mice rendered congenic homozygotes for NZM2410-derived Sle1z, a 37 centimorgan lupus susceptibility interval on chromosome 1, with termini at D1MIT101 and D1MI T155 (23). These mice are strongly seropositive for anti-chromatin and anti-histone/DNA Abs, but weakly positive for anti-dsDNA Abs (24), while the B6 controls were seronegative for these specificities. Mice used for studies were 6-9 mo old males and females, housed in a specific pathogen free colony at UT Southwestern Medical Center Department of Animal Resources, Dallas, TX.
Single cell PCR analysis
Single cell sorting was performed using a FACStar Plus machine with an automatic cell deposition unit (Becton Dickinson, Mountain View, CA). Calibrator beads were used to confirm the single-cell sorting efficiency of the machine. Splenic B220+ve, IgM+ve B-cells (i.e., total B-cells), B220+ve, IgM+ve, CD23+ve follicular B-cells, as well as IgM+ve, CD21+ve, CD23-ve marginal zone (MZ) B-cells were directly single-cell sorted into 96-well plates (Costar, Cambridge, MA), containing 19 ul of 1X PCR buffer (Promega, Madison, WI) and 1 ul of proteinase K (4 mg/ml, Sigma Chemicals, St. Louis, MO) per well. Care was taken to exclude T1 and T2 transitional B-cells, based on their expressions levels of CD21 and CD23 in all studies, and AA4.1 in some studies.
Single-cells were digested for 1 h at 55°C with proteinase K, which was subsequently inactivated for 10 minutes at 95°C. PCR amplification of Ig HC DNA was carried out in two rounds, following published protocols (26, 27, 30). Briefly, the first round of PCR was carried out over 40 cycles using a 5’ VH framework 1 primer (AGG T(C/G)(A/C) A(A/G)C TGC AG(C/G) AGT C(A/T)G G), and a 3’ primer specific for a sequence that lies 3’ of JH4 (GGG TCT AGA CTC TCG GCC GGC TCC CTC AGG), in a total reaction volume of 30 ul, using the following parameters: 60 s at 95°C, 60 s at 58°C, and 150 s at 72°C. One ul of the first round PCR product was used for a second round of PCR, using (AGG T(C/G)(A/C) A(A/G)C TGC AG(C/G) AGT C(A/T)G G) and (G(A/G)(G/C) CTG AGC T(G/T)G TGA (A/G)GC CTG G) as 5’ VH framework 1 primers, and four different 3’ JH-specific primers: JH1 (GAC GGT GAC CGT GGT CCC TGT), JH2 (GAC TGT GAG AGT GGT GCC TTG), JH3 (GAC AGT GAC CAG AGT CCC TTG), and JH4 (GAC GGT GAC TGA GGT TCC TTG), in 20 ul volume, over 35 cycles (60 s at 95°C, 60 s at 67°C, and 90 s at 72°C). PCR bands were excised and purified using Qiaquick Spin PCR purification kit (Qiagen, Chatsworth, CA), and direct-sequenced. The single-cell HC amplification efficiency was similar in the two strains studied, attaining 70-90% efficiency. Ambiguous sequences were re-sequenced. The Taq error rate was estimated to be about 1 in 4000 nucleotides, based on multiple rounds of sequencing of the Cu gene.
Antibody sequence analysis
Sequences were aligned using OMIGA 3.0 (Oxford Molecular, Oxford, UK), blasted against public databases of mouse Ig sequences (http://www.ncbi.nlm.nih.gov/igblast), assigned to their respective germline origins as described (8, 28, 29), and deposited into Genbank (accession numbers AY436820-AY436914). The control databases of non-ANA sequences described in this study represent recently assembled collections of non-ANA HC and LC sequences drawn from the Genbank (18, 34). Importantly, these abridged databases had no clonal replicates, and no 2 Abs shared the same antigen specificity. For the statistical comparisons, multi-member clones were represented by one member each so as to minimize the impact of clonal bias. Specifically, the single clone selected was the most mutated clone, so that as much of the mutational information was captured. The respective frequencies of VH and Vk gene usage, as well as the frequencies of individual amino acid residues at the different CDR positions were compared between the ANAs and control Abs using Chi square tests (with Yates correction, where appropriate), or Fisher's exact test. The Student's t-test was used for comparing group means. All statistical comparisons were performed using SigmaStat (Jandel Scientific). “Relative risk” and “odds ratio” were calculated as detailed elsewhere (http://www.childrensmercy.org/stats/definitions/or.htm).
Expression cloning and site directed mutagenesis
Ig HC and LC sequences were amplified from the cDNA of the B6.Sle1z hybridoma IDG5, and cloned into a human IgG1 and IgK expression vector (kind gift from Dr. Michel Nussenzweig, Rockefeller University, New York), using primers with the appropriate restriction sites: Age1VH: CTG CAA CCG GTG TAC ATT CCG AGG TGA AGC TGC AGC AGT CAG G and SalJH2: TGC GAA GTC GAC GCT GAG GAG ACT GTG AGA GTG GTG CC (for Ig HC), Age919: CTG CAA CCG GTG TAC ATT CTG AHA TCC AGA TGA TGA CHC AGW CT and BSJK2: GCC ACC GTA CGA CGT TTT ATT TCC AGC TTG G for Ig Vk LC. Ig HC was amplified with 0.5 ul cDNA in 20 ul reaction volume with 2 mM MgCl2 (60s at 95°C, 60s at 65°C, and 90s at 72°C). Ig kappa LC was likewise amplified using the following conditions: 60s at 95°C, 60s at 60°C, and 90s at 72°C. Site-directed mutagenesis was performed using appropriate oligonucleotide primers.
For example, the following oligonucleotides were used for PCR-based mutagenesis of Asp (D50) to Gly: 50GAS (5’-TCCAGGATAAATACCTCCAATCCACTCAAG-3’) and 50GS (5’-GAGTGGA TTGGAG GTATTTATCCTGGAAG -3’); Asp (D50) to Asn: 50NAS (5’-TCCAGGA TAAATA TTTCCAATCCAC TCAAG-3’) and 50NS (5’- GAGTGGATTGGAAATATTTATCCT GGAAGT-3’). Likewise, for the reverse mutagenesis of Gly (G97) to Arg, oligonucleotides 97RAS (5’-GTTTCTCCATATC CTCTTCGATC TTGCACA-3’) and 97RS (5’-GCAAGATCGAA GAG GATATGGAGAAACTCC-3’) were used. All induced mutations were confirmed by sequencing to ensure that the rest of the Ig sequence was unaltered. Mutated Abs were co-transfected into human embryonic kidney fibroblast 293A cells using FuGENE6 (Roche, Mannheim, Germany). Supernatants collected 7 days post-culture were purified using protein G Sepharose 4 Fast Flow (GE Healthcare, USA), neutralized with 1M Tris/Cl (pH=8), and tested for specificity by ELISA, using approaches described previously (7, 8).
Results
ANA-associated Ig HC sequence motifs in primary B-cell repertoire
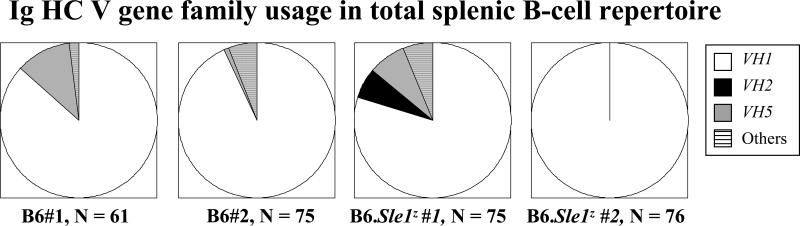
As reported recently, B6.Sle1z ANA HCs differ from non-ANA HC sequence in that they possess 3 distinct sequence motifs, including increased cationic residues in CDR3 (termed ‘motif A’), reduced anionic residues in CDR2 H52-H56 (termed ‘motif B’) and increased ‘D’ residues at H50 in CDR2 (termed ‘motif C’) (25). To elucidate the precise degree to which these 3 motifs might be encoded in the primary B-cell repertoire, the Ig HC of IgM+ve, B220+ve splenic B-cells were amplified from two B6 mice (B6 #1 and B6 #2, both being seronegative for ANAs) and 2 anti-chromatin sero-positive mice, B6.Sle1z #1 and B6.Sle1z #2, using single-cell PCR. The total B-cell repertoire from B6 and B6.Sle1z mice exhibited similar VH gene family usage profiles, comparable to that noted among B6.Sle1z-derived ANAs, with most of the Abs being encoded by the VH1/J558 germline gene (Fig. 1). As expected, the total B-cell HC repertoires in both strains were largely germline-encoded, with little evidence of somatic mutation (Table 1). Also, the B6.Sle1 HC CDR3 regions did not differ from their B6 counterparts in terms of CDR3 lengths, or in the frequencies of D-D fusions and inverted D gene usage (data not shown).
Figure 1. Ig HC gene usage by the different groups of antibodies examined in this study.
The Ig VH gene family usage among IgM+ve, B220+ve total splenic B cells drawn from 2 B6 mice (#1, and #2), and 2 B6.Sle1z mice (B6.Sle1#1 and B6.Sle1#2) was determined by single cell PCR. The numbers of Abs listed represent the final count of HC sequences after removal of clonal replicates. The depicted VH profiles did not differ significantly between the 2 strains.
Table 1.
Frequencies of non-germline amino acid residues in B6.Sle1z-derived and B6-derived splenic B-cell repertoires
| Total number of clonally independent HCs | Frequency of mutations in HC1 | Frequency of mutations in HC CDR1 | Frequency of mutations in HC CDR2 | |
|---|---|---|---|---|
| B6 #1, total B-cell repertoire | 61 | 0.8% | 1.5% | 1.1% |
| B6 #2, total B-cell repertoire | 75 | 0.5% | 0.4% | 0.4% |
| B6.Sle1 #1, total B-cell repertoire | 76 | 0.9% | 0.6% | 1.5% |
| B6.Sle1 #2, total B-cell repertoire | 75 | 1.0% | 2.0% | 1.2% |
Indicated are the frequencies of amino-acid residues that are non-germline, when compared to the NCBI/IgBlast database.
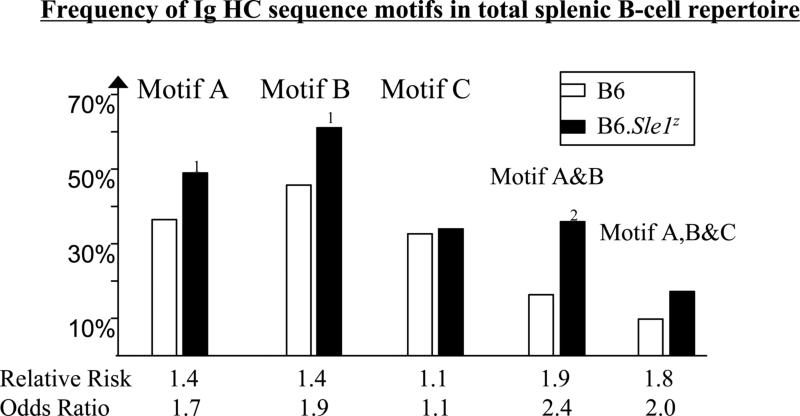
Interestingly, however, the B6.Sle1z primary Ig HC repertoire exhibited the ANA-associated sequence motifs “A” and “B” at a significantly higher frequency compared to the B6 controls (Fig. 2); the presence of these motifs conferred a 1.4-fold enhanced “risk” of nuclear antigen reactivity when acting separately, and 1.9-fold higher risk when operating in tandem, values that were similar to those noted previously when B6.Sle1z ANAs were compared to non-ANA mAbs (25). With respect to motif “A”, somatic recombination with N-nucleotide addition accounted for 71%, 77%, 78% and 79% of all CDR3 “R” residues in the HCs from B6 #1, B6 #2, B6.Sle1z #1, and B6.Sle1z #2 total splenic Ig repertoires, respectively (data not plotted). Thus, for example, “R” residues at H95 and H96 were predominantly the product of V:D joining, whereas “R” at H100 and beyond were largely due to D:D or D:J joining, with N-insertions, in all groups of Abs studied (data not shown). Thus, the molecular origins of CDR3 “R” residues did not differ between the two strains. With respect to motif “B”, it was of interest to note that the reduction in anionic residues seen in the CDR2 regions of B6.Sle1z B-cells was approximately compensated by the increased prevalence of “N”, “R”, or “Y” residues at H52–H54 (P < 0.08), and “G” or “S” residues at H56 (P < 0.005) (data not plotted).
Figure 2. The frequencies of ANA-associated HC sequence motifs, “A”, “B”, and “C” among B6 and B6.Sle1z primary B-cell repertoires.
The total or primary single-cell amplified HC repertoire of IgM+ve, B220+ve splenic B-cells from 2 B6 mice (represented by 61 + 75 = 136 HC sequences) and 2 B6.Sle1z mice (represented by 75 + 76 = 151 HC sequences) were examined for the frequencies of ANA-associated HC sequence motif “A” (i.e, the presence of at least 1 N/R/K residue within CDR3 H95-H100a), motif “B” (i.e, the absence of D/E residues across H52-H56), or motif “C” (i.e, D residue at H50) in the HC sequences. The association of these motifs with nuclear antigen reactivity has been reported recently (25). Clonal replicates have been removed from all datasets. Indicated P-values pertain to Chi-square test or Fisher's exact test comparisons of B6.Sle1z HC sequence versus B6 control sequences (1, P < 0.05; 2, P < 0.01). Indicated below each motif is the relative risk and odds ratio pertaining to the likelihood of the Ab being nuclear-antigen reactive if it were to possess that particular motif or combination of motifs. Note: the co-presence of motifs A and C, or motifs B and C, did not further augment the likelihood of the Ab being nuclear antigen reactive; hence these two motif combinations are not plotted.
There were no other sequence differences observed when the HC repertoires from the 2 strains were compared, with one exception. At the CDR3 position H95, B6.Sle1z HC possessed 3-fold more “N”, “R”, or “Y” residues compared to the corresponding frequencies in B6 CDR3 regions (P < 0.001). In contrast to motifs A and B, the primary B-cell repertoires from B6.Sle1z and B6 mice did not differ in their frequencies of the ANA-associated motif “C“ (Fig. 2), suggesting that this ANA-associated sequence motif may be the product of somatic mutation, at least in part.
Follicular and MZ B-cells differ in the frequency of ANA-associated HC sequence motifs
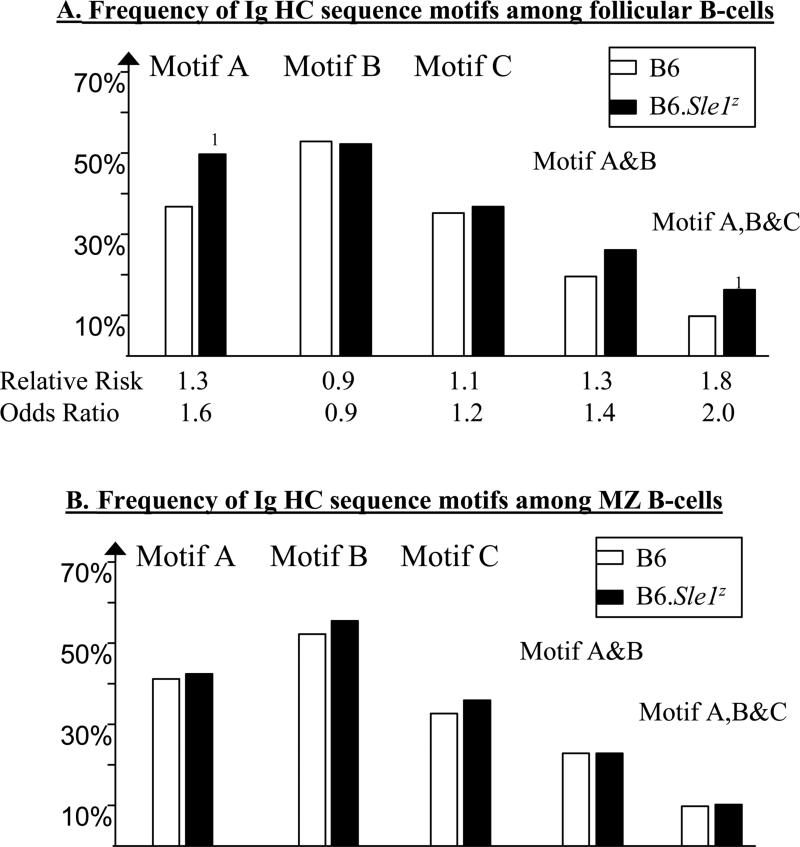
Although B6 and B6.Sle1z mice do not differ with respect to the frequencies of the 2 largest subsets of splenic B-cells (i.e., follicular and marginal zone B-cells), it became important to ascertain which particular B6.Sle1z B-cell subset was enriched for the above ANA-associated sequence motifs in the HC repertoire. As shown in Fig. 3, B6.Sle1z follicular B-cells, rather than the MZ B-cells exhibited increased presence of the ANA-associated HC sequence motif A, as well as motif A in combination with the other motifs. Importantly, in both compartments, these motifs, where present, were not the products of somatic mutations (Table 1 and data not shown). These studies indicate that the vast majority of the recirculating follicular B-cells in B6.Sle1z mice already possess some of the ANA-associated HC sequence motifs.
Figure 3. The frequencies of ANA-associated HC sequence motifs, “A”, “B”, and “C” among B6 and B6.Sle1z splenic follicular and MZ B-cell Ig repertoires.
The Ig HC repertoire was single-cell PCR amplified from splenic follicular (A) and MZ (B) B-cells from B6 (151 FO and 166 MZ HC sequences, all clonally independent, amplified from 2 mice) and B6.Sle1z (201 FO and 159 MZ HC sequences, all clonally independent, amplified from 7 mice), and examined for the frequencies of ANA-associated HC sequence motifs “A”, “B”, and “C”. Annotations are as detailed in Fig. 2. Indicated P-values pertain to Chi-square test or Fisher's exact test comparisons of B6.Sle1z HC sequence versus B6 control sequences (1, P < 0.05).
Given that the B6 and B6.Sle1z total Ig HC repertoires were largely encoded by unmutated germline genes (Table 1), one may infer that these Ab groups may differ with respect to the specific VH germline genes they utilized. Since different VH families differ significantly in their CDR2 sequences, and since most of the Abs in all study groups belonged to VH1/J558, we focused further analysis on just VH1/J558-encoded genes. For this analysis, all VH1/J558 germline genes were parsed into 3 categories, according to the number of anionic residues they possessed within H52-H56 (Table 2). Whereas ~70% of the B6.Sle1z total HC repertoire utilized motif B-bearing VH1/J558 germline genes (i.e., lacking anionic residues across H52-H56), only 48% of the control Abs used such germline genes, with the difference attaining statistical significance (Table 2). Conversely, whereas 35% of the total B6 B-cell HC repertoire was encoded by VH1/J558 germline genes possessing 2 anionic residues within H52-H56, this was half as frequent among the B6.Sle1z total Ig HC repertoire (Table 2). Collectively, the above observations suggest that the primary B-cell repertoire in lupus-prone mice may possess significantly increased frequencies of HCs with ANA-associated Ig sequence motifs that are either hardwired into germline VH genes (as is the case for motif B), or engineered through VDJ recombination early in B-cell development (as is the case with motif A), and then subjected to selective forces by self nuclear antigens.
Table 2.
Usage of VH1/J558 germline genes parsed according to the content of “D” residues in their HC CDR2 regions
| Type of VH1/J558 germline gene | Examples of VH1/J558 genes | B6 total B-cell repertoire | B6.Sle1z total B-cell repertoire |
|---|---|---|---|
| N = 1302 | N = 1632 | ||
| No“D”residues at H52-H561 | J558.53.146 | 48% | 70%*** |
| J558.26.116 | |||
| J558.55.149 | |||
| 1 “D” residue at H52-H561 | J558.36 | 17% | 15% |
| J558.75.177 | |||
| J558.88.194 | |||
| 2 “D” residues at H52-H561 | J558.72.173 | 35% | 15%*** |
| J558.83.189 | |||
| J558.85.191 |
The CDR2 sequences of VH1/J558 germline genes were examined for their content of “D” residues at H52-H56, and were then divided into 3 categories, according to whether they possessed 0, 1, or 2 “D” residues within this stretch. Note that VH genes lacking “D” residues at H52-H56 correspond to the ANA sequence motif “B”, as detailed in the text.
Only HCs that belonged to the VH1/J558 family were included in this analysis, as this constituted the major family in all Ab groups.
P < 0.001, Chi square test, comparing B6.Sle1z to B6 total B-cell repertoire
Site-directed mutagenesis reveals that ANA-associated Ig HC sequence motifs do influence nuclear antigen binding
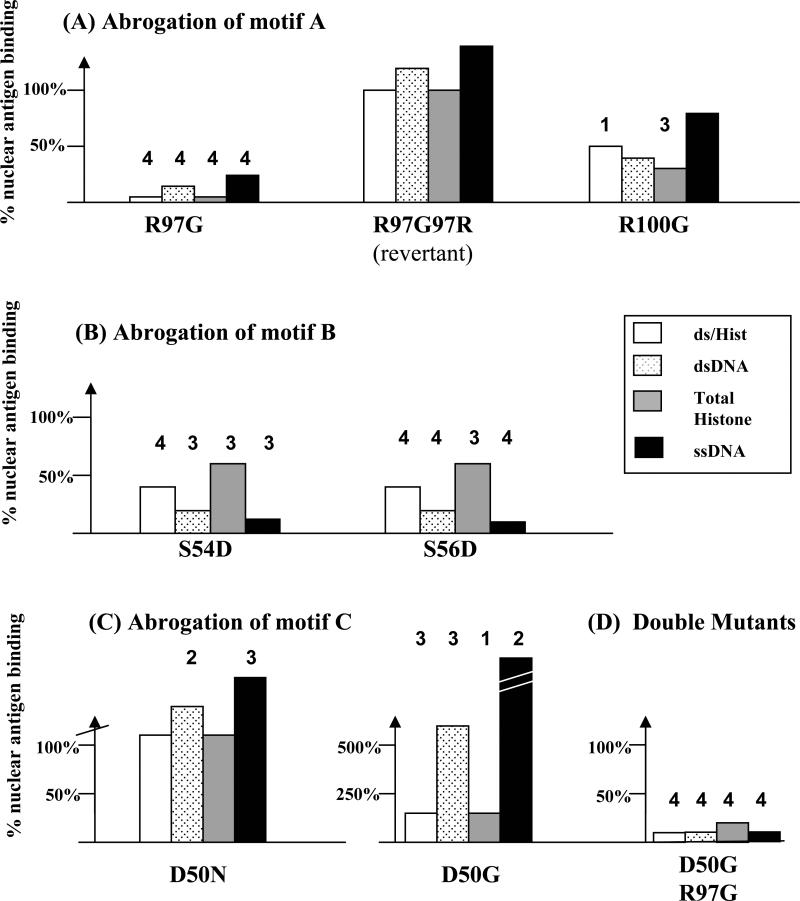
To validate if the ANA-associated Ig HC sequence motifs uncovered in this study do indeed influence nuclear antigen reactivity, site-directed mutagenesis targeting each of the 3 HC sequence motifs was carried out. In these studies, the Ig LC partner was kept constant, using an unmutated germline Vk4 gene, selected because it facilitates nuclear antigen binding (25). These studies utilized the HC of 1DG5, an oligoclonally expanded nuclear antigen reactive mAb which exhibited all 3 ANA-associated HC sequence motifs (25). Importantly, it possessed a specificity profile that was representative of the serology in B6.Sle1z mice, being strongly nucleosome-reactive but weakly dsDNA-reactive (24). The HC of 1DG5, belonging to the VH1/J558 family, possessed “R” residues at H97 and H100, constituting motif A (25). Mutating either of these residues to “G”, particularly “R” at H97, significantly reduced binding to all 4 nuclear antigens tested (Fig. 4A), compared to the unmutated 1DG5 Ab, and a double mutated revertant (R97 → G97 → R97). These studies revealed that nuclear antigen reactivity (nucleosome/histone binding, in particular) was absolutely dependent on “R” at H97 of the Ab HC.
Figure 4. Site-directed mutagenesis study to establish the relative contributions of Ig HC sequence motifs “A”, “B”, and “C”, in conferring reactivity to nucleosomes/histones and ss/dsDNA.
The B6.Sle1z-derived monoclonal ANA, 1DG5, possesses all 3 ANA-associated HC sequence motifs A, B, and C (25). The respective role of each sequence motif in conferring nuclear antigen reactivity and fine specificity was tested by site-directed mutagenesis of motif A (A), motif B (B) and motif C (C). All mutated HCs were co-expressed with a constant LC partner (a germline Vk4 gene, ai4), protein-G purified from the supernatant, adjusted to a fixed protein concentration, and then tested for nuclear antigen binding by ELISA. “R97G” means the “R” residue in the 1DG5 HC at CDR3 position #95 was mutated to “G”. The resulting degree of reactivity to the different nuclear antigens tested was expressed relative to the ELISA reactivity exhibited by the original unmutated 1DG5 Ab, expressed as a recombinant Ab using the same expression system. Supernatants from HC-only or LC-only transfectants were used as negative controls, yielding ELISA reactivities that were <10% of that seen with the positive control recombinant 1DG5 Ab. Each bar represents the mean of duplicate or triplicate ELISA reactivities, and all reactivity patterns shown have been reproduced in at least 2 independent experiments, 1 of which is illustrated._In (C), the D50N substitution resulted in 160% increase in dsDNA binding and _285% increase in ssDNA binding while the D50G substitution resulted in _604% increase in dsDNA binding and _4154% increase in ssDNA binding. Indicated P-values pertain to a Student's t-test comparison of the absolute reactivity of each recombinant Ab against the reactivity exhibited by 2 positive controls - the unmutated 1DG5 Ab and R97G→G97R double (revertant) mutant shown in (A) (1, P < 0.05; 2, P < 0.01; 3, P < 0.001; 4, P < 0.0001).
Likewise, when the Ig HC sequence motif B was abrogated by introducing aspartate residues at H54 or H56 in CDR2, nuclear antigen reactivity was also dampened (Fig. 4B), particularly the reactivity to ssDNA and dsDNA. The reactivity to nucleosomes and histones was also reduced, albeit to a lesser degree. Hence, nucleosome binding appears to be partly dependent on the HC sequence motif B whereas ss/dsDNA reactivity is heavily dependent on motif B. Interestingly, when the HC sequence motif C was abrogated by replacing aspartate at H50 with “N or “G” residues, nucleosome and histone binding were not affected, but ss/ds-DNA binding was augmented several fold (Fig. 4C), indicating that “D” at H50 may be an important determinant in skewing reactivity towards nucleosomes/histones, as opposed to DNA; these observations are consistent with the possibility that aspartate at H50 may be repelling DNA-binding. Although the abrogation of motifs “A” and “B” individually reduced nuclear antigen reactivity substantially (Fig. 4A, B), we were not able to demonstrate any further additive effects when these 2 mutations were combined (data not shown). On the other hand, double mutation studies indicated that the influence of motif A was dominant over the influence of motif C (Fig. 4D).
Discussion
These studies have important implications towards our understanding of the evolution of the anti-nuclear autoantibody response in lupus. First, they indicate that nuclear antigen binding motifs (and nuclear antigen reactivity) may already be encoded in the primary immunoglobulin repertoire of lupus-prone mice, with negligible contributions from somatic mutation. Remarkably, in transitioning from the total primary B-cell repertoire as detailed in this study to the expressed anti-nuclear Ab repertoire, as elucidated by our previous monoclonal ANA studies (25), the frequencies of motifs “A” (CDR3 stretches with at least one cationic residue) and motif “B” (CDR2s lacking D/E residues across H52-H56) do not appear to change substantially. In contrast, motif “C” was not encoded in the primary Ig HC repertoire of B6.Sle1z mice, suggesting that somatic mutation later in B cell development may indeed be responsible for this ANA-associated sequence motif.
The notion that the early Ig repertoire may be enriched with nuclear antigen reactive Abs (or at least the substrate for anti-nuclear reactivity) is boosted by recent observations by Nussenzweig and colleagues indicating that this also appears to be the case in humans (32, 33). In the latter study, the authors reported that up to 55-75% of the total Ig repertoire at the immature B-cell stage (in a normal healthy subject) may already be nuclear antigen reactive. Taken together with the findings reported in this manuscript, it's tempting to hypothesize that central B-cell tolerance mechanisms that function to “censor” such nuclear-antigen reactive specificities from the mature recirculating repertoire may be essential in thwarting autoimmunity, and that lupus-promoting genomes in mice and humans may harbor deficits in these early tolerance checkpoints. Indeed, we have recently reported that polymorphisms in Ly108, located within the Sle1b sublocus of the Sle1z disease interval, impair B-cell censoring (i.e., by deletion and BCR editing) at the immature stage of development (31), an observation that could account for the increased numbers of peripheral (including follicular) B-cells in these mice that are already “pre-wired” for anti-nuclear reactivity in their Ig heavy chains. It is certainly possible that deficits in checkpoints that censor newly emigrant B-cells in the periphery may also have contributed to the observed repertoire changes in the B6.Sle1z mice. Though it is tempting to attribute these tolerance deficits to polymorphisms in the Ly108 gene, further mechanistic studies to substantiate this hypothesis are clearly warranted.
A second important contribution of this work pertains to the structural requirements for nucleosome/histone and ssDNA/dsDNA binding. The presence of “R” residues in HC CDR3 (i.e., motif A) is perhaps the most commonly reported molecular signature among ANAs, particularly at H96 and H98-H100a (8-12, 35-38), and this observation is further bolstered by structural studies (39-44). Our observation that “R” residues in ANA HCs reach the highest usage frequencies at H96, H98, H100, and H100a in CDR3 (25), and the finding that they are necessary for maximal anti-nuclear reactivity is perfectly congruent with previously published reports (10-15). Interestingly, anti-dsDNA ANAs have been reported to exhibit relatively modest increases of “R” usage at H97, as well as H99 (8, 12, 45-47). Krishnan et al. have suggested that an “R” residue at H97 might actually destabilize key structural components of the antigen-binding groove, thus precluding dsDNA reactivity (45, 46). In our site-directed mutagenesis studies, “R” at H97 appears to be important for binding nucleosomes as well as DNA, though the former specificity appears to be absolutely dependent on this (Fig. 4A), again resonating well with earlier literature reports (45-47). Taking the previously published and current findings together, “R” residues at H96, H98, H100-100a in HC CDR3 appear to be particularly important for DNA-binding, while “R” at the intervening positions may serve to preferentially augment nucleosome-binding (over DNA-binding).
Charged residues have also been previously shown to be important in the HC CDR2 regions of ANAs (12, 14, 16, 17, 20, 25). Of note, deliberate mutagenesis of “D” at H56 to “R” profoundly enhanced DNA binding (20). Conversely, “D” residues at these positions have been suggested to augment nucleosome/histone binding (12, 48, 49). Our mutagenesis studies are consistent with the conclusion that “D” at H52-H56 may dampen nuclear antigen binding, with a particularly strong impact on ss/dsDNA reactivity (Fig. 4B). Given the modest “relative risk” this motif alone contributes towards nuclear antigen binding (Fig. 2), we postulate that this motif might influence nuclear antigen binding most effectively when operating in concert with motif A. In contrast to the above 2 motifs, no information is available in the literature concerning how motif “C” (i.e., “D” residues at H50) might impact anti-nuclear reactivity. Our mutagenesis studies are the first to indicate that aspartate residues at this position may serve to veto ss/ds-DNA binding, while still permitting nucleosome-binding. Besides directly influencing nuclear antigen contact, it is also possible that “D” residues at H50 may indirectly influence nuclear antigen specificity through potential intra-molecular salt bridges that could impact the 3-dimensional configuration of the antigen binding pocket.
Collectively, these findings provide novel insights on how the anti-nuclear Ab repertoire might evolve in lupus. To begin with, genetic defects in early B-cell tolerance may result in a primary Ig repertoire that already possesses substantial nuclear antigen binding capacity (or the substrate for such specificities), encoded by appropriately charged CDR2 and CDR3 HC sequence motifs. Violation of additional B-cell tolerance checkpoints in the periphery, such as those described to operate in germinal centers (50-52), may further shape the evolving ANA repertoire, either in terms of its affinity or fine specificity for nuclear antigens, or cross-reactivity to other target antigens. Strong selective pressure may then operate upon specific ANA clones bearing nuclear-antigen binding motifs pre-wired into the primary Ig repertoire (with superimposed somatic mutations at critical CDR positions), resulting in oligoclonal expansions of ANA-producing B-cells in lupus.
Acknowledgements
This work was supported in part by grants from the NIH P50 CORT AR 055503 and the Korea Science and Engineering Foundation (R11-2002-098-05001-0). We acknowledge the technical assistance provided by Kelvin Hsu. We also thank Dr. Michel Nussenzweig (Rockefeller University, New York) for generously providing us with the eukaryotic expression system used in this report.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hahn BH. Antibodies to DNA. N Engl J Med. 1998;338(19):1359–68. doi: 10.1056/NEJM199805073381906. [DOI] [PubMed] [Google Scholar]
- 2.Pisetsky DS. Anti-DNA and autoantibodies. Curr Opin Rheumatol. 2000;12(5):364–8. doi: 10.1097/00002281-200009000-00002. [DOI] [PubMed] [Google Scholar]
- 3.Kotzin BL. Systemic lupus erythematosus. Cell. 1996;85(3):303–6. doi: 10.1016/s0092-8674(00)81108-3. [DOI] [PubMed] [Google Scholar]
- 4.Lefkowith JB, Gilkeson GS. Nephritogenic autoantibodies in lupus: current concepts and continuing controversies. Arthritis Rheum. 1996;39(6):894–903. doi: 10.1002/art.1780390605. [DOI] [PubMed] [Google Scholar]
- 5.Chan OT, Hannum LG, Haberman AM, Madaio MP, Shlomchik MJ. A novel mouse with B cells but lacking serum antibody reveals an antibody-independent role for B cells in murine lupus. J Exp Med. 1999;189(10):1639–48. doi: 10.1084/jem.189.10.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Waters ST, McDuffie M, Bagavant H, Deshmukh US, Gaskin F, Jiang C, et al. Breaking tolerance to double stranded DNA, nucleosome, and other nuclear antigens is not required for the pathogenesis of lupus glomerulonephritis. J Exp Med. 2004;199(2):255–64. doi: 10.1084/jem.20031519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi X, Xie C, Chang S, Zhou XJ, Tedder T, Mohan C. CD19 hyperexpression augments Sle1-induced humoral autoimmunity but not clinical nephritis. Arthritis Rheum. 2007;56(9):3057–69. doi: 10.1002/art.22825. [DOI] [PubMed] [Google Scholar]
- 8.Liang Z, Xie C, Chen C, Kreska D, Hsu K, Li L, et al. Pathogenic profiles and molecular signatures of antinuclear autoantibodies rescued from NZM2410 lupus mice. J Exp Med. 2004;199(3):381–98. doi: 10.1084/jem.20030132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marion TN, Tillman DM, Jou NT, Hill RJ. Selection of immunoglobulin variable regions in autoimmunity to DNA. Immunol Rev. 1992;128:123–49. doi: 10.1111/j.1600-065x.1992.tb00835.x. [DOI] [PubMed] [Google Scholar]
- 10.Radic MZ, Weigert M. Genetic and structural evidence for antigen selection of anti-DNA antibodies. Annu Rev Immunol. 1994;12:487–520. doi: 10.1146/annurev.iy.12.040194.002415. [DOI] [PubMed] [Google Scholar]
- 11.Eilat D, Anderson WF. Structure-function correlates of autoantibodies to nucleic acids. Lessons from immunochemical, genetic and structural studies. Mol Immunol. 1994;31(18):1377–90. doi: 10.1016/0161-5890(94)90154-6. [DOI] [PubMed] [Google Scholar]
- 12.Chen L, Chang S, Mohan C. Molecular signatures of antinuclear antibodies-contributions of heavy chain CDR residues. Mol Immunol. 2002;39(56):333–47. doi: 10.1016/s0161-5890(02)00110-4. [DOI] [PubMed] [Google Scholar]
- 13.Martin T, Crouzier R, Weber JC, Kipps TJ, Pasquali JL. Structure-function studies on a polyreactive (natural) autoantibody. Polyreactivity is dependent on somatically generated sequences in the third complementarity-determining region of the antibody heavy chain. J Immunol. 1994;152(12):5988–96. [PubMed] [Google Scholar]
- 14.Radic MZ, Seal SN. Selection of recurrent V genes and somatic mutations in autoantibodies to DNA. Methods. 1997;11(1):20–6. doi: 10.1006/meth.1996.0383. [DOI] [PubMed] [Google Scholar]
- 15.Wloch MK, Clarke SH, Gilkeson GS. Influence of VH CDR3 arginine and light chain pairing on DNA reactivity of a bacterial DNA-induced anti-DNA antibody from a BALB/c mouse. J Immunol. 1997;159(12):6083–90. [PubMed] [Google Scholar]
- 16.Radic MZ, Mackle J, Erikson J, Mol C, Anderson WF, Weigert M. Residues that mediate DNA binding of autoimmune antibodies. J Immunol. 1993;150(11):4966–77. [PubMed] [Google Scholar]
- 17.Katz JB, Limpanasithikul W, Diamond B. Mutational analysis of an autoantibody: differential binding and pathogenicity. J Exp Med. 1994;180(3):925–32. doi: 10.1084/jem.180.3.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang Z, Chen C, Mohan C. Molecular signatures of anti-nuclear antibodies: contributions of specific light chain residues and a novel New Zealand Black V kappa 1 germline gene. J Immunol. 2003;171(7):3886–94. doi: 10.4049/jimmunol.171.7.3886. [DOI] [PubMed] [Google Scholar]
- 19.Ibrahim SM, Weigert M, Basu C, Erikson J, Radic MZ. Light chain contribution to specificity in anti-DNA antibodies. J Immunol. 1995;155(6):3223–33. [PubMed] [Google Scholar]
- 20.Li H, Jiang Y, Prak EL, Radic M, Weigert M. Editors and editing of anti-DNA receptors. Immunity. 2001;15(6):947–57. doi: 10.1016/s1074-7613(01)00251-5. [DOI] [PubMed] [Google Scholar]
- 21.Fitzsimons MM, Chen H, Foster MH. Diverse endogenous light chains contribute to basement membrane reactivity in nonautoimmune mice transgenic for an anti-laminin Ig heavy chain. Immunogenetics. 2000;51(1):20–9. doi: 10.1007/s002510050004. [DOI] [PubMed] [Google Scholar]
- 22.Spatz L, Saenko V, Iliev A, Jones L, Geskin L, Diamond B. Light chain usage in anti-double-stranded DNA B cell subsets: role in cell fate determination. J Exp Med. 1997;185(7):1317–26. doi: 10.1084/jem.185.7.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morel L, Mohan C, Yu Y, Croker BP, Tian N, Deng A, et al. Functional dissection of systemic lupus erythematosus using congenic mouse strains. J Immunol. 1997;158(12):6019–28. [PubMed] [Google Scholar]
- 24.Mohan C, Alas E, Morel L, Yang P, Wakeland EK. Genetic dissection of SLE pathogenesis. Sle1 on murine chromosome 1 leads to a selective loss of tolerance to H2A/H2B/DNA subnucleosomes. J Clin Invest. 1998;101(6):1362–72. doi: 10.1172/JCI728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang Zhiyan, Chang Sooghee, Youn Min So, Mohan Chandra. Molecular Hallmarks of Antinuclear Antibodies Exhibited by the Lupus Susceptibility Locus, Sle1. Mol Immunol. doi: 10.1016/j.molimm.2008.12.034. (manuscript in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ehlich A, Martin V, Muller W, Rajewsky K. Analysis of the B-cell progenitor compartment at the level of single cells. Curr Biol. 1994;4(7):573–83. doi: 10.1016/s0960-9822(00)00129-9. [DOI] [PubMed] [Google Scholar]
- 27.ten Boekel E, Melchers F, Rolink A. The status of Ig loci rearrangements in single cells from different stages of B cell development. Int Immunol. 1995;7(6):1013–9. doi: 10.1093/intimm/7.6.1013. [DOI] [PubMed] [Google Scholar]
- 28.Gu H, Tarlinton D, Muller W, Rajewsky K, Forster I. Most peripheral B cells in mice are ligand selected. J Exp Med. 1991;173(6):1357–71. doi: 10.1084/jem.173.6.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haines BB, Angeles CV, Parmelee AP, McLean PA, Brodeur PH. Germline diversity of the expressed BALB/c VhJ558 gene family. Mol Immunol. 2001;38(1):9–18. doi: 10.1016/s0161-5890(01)00049-9. [DOI] [PubMed] [Google Scholar]
- 30.Kantor AB, Merrill CE, Hillson JL. Construction of cDNA from single unstimulated mouse B lymphocytes: method and application to the study of expressed antibody repertoire in FACS-sorted murine B cell subsets. Blackwell Scientific. 1996 [Google Scholar]
- 31.Kumar KR, Li L, Yan M, Bhaskarabhatla M, Mobley AB, Nguyen C, et al. Regulation of B cell tolerance by the lupus susceptibility gene Ly108. Science. 2006;312(5780):1665–9. doi: 10.1126/science.1125893. [DOI] [PubMed] [Google Scholar]
- 32.Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant autoantibody production by early human B cell precursors. Science. 2003;301(5638):1374–7. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- 33.Yurasov S, Wardemann H, Hammersen J, Tsuiji M, Meffre E, Pascual V, et al. Defective B cell tolerance checkpoints in systemic lupus erythematosus. J Exp Med. 2005;201(5):703–11. doi: 10.1084/jem.20042251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sedrak P, Hsu K, Mohan C. Molecular signatures of anti-nuclear antibodies--contribution of heavy chain framework residues. Mol Immunol. 2003;40(8):491–9. doi: 10.1016/s0161-5890(03)00223-2. [DOI] [PubMed] [Google Scholar]
- 35.Kieber-Emmons T, Foster MH, Williams WV, Madaio MP. Structural properties of a subset of nephritogenic anti-DNA antibodies. Immunol Res. 1994;13(23):172–85. doi: 10.1007/BF02918278. [DOI] [PubMed] [Google Scholar]
- 36.Shlomchik M, Mascelli M, Shan H, Radic MZ, Pisetsky D, Marshak-Rothstein A, et al. Anti-DNA antibodies from autoimmune mice arise by clonal expansion and somatic mutation. J Exp Med. 1990;171(1):265–92. doi: 10.1084/jem.171.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koelsch K, Zheng NY, Zhang Q, Duty A, Helms C, Mathias MD, et al. Mature B cells class switched to IgD are autoreactive in healthy individuals. J Clin Invest. 2007;117(6):1558–65. doi: 10.1172/JCI27628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koutouzov S, Jovelin F, Brard F, Raux G, Tron F, Gilbert D. Comparison of structural characteristics of antisubnucleosome and anti-DNA monoclonal antibodies derived from lupus mice. Ann N Y Acad Sci. 1997;815:327–30. doi: 10.1111/j.1749-6632.1997.tb52076.x. [DOI] [PubMed] [Google Scholar]
- 39.Seeman NC, Rosenberg JM, Rich A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc Natl Acad Sci U S A. 1976;73(3):804–8. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barry MM, Mol CD, Anderson WF, Lee JS. Sequencing and modeling of anti-DNA immunoglobulin Fv domains. Comparison with crystal structures. J Biol Chem. 1994;269(5):3623–32. [PubMed] [Google Scholar]
- 41.Cygler M, Boodhoo A, Lee JS, Anderson WF. Crystallization and structure determination of an autoimmune anti-poly(dT) immunoglobulin Fab fragment at 3.0 A resolution. J Biol Chem. 1987;262(2):643–8. [PubMed] [Google Scholar]
- 42.Herron JN, He XM, Ballard DW, Blier PR, Pace PE, Bothwell AL, et al. An autoantibody to single-stranded DNA: comparison of the three-dimensional structures of the unliganded Fab and a deoxynucleotide-Fab complex. Proteins. 1991;11(3):159–75. doi: 10.1002/prot.340110302. [DOI] [PubMed] [Google Scholar]
- 43.Pokkuluri PR, Bouthillier F, Li Y, Kuderova A, Lee J, Cygler M. Preparation, characterization and crystallization of an antibody Fab fragment that recognizes RNA. Crystal structures of native Fab and three Fab-mononucleotide complexes. J Mol Biol. 1994;243(2):283–97. doi: 10.1006/jmbi.1994.1654. [DOI] [PubMed] [Google Scholar]
- 44.Mol CD, Muir AK, Cygler M, Lee JS, Anderson WF. Structure of an immunoglobulin Fab fragment specific for triple-stranded DNA. J Biol Chem. 1994;269(5):3615–22. [PubMed] [Google Scholar]
- 45.Krishnan MR, Jou NT, Marion TN. Correlation between the amino acid position of arginine in VHCDR3 and specificity for native DNA among autoimmune antibodies. J Immunol. 1996;157(6):2430–9. [PubMed] [Google Scholar]
- 46.Krishnan MR, Marion TN. Comparison of the frequencies of arginines in heavy chain CDR3 of antibodies expressed in the primary B-cell repertoires of autoimmune-prone and normal mice. Scand J Immunol. 1998;48(3):223–32. doi: 10.1046/j.1365-3083.1998.00426.x. [DOI] [PubMed] [Google Scholar]
- 47.Tillman DM, Jou NT, Hill RJ, Marion TN. Both IgM and IgG anti-DNA antibodies are the products of clonally selective B cell stimulation in (NZB x NZW)F1 mice. J Exp Med. 1992;176(3):761–79. doi: 10.1084/jem.176.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Monestier M, Fasy TM, Losman MJ, Novick KE, Muller S. Structure and binding properties of monoclonal antibodies to core histones from autoimmune mice. Mol Immunol. 1993;30(12):1069–75. doi: 10.1016/0161-5890(93)90153-3. [DOI] [PubMed] [Google Scholar]
- 49.Monestier M. Variable region genes of anti-histone autoantibodies from a MRL/Mp-lpr/lpr mouse. Eur J Immunol. 1991;21(7):1725–31. doi: 10.1002/eji.1830210721. [DOI] [PubMed] [Google Scholar]
- 50.Cappione A, 3rd, Anolik JH, Pugh-Bernard A, Barnard J, Dutcher P, Silverman G, et al. Germinal center exclusion of autoreactive B cells is defective in human systemic lupus erythematosus. J Clin Invest. 2005;115(11):3205–16. doi: 10.1172/JCI24179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shokat KM, Goodnow CC. Antigen-induced B-cell death and elimination during germinal-centre immune responses. Nature. 1995;375(6529):334–8. doi: 10.1038/375334a0. [DOI] [PubMed] [Google Scholar]
- 52.Paul E, Lutz J, Erikson J, Carroll MC. Germinal center checkpoints in B cell tolerance in 3H9 transgenic mice. Int Immunol. 2004;16(2):377–84. doi: 10.1093/intimm/dxh035. [DOI] [PubMed] [Google Scholar]