Abstract
Immunogenicity of non-human proteins with useful therapeutic properties has prevented their development for use in the therapy of disease. However, this class of proteins could be very useful, if their immunogenicity could be markedly reduced so that many treatment cycles could be administered. One approach to reduce the immunogenicity of foreign proteins is to identify B-cell epitopes on the protein and eliminate them by mutagenesis. In this article, theoretical aspects and experimental evidence for the feasibility of B cell epitope removal is reviewed. A special focus is given to our results with deimmunization of recombinant immunotoxins in which Fvs are fused to a 38 kDa portion of the bacterial protein, Pseudomonas exotoxin A (PE38). Immunotoxins targeting CD22 and CD25 have produced complete remissions in many patients with drug resistant Hairy Cell Leukemia and are being evaluated in other malignancies. Experimental data summarized in this review indicates that removal of B-cell epitopes is a practical approach for making less immunogenic protein therapeutics from non-human functional proteins. This approach requires grouping of the epitopes to identify targets for de-immunization followed by quantitative analysis of the decrease in affinity produced by the mutations in B cell epitopes.
Keywords: Deimmunization, mutagenesis, antigen, antigenicity, antibody, biopharmaceutical
1. Introduction
Biologically useful proteins originating from non-human species are an attractive source of biopharmaceuticals, due to their high selectivity and potency. However, because they are recognized as foreign by the human immune system, the number of treatment cycles that can be administered to patients is limited by the formation of antibodies [1–5]. Nevertheless a few foreign proteins have proven to be clinically useful. Streptokinase is a bacterial protein secreted by hemolytic streptococci that is an effective clot-dissolving medication for myocardial infarction and pulmonary embolism [6]. Although streptokinase is highly immunogenic and inactivating antibodies can be present from prior streptococcal infections, the levels of antibodies are generally of little clinical significance, when streptokinase is used in the large doses recommended. Streptokinase has relatively less bleeding risk for patients than newer agents and is still used for the first line treatment of acute myocardial infarction. Another example of a foreign protein in clinical use is Botulinum toxin, a neurotoxic protein produced by the bacterium Clostridium botulinum [7]. Botulinum toxin is a very potent toxin and minute doses are used to treat muscle spasms. The very small protein load (usually less than 100 ng) needed for its medical effect does not usually induce significant antibody responses; only 5–15% of patients injected serially with Botulinum toxin became unresponsive due to the production of neutralizing antibodies [8]. These examples indicate that highly immunogenic foreign proteins can be used for medical purposes. Another important factor is that non-human proteins are unlikely to produce auto-immunity that could neutralize endogenous protein function. This suggests that it is not necessary to aim for the complete elimination of the immunogenicity for medical benefit [9].
We have successfully used a 38 kDa portion of Pseudomonas exotoxin A (PE38) as a cytotoxic moiety in recombinant immunotoxins for the therapy of cancer [10–12]. In these immunotoxins, PE38 is genetically linked to the Fv portion of a monoclonal antibody, guiding the PE38 toxin to cancer cells that express the antigen on their cell surface. We have been actively pursuing the reduction of immunogenicity of recombinant immunotoxins to expand their usefulness in cancer treatment. Clinical trials revealed that over half of the patients with life threatening drug resistant Hairy Cell Leukemia achieved a complete remission after 3 to 10 cycles of treatment with BL22, a recombinant immunotoxin containing PE38 [13, 14]. However, such multiple cycles of treatment are not possible, in patients with normal immune systems, because neutralizing antibodies usually develop within three weeks. These antibodies almost always react with the bacterial toxin and very infrequently with the Fv, and limit the number of cycles of therapy that can be given. Fortunately, patients with leukemias and lymphomas make antibodies to the immunotoxin relatively infrequently, because the chemotherapy used to treat this disease is toxic to the immune system and because leukemias and lymphomas infiltrate and damage the immune system. The success in treating drug resistant leukemia suggests that immunotoxin therapy can be useful in the treatment of other types of cancer, if we can reduce immunogenicity to a level, which permits multiple cycles of treatment to be given.
One approach to de-immunize a protein is to identify B-cell epitopes on the protein and eliminate them by mutagenesis [15, 16]. PE38 is a highly immunogenic protein and de-immunizing appeared to present a formidable task. Our success over the last 5 years in substantially reducing the immunogenicity of PE38 containing immunotoxins [17, 18] suggests that B cell epitope removal can also be accomplished for other foreign proteins. In this review, both theoretical aspects and experimental evidence on the reduction of immunogenicity by B cell epitope removal will be discussed.
2. Theoretical basis of B cell epitope removal for reducing immunogenicity
There are several essential prerequisites for deimmunization by B cell epitope removal. They are: (1) the presence of antigenic hot spots on a protein surface that more frequently serve as epitopes to the antibodies than other surface regions of the protein; (2) the antigenic structural signatures of the hot spots can be altered so that they are less immunogenic by point mutations in amino acids located at these sites; and (3) these point mutations can be combined into a functional protein that induces a diminished antibody response vivo. There are no immunologic theories to support these prerequisites due to the heterogeneity of the mechanisms of the antibody generation. The next section analyzes the properties of B cell epitopes as targets for reducing immunogenicity.
2.1. High affinity is required for antibody binding in patients
It is well known that the affinities of antibodies generated against an antigen vary over 10,000-fold [19] . Repeated immunizations does not always increase affinity, possibly depending on the epitope [20]. As expected, higher affinity antibodies have a greater impact on antigen function and elimination through antigen-antibody complex formation [21, 22]. For foreign proteins used at a relatively low dose, low affinity antibodies may not inhibit activity, because the immune complexes are short lived at the concentration levels of the protein and the antibodies in patients’ body fluids [23]. Binding assays and neutralization assays currently used in the evaluation of antibody induction to biopharmaceuticals are biased toward the detection of higher affinity antibodies [24, 25]. This correlation of the affinity with the severity of the immunogenicity problem should be taken into consideration in a deimmunization plan, because it emphasizes the importance of ranking of epitopes in terms of the affinity of the induced antibody.
B cells that produce high affinity antibodies have undergone affinity maturation in germinal centers in secondary lymphoid organs such as lymph nodes [26]. Helper T cell support is absolutely necessary to enable a B cell to produce a high affinity antibody [27, 28]. In addition, the helper T-cell must be antigen-specific and must physically contact the B cell specific to a B cell epitope of the same antigen [29]. When a B-cell encounters an antigen, the antigen is trapped by the surface immunoglobulin specific to a B cell epitope on the antigen. The B cell then internalizes the antigen by receptor-mediated endocytosis and digests it into peptides, some of which bind to major histocompatibility complex (MHC) class II molecules (e.g. T cell epitope), providing the binding site on the B cell surface for the specific T cell receptor expressed on the helper T cell [30]. The combined B and T cells then collaborate to promote affinity maturation and production of high affinity antibody by mutual stimulation. After the initial binding of the B cell to the T cell, subsequent binding of the co-receptor ligand (e.g. CD40L) on the T cell to a co-receptor on the B cell (e.g. CD40) initiates the supplemental intracellular signal for differentiation of the B-cell to become a high affinity antibody-producing cell. The CD40-CD40L interaction is critical for production of high affinity antibody as demonstrated by the impairment of T cell-dependent antibody responses in mice deficient in either CD40 or CD40L [31, 32]. The B cell also signals the T cell to proliferate through binding between the co-receptor molecules on the T cell (such as CD28) and the ligands on the B cell (such as B7).
As indicated from this mechanism, the repertory of high affinity antibodies produced in a patient is restricted by the epitopes displayed on the surface of the biopharmaceutical molecule. In addition, the location of B cell epitopes is unconnected to that of the T cell epitopes [33, 34]. For effective deimmunization, B cell epitopes for high affinity antibodies should be given a high priority in the removal process.
2.2. Foreignness as a secondary factor for determining epitope locations of proteins: antibody response in different animals
To date the most success in making non-immunogenic proteins has been achieved with the humanization of mouse antibodies [35]. This was initially carried out by replacing the mouse Fc with human constant regions to make chimeric antibodies [36, 37], and later by genetically grafting complementarity-determining regions (CDRs) (antigen binding sites of the rodent antibody) into human immunoglobulin frameworks to further minimize the mouse component [38]. Foreignness, by which we mean the evolutionary distance between humans and the organism producing the protein to be used as a therapeutic agent, is considered as the primary factor determining the immunogenicity of protein biopharmaceuticals [39–41]. The importance of foreignness is supported by the fact that immune systems have been educated not to react with the self proteins [42]. This immunological tolerance is acquired through epitope-specific mechanisms such as clonal deletion, receptor editing and clonal anergy [43, 44], even though the initial antibody repertory before the education has a very wide potential for diversity through gene recombination and mutation events. Although the education by a different panel of self antigens indicates that different species have a unique deficiency in the antibody repertoire so as not to react with self antigens, the acquired immune systems in vertebrates have developed so that they can efficiently react with previously unseen foreign proteins such as those produced by bacteria that can injure the host. Since there is no biochemical reason to indicate that there is a unique antibody repertoire in each species, we think it is reasonable to expect that the B-cell epitopes in different animals for foreign antigens are similar and based on properties of the foreign protein [45–49].
In fact, characterizations of repertoires of B cell epitopes indicate that epitope profiles of foreign proteins are overlap each other in different animals [45]. For example, characterization of linear B cell epitopes of Botulinum toxin show good agreement in different animals, including mice with different MHCs utilizing different T-cell epitopes [50, 51]. Another example is a significant overlap of mouse and human B-cell epitopes on HIV gp160 recorded in the HIV molecular immunology database [52]. In an analysis of B-cell epitopes of PE38, 7 epitopes on PE38 that were identified by a panel of mouse monoclonal antibodies were also recognized by human antibodies in the sera of patients treated with immunotoxins containing PE38 [17].
Therefore, for deimmunization purposes, the B cell epitope positions and the structural factors that determine B cell epitope locations (virtually free from host-dependent factors) can be analyzed by the use of antibodies produced in different species. Moreover, this suggests that the immunogenicity of a foreign protein (the property inducing antibody response in vivo) can be predicted by its antigenicity (reactivity with existing antibody) [53, 54]. The original and modified proteins after the epitope removal can be assessed for immunogenicity by measuring their binding to existing antibodies.
2.3. Heterogeneity of binding mode and strength between antigen and antibody
There are two contrasting views on the nature of the B cell epitopes on proteins [55]: one is that certain parts of proteins are inherently antigenic and this property is intrinsic to the nature of the protein [56]. The other is that virtually any accessible part of a protein is potentially an antigenic site [55, 57]. This dispute remains unsolved. In contrast to T cell epitopes that require appropriate lengths and anchor residues for binding to the MHC, B cell epitopes do not need the support of other molecules to bind to antibody. Moreover, epitope is a functional term, only defined by its binding activity to the antibody and cannot be simply deduced from the antigen structure itself [58]. Using crystal structures, the analysis of antigen-antibody complexes have not yet led to the identification of a specific mode of interaction or identified specific amino acids that are uniformly important in the interaction, although the presence of hydrophilic and large amino acid residues are often observed in antigen-antibodies interfaces [59–64]. Their presence could be the result of the higher area exposure on the surface of the antigen combined with hydrophilicity leading to an increased potential to provide interactions with the antibody. Turn regions of the secondary structure are also relatively frequently found in interfaces [65].
Many methods of predicting the location of B cell epitopes have been developed based on the amino acid usage in antigen-antibody interfaces, but the success rate is not very high [66, 67]. Most widely used prediction methods use amino acid sequence information and calculate the probability of exposure [68–71]. Bioinformatics has also been used to predict B cell epitopes [72–75]. However the usefulness of these approaches has not been fully experimentally confirmed.
The unique mode of binding for each antigen-antibody pair makes it difficult to predict the effect of a mutation on the binding strength. The modification of epitopes in deimmunized mutants must be evaluated quantitatively and related to the biologic function of the protein. For example, neutralizing antibodies in the blood of patients treated with PE38-based immunotoxins are usually converted to positive in the neutralization assay at about a 10 nM concentration of PE38-specific antibody (unpublished data), but a low affinity antibody (> 20 nM of KD) showed very little binding at this concentration. Practically, the degree of reduction of immunogenicity that is needed is dependent on the nature of the protein and how it is used.
2.4. Dominance of discontinuous epitopes (conformational epitopes) over continuous epitopes (linear epitopes)
Epitopes on the surface of proteins are almost always discontinuous and conformation dependent [58, 76, 77]. It is very unusual for an area on the surface of a protein to be made up of a continuous string of amino acids and to serve as an epitope in the native form of the protein. The common use of synthetic peptide fragments for the production of antibody reagents has focused attention on linear epitopes. Such antibody reagents, although useful in research for western blotting, etc., often do not react with the protein in immunoprecipitation, indicating the epitope is not located on the accessible surface of the native form of the antigen. We analyzed the reactivity of anti-sera from patients, who have made antibodies to immunotoxins containing PE38, and found that the antibody titers to conformational epitopes usually relate to the neutralizing activity, indicating the importance of the discontinuous epitopes in the neutralization of immunotoxins in patients. Therefore, effective deimmunization strategies for biopharmaceuticals need to address discontinuous epitopes on the surface of the native proteins.
Although crystallography of an antigen-antibody complex is the ultimate method to locate an epitope, this technique is too laborious to be used to analyze numerous antigen-antibody complexes. One widely used approach to identify B-cell epitopes is the use of small fragments or peptides derived from the protein [78–80], but this approach cannot be applied to discontinuous B-cell epitopes. An important technical difficulty in discontinuous epitope analysis is the fragileness of the native conformational structure of proteins in widely used solid phase assays measuring antigen-antibody binding [81]. For example, passive adsorption of proteins onto the plastic surfaces of ELISA plates often alters the protein conformation, causes denaturation of conformational epitopes and reveals cryptic epitopes [82, 83]. To determine the location of an epitope on the surface of a protein, it is necessary to employ an assay that monitors antigen-antibody complex formation in solution (under native conditions) [17, 84].
2.5. Boundaries, overlap and grouping of B cell epitopes
For deimmunization purposes, the number of epitope groups must be established and there must be a limited number of epitopes on a protein so they all can be modified and eliminated by a small number of mutations. As we have discussed above, peptide-scanning techniques are not useful, because they usually fail to detect conformational epitopes. Polyclonal antibodies cannot be used for epitope analysis, because there is a limitation of the number of antibody molecules that can bind to the antigen at one time and epitopes can be silent in the presence of a large amount of antibodies to other epitopes until the main epitopes are eliminated by mutation [85, 86]. The best solution is the production of a panel of monoclonal antibodies so that the binding of each can be analyzed and studied [87]. Then competition assays using pairs of monoclonal antibodies enable one to group the epitopes appropriate for deimmunization [88–90], because two antibodies showing competitive binding should share at least one amino acid residue in their epitopes and this then could be modified by a single point mutation. In previous reports, analyses using 103 different monoclonal antibodies against horse cytochrome C (14kD) revealed three major non-overlapping epitope groups, although 22 distinct reactive patterns were detected to various mutants and fragments of the antigen [91]. Another example is that 49 anti-lysozyme monoclonal antibodies identified three major non-overlapping epitope groups [92]. As such, a moderately large panel of monoclonal antibodies should be comprehensive enough to map all epitope groups on the protein surface.
2.6. Strategy of mutagenesis to remove B cell epitopes
The location of an epitope requires knowledge of the structure of a protein and the availability of a panel of monoclonal antibodies to it. Epitope locations are defined by specific mutations that destroy the binding of several antibodies that also compete with each other for binding to the protein [85, 93]. Alanine is usually chosen as the replacement residue, because alanine removes all side chain atoms after the β-carbon that can react with antibodies and alanine usually maintains the conformation of antigen [94, 95]. Alanine is commonly distributed in buried and exposed positions and in all manner of secondary structures and also does not impose new hydrogen bonding. For these reasons, a series of mutants with alanine substitution of a single amino acid (alanine scanning) has been successfully used to analyze many protein-protein interactions including antigen-antibody interactions [85, 96].
Because of the large number of exposed residues, it is necessary to develop criteria that will identify residues likely for antibody binding that can be mutated to alanine. The most likely candidates are amino acids with bulky side chains that are highly exposed with a high ASA accessible surface area [97]. Using alanine several point mutations can be combined in a single mutant molecule with minimum risk of the creation of new epitopes and loss of protein function. Our previous results on PE38 deimmunization indicate that alanine is generally applicable for substitution of most residues. However, we occasionally found that glycine or serine was a better replacement, probably because of the need for flexibility or hydrophilicity [17, 18].
2.7. Removal of B cell epitopes reduces antibody production: presence of hot spots of B cell epitopes
There are a few reports of successful B cell epitope removal for reducing immunogenicity of foreign proteins used for biopharmaceuticals. A significant reduction in the antigenicity of streptokinase was achieved by site directed mutagenesis of antibody epitopes [15, 98]. Neutralizing antibodies were observed following a single treatment in 81% of patients administered wild-type streptokinase, but in only 47% of those treated with the best variant. Another example is a less immunogenic mutated form of MFECP (a bacterial carboxypeptidase G2 [CP]-based reagent for antibody-directed enzyme pro-drug therapy [ADEPT]) [16]. Although the B cell epitope removal did not eliminate the antibody response completely, a mutant with the modification of an epitope for an antibody (CM79) that showed a delayed polyclonal antibody response in both mice and patients. We recently carried out comprehensive studies using many monoclonal antibodies and mutants on the impact of B cell epitope removal upon immunogenicity [17, 18]. The analysis showed a statistically significant bias toward hot spots in the distribution B cell epitopes on the surface of PE38. We further showed that 8 point mutations that are located in each of the major epitopes can be combined in a new molecule that shows reduced antibody responses in mice with different MHC haplotypes. This result demonstrated that it is feasible to identify epitope hot spots using existing antibodies and to alter the hot spots to be less immunogenic for inducing fewer antibodies in vivo.
3. Practice of B cell epitope removal: a lesson from the deimmunization of a Pseudomonas exotoxin-based immunotoxin
We have treated more than 300 patients with immunotoxins containing several different Fvs but the same 38 kDa fragment of Pseudomonas exotoxin A .We observed that when antibodies were produced they were directed at PE38 and rarely against the Fv. Such anti-PE38 antibodies neutralize the immunotoxins and reduce their efficacy [10–12]. The clinical success of the immunotoxins encouraged us in trying practical level of the reduction of the immunogenicity of PE38. In this section, our recent accomplishment of the deimmunization of a PE38-based immunotoxin will be described [17, 18]. This example illustrates critical points that can be generalized for the deimmunization of foreign proteins by B cell epitope removal.
3.1. Use of a conformation-dependent assay to assess antigen-antibody binding
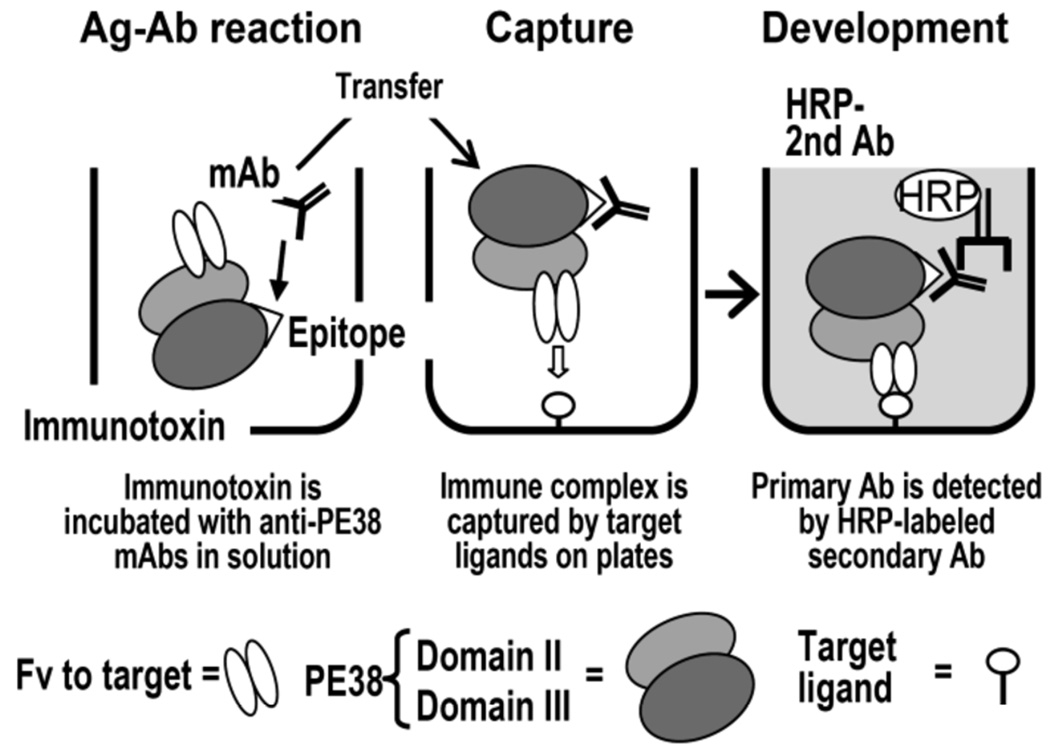
In clinical trials of immunotoxins, we have used a biological neutralization assay to make decisions to continue or stop the therapy due to the immune response, and found that the neutralization activity is a good index in predicting blood levels and the benefit of the therapy for patients. We have also measured antibody levels by ELISA, which is frequently used as a standard for the assessment of antibodies to biopharmaceuticals in patients. We have observed that antibody titers in ELISA largely depend on the assay format and a special ELISA format is required to accurately estimate neutralizing antibody levels in patients [17, 18]. In an ELISA called immune complex capture ELISA (ICC-ELISA), the antigen-antibody interaction takes place in solution, where the protein is in its native form, and the immune complexes are captured by ligands for the immunotoxin (cancer antigens for the Fv part) coated on the plate (Fig. 1). When compared to a general ELISA in which immunotoxins are directly coated on the plate for later challenge by antibody from patients (DC-ELISA), ICC-ELISA generally gives much higher titers in sera from patients who developed neutralizing antibodies to the immunotoxin. To investigate the difference between the two ELISAs, we performed an adsorption experiment of antibody from patients by native immunotoxin in solution. As expected, the antibody detected in DC-ELISA was not inhibited by native PE38 for its binding to PE38 coated on a plate. In contrast, antibody binding detected in ICC-ELISA was largely competed with native PE38. Based on these findings, we use ICC-ELISA that measures antigen-antibody interactions in solution for the deimmunization procedure.
Fig. 1.
Principle of the ICC-ELISA. Immunotoxins, comprising the 38 kDa portion (domain II and III) of Pseudomonas exotoxin A fused to an Fv specific for target antigens on cancer cells, are incubated with candidate mAbs. Because the reaction takes place in solution, all epitopes on native PE38 are theoretically available. Complexes are captured to the plate with immobilized target ligands such as CD22-Fc or similar molecules. The primary antibody is then detected with an HRP-labeled secondary antibody. Reproduced with permission from The Journal of Immunology [published 2006, Vol. 177, 8822–8834] Permission pending.
3.2. Concept of a B cell epitope group and its identification
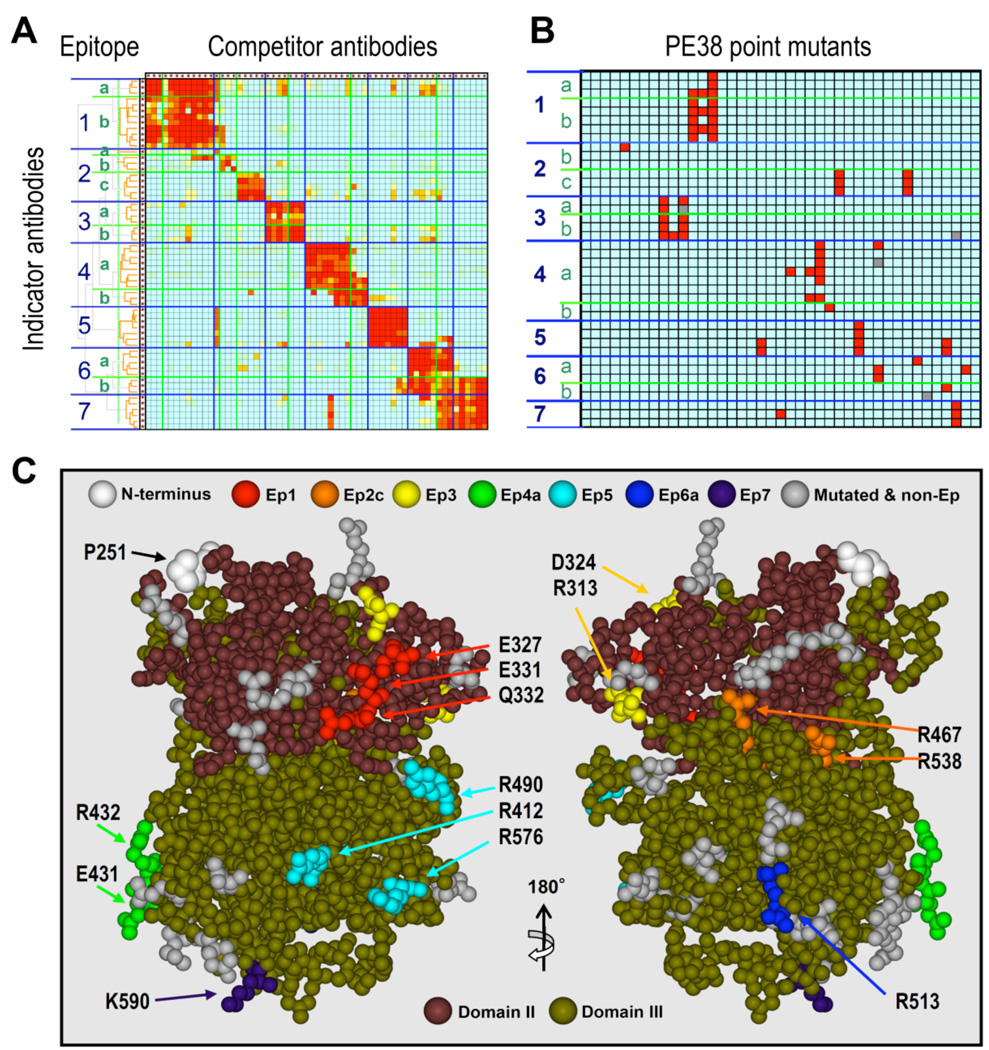
We have used a mutual competition assay employing 60 monoclonal antibodies in a pairwise manner to elucidate the topological epitope map on PE38. The results indicate the presence of 7 major epitope groups and 13 subgroups (Fig. 2A) [90]. This method is extremely useful for deimmunization because: (1) the mutual competition experiments can be carried out with native antigen in solution without fragmentation or adsorption of the antigen on solid phases that often destroy conformational structure important for antibody binding; (2) antibodies that compete with each other must share at least one amino acid residue providing an interaction for the binding and serving as the mutation candidate; and (3) the use of a pair of selected antibodies organizes the data by focusing on the relationship of the selected two epitope locations and minimizes the possibility of artifacts caused by a collision of multiple antibody molecules. For this approach to succeed, objective criteria must be established for grouping of the epitopes. We employed several contrivances in our topographical mapping method that enabled us to accurately map epitopes. These include the development of a label-free competition assay that does not require the purification or labeling of the antibodies [90] and the use of objective statistical cluster analysis to derive the epitope grouping from the competition data matrix (Fig. 2A) [90]. The method has since been validated for its usefulness for other proteins [99, 100]. This improved competition assay made it possible to evaluate the distribution of epitope groups on the whole surface of PE38 without modifying the antigen or the antibodies.
Fig. 2.
Summary of steps to identify and remove B cell epitopes in PE38. A. Mutual competitive binding assay of 60 mouse monoclonal antibodies against PE38 produced for B cell epitope analysis for the de-immunization of PE38 The binding of the indicator antibodies (in rows) in the presence of competitor antibodies (in columns) was measured by ICC-ELISA. Color code (red to blue) shows the degree of competition from 100 to 0%. B. Binding of each antibody (in rows) to each point mutant of PE38 (in columns). Mutations were made in highly exposed amino acids in a crystal structure. The binding assay measured the affinity difference between the mutant and the wild type PE38; mutations that reduced the affinity to the antibody under 10% compared to wild type are shown in red. C. The locations of mutated amino acids that decrease binding of PE38 to antibodies in an epitope-specific manner (from the data shown in Fig. 2B) are displayed in different colors on a structural model of PE38. The clustered residues shown in different colors indicate the location of each epitope. The two different approaches (topographical epitope mapping and the location of the epitopes on the crystal structure determined by mutagenesis) are in close agreement. This concordance verifies the epitope locations without analyzing crystal structures of the antigen-antibody complexes. Adapted with permission from The Journal of Immunology [published 2006, Vol. 177, 8822–8834] Permission pending.
3.3. Presence of hot spots of B cell epitopes on a foreign protein
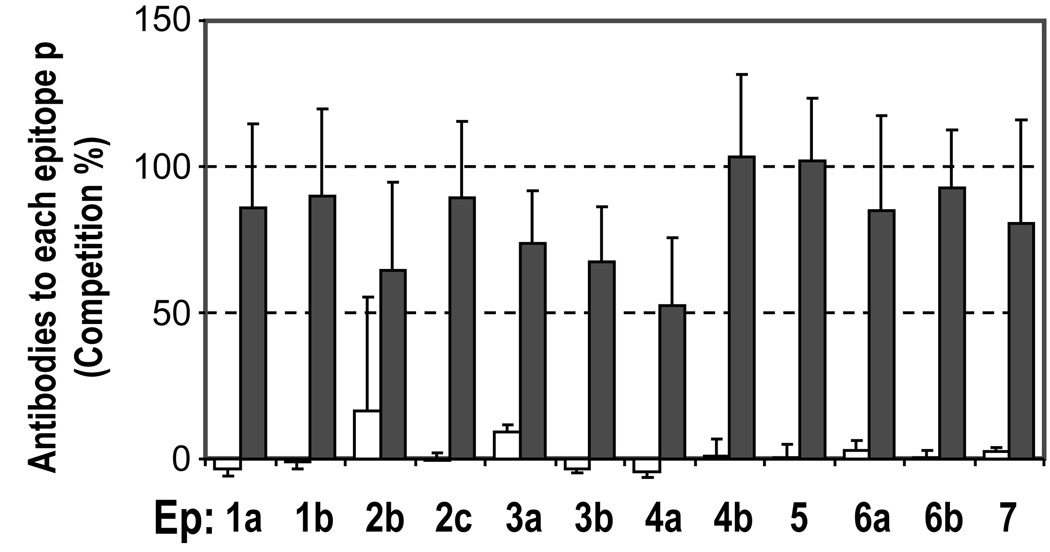
We found that the B cell epitopes are located in discrete clusters that could not be reproduced in a computer simulation that created artificial epitopes in random residues on the PE38 surface [17]. This confirmed that the B-cell epitopes are preferentially distributed on hot spots on the surface of PE38 that more frequently serve as epitopes than other surface areas of the protein. Using sera from immunotoxin treated patients, the formation of human antibodies to each of the identified hot spots was also demonstrated by a competition assay (Fig. 3). We believe that this study is the first comprehensive analysis that provides objective evidence for the presence of antigenic hot spots on a foreign protein. This important finding strongly supports the idea that deimmunization by B cell epitope removal is possible, if we can alter the hot spot structures to less immunogenic ones by mutations that remove the feature causing the high immunogenicity. We suggest that similar computer-based analyses can be performed to examine the competitive pattern of antibodies to other antigens. Such an examination is important to judge if the identified epitope groups indicate the location of true hot spots, which can be modified to make the antigen less immunogenic.
Fig. 3.
Generation of antibodies in patients against epitope hot spots identified by the mouse monoclonal antibodies. Human serum samples of patients before (open bars) and after (closed bars) treatment with anti-mesothelin immunotoxin SS1P were analyzed in the competition assay used for mapping topographical epitopes (Fig. 2A). Averages and standard deviations of 4 different patient samples are shown. Adapted with permission from The Journal of Immunology Vol. 177, 8822–8834, 2006; copyright 2006 The American Association of Immunologists, Inc.
3.4. Assessment of affinity of antigen-antibody binding
We determined the location of the epitopes on the structure of PE38 by making 41 point mutations in which alanine replaced amino acids with large ASAs and then assessing loss of binding to our panel of monoclonal antibodies (Fig. 2B). Our assay is based on the competition of the antibody binding by a series of concentrations of native protein or mutant protein, and measuring the difference of the affinities of the antibody to different mutants in solution (native condition). Such quantitative assessment of the binding is essential for B cell epitope removal, because the large variation of the affinity of antibodies does not allow the use of a uniform cutoff value to choose the best mutant.
3.5. Epitope core: a point mutation affects the binding of a group of antibodies
In the deimmunization of PE38, the location of the epitopes on the PE38 structure obtained by mutagenesis of specific residues show close agreement with the topographical epitope mapping data (Fig. 2C). Each antibody bound to most of the mutants as well as to wild type PE38, but failed to bind a few mutants (95 out of 1542). When mutants bound to most of the antibodies, but lost reactivity to a few antibodies they were assigned to the same epitope group. These results indicate that the structural alterations of these mutants are restricted within a limited area recognized by the antibodies assigned for the same epitope without changing the structure of other areas.
The data also showed that there are a small number of key residues that primarily contribute to epitope hot spot formation (epitope cores), based on their elimination by the alanine mutagenesis strategy. This is important for deimmunization, because a single mutation can abolish binding of many antibodies associated with the epitope group and the immunogenicity of the whole molecule can be reduced by the introduction of a handful of mutations. Sites to be mutated were mainly selected because they were hydrophilic and had high ASAs [97], properties important for antibody binding. Our mutant panel consists of only 41 mutant residues out of 369 residues (12%) yet has so far successfully identified 10 of the 13 sub-epitopes on PE38. Such a high success rate suggests that our strategy of amino acid selection for mutation is practical, even though we cannot predict the exact binding mode between antigen and antibody.
3.6. Successful combination of each point mutation into one mutant molecule
We then incrementally combined the point mutations at the epitope cores of PE38 into one molecule [18]. The elimination of the epitopes was confirmed by the loss of reactivity to the corresponding antibodies. The final molecule contained eight mutations (designated as 8X) with a mutation targeting each of the major epitope groups. Most of the mutants, including 8X, showed full cytotoxic activity. We were concerned that the replacement of all eight surface residues with alanine might alter protein folding or cause instability and we ultimately used alanine for four mutations, serine for three others, and glycine for one. The final immunotoxin, HA22-8X with 8X mutated PE38, targets CD22 on leukemia cells and shows similar anti-tumor activity as the non-mutated HA22 in a xenograft model of CD22-positive cancers in mice.
3.7. A less antigenic molecule is less immunogenic: no generation of new epitopes
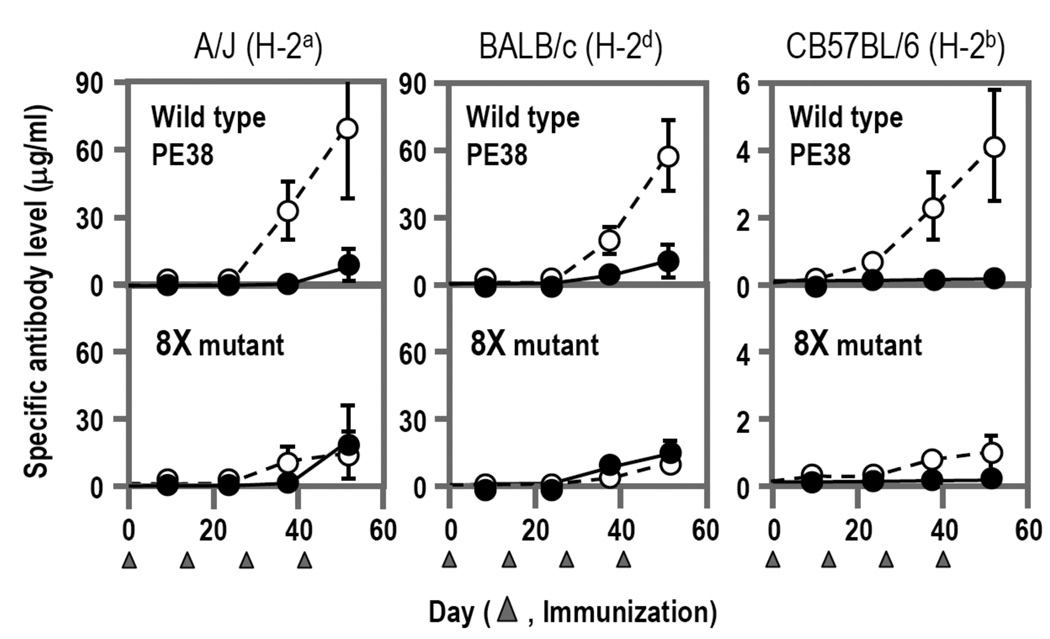
When tested in mice by a series of intravenous injections, we found that the HA22-8X mutant protein induced much lower and slower antibody responses compared to the HA22 containing the original PE38 (Fig. 4). We also examined the mutant anti-sera for the presence of new epitopes and could not detect any new epitopes. This result indicates that the hot spots identified by the strategy we developed are critical for immunogenicity of a whole protein and their elimination by alanine mutagenesis does not create new epitopes.
Fig. 4.
Reduced Immunogenicity of an Immunotoxin with 8 mutations in PE38 in 3 strains of mice. Immunotoxins containing native PE38 or mutant PE38 (8X mutant) were injected intravenously every 14 days into 3 different strains of mice. Specific antibody responses were measured by an ELISA, which only detects antibodies to conformational epitopes on the surface of the protein. Open circles show reactivity with native PE38 and closed circles with the mutant with 8X mutant. In all strains of mice the mutant was less immunogenic and less antigenic. Adapted from The Proceedings of the National Academy of Sciences of the United States of America [published 2008, Vol. 105, 11311–11316].
3.8. T cell epitope-independent deimmunization
To examine the possible role of T cell help we immunized three different strains of mice with different MHC haplotypes (H-2k, H-2b, and H-2d). Although different MHC molecules are supposed to load different regions of peptide fragments for presentation as T cell epitopes, we observed a significant decrease in antibody production for 8X compared to un-mutated PE38 in all three strains of mice (Fig. 4). Thus, the reduced immunogenicity we observed is due to alterations in B cell epitopes, as expected by the non-linkage between the locations of B cell epitopes and T cell epitopes [33, 34]. We suggest that removal of B cell epitopes reduces the immunogenicity likely independent of T cell epitopes and could be a robust deimmunization method that tolerates the high polymorphism of MHC.
4. Concluding remarks
Theoretical considerations and experimental data show that B cell epitope removal is a feasible and effective method to reduce the immunogenicity of a foreign protein. Each step of deimmunization should be carefully designed to take into account the nature of a B cell epitope and its interactions with antibody. B cell epitope-based deimmunization has the advantages that it also removes binding to pre-existing antibodies and is independent of T cell epitopes. Based on these mouse experiments and the theory behind our approach, the method we have developed may be useful for de-immunizing other foreign proteins. We plan to analyze and identify antibody responses in humans with a similar approach.
Acknowledgement
This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Leader B, Baca QJ, Golan DE. Protein therapeutics: a summary and pharmacological classification. Nat. Rev. Drug Discov. 2008;7:21–39. doi: 10.1038/nrd2399. [DOI] [PubMed] [Google Scholar]
- 2.Johnson-Leger C, Power CA, Shomade G, Shaw JP, El Proudfoot A. Protein therapeutics - lessons learned and a view of the future. Expert Opin. Biol. Ther. 2006;6:1–7. doi: 10.1517/14712598.6.1.1. [DOI] [PubMed] [Google Scholar]
- 3.Crommelin DJ, Storm G, Verrijk R, de Leede L, Jiskoot W, Hennink WE. Shifting paradigms: biopharmaceuticals versus low molecular weight drugs. Int. J. Pharm. 2003;266:3–16. doi: 10.1016/s0378-5173(03)00376-4. [DOI] [PubMed] [Google Scholar]
- 4.Subramanyam M. Immunogenicity of biotherapeutics-an overview. J. Immunotoxicol. 2006;3:151–156. doi: 10.1080/15476910600845740. [DOI] [PubMed] [Google Scholar]
- 5.Schellekens H. Immunogenicity of therapeutic proteins: Clinical implications and future prospects. Clin. Ther. 2002;24:1720–1740. doi: 10.1016/s0149-2918(02)80075-3. [DOI] [PubMed] [Google Scholar]
- 6.Khan IA, Gowda RM. Clinical perspectives and therapeutics of thrombolysis. Int. J. Cardiol. 2003;91:115–127. doi: 10.1016/s0167-5273(03)00019-6. [DOI] [PubMed] [Google Scholar]
- 7.Mahajan ST, Brubaker L. Botulinum toxin: from life-threatening disease to novel medical therapy. Am. J. Obstet. Gynecol. 2007;196:7–15. doi: 10.1016/j.ajog.2006.03.108. [DOI] [PubMed] [Google Scholar]
- 8.Yablon SA, Brashear A, Gordon MF, Elovic EP, Turkel CC, Daggett S, Liu JY, Brin MF. Formation of neutralizing antibodies in patients receiving botulinum toxin type A for treatment of poststroke spasticity: A pooled-data analysis of three clinical trials. Clin. Ther. 2007;29:683–690. doi: 10.1016/j.clinthera.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 9.Shankar G, Pendley C, Stein KE. A risk-based bioanalytical strategy for the assessment of antibody immune responses against biological drugs. Nat. Biotechnol. 2007;25:555–561. doi: 10.1038/nbt1303. [DOI] [PubMed] [Google Scholar]
- 10.Pastan I, Hassan R, FitzGerald DJ, Kreitman RJ. Immunotoxin treatment of cancer. Annu. Rev. Med. 2007;58:221–237. doi: 10.1146/annurev.med.58.070605.115320. [DOI] [PubMed] [Google Scholar]
- 11.Pastan I, Hassan R, Fitzgerald DJ, Kreitman RJ. Immunotoxin therapy of cancer. Nat. Rev. Cancer. 2006;6:559–565. doi: 10.1038/nrc1891. [DOI] [PubMed] [Google Scholar]
- 12.FitzGerald DJ, Kreitman R, Wilson W, Squires D, Pastan I. Recombinant immunotoxins for treating cancer. Int. J. Med. Microbiol. 2004;293:577–582. doi: 10.1078/1438-4221-00302. [DOI] [PubMed] [Google Scholar]
- 13.Kreitman RJ, Squires DR, Stetler-Stevenson M, Noel P, FitzGerald DJ, Wilson WH, Pastan I. Phase I trial of recombinant immunotoxin RFB4(dsFv)-PE38 (BL22) in patients with B-cell malignancies. J. Clin. Oncol. 2005;23:6719–6729. doi: 10.1200/JCO.2005.11.437. [DOI] [PubMed] [Google Scholar]
- 14.Kreitman RJ, Wilson WH, Bergeron K, Raggio M, Stetler-Stevenson M, FitzGerald DJ, Pastan I. Efficacy of the anti-CD22 recombinant immunotoxin BL22 in chemotherapy-resistant hairy-cell leukemia. N. Engl. J. Med. 2001;345:241–247. doi: 10.1056/NEJM200107263450402. [DOI] [PubMed] [Google Scholar]
- 15.Laroche Y, Heymans S, Capaert S, De Cock F, Demarsin E, Collen D. Recombinant staphylokinase variants with reduced antigenicity due to elimination of B-lymphocyte epitopes. Blood. 2000;96:1425–1432. [PubMed] [Google Scholar]
- 16.Mayer A, Sharma SK, Tolner B, Minton NP, Purdy D, Amlot P, Tharakan G, Begent RH, Chester KA. Modifying an immunogenic epitope on a therapeutic protein: a step towards an improved system for antibody-directed enzyme prodrug therapy (ADEPT) Br J Cancer. 2004;90:2402–2410. doi: 10.1038/sj.bjc.6601888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Onda M, Nagata S, FitzGerald DJ, Beers R, Fisher RJ, Vincent JJ, Lee B, Nakamura M, Hwang J, Kreitman RJ, Hassan R, Pastan I. Characterization of the B cell epitopes associated with a truncated form of Pseudomonas exotoxin (PE38) used to make immunotoxins for the treatment of cancer patients. J. Immunol. 2006;177:8822–8834. doi: 10.4049/jimmunol.177.12.8822. [DOI] [PubMed] [Google Scholar]
- 18.Onda M, Beers R, Xiang L, Nagata S, Wang QC, Pastan I. An immunotoxin with greatly reduced immunogenicity by identification and removal of B cell epitopes. Proc. Natl. Acad. Sci. U.S.A. 2008;105:11311–11316. doi: 10.1073/pnas.0804851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eisen HN, Siskind GW. Variations in affinities of antibodies during the immune response. Biochemistry. 1964;3:996–1008. doi: 10.1021/bi00895a027. [DOI] [PubMed] [Google Scholar]
- 20.Tai Kim Y, Greenbaum D, Davis P, Fink SA, Werblin TP, Siskind GW. Studies on the control of antibody synthesis. IX. Effect of boosting on antibody affinity. J. Immunol. 1975;114:1302–1306. [PubMed] [Google Scholar]
- 21.Marzocchi-Machado CM, Polizello AC, Azzolini AE, Lucisano-Valim YM. The influence of antibody functional affinity on the effector functions involved in the clearance of circulating immune complexes anti-BSA IgG/BSA. Immunol. Invest. 1999;28:89–101. doi: 10.3109/08820139909061139. [DOI] [PubMed] [Google Scholar]
- 22.Kawade Y, Finter N, Grossberg SE. Neutralization of the biological activity of cytokines and other protein effectors by antibody: theoretical formulation of antibody titration curves in relation to antibody affinity. J. Immunol. Methods. 2003;278:127–144. doi: 10.1016/s0022-1759(03)00203-5. [DOI] [PubMed] [Google Scholar]
- 23.Alpers JH, Steward MW, Soothill JF. Differences in immune elimination in inbred mice. The role of low affinity antibody. Clin. Exp. Immunol. 1972;12:121–132. [PMC free article] [PubMed] [Google Scholar]
- 24.Liang M, Klakamp SL, Funelas C, Lu H, Lam B, Herl C, Umble A, Drake AW, Pak M, Ageyeva N, Pasumarthi R, Roskos LK. Detection of high- and low-affinity antibodies against a human monoclonal antibody using various technology platforms. Assay Drug Dev. Technol. 2007;5:655–662. doi: 10.1089/adt.2007.089. [DOI] [PubMed] [Google Scholar]
- 25.Shankar G, Devanarayan V, Amaravadi L, Barrett YC, Bowsher R, Finco-Kent D, Fiscella M, Gorovits B, Kirschner S, Moxness M, Parish T, Quarmby V, Smith H, Smith W, Zuckerman LA, Koren E. Recommendations for the validation of immunoassays used for detection of host antibodies against biotechnology products. J. Pharm. Biomed. Anal. 2008;48:1267–1281. doi: 10.1016/j.jpba.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 26.MacLennan IC. Germinal centers. Annu. Rev. Immunol. 1994;12:117–139. doi: 10.1146/annurev.iy.12.040194.001001. [DOI] [PubMed] [Google Scholar]
- 27.Parker DC. T cell-dependent B cell activation. Annu. Rev. Immunol. 1993;11:331–360. doi: 10.1146/annurev.iy.11.040193.001555. [DOI] [PubMed] [Google Scholar]
- 28.Bernard A, Coitot S, Bremont A, Bernard G. T and B cell cooperation: a dance of life and death. Transplantation. 2005;79:S8–S11. doi: 10.1097/01.tp.0000153290.75695.31. [DOI] [PubMed] [Google Scholar]
- 29.Abbas AK. Antigen presentation by B lymphocytes: mechanisms and functional significance. Semin. Immunol. 1989;1:5–12. [PubMed] [Google Scholar]
- 30.Pieters J. MHC class II-restricted antigen processing and presentation. Adv. Immunol. 2000;75:159–208. doi: 10.1016/s0065-2776(00)75004-8. [DOI] [PubMed] [Google Scholar]
- 31.Kawabe T, Naka T, Yoshida K, Tanaka T, Fujiwara H, Suematsu S, Yoshida N, Kishimoto T, Kikutani H. The immune responses in CD40-deficient mice: impaired immunoglobulin class switching and germinal center formation. Immunity. 1994;1:167–178. doi: 10.1016/1074-7613(94)90095-7. [DOI] [PubMed] [Google Scholar]
- 32.Renshaw BR, Fanslow WC, Armitage RJ, Campbell KA, Liggitt D, Wright B, Davison BL, Maliszewski CR. Humoral immune-responses in CD40 ligand-deficient mice. J. Exp. Med. 1994;180:1889–1900. doi: 10.1084/jem.180.5.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brons NH, Blaich A, Wiesmuller KH, Schneider F, Jung G, Muller CP. Hierarchic T-cell help to non-linked B-cell epitopes. Scand. J. Immunol. 1996;44:478–484. doi: 10.1046/j.1365-3083.1996.d01-336.x. [DOI] [PubMed] [Google Scholar]
- 34.Rosenberg JS, Atassi MZ. Intersite helper function of T cells specific for a protein epitope that is not recognized by antibodies. Immunol Invest. 1997;26:473–489. doi: 10.3109/08820139709022703. [DOI] [PubMed] [Google Scholar]
- 35.Mukovozov I, Sabljic T, Hortelano G, Ofosu FA. Factors that contribute to the immmunogenicity of therapeutic recombinant human proteins. Thromb. Haemost. 2008;99:874–882. doi: 10.1160/TH07-11-0654. [DOI] [PubMed] [Google Scholar]
- 36.Boulianne GL, Hozumi N, Shulman MJ. Production of functional chimaeric mouse/human antibody. Nature. 1984;312:643–646. doi: 10.1038/312643a0. [DOI] [PubMed] [Google Scholar]
- 37.Morrison SL, Johnson MJ, Herzenberg LA, Oi VT. Chimeric human antibody molecules: mouse antigen-binding domains with human constant region domains. Proc. Natl. Acad. Sci. U.S.A. 1984;81:6851–6855. doi: 10.1073/pnas.81.21.6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones PT, Dear PH, Foote J, Neuberger MS, Winter G. Replacing the complementarity-determining regions in a human antibody with those from a mouse. Nature. 1986;321:522–525. doi: 10.1038/321522a0. [DOI] [PubMed] [Google Scholar]
- 39.Kanduc D. Immunogenicity in peptide-immunotherapy: from self/nonself to similar/dissimilar sequences. Adv. Exp. Med. Biol. 2008;640:198–207. doi: 10.1007/978-0-387-09789-3_15. [DOI] [PubMed] [Google Scholar]
- 40.Shankar G, Shores E, Wagner C, Mire-Sluis A. Scientific and regulatory considerations on the immunogenicity of biologics. Trends Biotechnol. 2006;24:274–280. doi: 10.1016/j.tibtech.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 41.van den Berg HA, Rand DA. Foreignness as a matter of degree: the relative immunogenicity of peptide/MHC ligands. J. Theor. Biol. 2004;231:535–548. doi: 10.1016/j.jtbi.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 42.Paul WE. Fundamental Immunology. Lippincott Williams & Wilkins Philadelphia; 2003. [Google Scholar]
- 43.Goodnow CC. Balancing immunity and tolerance: deleting and tuning lymphocyte repertoires. Proc. Natl. Acad. Sci. U.S.A. 1996;93:2264–2271. doi: 10.1073/pnas.93.6.2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ferry H, Leung JC, Lewis G, Nijnik A, Silver K, Lambe T, Cornall RJ. B-cell tolerance. Transplantation. 2006;81:308–315. doi: 10.1097/01.tp.0000203830.79357.39. [DOI] [PubMed] [Google Scholar]
- 45.Bugelski PJ, Treacy G. Predictive power of preclinical studies of recombinant therapeutic proteins in in animals for the immunogenicity humans. Curr. Opin. Mol. Ther. 2004;6:10–16. [PubMed] [Google Scholar]
- 46.Vanderschueren SM, Stassen JM, Collen D. On the immunogenicity of recombinant staphylokinase in patients and in animal models. Thromb. Haemost. 1994;72:297–301. [PubMed] [Google Scholar]
- 47.McCabe WR, DeMaria A, Johns M. Potential use of shared antigens for immunization against gram-negative bacillary infections. Prog. Clin. Biol. Res. 1980;47:107–117. [PubMed] [Google Scholar]
- 48.Wallach S. The role of calcitonin treatment in postmenopausal osteoporosis. Orthop. Rev. 1992;21:1034–1042. 1130-1031. [PubMed] [Google Scholar]
- 49.Wang ZG, Williams LJ, Zhang XF, Zatorski A, Kudryashov V, Ragupathi G, Spassova M, Bornmann W, Slovin SF, Scher HI, Livingston PO, Lloyd KO, Danishefsky SJ. Polyclonal antibodies from patients immunized with a globo H-keyhole limpet hemocyanin vaccine: isolation, quantification, and characterization of immune responses by using totally synthetic immobilized tumor antigens. Proc. Natl. Acad. Sci. U.S.A. 2000;97:2719–2724. doi: 10.1073/pnas.97.6.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dolimbek BZ, Aoki KR, Steward LE, Jankovic J, Atassi MZ. Mapping of the regions on the heavy chain of botulinum neurotoxin A (BoNT/A) recognized by antibodies of cervical dystonia patients with immunoresistance to BoNT/A. Mol. Immunol. 2007;44:1029–1041. doi: 10.1016/j.molimm.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 51.Atassi MZ, Dolimbek BZ. Mapping of the antibody-binding regions on the H-N-domain (residues 449–859) of botulinum neurotoxin A with antitoxin antibodies from four host species. Full profile of the continuous antigenic regions of the H-chain of botulinum neurotoxin A. Protein J. 2004;23:39–52. doi: 10.1023/b:jopc.0000016257.91979.06. [DOI] [PubMed] [Google Scholar]
- 52.Korber BTM, Brander C, Haynes BF, Koup R, Moore JP, Walker BD, Watkins DI Theoretical Biology and Biophysics Group. Los Alamos, NM: Los Alamos National Laboratory; 2007. HIV Molecular Immunology 2006/2007. [Google Scholar]
- 53.Van Regenmortel MH. Antigenicity and immunogenicity of synthetic peptides. Biologicals. 2001;29:209–213. doi: 10.1006/biol.2001.0308. [DOI] [PubMed] [Google Scholar]
- 54.Selvarajah S, Puffer B, Pantophlet R, Law M, Doms RW, Burton DR. Comparing antigenicity and immunogenicity of engineered gp120. J. Virol. 2005;79:12148–12163. doi: 10.1128/JVI.79.19.12148-12163.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Berzofsky JA. Intrinsic and extrinsic factors in protein antigenic structure. Science. 1985;229:932–940. doi: 10.1126/science.2410982. [DOI] [PubMed] [Google Scholar]
- 56.Van Regenmortel MH. Reductionism and the search for structure-function relationships in antibody molecules. J. Mol. Recognit. 2002;15:240–247. doi: 10.1002/jmr.584. [DOI] [PubMed] [Google Scholar]
- 57.Geysen HM, Tainer JA, Rodda SJ, Mason TJ, Alexander H, Getzoff ED, Lerner RA. Chemistry of antibody binding to a protein. Science. 1987;235:1184–1190. doi: 10.1126/science.3823878. [DOI] [PubMed] [Google Scholar]
- 58.Van Regenmortel MH. What is a B-cell epitope? Methods Mol. Biol. 2009;524:3–20. doi: 10.1007/978-1-59745-450-6_1. [DOI] [PubMed] [Google Scholar]
- 59.Davies DR, Padlan EA, Sheriff S. Antibody-antigen complexes. Annu. Rev. Biochem. 1990;59:439–473. doi: 10.1146/annurev.bi.59.070190.002255. [DOI] [PubMed] [Google Scholar]
- 60.van Regenmortel MHV. Structure of Antigens Volume I. Florida: CRC Press; 1992. [Google Scholar]
- 61.van Regenmortel MHV. Structure of Antigens Volume II. Florida: CRC Press; 1992. [Google Scholar]
- 62.Padlan EA. Antibody-Antigen Complex. Florida: CRC Press; 1994. [Google Scholar]
- 63.Almagro JC. Identification of differences in the specificity-determining residues of antibodies that recognize antigens of different size: implications for the rational design of antibody repertoires. J. Mol. Recognit. 2004;17:132–143. doi: 10.1002/jmr.659. [DOI] [PubMed] [Google Scholar]
- 64.Schlessinger A, Ofran Y, Yachdav G, Rost B. Epitome: database of structure-inferred antigenic epitopes. Nucleic Acids Res. 2006;34:D777–D780. doi: 10.1093/nar/gkj053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Westhof E, Altschuh D, Moras D, Bloomer AC, Mondragon A, Klug A, Van Regenmortel MH. Correlation between segmental mobility and the location of antigenic determinants in proteins. Nature. 1984;311:123–126. doi: 10.1038/311123a0. [DOI] [PubMed] [Google Scholar]
- 66.Blythe MJ, Flower DR. Benchmarking B cell epitope prediction: underperformance of existing methods. Protein Sci. 2005;14:246–248. doi: 10.1110/ps.041059505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Greenbaum JA, Andersen PH, Blythe M, Bui HH, Cachau RE, Crowe J, Davies M, Kolaskar AS, Lund O, Morrison S, Mumey B, Ofran Y, Pellequer JL, Pinilla C, Ponomarenko JV, Raghava GP, van Regenmortel MH, Roggen EL, Sette A, Schlessinger A, Sollner J, Zand M, Peters B. Towards a consensus on datasets and evaluation metrics for developing B-cell epitope prediction tools. J. Mol. Recognit. 2007;20:75–82. doi: 10.1002/jmr.815. [DOI] [PubMed] [Google Scholar]
- 68.Pellequer JL, Westhof E, Van Regenmortel MH. Predicting location of continuous epitopes in proteins from their primary structures. Methods Enzymol. 1991;203:176–201. doi: 10.1016/0076-6879(91)03010-e. [DOI] [PubMed] [Google Scholar]
- 69.Chen J, Liu H, Yang J, Chou K. Prediction of linear B-cell epitopes using amino acid pair antigenicity scale. Amino Acids. 2007;33:423–428. doi: 10.1007/s00726-006-0485-9. [DOI] [PubMed] [Google Scholar]
- 70.Haste Andersen P, Nielsen M, Lund O. Prediction of residues in discontinuous B-cell epitopes using protein 3D structures. Protein Sci. 2006;15:2558–2567. doi: 10.1110/ps.062405906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Reimer U. Prediction of linear B-cell epitopes. Methods Mol. Biol. 2009;524:335–344. doi: 10.1007/978-1-59745-450-6_24. [DOI] [PubMed] [Google Scholar]
- 72.Rubinstein ND, Mayrose I, Pupko T. A machine-learning approach for predicting Bcell epitopes. Mol. Immunol. 2009;46:840–847. doi: 10.1016/j.molimm.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 73.Ponomarenko J, Bui HH, Li W, Fusseder N, Bourne PE, Sette A, Peters B. ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinformatics. 2008;9:8. doi: 10.1186/1471-2105-9-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Peters B, Sette A. Integrating epitope data into the emerging web of biomedical knowledge resources. Nat. Rev. Immunol. 2007;7:485–490. doi: 10.1038/nri2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Peters B, Sidney J, Bourne P, Bui HH, Buus S, Doh G, Fleri W, Kronenberg M, Kubo R, Lund O, Nemazee D, Ponomarenko JV, Sathiamurthy M, Schoenberger SP, Stewart S, Surko P, Way S, Wilson S, Sette A. The design and implementation of the immune epitope database and analysis resource. Immunogenetics. 2005;57:326–336. doi: 10.1007/s00251-005-0803-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Barlow DJ, Edwards MS, Thornton JM. Continuous and discontinuous protein antigenic determinants. Nature. 1986;322:747–748. doi: 10.1038/322747a0. [DOI] [PubMed] [Google Scholar]
- 77.Laver WG, Air GM, Webster RG, Smithgill SJ. Epitopes on protein antigens - misconceptions and realities. Cell. 1990;61:553–556. doi: 10.1016/0092-8674(90)90464-p. [DOI] [PubMed] [Google Scholar]
- 78.Harlow E, Lane P. Epitope Mapping, Using Antibodies: A Laboratory Manual. NY: Cold Spring Harbor Laboratory Press; 1999. pp. 381–405. [Google Scholar]
- 79.Carter JM, Loomis-Price L. B cell epitope mapping using synthetic peptides, Chapter 9. Curr. Protoc. Immunol. 2004 doi: 10.1002/0471142735.im0904s60. Unit 9 4. [DOI] [PubMed] [Google Scholar]
- 80.Fack F, Hugle-Dorr B, Song D, Queitsch I, Petersen G, Bautz EK. Epitope mapping by phage display: random versus gene-fragment libraries. J. Immunol. Methods. 1997;206:43–52. doi: 10.1016/s0022-1759(97)00083-5. [DOI] [PubMed] [Google Scholar]
- 81.Vijayendran RA, Leckband DE. A quantitative assessment of heterogeneity for surface-immobilized proteins. Anal. Chem. 2001;73:471–480. doi: 10.1021/ac000523p. [DOI] [PubMed] [Google Scholar]
- 82.Schwab C, Bosshard HR. Caveats for the use of surface-adsorbed protein antigen to test the specificity of antibodies. J. Immunol. Methods. 1992;147:125–134. doi: 10.1016/s0022-1759(12)80037-8. [DOI] [PubMed] [Google Scholar]
- 83.Butler JE, Ni L, Nessler R, Joshi KS, Suter M, Rosenberg B, Chang J, Brown WR, Cantarero LA. The physical and functional behavior of capture antibodies adsorbed on polystyrene. J. Immunol. Methods. 1992;150:77–90. doi: 10.1016/0022-1759(92)90066-3. [DOI] [PubMed] [Google Scholar]
- 84.Rabbany SY, Piervincenzi R, Judd L, Kusterbeck AW, Bredehorst R, Hakansson K, Ligler FS. Assessment of heterogeneity in antibody-antigen displacement reactions. Anal. Chem. 1997;69:175–182. doi: 10.1021/ac960680h. [DOI] [PubMed] [Google Scholar]
- 85.Jin L, Wells JA. Mutational analysis of antibody binding sites. In: Van Regenmortel M, editor. Structure of Antigens. Vol. 3. Boca Raton: CRC Press; 1995. pp. 21–36. [Google Scholar]
- 86.Mitchell AJ, Edwards MR, Collins AM. Valency or wahlency: is the epitope diversity of the B-cell response regulated or chemically determined? Immunol. Cell Biol. 2001;79:507–511. doi: 10.1046/j.1440-1711.2001.01021.x. [DOI] [PubMed] [Google Scholar]
- 87.Mueller CM, Minnerath JM, Jemmerson R. B lymphocyte recognition of the self antigen mouse cytochrome C in different mouse strains: targeting of the same dominant epitope by naturally-occurring cells expressing distinct VH genes. Mol. Immunol. 1997;34:843–853. doi: 10.1016/s0161-5890(97)00109-0. [DOI] [PubMed] [Google Scholar]
- 88.Tzartos SJ. Epitope mapping by antibody competition. Methodology and evaluation of the validity of the technique. Methods Mol. Biol. 1996;66:55–66. doi: 10.1385/0-89603-375-9:55. [DOI] [PubMed] [Google Scholar]
- 89.Kuroki M, Wakisaka M, Murakami M, Haruno M, Arakawa F, Higuchi H, Matsuoka Y. Determination of epitope specificities of a large number of monoclonal antibodies by solid-phase mutual inhibition assays using biotinylated antigen. Immunol. Invest. 1992;21:523–538. doi: 10.3109/08820139209069388. [DOI] [PubMed] [Google Scholar]
- 90.Nagata S, Numata Y, Onda M, Ise T, Hahn Y, Lee B, Pastan I. Rapid grouping of monoclonal antibodies based on their topographical epitopes by a label-free competitive immunoassay. J. Immunol. Methods. 2004;292:141–155. doi: 10.1016/j.jim.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 91.Jemmerson R. Multiple overlapping epitopes in the three antigenic regions of horse cytochrome c1. J. Immunol. 1987;138:213–219. [PubMed] [Google Scholar]
- 92.Newman MA, Mainhart CR, Mallett CP, Lavoie TB, Smith-Gill SJ. Patterns of antibody specificity during the BALB/c immune response to hen eggwhite lysozyme. J. Immunol. 1992;149:3260–3272. [PubMed] [Google Scholar]
- 93.Jin L, Fendly BM, Wells JA. High resolution functional analysis of antibody-antigen interactions. J. Mol. Biol. 1992;226:851–865. doi: 10.1016/0022-2836(92)90636-x. [DOI] [PubMed] [Google Scholar]
- 94.Jin L, Wells JA. Dissecting the energetics of an antibody-antigen interface by alanine shaving and molecular grafting. Protein Sci. 1994;3:2351–2357. doi: 10.1002/pro.5560031219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cunningham BC, Wells JA. High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science. 1989;244:1081–1085. doi: 10.1126/science.2471267. [DOI] [PubMed] [Google Scholar]
- 96.Meyer DL, Schultz J, Lin YK, Henry A, Sanderson J, Jackson JM, Goshorn S, Rees AR, Graves SS. Reduced antibody response to streptavidin through site-directed mutagenesis. Protein Sci. 2001;10:491–503. doi: 10.1110/ps.19901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lee B, Richards FM. The interpretation of protein structures: estimation of static accessibility. J. Mol. Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 98.Collen D, Bernaerts R, Declerck P, De Cock F, Demarsin E, Jenne S, Laroche Y, Lijnen HR, Silence K, Verstreken M. Recombinant staphylokinase variants with altered immunoreactivity. I: Construction and characterization. Circulation. 1996;94:197–206. doi: 10.1161/01.cir.94.2.197. [DOI] [PubMed] [Google Scholar]
- 99.Nagata S, Ise T, Onda M, Nakamura K, Ho M, Raubitschek A, Pastan I. Cell membrane-specific epitopes on CD30: potentially superior targets for immunotherapy. Proc. Natl. Acad. Sci. U.S.A. 2005;102:7946–7951. doi: 10.1073/pnas.0502975102. [Erratum, ibid 103 (2006) 3007] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Du X, Nagata S, Ise T, Stetler-Stevenson M, Pastan I. FCRL1 on chronic lymphocytic leukemia, hairy cell leukemia, and B-cell non-Hodgkin lymphoma as a target of immunotoxins. Blood. 2008;111:338–343. doi: 10.1182/blood-2007-07-102350. [DOI] [PMC free article] [PubMed] [Google Scholar]