Abstract
Computerized Clinical Decision Support (CDS) aims to aid decision making of health care providers and the public by providing easily accessible health-related information at the point and time it is needed. Natural Language Processing (NLP) is instrumental in using free-text information to drive CDS, representing clinical knowledge and CDS interventions in standardized formats, and leveraging clinical narrative. The early innovative NLP research of clinical narrative was followed by a period of stable research conducted at the major clinical centers and a shift of mainstream interest to biomedical NLP. This review primarily focuses on the recently renewed interest in development of fundamental NLP methods and advances in the NLP systems for CDS. The current solutions to challenges posed by distinct sublanguages, intended user groups, and support goals are discussed.
Keywords: Natural Language Processing, Decision Support Techniques, Clinical Decision Support Systems, Review
1. Introduction
The goal of Clinical Decision Support (CDS) is to “help health professionals make clinical decisions, deal with medical data about patients or with the knowledge of medicine necessary to interpret such data” [1]. Clinical Decision Support Systems are defined as “any software designed to directly aid in clinical decision making in which characteristics of individual patients are matched to a computerized knowledge base for the purpose of generating patient-specific assessments or recommendations that are then presented to clinicians for consideration” [2]. A CDS system structure could be envisioned as a neural reflex arc: its receptors reside in, and are activated by patient data; its integration center contains decision rules and the knowledge base; and its effectors are the patient-specific assessments and recommendations.
Patient data can be manually entered into a CDS system by clinicians seeking support, but then they only get support when they recognize the need and have time to find and enter the requisite data. Support is thus much more effective when the computerized system has access to Electronic Heath Record (EHR) data that can trigger reminders or alerts automatically as situations arise that require physician action1. The EHR will often carry some clinical information, for example, laboratory results, pharmacy orders, and discharge diagnoses in a structured and coded form. Today, a major portion of the patients clinical observations, including radiology reports, operative notes, and discharge summaries are recorded as narrative text (dictated and transcribed, or directly entered into the system by care providers). And in some systems even laboratory and medication records are only available as part of the physician’s notes. Moreover, in some cases the facts that should activate a CDS system can be found only in the free text. For example, the CDC Technical Instructions for Tuberculosis Screening and Treatment2 define a complete screening medical examination for tuberculosis as consisting “of a medical history, physical examination, chest radiography (when required), determination of immune response to Mycobacterium tuberculosis antigens (i.e., tuberculin skin testing, when required), and laboratory testing for human immunodeficiency virus infection … and M. tuberculosis (when required.)” Notably, medical history, physical examination, and chest radiography results are routinely obtained in free-text form. Indications for further tuberculosis screening could be identified in these clinical notes using NLP methods [3] at no additional cost.
In principle, natural language processing could extract the facts needed to actuate many kinds of decisions rules. In theory, NLP systems might also be able to represent clinical knowledge and CDS interventions in standardized formats [4, 5, 6]. That is, NLP could potentially enrich all three major components of CDS systems. The goal of this review is to present the current state of clinical NLP, its contributions to CDS, and the development of biomedical NLP towards processing of clinical narrative and information resources required for further and more involved participation in the CDS process.
Since their introduction about 40 years ago, CDS systems have improved practitioner performance in approximately 60% of the reviewed cases [7]. Several features are correlated with decision support systems’ ability to improve patient care: automatically and proactively providing decision support as part of clinician workflow; providing recommendations rather than assessments; and providing decision support at the time and location of decision making [8]. If CDS systems were to depend upon NLP, it would require reliable, high-quality NLP performance and modular, flexible, and fast systems. Some NLP applications have been integrated in both active and passive CDS. Active NLP CDS applications leverage existing information and push patient-specific information to users. Passive NLP CDS applications require input by the user to generate output.
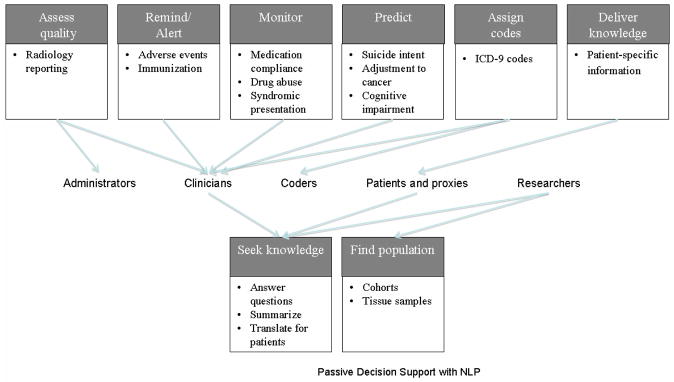
Active NLP CDS includes alerting, monitoring, coding, and reminding. Active NLP CDS is considered quintessential by most interested parties. Passive NLP CDS has focused on providing knowledge and finding patient populations. This review presents the view of NLP CDS which includes active and passive support, and leaves out the related burgeoning areas of tutoring [9] and clinical text mining [10]. Both active and passive NLP CDS systems process a variety of textual sources, such as clinical records, biomedical literature, web pages, and suicide notes. A variety of NLP CDS systems have been targeted to clinicians, but other users are researchers, patients, administrators, students, and coders. Although the application types targeted to clinicians represent a diverse set of active tools, there appears to be a recent trend toward a higher volume of passive tools being targeted to clinicians and researchers. Figure 1 demonstrates the types of active and passive CDS applications to which NLP tools have contributed and have potential to contribute in the future.
Figure 1.
NLP CDS applications
The scope and workflow of an idealized NLP system capable of supporting various clinical decisions and text types are discussed in Section 3. The rest of the review is organized as follows: Section 2 presents a short overview of CDS systems. Section 4 provides an overview of the fundamental NLP methods applied to clinical text. Section 5 describes integrated and end-to-end medical natural language processing systems. Section 6 presents methods and systems for generation of bottom-line advice to clinicians and the public, which are likely to be integrated with CDS systems in the near future. Section 7 presents research on direct applications of NLP in diagnosis, treatment, and other healthcare decisions. The review concludes with potential future directions in natural language processing for clinical decision support.
2. Current state of Clinical Decision Support Systems
Clinical decision support systems can be described along several axes [11, 12]:
access (integrated with an EHR or standalone systems)
setting (used in ambulatory care or inpatient setting)
task (targeting a specific clinical or administrative task such as diagnosis, immunization, or quality control)
scope (general or targeting a specialty)
timing (before, during, or after the clinical decision is made)
output (active, for example, reminders, or passive)
implementation (knowledge-based, statistical)
A thorough review of the systems is beyond the scope of this paper, but further information on CDS and CDS systems can be found in [1, 2, 4, 5, 11, 12].
Much of the data that could support CDS is textual and therefore cannot be leveraged by a CDS system without natural language processing. For example, Aronsky and colleagues studied the usefulness of NLP for a CDS system that identified community-acquired pneumonia in emergency department patients and showed that performance was significantly better with the NLP output [13]. The following examples of integrated CDS systems demonstrate the possibilities for NLP in the context of CDS.
2.1 An outpatient reminder system
In the early 1970s, the Regenstrief Institute introduced prospective, protocol-driven reminders in the outpatient clinics of Wishard Memorial Hospital. The reminder system searched patients’ charts for conditions specified initially in about 300 rules in two categories: ordering specific tests after starting certain drug therapy; and changing drug therapy in response to abnormal test results [11]. For example, if a patient’s serum aminophylline level had not been measured within a certain time after starting aminophylline therapy, the system generated a reminder for the responsible physician suggesting to order the test. The messages suggesting specific actions and explaining the rationale with references to research articles were printed on paper before each patient encounter and attached to the front of the patient’s chart [11, 15, 16].
In a summative two-year evaluation of the expanded system, the clinicians who received reminders undertook the expected actions at a significantly higher rate (44–49% vs. 29%) than those who did not receive the reminders. Interestingly, the study participants never requested articles referenced in the reminders because of time pressures or because they knew the evidence that justified the reminder [11, 15]. Reduction of time required for analysis of evidence presented in research articles is the goal of text summarization and question answering methods discussed in Section 6. Further details on over a quarter century of CDS experience at the Regenstrief Institute can be found in [11, 15, 16].
2.2 Inpatient reminder and diagnostic decision support systems
The HELP (Health Evaluation through Logical Processing) Hospital Information System installed in the Intermountain Healthcare hospitals provides examples of CDS in the inpatient setting. In addition to alert, critique, and suggestion systems similar to the Regenstrief reminder system, the HELP system provides diagnostic decision support. For example, a rule-based subsystem helps diagnose adverse drug events (ADE) through identifying patients with specific chemistry test results, drug level tests, orders for drugs that are commonly used to treat ADEs, and a program in which providers choose symptoms that may be caused by ADEs (e.g., rash, change in heart rate, respiratory rate, mental status, etc.) [17]. Manual entry of the symptoms that may be caused by ADEs could be facilitated or replaced by an NLP tool for extraction of symptoms from free text (for example, from patients’ progress notes), particularly because several NLP systems (discussed in section 5) are already in use at Intermountain Healthcare.
The Antibiotic Assistant [18], initially developed and implemented at LDS Hospital, identifies patients with potential nosocomial infections, alerts physicians to the possible need for anti-infective therapy, and suggests dose and regiment for individual patients. Evans et al. have shown that use of the Antibiotic Assistant decreased the number of ADEs, length of stay, morbidity, and significant reductions in adverse drug events and cost [18]. Recently, the Antibiotic Assistant has been deployed in multiple IHC hospitals and a commercialized version is being marketed across the U.S.3 A key element in the diagnostic reasoning for the Antibiotic Assistant is whether there is radiographic evidence of bacterial pneumonia. Fiszman et al. [19] showed that application of an NLP system to identification of pneumonia performed better than the simple keyword-based method implemented in the Antibiotic Assistant, but integrating the NLP system in a production-level system proved too complex, and the Antibiotic Assistant still uses the internal keyword-based algorithm. If the NLP system were deployed, processing clinical reports in real-time, and storing the NLP annotations, as is the case at Columbia University (see section 5.1), making use of the NLP system’s output would be more straightforward for existing CDSs.
2.3 Decision support centered on CPOE
Many computerized provider order entry (CPOE) systems use controlled vocabularies to avoid unstructured narrative that can “result in confusion among the lab technicians and pharmacists who receive completed orders” [20]. This, however, does not preclude NLP contributions to CDS systems. A passive CPOE-centered NLP CDS system would have the advantage of receiving structured input in the form of patient-specific variables plugged into a text template. It would still need to formally represent knowledge in textual resources and find matching representations. For example, an NLP CDS system could contribute to educational information provided by the Vanderbilt University Medical Center HEO (Horizon Expert Orders, McKesson®), formerly the WizOrder system, which (in addition to recommendations about patient safety and quality of care issues traditionally provided by CPOE systems [16, 17]) provides summaries of disease-specific national guidelines [21] by matching formal representations of guidelines relevant to a specific order. An NLP system could also contribute by processing free text collected by the HEO system, which ranges from comments to structured fields such as orderables for baby aspirin to completely free-text nursing orders (personal communication with Dr. Dominik Aronsky and Dr. Russ Waitman). For example, if the system can process a skin-care related order “turn every three hours”, it can then search the EHR for the last documented turn and issue a reminder, if the elapsed time exceeds the ordered time.
Centering CDS on CPOE allows decision support at various stages of order entry (initiation, patient selection, order selection, order construction, and order completion) [21]. For example, decision support at Partners Healthcare originated at the Brigham and Women’s Hospital as a set of passive tools focused on referential knowledge, anticipated order sets, guidelines and feedback and evolved into an active system integrated with the workflow. For example, if a physician prescribes a drug that lowers potassium, the system displays the patient’s potassium lab values [22].
CPOE systems provide a mechanism for complex, interactive decision support based on protocols and guidelines [21, 23]. Only decidable guidelines shown to be useful in clinical trials and tested against patients’ cases should be provided for CDS in the form of preprogrammed suggestions [23]. Although CPOE systems often collect patient data in structured form, NLP systems could enrich CDS through linking data collected through CPOE with additional information contained in the free-text fields of the EHR and providing assistance in guideline generation, monitoring programmed guidelines for applicability, and generating updates.
3. NLP for CDS: scope and models
The existing NLP and CDS systems provide a solid foundation for the generalized models presented in this section. The models of NLP-CDS systems range from specialized systems dedicated to a specific task, to a set of NLP modules run by a CDS system, to stand-alone systems/services that take clinical text as input and generate output to be used in a CDS system. The implementation and expansion or retargeting of these models differ along the axes shown in Figure 2. The three axes represent relationship types between the NLP and CDS modules: whether the NLP system (1) is integrated within a CDS or coupled with it to various degrees of tightness, (2) is governed by a CDS or implements knowledge and logic necessary to support decisions, or (3) has been developed for a specific task or as a generalized tool that could be customized for different tasks.
Figure 2.
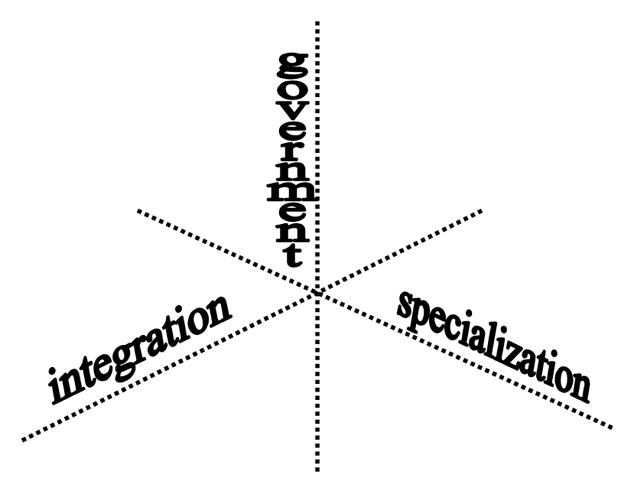
Axes of NLP-CDS relations in clinical NLP models
Particular combinations of system features along the three axes will result in specific NLP-CDS models. Some of those are described in the next section and illustrated in Figures 3 and 4.
Figure 3.
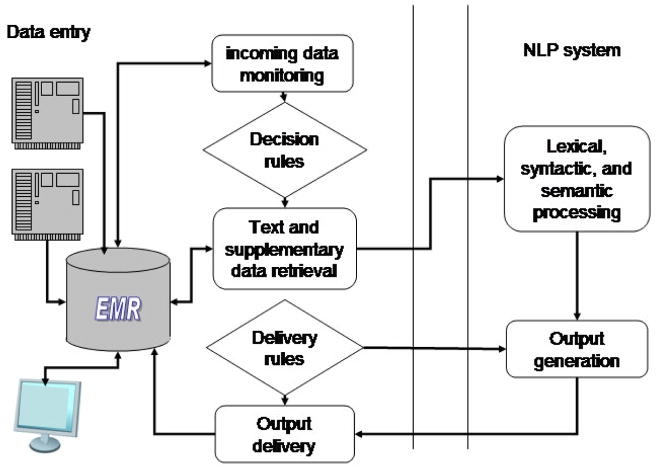
A coupled task-specific NLP system governed by the CDS module
Figure 4.
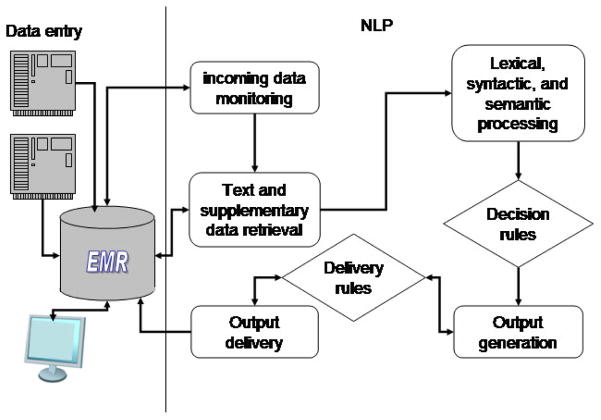
An integrated self-governed multi-task NLP-CDS system
3.1 NLP models
A coupled or integrated NLP system that performs one specific task can, for example, determine whether a chest radiograph report shows evidence of pneumonia or assign a pre-defined subset of ICD-9-CM codes to radiology reports for billing purposes [24]. Such a system might be activated when a new radiology report is submitted to an EHR. The coupled NLP system will be invoked by the EHR (See Figure 3), whereas the integrated system will monitor the incoming reports and start the task as needed (See Figure 4). The NLP system might be self-contained and resort to searching phrases and regular expressions associated with each code or use some of the basic tools described in Section 4. The ICD-9-CM codes obtained by the system and submitted to an EHR could be used to assist human coders while assigning codes or to enable quality control after code assignment. For example, a sophisticated NLP engine is used in a successful, widely-deployed, commercial computer-assisted coding solution, which employs human review for quality assurance of the system output and in cases of low confidence in automatic coding.4
An NLP system governed by and in support of a CDS system comprises a suite of modules that can be selected from and aligned into a pipeline customizable for a variety of tasks. In this model, the CDS system drives and monitors the tightly integrated set of NLP modules and ensures application-specific workflow. Software systems such as the National Library of Medicine’s Unified Medical Language System (UMLS), developed to facilitate clinical data processing and linking to biomedical knowledge [25], General Architecture for Text Engineering (GATE)5, systems based on the Unstructured Information Management applications (UIMA)6 and provided by the Open Health Natural Language Processing (OHNLP) Consortium7, and LingPipe8 can be used in this type of customizable NLP-CDS system coupling.
A specialized NLP system is provided with information about the tasks and takes over the management of the process. The NLP system could be loosely coupled with a CDS system. For example, it may get a signal and text for performing a certain task from a CDS system, perform the task independently, and deliver the pre-specified output to the CDS system, which then incorporates the results into an EHR. Or the NLP system could be used as a module integrated directly in an EHR. Such a system could also use readily available software systems (UMLS, UIMA, GATE, etc.) for basic NLP tasks. A schematic representation of such a system, which seems to be the current model partially implemented at the leading clinical centers, is shown in Figure 4.
The idealized system in Figure 4 will have a module that monitors an EHR for insertion of new data into specific fields. When a radiology report of a patient admitted to a hospital after a pedestrian accident, for example, is entered into the EHR, the NLP system could activate the basic processing pipeline. Processing the following impression section: “Right lower lung opacity, which could be contusion or pneumonia,” the system will extract information about potential pneumonia or pulmonary contusion. The system will look up decision rules for suspected pneumonia that might, for example, contain instructions to retrieve the structured results of blood tests and evaluate the white blood cell count. If the count were high, the reminder message generated by the system would say the patient is more likely to have pneumonia than pulmonary contusion. The system could use the results of the text analysis to solicit more information (for example, find evidence for best approaches to management of both disorders) and present succinct summaries of the information. At this point, the NLP system can hand off the reminder text and summaries to the CDS system, or issue the reminder directly and insert the summaries into designated EHR fields.
The above idealized systems would have to deal with unique challenges faced by existing NLP systems that have to process text and document types ranging from informal notes typed into a patient’s record by various caregivers to highly structured peer reviewed publications in scientific journals. It is not clear whether an NLP system can be designed to handle all text types, applying common modules to both clinical notes and literature, or whether specialized clinical text processing systems should communicate with specialized biomedical literature processing systems using common representation and messaging.
3.2 Text and document types encountered in CDS
Successful processing of clinical narrative is the key to overall success of any NLP-CDS system. This type of text is particularly challenging, because clinical notes are often entered by healthcare providers who have limited time and therefore frequently use domain-specific abbreviations and do not check spelling. Development of NLP processors of clinical text requires access to large volumes of such text, but privacy considerations present barriers to such access Privacy issues have stimulated research in de-identification and anonymization of clinical records [26, 27], in order to reduce the privacy constrains and generate the corpora needed to advance the field. Some modest number of de-identified clinical narratives has been made available to the community [24], however, much larger sets are needed to unleash the potential of NLP and provide access to clinical narratives to NLP researchers working outside of medical centers.
Because CDS involves not only patient-specific information from the clinical record but also general medical knowledge regarding best practices in diagnosing or treating conditions experienced by the patient, NLP beyond current capabilities is needed to find and formally represent publications containing guidelines, CDS rules, and actionable recommendations offered in free text in publicly available online databases (such as MEDLINE/PubMed9, BioMed Central10, and PubMedCentral11) that provide access to scientific literature. Another publicly available resource is “gray literature,” which is more likely to report preliminary, non-significant or negative results than peer-reviewed, commercially published literature. Taking grey literature into account when analyzing and summarizing best available evidence may provide a more complete and objective answer to the question under consideration [28]. For example, averaging over 39 manually conducted meta-analyses alone, treatment was shown to be more effective for preventing an undesirable health outcome compared to manual analysis which including abstracts of conference proceedings, Food and Drug Administration (FDA) documents, and unpublished resources in addition to published literature [28]. Bringing together evidence from formal studies and grey literature is a promising venue for NLP CDS.
CDS-specific language processing builds upon the fundamental clinical text processing briefly described in the next section. The biomedical natural language processing of different document and text types is discussed in parallel throughout the remainder of this review.
4. NLP Building Blocks
Even sophisticated NLP systems are built on the foundation of recognizing words or phrases as medical terms that represent the domain concepts (named entity recognition) and understanding the relations between the identified concepts.
4.1 Text pre-processing
The pre-processing steps leading to term and relation identification usually include tokenization, part-of-speech tagging, and syntactic parsing. Processing of clinical notes often starts with spelling correction and context specific expansion of abbreviations. Unlike many abbreviations in the literature, abbreviations in clinical notes are not often expressed in parenthetical phrases following the expansion; therefore, researchers sometimes treat abbreviation expansion as a word sense disambiguation problem. Similarly, spell checking algorithms that use NLP techniques, such as word sense disambiguation and named entity recognition, perform better than traditional string edit distance algorithms at correcting spelling in clinical notes [29, 30, 31, 32].
Part-of-speech tagging is an essential step in natural language understanding. For example, the following phrase found in a patient’s record can be interpreted differently depending on the assigned part-of-speech tag (the contextually correct tag in square brackets follows the word, the wrong tag assigned by the part-of-speech-tagger is shown in parenthesis): “hemorrhagic [adjective] corpus [adjective] (noun) luteum [adjective](noun) cyst[noun], left [adjective](verb) ovary [noun](adjective).” Given the tagging provided by the part-of-speech-tagger, we should interpret the phrase as a report about the cyst disappearing from the ovary, as opposed to a correct interpretation of a specific cyst found in the left ovary.
Several part-of-speech taggers (POS-taggers) were developed specifically for biomedical domain: the MedPost tagger, based on a hidden Markov model (HMM) and the Viterbi algorithm, was trained and tested on 5700 manually tagged sentences, achieving 97% accuracy on sentences extracted from various thematic MEDLINE subsets [33]. Despite an opinion that at the part-of-speech tagging level, sublanguage differences seem to vanish [34], there is evidence that POS-taggers trained and tested on formal text that does not include clinical documents do not achieve state-of-the-art performance. For example, training a POS-tagger on a relatively small set of clinical notes improves the performance of the POS-tagger trained on Penn Treebank from 90% to 95% in one study [35] and from 79% to 94% in another study [36].
One of the main causes of errors when porting part-of-speech taggers to new domains is assignment of tags to out-of-vocabulary words in the new domain. Errors at the part-of-speech level can propagate upward to create more errors at the syntactic processing level. Errors in syntactic analysis, which provides information necessary for semantic interpretation of both the clinical narrative and the biomedical literature text, can, in turn, cause errors in text understanding.
4.2 NER
Named entity recognition (NER) involves identifying the boundaries of the name in the text and understanding (and disambiguating) its meaning, often through mapping the entity to a unique concept identifier in an appropriate ontology [37].
4.2.1 Dictionary-based NER
By its nature, dictionary-based NER needs resources that at a minimum provide a list of names for a given entity type. For example, the NCI Dictionary of Genetics Terms12, contains about 100 terms and their definitions that support genetics cancer information summaries. This and other dictionaries and thesauri are included in the UMLS that preserves information from the original contributing sources and enriches it through linking and adding meta-information, such as semantic types. The UMLS 2009AA version13 includes 2,125,395 concepts with 8,006,171 distinct names contributed by 152 sources and merged into the Metathesaurus (UMLS Meta). Many NER methods (applied to both the clinical narrative and the biomedical literature text) utilize UMLS Meta and tools developed within the UMLS.
The expanse and origins of the UMLS Metathesaurus present the need for customization to address specific needs and sublanguages, as demonstrated in the comparative study of UMLS content views to support NLP processing of biomedical literature and clinical text [38]. One of the issues faced by dictionary-based methods is whether the lexicon coverage is sufficient for specific sub-domains [39]. The coverage of the semantic lexicon could be increased employing morphosemantics-based systems capable of generating definitions for unknown terms [40]. In clinical NLP, it might be desirable to use a local lexicon instead of or in addition to significant domain knowledge captured in the UMLS. Manual integration of local terminologies and the UMLS can be aided through WordNet-based mapping methods [41].
4.2.2 Statistical NER
One of the successful alternatives to dictionary-based methods is the supervised machine learning approach to biomedical NER. This approach usually requires a substantial manual annotation effort. For example, in one effort 1,442 MEDLINE abstracts were manually tokenized and annotated for automatic recognition of malignancy named entities [42]. Reducing annotation effort for NER can be achieved through dynamic selection of sentences to be annotated [43] or through active learning [44]. These methods could be of value for annotation of clinical notes because the existing collections of annotated clinical notes are significantly smaller than those of the medical literature (most dataset owners report gold standards around 160 notes [45, 46]. The F-scores achieved for statistical NER on these collections range from low 70s [46] to 86% [47]. It remains to be seen if a larger annotated collection of clinical notes will prove beneficial for statistical NER.
An in-depth overview of the dictionary-based, rule-based, statistical, and hybrid approaches to automatic named entity recognition in biomedical literature is provided in Krauthammer and Nenadic [48]. Meystre et al. review information extraction from clinical narrative [49].
4.3 Context extraction
The key functions of clinical decision support systems require understanding the context from which an event or a named entity is extracted. For example, supporting clinical diagnosis and treatment processes with best evidence will require not only recognizing a clinical condition, but determining whether the condition is present or absent. Several algorithms have been developed for negation identification [50, 51, 52, 53]. Chapman et al. [54] developed a stand-alone algorithm, ConText, for identifying three contextual features: Negation (for example, no pneumonia); Historicity (the condition is recent, occurred in the past, or might occur in the future); and Experiencer (the condition occurs in the patient or in someone else, such as parents abuse alcohol). In many cases it is desirable to detect the degree of certainty in the context (for example, suspected pneumonia). Solt and colleagues described an algorithm for determining whether a condition is absent, present, or uncertain [55], and Uzuner and colleagues compared rule-based and machine learning approaches to assertion classification [56]. Aramaki et al. developed an application for creating tables from clinical texts that places information in the context of whether the information is negated, may occur in the future, and is needed, planned, or recommended [57]. Denny and colleagues developed an application for identifying timing and status of colonscopy testing from reports, including whether a test was described in the context of being refused or scheduled [58]. A comprehensive review of temporal reasoning with medical data can be found in [59].
4.3 Associations and relations extraction
A better understanding of the clinical narrative text might be gained through identification and extraction of meaningful relationships between the identified entities and events. Similarly to NER, relation extraction can be decomposed into relation detection and determination of the relation type. Several resources contain relation types for the biomedical domain, for example, the UMLS Semantic Network14 that defines binary relations allowed between the UMLS semantic types. Although annotation efforts sometimes include relations [46, 60], it remains to be seen whether explicitly stated relations occur in clinical narrative regularly and frequently enough to be necessary or useful for clinical decision support and whether experience in relation extraction from the literature [61, 62] can be leveraged in clinical text processing.
In the absence of explicitly stated relations, researchers rely on co-occurrence statistics of certain semantic types to infer a relationship. For example, Chen et al. [10] applied the Medical Language Extraction and Encoding System (MedLEE) and BioMedLEE [63] to discharge summaries and MEDLINE articles to identify disease and drug entities and the chi-square statistic to measure the significance of associations between diseases and drugs. The subsequent overview of the top five disease-drug associations by a medical expert across eight diseases confirmed the appropriateness of the method for extracting disease-drug associations from both text sources [10].
To date, the bulk of the relation extraction experience stems from processing of the literature. One of the leading systems in this research, SemRep, is a rule-based, symbolic natural language processing system developed to identify relations defined in the UMLS Semantic Network. SemRep relies on its “indicator” rules to map syntactic elements (such as verbs and nominalizations) to predicates in the Semantic Network, such as TREATS, CAUSES, and LOCATION_OF. Argument identification rules (which take into account coordination, relativization, and negation) then find syntactically allowable noun phrases to serve as arguments for indicators [64]. Propositional representation (predications) of the TREATS(DISORDER, PHARMACOLOGICAL_SUBSTANCE) relation identified using SemRep significantly outperformed the co-occurrence frequency method in finding evidence for treatment suggestions for over 50 diseases, gaining 0.17 in mean average precision [65].
5. Current state of clinical NLP systems
The currently existing systems roughly fall into two categories: general-purpose clinical NLP architectures (increasingly publicly available), and specialized systems developed for specific tasks.
5.1 General-purpose clinical NLP systems
The early vision of medical NLP was implemented in the Linguistic String Project (LSP) system that developed the basic components and the formal representation of clinical narrative, and implemented the transformation of the free-text clinical documents into a formal representation [66]. The LSP system evolved into the Medical Language Processor (MLP) that includes the English healthcare syntactic lexicon and medically tagged lexicon, the MLP parser, parsing with English medical grammar, selection with medical co-occurrence patterns, English transformation, syntactic regularization, mapping into medical information format structures, and a set of XML tools for browsing and display15.
MedLEE is an NLP system that extracts information from clinical narratives and presents this information in structured form using a controlled vocabulary. MedLEE uses a lexicon to map terms into semantic classes and a semantic grammar to generate formal representation of sentences. It is in use at Columbia University Medical Center, and is one of the few natural language processing systems integrated with clinical information systems. MedLEE has been successfully used to process radiology reports, discharge summaries, sign-out notes, pathology reports, electrocardiogram reports, and echocardiogram reports [10, 67, 68, 69, 70]. An in-depth overview of the system and a case scenario are provided in [71].
The Text Analytics architecture developed in collaboration between the Mayo Clinic and IBM is using Unstructured Information Management Architecture (UIMA) to identify clinically relevant entities in clinical notes. The entities are subsequently used for information retrieval and data mining [72]. The ongoing development of this architecture resulted in two specialized pipelines: medKAT/P, which extracts cancer characteristics from pathology reports, and cTAKES, which identifies disorders, drugs, anatomical sites, and procedures in clinical notes. Evaluated on a set of manually annotated colon cancer pathology reports, MedTAS/P achieved F1-scores in the 90% range in extraction of histology, anatomical entities, and primary tumors [73]. A lower score achieved for metastatic tumors was attributed to the small number of instances in the training and test sets [73]. cTAKES and HiTEx, described below, are the first generalized clinical NLP systems to be made publicly available.
Developed at the National Center for Biomedical Computing, Informatics for Integrating Biology & the Bedside (I2B2), the Health Information Text Extraction (HiTEx) tool based on GATE is a modular system that assembles a different pipeline for extracting specific findings from clinical narrative. For example, a pipeline to extract diagnoses is formed by applying sequentially a section splitter, section filter, sentence splitter, sentence tokenizer, POS tagger, noun phrase finder, UMLS concept mapper, and negation finder [74]. A pipeline for extraction of family history from discharge summaries and outpatient clinic notes evaluated on 350 sentences achieved 85% precision and 87% recall in identifying diagnoses; 96% precision and 93% recall differentiating family history from patient history; and 92% precision and recall exactly assigning diagnoses to family members [75].
The MediClass (a “Medical Classifier”) system was designed to automatically detect clinical events in any electronic medical record by analyzing the coded and free-text portions of the record. It was assessed in detecting care delivery for smoking cessation; immunization adverse events; and subtypes of diabetic retinopathy. Although the system architecture remained constant for each clinical event detection task, new classification rules and terminology were defined for each task [76]. For example, to detect possible vaccine reactions in the clinical notes, MediClass developers identified the relevant concepts and the linguistic structures used in clinical notes to record and attribute an adverse event to an immunization or vaccine [77]. The identified terms and structures were encoded into rules of a MediClass knowledge module that defines the classification scheme for automatic detection of possible vaccine reactions. The scheme requires detecting an explicit mention of an immunization event and detecting or inferring at least one finding of an adverse event [77]. In 227 of 248 cases (92%), MediClass correctly detected a possible vaccine reaction [77].
5.2 Specialized clinical NLP systems
The evaluation of the general-purpose architectures in specific tasks and the use of the general-purpose systems as components or foundation of many task- or document-specific systems, make the line between these system types somewhat fuzzy. The differences are most probably not in the end-results but in the initial goals of developing a system to process free-text for any task versus solving a specific clinical task. Independent of the starting point and reuse of the general-purpose components, solving a specific task at minimum requires developing a task-specific database and decision rules. Some examples of task-specific systems are provided in this section.
5.2.1 Clinical events monitoring
Clinical events monitoring is one of the most common and essential tasks of CDS systems. Particularly important are detection and prevention of adverse events. Murff et al. found the electronic discharge summaries to be an excellent source for detecting adverse events; however, practically useful automatic detection of those events could not be achieved using simple keyword queries to trigger an alert [78]. Building upon rule-based extraction of clinical conditions from radiology reports [67], Hripcsak et al. describe a NLP-based framework for adverse event discovery [79]. The event discovery process involved seven steps: 1) identification of the target event, for example, drug interactions; 2) selection of a clinical data repository for monitoring; 3) natural language processing to formally represent clinical narrative. The formal representation was generated by MedLEE [68]; 4) query generation for event detection and classification; 5) verification of the accuracy of detection and classification; 6) error analysis; 7) iterative improvement of steps 1, 3, and 4 based on findings in steps 5 and 6. The framework sensitivity ranged from 0.15 to 0.37 at 0.99 specificity levels in identifying 45 types of adverse events such as pulmonary embolism, wound dehiscence requiring repair, medication errors, and other serious adverse events [79].
5.2.2 Processing radiology reports
Radiology reports are probably the most studied type of clinical narrative. This extremely important source of clinical data provides information not otherwise available in the coded data and allows performing tasks from coding of the findings and impressions [67, 80], to detection of imaging technique suggested for follow-up or repeated examinations [81], to decision support for nosocomial infections [19], to biosurveillance [82]. The complete description of systems developed for processing of radiology reports is beyond the scope of this review. This section outlines the scope and research directions in processing of radiology reports and omits most of the radiology report studies based on the described above general-purpose systems.
A family of systems with the initial goal of processing radiology reports was developed at the LDS Hospital (Intermountain Healthcare). The Special Purpose Radiology Understanding System (SPRUS) extracts and encodes the findings and the radiologists’ interpretations using information from a diagnostic expert system [80]. SPRUS was followed by the Natural language Understanding Systems (NLUS) and Symbolic Text Processor (SymText) systems that combine semantic knowledge stored and applied in the form of a Bayesian Network with syntactic analysis based on a set of augmented transition network (ATN) grammars [83, 84]. SymText was deployed at LDS Hospital for semi-automatic coding of admit diagnoses to ICD-9 codes [85]. SymText was also used to automatically extract interpretations from Ventilation/Perfusion lung scan reports for monitoring diagnostic performance of radiologists [86]. The accuracy of the system in identifying pneumonia-related concepts and inferring the presence or absence of acute bacterial pneumonia was evaluated using 292 chest x-ray reports annotated by physicians and lay persons. The 95% recall, 78% precision, and 85% specificity achieved by the system were comparable to that of physicians and better than that of lay persons [19]. SymText evolved to MPLUS (M+), which also uses a semantic model based on Bayesian Networks (BNs), but differs from SymText in the size and modularity of its semantic BNs and in its use of a chart parser [87]. M+ was evaluated for the extraction of American College of Radiology utilization review codes from 600 head CT reports. The system achieved 87 % recall, 98% specificity and 85% precision in classifying reports as positive (containing brain conditions) [87]. M+ was also evaluated for classifying chief complaints into syndrome categories [88]. Currently, M+ has been redesigned as Onyx and is being applied to spoken dental exams [89]. These evolving NLP systems provide examples of successful retargeting to coding of other types of clinical reports.
Elkin et al. [82] presented a specialized tool based on the general-purpose architecture, which was used to code radiology reports into the SNOMED CT reference terminology. The subsequent processing was based on the SNOMED CT encoded rule for the identification of Pneumonias, Infiltrates or Consolidations or other types of pulmonary densities [82]. The rule consisted of 17 increasingly complex clauses, starting with “if pneumonia 233604007 Positive Assertion Explode Impression Section --> Positive Assertion Pneumonia”. Identification of pneumonias was evaluated on 400 reports and resulted in 100% recall (sensitivity), 97% precision (positive predictive value), and 98% specificity [82].
The REgenstrief data eXtraction tool (REX) coded raw version 2.x Health Level 7 (HL7) messages to a targeted small to medium sized sets of concepts for a particular purpose in a given kind of narrative text [90]. REX was applied to 39,000 chest x-rays performed at Wishard Hospital in a 21-month period to identify findings related to CHF, tuberculosis, pneumonia, suspected malignancy, compression fractions, and several other disorders. REX achieved 100% specificity for all conditions, 94% to 100% sensitivity, and 95% to 100% positive predictive value, outperforming human coders in sensitivity [90]. In contrast, mapping six types of radiology reports to a UMLS subset and then selectively recognizing most salient concepts using information retrieval techniques, resulted in 63% recall and 30% precision [91].
5.2.3 Processing emergency department reports
Topaz targets 55 clinical conditions relevant for detecting patients with an acute lower respiratory syndrome [92]. Topaz uses three methods for mapping text to the 55 conditions: index UMLS concepts with MetaMap [93]; create compound concepts from UMLS concepts or keywords and section titles (e.g., Section:Neck + UMLS concept for lymphadenopathy = Cervical Lymphadenopathy); and identify measurement-value pairs (e.g., “temp” + number > 38 degrees Celsius = Fever). Topaz is built on the GATE platform and implements ConText as a GATE module for determining whether indexed conditions are present or absent, experienced by the patient or someone else, and historical, recent, or hypothetical. After integrating potentially multiple mentions of a condition from a report, agreement between Topaz and physicians reading the report was 0.85 using weighted kappa.
5.2.4 Processing pathology reports
Surgical pathology reports are another trove of clinical data for locating information about appropriate human tissue specimens [94] and supporting cancer research. For example, a preprocessor integrated with MedLEE to abstract 13 types of findings related to risks of developing breast cancer achieved a sensitivity of 90.6% and a precision of 91.6% [69].
In MEDSYNDIKATE, an NLP system for extraction of medical information from pathology reports, the basic sentence-level understanding of the clinical narrative (that takes into consideration grammatical knowledge, conceptual knowledge, and the link between syntactic and conceptual representations) is followed by the text-level analysis that tracks reference relations to eliminate representation errors [95].
Liu et al. assessed the feasibility of utilizing an existing GATE pipeline for extraction of the Gleason score (a measure of tumor grade), tumor stage, and status of lymph node metastasis from free-text pathology reports [96]. The pipeline was evaluated on committing errors related to the text processing and extraction of values from the report, and errors related to semantic disagreement between the report and the gold standard. Each variable had a different profile of errors. Numerous system errors were observed for Gleason Score extraction that requires fine distinctions and TNM stage extraction requiring multiple discrete decisions. The authors conclude that the existing system could be used to aid manual annotation or could be extended for automatic annotation [96]. These findings second observations of Schadow and McDonald that general-purpose tools and vocabularies need to be adapted to the specific needs of surgical pathology reports [94]. The Cancer Text Information Extraction System (caTIES) system, built on a GATE framework, uses MetaMap [93] and NegEx [51] to annotate findings, diagnoses, and anatomic locations in pathology reports. caTIES provides researchers with the ability to query, browse and create orders for annotated tissue data and physical material across a network of federated sources using automatically annotated pathology reports16.
5.2.5 Processing a mixture of clinical note types
The above studies indicate that natural language processing acceptable for clinical decision support is better achieved using tools developed for specific tasks and document types. It is therefore not surprising that processing of a mixture of clinical notes is successful when the task is well-defined and a small knowledge base is developed specifically for the task. For example, Meystre and Haug created a subset of the UMLS Metathesaurus for 80 problems of interest to their longitudinal Electronic Medical Record and evaluated extraction of these problems from 160 randomly selected discharge summaries, radiology reports, pathology reports, progress notes, and other document types [97]. The evaluation demonstrated that using a general purpose entity extraction tool with a custom data subset, disambiguation, and negation detection achieves 89.2% recall and 75.3% precision [97].
The MediClass system [76] was configured to automatically assess delivery of evidence-based smoking-cessation care [98]. A group of clinicians and tobacco-cessation experts met over several weeks to encode the recommended treatment model using the concepts and the types of phrases that provide evidence for smoking-cessation medications, discussions, referral activities, quitting activities, smoking and readiness-to-quit assessments. The treatment model involves five steps, “5A’s”: (1) ask about smoking status; (2) advise to quit; (3) assess a patient’s willingness to quit; (4) assist the patient’s quitting efforts; and (5) arrange follow-up. Evaluated on 500 patient records containing structured data in addition to progress notes, patient instructions, medications, referrals, reasons for visit, and other smoking-related data, MediClass performance was judged adequate to replace human coders of the 5A’s of smoking-cessation care [98].
The InfoBot system under development at the National Library of Medicine identifies the elements of a well-formed clinical question [99] in clinical notes. It subsequently invokes a question answering module (the CQA 1.0 system described in section 6.3) that extracts answers to the question about the best care plan for a given patient with the identified problems from the literature, and delivers documents containing the answers [100]. In a pilot evaluation by 16 NIH Clinical Center nurses, each evaluating 15 patient cases, documents containing answers were found to be relevant and useful in the majority of cases [101]. It remains to be seen if such automated methods of linking evidence to a patient’s record can achieve the accuracy of more controlled delivery implemented in Infobuttons, decision support tools that deliver information based on the context of the interaction between a clinician and an EHR [102]. Automatic linking of external knowledge bases and patients’ records will be useful if the NLP systems achieve acceptable accuracy in extraction of bottom-line advice and presentation of this information in an easily comprehensible form. Extraction of the bottom-line advice and answers to clinical questions are presented in the next section.
6. Providing evidence: Personalized context-sensitive summarization and question answering
The need to link evidence to patients’ records was stated in the 1977 assessment of computer-based medical information systems undertaken because of increased concern over the quality and rising costs of medical care [103]. The assessment concluded that the quality and cost concerns could be addressed by medical information systems that will supply physicians with information and incorporate valid findings of medical research [103]. The results of medical research might soon become directly available through querying clinical research databases, however to date, findings of medical research can be primarily found in the literature. Following the 1977 report, medical informatics research focused on understanding physicians’ information needs and enabling physicians’ access to the published results of clinical studies. This research provides a solid foundation for NLP aimed at satisfying physicians’ desiderata. The most desired features include comprehensive specific bottom-line recommendations that anticipate and directly answer clinical questions, rapid access, current information, and evidence-based rationale for recommendations [104].
One important summarization task is to provide an overview of the latest scientific evidence pertaining to a specific clinical situation. The secondary sources that compile evidence found in the scientific publication (such as Family Physicians Inquiry Network17 and BMJ Clinical Evidence18) deliver expert-generated support in the form of short answers to clinical questions followed by summaries. This model is partially implemented in several systems described in this section.
6.1 Clinical data and evidence summarization for clinicians
Unlike the comparatively better researched summarization and visualization of structured clinical data [105, 106, 107, 108], summarization of clinical narrative is an evolving area of research. Afantenos et al surveyed the potential of summarization technology in the medical domain [109]. Van Vleck et al identified information physicians consider relevant to summarizing a patient’s medical history in the medical record. The following categories were identified as necessary to capturing patient’s history: Labs and Tests, Problem and Treatment, History, Findings, Allergies, Meds, Plan, and Identifying Info [110]. Meng et al. approached generation of clinical notes as an extractive summarization problem [111]. In this approach, sentences containing patient information that needs to be repeated are extracted based on their rhetorical categories determined using semantic patterns. This extraction method compares favorably to the baseline extraction method (the position of a sentence in the note) on a test set of 162 sentences in urological clinical notes [111]. Cao et al summarized patients’ discharge summaries into problem lists [70].
The PERSIVAL project (a prototype system, not currently in use) summarized medical scientific publications [112, 113]. The summarization module of the PERSIVAL system generated summaries tailored for physicians and patients. Summaries generated for a physician contained information relevant to a specific patient’s record. Each publication was represented using a set of templates. Templates were then clustered into semantically related units in order to generate a summary [112, 113].
Based on the semantic abstraction paradigm, Fiszman et al. are developing a summarization system that relies on SemRep for semantic interpretation of the biomedical literature. The system condenses SemRep predications and presents them in graphical format [114]. We hope to see in the future if the above method holds promise for summarization and visual presentation of clinical notes
6.2 Clinical data and evidence summarization for patients
The online access to personal health and medical records and the overwhelming amount of health-related information available to patients (alternatively called health care consumers and lay users) pose many interesting questions. Hardcastle and Hallet studied which text segments of a patient record require explanation before being released to patients and what types of explanation are appropriate [115]. Elhadad and Sutaria presented an unsupervised method for building a lexicon of semantically equivalent pairs of technical and lay medical terms [116].
Ahlfeldt et al surveyed issues related to communicating technical medical terms in everyday language for patients and generating patient-friendly texts [117]. The survey presents research on alleviating the lack of understanding of clinical documents caused by medical terminology. This research includes generation of patient vocabularies and matching those vocabularies and problem lists with standard terminologies; generation of terminological resources, corpora and annotation tool; development of natural consumer language generation systems; and customization of patient education materials [117]. Green presents the design of a discourse generator that plans the content and organization of lay-oriented genetic counseling documents to assist drafting letters that summarize the results for patients [118].
6.3 Clinical question answering
One of the principal purposes of CDS is answering questions [14]. Questions occurring in clinical situations could pertain to “information on particular patients; data on health and sickness within the local population; medical knowledge; local information on doctors available for referral; information on local social influences and expectations; and information on scientific, political, legal, social, management, and ethical changes affecting both how medicine is practiced and how doctors interact with individual patients” [119]. Some questions do not need NLP and can be answered directly by a known recourse. For example, the NLM Go Local service19 (which connects users to health services in their local communities and directs users of the Go Local sites to MedlinePlus health information) was established to answer logistics questions by providing access to local information. Questions about particular patients are currently answered by manually browsing or searching the EHR. Answering these questions can be facilitated by summarization (which requires NLP if information is extracted from free text fields) and visualization tools [105, 106, 107, 108]. Facilitating access to medical knowledge by providing answers to clinical questions is an area of active NLP research [120]. The goal of clinical question answering systems is to satisfy medical knowledge questions providing answers in the form of short action items supported by strong evidence.
Jacquemart and Zweigenbaum studied the feasibility of answering students’ questions in the domain of oral pathology using Web resources. Questions involving pathology, procedures, treatments, examinations, indications, diagnosis and anatomy were used to develop eight broad semantic models comprised of 66 different syntactico-semantic patterns representing the questions. The triple-based model ([concept]–(relation)–[concept]) combined with which, why, and does modalities accounted for a vast majority of questions. The formally represented questions were used to query 10 different search engines. Search results were checked manually to find a passage answering the question in a consistent context [121].
The [concept]–(relation)–[concept] triples generated by SemRep can be used to generate conceptual condensates that summarize a set of documents [114], or answer specific questions, for example, finding the best pharmacotherapy for a given disease [65]. Within the EpoCare project, the same question type is answered by using an SVM to classify MEDLINE abstract sentences as containing an outcome (answer) or not and extracting the high-ranking sentences [122]. The CQA-1.0 system also implements an Evidence Based Medicine (EBM)-inspired approach to outcome extraction [120]. In addition to extracting outcomes from individual MEDLINE abstracts to answer a wide range of questions, the CQA-1.0 system aggregates answers to questions about the best drug therapy into 5–6 drug classes generated based on the individual pharmaceutical treatments extracted from each abstract. Each class is supported by the strongest patient-oriented outcome pertaining to each drug in the class.. The EpoCare and CQA-1.0 systems rely on the Patient-Intervention-Comparison-Outcome (PICO) framework developed to help clinicians formulate clinical questions [99]. The MedQA system answers definitional questions by integrating information retrieval, extraction, and summarization techniques to automatically generate paragraph-level text [123].
7. Clinical NLP: Direct applications of NLP in healthcare
In addition to processing text pertaining to patients and generated by clinicians and researchers, NLP methods have been applied directly to patients’ narratives for diagnostic and prognostic purposes.
The Linguistic Inquiry and Word Count (LIWC)20 tool was used to explore personality expressed through a person’s linguistic style [124]. The LIWC tool (which calculates the percentage of words in written text that match up to 82 language dimensions) was evaluated in predicting post-bereavement improvements in mental and physical health [125], predicting adjustment to cancer [126], differentiating between the Internet message board entries and homepages of pro-anorexics or recovering anorexics [127], and recognizing suicidal and non-suicidal individuals [128]. Pestian et al. demonstrated that the sequential minimization optimization algorithm can classify completer and simulated suicide notes as well as mental health professionals [129].
Another potential clinical NLP application is assessment of neurodegenerative impairments. Roark et al studied automation of NLP methods for diagnosis of mild cognitive impairment (MCI). Automatic psychometric evaluation included syntactic annotation and analysis of spoken language samples elicited during neuropsychological exams of elderly subjects. Evaluation of syntactic complexity of the narrative was based on analysis of dependency structures and deviations from the standard (for English) right-branching trees in parse trees of subjects’ utterances. Measures derived from automatic parses highly correlated with manually derived measures, indicating that automatically derived measures may be useful for discriminating between healthy and MCI subjects. [130].
Clinical NLP is also used for medication compliance and drug abuse monitoring. Butler et al explored usefulness of content analysis of Internet message board postings for detection of potentially abusable opioid analgesics [131]. In this study, attractiveness for abuse of OxyContin®, Vicodin®, and Kadian® determined automatically (using the total number of posts by product, total number of mentions by product (including synonyms and misspellings), total number of posts containing at least one mention of each product, total number of unique authors, and the number of unique authors of posts referencing any of the 3 target products) was compared to the known attractiveness of the products. The numbers of mentions of the products were significantly different and corresponded to the product attractiveness. Based on this and other metrics, the authors conclude that a systematic approach to post-marketing surveillance of Internet chatter related to pharmaceutical products is feasible [131]. Understanding patient compliance issues could help in clinical decisions. This understanding could be gained through processing of informal textual communications found in the publicly available blog postings and e-mail archives. For example, Malouf et al. analyzed 316,373 posts to 19 Internet discussion groups and other websites from 8,731 distinct users and found associations (such as cognitive side effects, risks, and dosage related issues) the epilepsy patients and their caregivers have for different medications [132].
To the best of our knowledge, the applications described in this section are experimental rather than deployed and regularly used in clinical setting. The difficulties in translation of clinical NLP research into clinical practice and obstacles in determining the level of practical engagement of NLP systems are discussed in the next section.
8. Conclusions and thoughts on Future Work
Discussing the road ahead for the clinical decision support systems, Greenes notes that despite the demonstrated benefits and local successes of CDS systems, the past 45 years of CDS research have not been translated into widespread use and daily practice [14]. This observation can be expanded to NLP systems and methods for CDS. The strong foundation and local successes combined with the renewed community-wide interest to medical language processing provide hope that mature NLP systems for CDS will become available to the wider community in the near future.
Most of the above presented methods and systems were developed for specific users, document types and CDS goals. Future research might indicate if such systems could be easily retargeted for new users and goals and whether the retargeted systems can compete with those designed for specific tasks and clinical systems. Evaluation methods for measuring the impact of NLP methods on healthcare in addition to reliable standardized evaluation of NLP systems need to be developed.
For several issues very important to the future development of NLP for CDS, there is currently only anecdotal evidence and sparse publications. For example, with few exceptions, we do not know which of the reviewed NLP-CDS systems are actually implemented or deployed, and what makes these systems worthwhile. We might speculate that, for example, MedLEE is successfully integrated with a clinical information system because it was developed and adapted, as needed, for specific users and CDS goals, but the reason for its success could also be its sophisticated NLP. We could better judge which features determine whether NLP-CDS systems are applied outside of the experimental setting if we had more data points. We believe it would be valuable to have a special venue for presenting case studies and analysis of applied NLP systems in the near future.
Priorities in NLP development will be determined by the readiness of intended users to adopt NLP. The early successes in NLP and CDS led to high user expectations that were not always met. NLP researchers need to re-gain clinicians’ trust, which is achievable based on better understanding of the NLP strengths and weaknesses by clinicians, as well as significant progress in biomedical NLP. Reacquainting clinicians with NLP can be facilitated by NLP training, well-planned NLP experiments, careful and thoughtful evaluation of the results, high quality implementation of NLP modules, semi-automated and easier methods for adapting NLP for other domains, and evaluations of NLP-CDS adequacy in satisfying user needs.
We believe NLP can contribute to decision support for all groups involved in the clinical process, but the development will probably focus on the areas for which there is higher demand. For example, if researchers are more eager consumers of NLP than clinicians, NLP research into text mining and literature summarization will continue dominating the field.
The NLP CDS tasks are so numerous and complex that this area of research will succeed in making practical impact only as a result of coordinated community-wide effort.
Acknowledgments
This work was partially supported by the Intramural Research Program of the National Library of Medicine, National Institutes of Health. We also thank Kevin Bretonnel Cohen for inspiration and valuable comments, and the anonymous reviewers for the detailed analysis and helpful comments.
Footnotes
According to the Personalized Health Care (PHC) Initiative team at the Department of Health and Human Services data collection supported by CDS tools is either active or passive. http://www.hhs.gov/healthit/ahic/materials/09_07/phc/background.pdf
https://cabig-kc.nci.nih.gov/Vocab/KC/index.php/OHNLP_Documentation_and_Downloads
The system, provided by Colorado-based Medical Language Processing, L.L.C. corporation, can be downloaded from http://mlp-xml.sourceforge.net/.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shortliffe EH. Computer programs to support clinical decision making. JAMA. 1987 Jul 3;258(1):61–6. [PubMed] [Google Scholar]
- 2.Hunt DL, Haynes RB, Hanna SE, Smith K. Effects of computer-based clinical decision support systems on physician performance and patient outcomes: a systematic review. JAMA. 1998 Oct 21;280(15):1339–46. doi: 10.1001/jama.280.15.1339. [DOI] [PubMed] [Google Scholar]
- 3.Hripcsak G, Knirsch CA, Jain NL, Pablos-Mendez A. Automated tuberculosis detection. J Am Med Inform Assoc. 1997 Sep;4(5):376–81. doi: 10.1136/jamia.1997.0040376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Osheroff JA, Teich JM, Middleton B, Steen EB, Wright A, Detmer DE. A roadmap for national action on clinical decision support. J Am Med Inform Assoc. 2007 Mar–Apr;14(2):141–5. doi: 10.1197/jamia.M2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sittig DF, Wright A, Osheroff JA, Middleton B, Teich JM, Ash JS, Campbell E, Bates DW. Grand challenges in clinical decision support. J Biomed Inform. 2008 Apr;41(2):387–92. doi: 10.1016/j.jbi.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aspden P, Corrigan JM, Wolcott J, Erickson SM, editors. Patient Safety: Achieving a New Standard for Care. Washington, DC: The National Academies Press; 2004. [PubMed] [Google Scholar]
- 7.Garg AX, Adhikari NK, McDonald H, Rosas-Arellano MP, Devereaux PJ, Beyene J, Sam J, Haynes RB. Effects of computerized clinical decision support systems on practitioner performance and patient outcomes: a systematic review. JAMA. 2005 Mar 9;293(10):1223–38. doi: 10.1001/jama.293.10.1223. [DOI] [PubMed] [Google Scholar]
- 8.Kawamoto K, Houlihan CA, Balas EA, Lobach DF. Improving clinical practice using clinical decision support systems: a systematic review of trials to identify features critical to success. BMJ. 2005 Apr 2;330(7494):765. doi: 10.1136/bmj.38398.500764.8F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crowley RS, Tseytlin E, Jukic D. ReportTutor - an intelligent tutoring system that uses a natural language interface. Proc AMIA Symp. 2005:171–175. [PMC free article] [PubMed] [Google Scholar]
- 10.Chen ES, Hripcsak G, Xu H, Markatou M, Friedman C. Automated acquisition of disease drug knowledge from biomedical and clinical documents: an initial study. J Am Med Inform Assoc. 2008 Jan–Feb;15(1):87–98. doi: 10.1197/jamia.M2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prokosch HU, McDonald CJ. The Effect of Computer Reminders on the Quality of Care and Resource Use. Hospital Information Systems: Design and Development Characteristics; Impact and Future Architecture. In: Prokosch HU, Dudeck J, editors. Elsevier Science. 1995. pp. 221–40. [Google Scholar]
- 12.Berner ES, editor. Clinical Decision Support Systems, Theory and Practice. 2. Springer; New York: 2007. [Google Scholar]
- 13.Aronsky D, Fiszman M, Chapman WW, Haug PJ. Combining decision support methodologies to diagnose pneumonia. Proc AMIA Symp. 2001:12–6. [PMC free article] [PubMed] [Google Scholar]
- 14.Greenes RA, editor. Clinical Decision Support: The road ahead. Burlington, MA: Elsevier, Inc; 2007. [Google Scholar]
- 15.Mamlin BW, Overhage JM, Tierney W, Dexter P, McDonald CJ. Clinical decision support within the Regenstrief medical record system. In: Berner ES, editor. Clinical Decision Support Systems, Theory and Practice. 2. Springer; New York: 2007. pp. 190–314. [Google Scholar]
- 16.Biondich P, Mamlin BW, Tierney W, Overhage JM, McDonald CJ. Regenstrief medical informatics: Experiences with clinical decision support systems. In: Greenes RA, editor. Clinical Decision Support: The road ahead. Burlington, MA: Elsevier, Inc; 2007. pp. 111–126. [Google Scholar]
- 17.Haug PJ, Gardner RM, Evans RS, Rocha BH, Rocha RA. Clinical decision support at Intermountain Healthcare. In: Berner ES, editor. Clinical Decision Support Systems, Theory and Practice. 2. Springer; New York: 2007. pp. 159–189. [Google Scholar]
- 18.Evans RS, Pestotnik SL, Classen DC, Clemmer TP, Weaver LK, Orme JF, Jr, Lloyd JF, Burke JP. A computer-assisted management program for antibiotics and other antiinfective agents. N Engl J Med. 1998 Jan 22;338(4):232–8. doi: 10.1056/NEJM199801223380406. [DOI] [PubMed] [Google Scholar]
- 19.Fiszman M, Chapman WW, Aronsky D, Evans RS, Haug PJ. Automatic detection of acute bacterial pneumonia from chest X-ray reports. J Am Med Inform Assoc. 2000 Nov–Dec;7(6):593–604. doi: 10.1136/jamia.2000.0070593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dixon BE, Zafar A. Inpatient Computerized Provider Order Entry: Findings from the AHRQ Health IT Portfolio (Prepared by the AHRQ National Resource Center for Health IT) Rockville, MD: Agency for Healthcare Research and Quality; Jan, 2009. AHRQ Publication No. 09-0031-EF. [Google Scholar]
- 21.Miller RA, Waitman LR, Chen S, Rosenbloom ST. Decision support during inpatient care provider order entry: The Vanderbilt Experience. In: Berner ES, editor. Clinical Decision Support Systems, Theory and Practice. 2. Springer; New York: 2007. pp. 215–248. [Google Scholar]
- 22.Bates DW, Lo HG. Patients, doctors, and information technology: Clinical Decision support at Brigham and Women’s Hospital and Partners Healthcare. In: Greenes RA, editor. Clinical Decision Support: The road ahead. Burlington, MA: Elsevier, Inc; 2007. pp. 127–141. [Google Scholar]
- 23.McDonald CJ, Overhage JM. Guidelines you can follow and can trust: An ideal and an example. JAMA. 1994 Mar 16;271(11):872–3. [PubMed] [Google Scholar]
- 24.Pestian J, Brew C, Matykiewicz P, Hovermale DJ, Johnson N, Cohen KB, et al. A shared task involving multi-label classification of clinical free text. ACL’07 workshop on Biological, translational, and clinical language processing (BioNLP’07); Prague, Czech Republic. 2007. pp. 36–40. [Google Scholar]
- 25.Humphreys BL, Lindberg DA. The UMLS project: making the conceptual connection between users and the information they need. Bull Med Libr Assoc. 1993 Apr;81(2):170–7. [PMC free article] [PubMed] [Google Scholar]
- 26.Friedlin FJ, McDonald CJ. A software tool for removing patient identifying information from clinical documents. J Am Med Inform Assoc. 2008 Sep–Oct;15(5):601–10. doi: 10.1197/jamia.M2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neamatullah I, Douglass MM, Lehman LW, Reisner A, Villarroel M, Long WJ, Szolovits P, Moody GB, Mark RG, Clifford GD. Automated de-identification of free-text medical records. BMC Med Inform Decis Mak. 2008 Jul 24;:8–32. doi: 10.1186/1472-6947-8-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McAuley L, Pham B, Tugwell P, Moher D. Does the inclusion of grey literature influence estimates of intervention effectiveness reported in meta-analyses? Lancet. 2000 Oct 7;356(9237):1228–31. doi: 10.1016/S0140-6736(00)02786-0. [DOI] [PubMed] [Google Scholar]
- 29.Ruch P, Baud R, Geissbuhler A. Using lexical disambiguation and named-entity recognition to improve spelling correction in the electronic patient record. Artif Intell Med. 2003 Sep–Oct;29(1–2):169–84. doi: 10.1016/s0933-3657(03)00052-6. [DOI] [PubMed] [Google Scholar]
- 30.Xu H, Friedman C, Stetson PD. Methods for building sense inventories of abbreviations in clinical notes. AMIA Annu Symp Proc. 2008:819. [PMC free article] [PubMed] [Google Scholar]
- 31.Pakhomov S, Pedersen T, Chute CG. Abbreviation and acronym disambiguation in clinical discourse. AMIA Annu Symp Proc. 2005:589–93. [PMC free article] [PubMed] [Google Scholar]
- 32.Joshi M, Pakhomov S, Pedersen T, Chute CG. A comparative study of supervised learning as applied to acronym expansion in clinical reports. AMIA Annu Symp Proc. 2006:399–403. [PMC free article] [PubMed] [Google Scholar]
- 33.Smith L, Rindflesch T, Wilbur WJ. MedPost: a part-of-speech tagger for bioMedical text. Bioinformatics. 2004 Sep 22;20(14):2320–1. doi: 10.1093/bioinformatics/bth227. [DOI] [PubMed] [Google Scholar]
- 34.Wermter J, Hahn U. Really, is medical sublanguage that different? Experimental counter-evidence from tagging medical and newspaper corpora. Stud Health Technol Inform. 2004;107(Pt 1):560–4. [PubMed] [Google Scholar]
- 35.Pakhomov SV, Coden A, Chute CG. Developing a corpus of clinical notes manually annotated for part-of-speech. Int J Med Inform. 2006 Jun;75(6):418–29. doi: 10.1016/j.ijmedinf.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 36.Liu K, Chapman W, Hwa R, Crowley RS. Heuristic sample selection to minimize reference standard training set for a part-of-speech tagger. J Am Med Inform Assoc. 2007 Sep–Oct;14(5):641–50. doi: 10.1197/jamia.M2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ananiadou S, Friedman C, Tsujii J. Introduction: named entity recognition in biomedicine. J Biomed Inform. 2004 Dec;37(6):393–95. [Google Scholar]
- 38.Demner-Fushman D, Mork JG, Shooshan SE, Aronson AR. UMLS Content Views Appropriate for NLP Processing of the Biomedical Literature vs. Clinical Text. AMIA Annu Symp Proc. 2009 Nov 14; doi: 10.1016/j.jbi.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson SB. A semantic lexicon for medical language processing. J Am Med Inform Assoc. 1999 May–Jun;6(3):205–18. doi: 10.1136/jamia.1999.0060205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deléger L, Namer F, Zweigenbaum P. Morphosemantic parsing of medical compound words: Transferring a French analyzer to English. Int J Med Inform. 2008 Sep 16; doi: 10.1016/j.ijmedinf.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 41.Mougin F, Burgun A, Bodenreider O. Using WordNet to Improve the Mapping of Data Elements to UMLS for Data Sources Integration. AMIA Annu Symp Proc. 2006 Nov 6;:574–8. [PMC free article] [PubMed] [Google Scholar]
- 42.Jin Y, McDonald RT, Lerman K, Mandel MA, Carroll S, Liberman MY, Pereira FC, Winters RS, White PS. Automated recognition of malignancy mentions in biomedical literature. BMC Bioinformatics. 2006 Nov 7;:7–492. doi: 10.1186/1471-2105-7-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsuruoka Y, Tsujii J, Ananiadou S. Accelerating the Annotation of Sparse Named Entities by Dynamic Sentence Selection. Proceedings of the Workshop on Current Trends in Biomedical Natural Language Processing (BioNLP’08); Columbus, Ohio. 2008. Jun 19, pp. 30–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tomanek K, Wermter J, Hahn U. An approach to text corpus construction which cuts annotation costs and maintains reusability of annotated data. Proceedings of the 2007 Joint Conference on Empirical Methods in Natural Language Processing and Computational Natural Language Learning (EMNLP-CoNLL 2007); Jun 28–30; Prague, Czech Republic. 2007. pp. 486–95. [Google Scholar]
- 45.Ogren P, Savova G, Chute C. Constructing Evaluation Corpora for Automated Clinical Named Entity Recognition. Proceedings of the Sixth International Language Resources and Evaluation (LREC’08); May 28–30; Marrakech, Morocco. 2008. [Google Scholar]
- 46.Roberts A, Gaizauskas R, Hepple M, Demetriou G, Yikun Guo Y, Ian Roberts I, Setzer A. Building a semantically annotated corpus of clinical texts. J Biomed Inform. doi: 10.1016/j.jbi.2008.12.013. this issue. [DOI] [PubMed] [Google Scholar]
- 47.Li D, Savova G, Kipper-Schuler K. Conditional Random Fields and Support Vector Machines for Disorder Named Entity Recognition in Clinical Texts. Proceedings of the Workshop on Current Trends in Biomedical Natural Language Processing (BioNLP’08); Jun 19; Columbus, Ohio. 2008. pp. 94–95. [Google Scholar]
- 48.Krauthammer M, Nenadic G. Term identification in the biomedical literature. J Biomed Inform. 2004 Dec;37(6):512–26. doi: 10.1016/j.jbi.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 49.Meystre SM, Savova GK, Kipper-Schuler KC, Hurdle JF. Extracting information from textual documents in the electronic health record: a review of recent research. Yearb Med Inform. 2008:128–44. [PubMed] [Google Scholar]
- 50.SMutalik PG, Deshpande A, Nadkarni PM. Use of general-purpose negation detection to augment concept indexing of medical documents: a quantitative study using the UMLS. J Am Med Inform Assoc. 2001 Nov–Dec;8(6):598–609. doi: 10.1136/jamia.2001.0080598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform. 2001 Oct;34(5):301–10. doi: 10.1006/jbin.2001.1029. [DOI] [PubMed] [Google Scholar]
- 52.Elkin PL, Brown SH, Bauer BA, Husser CS, Carruth W, Bergstrom LR, et al. A controlled trial of automated classification of negation from clinical notes. BMC Med Inform Decis Mak. 2005;5(1):13. doi: 10.1186/1472-6947-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang Y, Lowe HJ. A Novel Hybrid Approach to Automated Negation Detection in Clinical Radiology Reports. J Am Med Inform Assoc. 2007 Feb 28; doi: 10.1197/jamia.M2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chapman WW, Chu D, Dowling JN. ConText: An Algorithm for Identifying Contextual Features from Clinical Text. ACL’07 workshop on Biological, translational, and clinical language processing (BioNLP’07); Prague, Czech Republic. 2007. pp. 81–8. [Google Scholar]
- 55.Solt I, Tikk D, Gal V, Kardkovacs ZT. Semantic classification of diseases in discharge summaries using a context-aware rule-based classifier. J Am Med Inform Assoc. 2009 Jul–Aug;16(4):580–4. doi: 10.1197/jamia.M3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Uzuner O, Zhang X, Sibanda T. Machine learning and rule-based approaches to assertion classification. J Am Med Inform Assoc. 2009 Jan–Feb;16(1):109–15. doi: 10.1197/jamia.M2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aramaki E, Miurua Y, Tonoike M, Ohkuma T, Mashuichi H, Ohe Kazuhiko. TEXT2TABLE: Medical text summarization system based on named entity recognition and modality identification. Proc of the Workshop on BioNLP2009 at the NAACL Symposium; 2009. pp. 185–192. [Google Scholar]
- 58.Denny J, Peterson J, Choma N, Xu H, Miller R, Bastarache L, Peterson N. Development of a natural language processing system to identify timing and status of colonoscopy testing in electronic medical records. AMIA Annu Symp Proc. 2009 in press. [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou L, Hripcsak G. Temporal reasoning with medical data--a review with emphasis on medical natural language processing. J Biomed Inform. 2007 Apr;40(2):183–202. doi: 10.1016/j.jbi.2006.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roberts A, Gaizauskas R, Hepple M. Extracting Clinical Relationships from Patient Narratives. Proceedings of the Workshop on Current Trends in Biomedical Natural Language Processing (BioNLP’08); Jun 19; Columbus, Ohio. 2008. pp. 10–8. [Google Scholar]
- 61.Cohen KB, Palmer M, Hunter L. Nominalization and alternations in biomedical language. PLoS ONE. 2008 Sep 9;3(9):e3158. doi: 10.1371/journal.pone.0003158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cohen KB, Hunter L. A critical review of PASBio’s argument structures for biomedical verbs. BMC Bioinformatics. 2006 Nov 24;7(Suppl 3):S5. doi: 10.1186/1471-2105-7-S3-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen L, Friedman C. Extracting phenotypic information from the literature via natural language processing. Stud Health Technol Inform. 2004;107(Pt 2):758–62. [PubMed] [Google Scholar]
- 64.Rindflesch TC, Bodenreider O. Advanced Library Services: Developing a Biomedical Knowledge Repository to Support Advanced Information Management Applications. The Lister Hill National Center for Biomedical Communications. 2006 September; LHNCBC-TR-2006-001. Available from: http://lhncbc.nlm.nih.gov/lhc/docs/reports/2006/tr2006001.pdf.
- 65.Fiszman M, Demner-Fushman D, Kilicoglu H, Rindflesch TC. Automatic summarization of MEDLINE citations for evidence-based medical treatment: A topic-oriented evaluation. J Biomed Inform. 2008 doi: 10.1016/j.jbi.2008.10.002. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sager N, Lyman M, Bucknall C, Nhan N, Tick LJ. Natural language processing and the representation of clinical data. J Am Med Inform Assoc. 1994 Mar–Apr;1(2):142–60. doi: 10.1136/jamia.1994.95236145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Friedman C, Alderson PO, Austin JH, Cimino JJ, Johnson SB. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc. 1994 Mar–Apr;1(2):161–74. doi: 10.1136/jamia.1994.95236146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Friedman C, Shagina L, Lussier Y, Hripcsak G. Automated encoding of clinical documents based on natural language processing. J Am Med Inform Assoc. 2004;11(5):392–402. doi: 10.1197/jamia.M1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu H, Anderson K, Grann VR, Friedman C. Facilitating cancer research using natural language processing of pathology reports. Stud Health Technol Inform. 2004;107(Pt 1):565–72. [PubMed] [Google Scholar]
- 70.Cao H, Chiang MF, Cimino JJ, Friedman C, Hripcsak G. Automatic Summarization of Patient Discharge Summaries to Create Problem Lists using Medical Language Processing. Stud Health Technol Inform. 2004;107(Pt 2):1540. [Google Scholar]
- 71.Friedman C. Semantic text parsing for patient records. In: Chun H, Fuller SS, Friedman C, Hersh W, editors. Knowledge Management and Data Mining in Biomedicine. Springer; NY: 2005. pp. 423–48. [Google Scholar]
- 72.Pakhomov S, Buntrock J, Duffy P. High Throughput Modularized NLP System for Clinical Text. Proceedings of the 43rd Annual Meeting on Association for Computational Linguistics (ACL’05). Interactive Poster and Demonstration Sessions; Jun 25 – 30, 2005; Ann Arbor, Michigan. pp. 25–8. [Google Scholar]
- 73.Coden A, Savova G, Sominsky I, Tanenblatt M, Masanz J, Schuler K, Cooper J, Guan W, de Groen PC. Automatically extracting cancer disease characteristics from pathology reports into a Disease Knowledge Representation Model. Journal of Biomedical Informatics. 2009 doi: 10.1016/j.jbi.2008.12.005. this issue. [DOI] [PubMed] [Google Scholar]
- 74.Zeng QT, Goryachev S, Weiss S, Sordo M, Murphy SN, Lazarus R. Extracting principal diagnosis, co-morbidity and smoking status for asthma research: evaluation of a natural language processing system. BMC Med Inform Decis Mak. 2006 Jul 26;6:30. doi: 10.1186/1472-6947-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Goryachev S, Kim H, Zeng-Treitler Q. Identification and Extraction of Family History Information from Clinical Reports. AMIA Annu Symp Proc. 2008:247–251. [PMC free article] [PubMed] [Google Scholar]
- 76.Hazlehurst B, Frost HR, Sittig DF, Stevens VJ. MediClass: A system for detecting and classifying encounter-based clinical events in any electronic medical record. J Am Med Inform Assoc. 2005 Sep–Oct;12(5):517–29. doi: 10.1197/jamia.M1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hazlehurst B, Mullooly J, Naleway A, Crane B. Detecting possible vaccination reactions in clinical notes. AMIA Annu Symp Proc. 2005:306–10. [PMC free article] [PubMed] [Google Scholar]
- 78.Murff HJ, Patel VL, Hripcsak G, Bates DW. Detecting adverse events for patient safety research: a review of current methodologies. J Biomed Inform. 2003 Feb–Apr;36:1–2. 131–43. doi: 10.1016/j.jbi.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 79.Hripcsak G, Bakken S, Stetson PD, Patel VL. Mining complex clinical data for patient safety research: a framework for event discovery. J Biomed Inform. 2003 Feb–Apr;36(1–2):120–30. doi: 10.1016/j.jbi.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 80.Haug PJ, Ranum DL, Frederick PR. Computerized extraction of coded findings from free-text radiologic reports. Work in progress. Radiology. 1990 Feb;174(2):543–8. doi: 10.1148/radiology.174.2.2404321. [DOI] [PubMed] [Google Scholar]
- 81.Dang PA, Kalra MK, Blake MA, Schultz TJ, Halpern EF, Dreyer KJ. Extraction of recommendation features in radiology with natural language processing: exploratory study. AJR Am J Roentgenol. 2008 Aug;191(2):313–20. doi: 10.2214/AJR.07.3508. [DOI] [PubMed] [Google Scholar]
- 82.Elkin PL, Froehling D, Wahner-Roedler D, Trusko B, Welsh G, Ma H, Asatryan AX, Tokars JI, Rosenbloom ST, Brown SH. NLP-based identification of pneumonia cases from free-text radiological reports. AMIA Annu Symp Proc. 2008 Nov 6;:172–6. [PMC free article] [PubMed] [Google Scholar]
- 83.Haug P, Koehler S, Lau LM, Wang P, Rocha R, Huff S. A natural language understanding system combining syntactic and semantic techniques. Proc Annu Symp Comput Appl Med Care. 1994:247–51. [PMC free article] [PubMed] [Google Scholar]
- 84.Haug PJ, Koehler S, Lau LM, Wang P, Rocha R, Huff SM. Experience with a mixed semantic/syntactic parser. Proc Annu Symp Comput Appl Med Care. 1995:284–8. [PMC free article] [PubMed] [Google Scholar]
- 85.Haug PJ, Christensen L, Gundersen M, Clemons B, Koehler S, Bauer K. A natural language parsing system for encoding admitting diagnoses. Proc AMIA Annu Fall Symp. 1997:814–8. [PMC free article] [PubMed] [Google Scholar]
- 86.Fiszman M, Haug PJ, Frederick PR. Automatic extraction of PIOPED interpretations from ventilation/perfusion lung scan reports. Proc AMIA Symp. 1998:860–4. [PMC free article] [PubMed] [Google Scholar]
- 87.Christensen LM, Haug PJ, Fiszman M. MPLUS: A Probabilistic Medical Language Understanding System. Proceedings of the Workshop on Natural Language Processing in the Biomedical Domain; Philadelphia. July 2002; Association for Computational Linguistics; pp. 29–36. [Google Scholar]
- 88.Chapman WW, Christensen LM, Wagner MM, Haug PJ, Ivanov O, Dowling JN, et al. Classifying free-text triage chief complaints into syndromic categories with natural language processing. Artif Intell Med. 2005 Jan;33(1):31–40. doi: 10.1016/j.artmed.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 89.Christensen L, Harkema H, Haug P, Irwin J, Chapman WW. ONYX: A System for the Semantic Analysis of Clinical Text. BioNLP Workshop of the Association for Computational Linguistics Conference; Boulder, CO. 2009. [Google Scholar]
- 90.Friedlin J, McDonald CJ. A natural language processing system to extract and code concepts relating to congestive heart failure from chest radiology reports. AMIA Annu Symp Proc. 2006:269–73. [PMC free article] [PubMed] [Google Scholar]
- 91.Hersh W, Mailhot M, Arnott-Smith C, Lowe H. Selective automated indexing of findings and diagnoses in radiology reports. J Biomed Inform. 2001 Aug;34(4):262–73. doi: 10.1006/jbin.2001.1025. [DOI] [PubMed] [Google Scholar]
- 92.Chapman WW, Fiszman M, Dowling JN, Chapman BE, Rindflesch TC. Identifying respiratory findings in emergency department reports for biosurveillance using MetaMap. Medinfo. 2004;2004:487–91. [PubMed] [Google Scholar]
- 93.Aronson AR. Effective mapping of biomedical text to the UMLS metathesaurus: the MetaMap program. Proc AMIA Symp AMIA. 2001:17–21. [PMC free article] [PubMed] [Google Scholar]
- 94.Schadow G, McDonald CJ. Extracting structured information from free text pathology reports. AMIA Annu Symp Proc. 2003:584–8. [PMC free article] [PubMed] [Google Scholar]
- 95.Hahn U, Romacker M, Schulz S. MEDSYNDIKATE--a natural language system for the extraction of medical information from findings reports. Int J Med Inform. 2002 Dec 4;67(1–3):63–74. doi: 10.1016/s1386-5056(02)00053-9. [DOI] [PubMed] [Google Scholar]
- 96.Liu K, Mitchell KJ, Chapman WW, Crowley RS. Automating tissue bank annotation from pathology reports - comparison to a gold standard expert annotation set. AMIA Annu Symp Proc. 2005:460–4. [PMC free article] [PubMed] [Google Scholar]
- 97.Meystre S, Haug PJ. Natural language processing to extract medical problems from electronic clinical documents: performance evaluation. J Biomed Inform. 2006;39(6):589–99. doi: 10.1016/j.jbi.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 98.Hazlehurst B, Sittig DF, Stevens VJ, Smith KS, Hollis JF, Vogt TM, et al. Natural language processing in the electronic medical record: assessing clinician adherence to tobacco treatment guidelines. Am J Prev Med. 2005:434–9. doi: 10.1016/j.amepre.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 99.Richardson W, Wilson MC, Nishikawa J, Hayward RSA. The well-built clinical question: a key to evidence-based decisions. ACP Journal Club. 1995;123:A-12. [PubMed] [Google Scholar]
- 100.Demner-Fushman D, Seckman C, Fisher C, Hauser S, Clayton J, Thoma G. A Prototype System to Support Evidence-based Practice. AMIA Annu Symp Proc. 2008 Nov 6;:151–5. [PMC free article] [PubMed] [Google Scholar]
- 101.Seckman C, Demner-Fushman D, Fisher C, Hauser SE, Thoma GR. InfoBot: A prototype system to support evidenced based practice. Building Connections for Patient Centered Records. University of Maryland School of Nursing 18th Annual Summer Institute in Nursing Informatics; July 2008; Baltimore, MD. [Google Scholar]
- 102.Cimino JJ, Del Fiol G. Infobuttons and point of care access to knowledge. In: Greenes RA, editor. Clinical decision support: the road ahead. Academic Press; New York: [Google Scholar]
- 103.Policy implications of medical information systems. Washington, DC: U.S. Government Printing Office; 1977. [Google Scholar]
- 104.Ely JW, Osheroff JA, Chambliss ML, Ebell MH, Rosenbaum ME. Answering physicians’ clinical questions: obstacles and potential solutions. J Am Med Inform Assoc. 2005 Mar–Apr;12(2):217–24. doi: 10.1197/jamia.M1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Downs S, Walker MG, Blum RL. Proceedings of the Fifth World Congress on Medical lnformatics (Medinfo) Elsevier Science Publishers; 1986. Automated Summarization of On-line Medical Records; pp. 800–4. [Google Scholar]
- 106.Shahar Y, Goren-Bar D, Boaz D, Tahan G. Distributed, intelligent, interactive visualization and exploration of time-oriented clinical data and their abstractions. Artif Intell Med. 2006 Oct;38(2):115–35. doi: 10.1016/j.artmed.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 107.Plaisant C, Lam SJ, Shneiderman B, Smith MS, Roseman DH, Marchand G, Gillam M, Feied C, Handler J, Rappaport H. Searching electronic health records for temporal patterns in patient histories: a case study with microsoft amalga. AMIA Annu Symp Proc. 2008 Nov 6;:601–5. [PMC free article] [PubMed] [Google Scholar]
- 108.Stacey M, McGregor C. Temporal abstraction in intelligent clinical data analysis: A survey. Artif Intell Med. 2007 Jan;39(1):1–24. doi: 10.1016/j.artmed.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 109.Afantenos S, Karkaletsis V, Stamatopoulos P. Summarization from medical documents: a survey. Artif Intell Med. 2005 Feb;33(2):157–77. doi: 10.1016/j.artmed.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 110.Van Vleck TT, Stein DM, Stetson PD, Johnson SB. Assessing data relevance for automated generation of a clinical summary. AMIA Annu Symp Proc. 2007 Oct 11;:761–5. [PMC free article] [PubMed] [Google Scholar]
- 111.Meng F, Taira RK, Bui AA, Kangarloo H, Churchill BM. Automatic generation of repeated patient information for tailoring clinical notes. Int J Med Inform. 2005 Aug;74(7–8):663–73. doi: 10.1016/j.ijmedinf.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 112.McKeown K, Chang SF, Cimino JJ, Feiner S, Friedman C, Gravano L, et al. PERSIVAL, a system for personalized search and summarization over multimedia healthcare information. Proceedings of the Joint Conference on Digital Libraries (JCDL) 2001:331–40. [Google Scholar]
- 113.Elhadad N, Kan MY, Klavans J, McKeown K. Customization in a Unified Framework for Summarizing Medical Literature. Artif Intell Med. 2005;33(2):179–98. doi: 10.1016/j.artmed.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 114.Fiszman M, Rindflesch TC, Kilicoglu H. Summarization of an Online Medical Encyclopedia. Stud Health Technol Inform. 2004;107(Pt 2):506–10. [PubMed] [Google Scholar]
- 115.Hardcastle D, Hallett C. Exploring the Use of NLP in the Disclosure of Electronic Patient Records; ACL’07 workshop on Biological, translational, and clinical language processing (BioNLP’07); Prague, Czech Republic. 2007. pp. 161–2. [Google Scholar]
- 116.Elhadad N, Sutaria K. Mining a Lexicon of Technical Terms and Lay Equivalents. ACL’07 workshop on Biological, translational, and clinical language processing (BioNLP’07); Prague, Czech Republic. 2007. pp. 49–56. [Google Scholar]
- 117.Åhlfeldt H, Borin L, Daumke P, Grabar N, Hallett C, Hardcastle D, et al. Literature Review on Patient-Friendly Documentation Systems. The Open University; United Kingdom: May, 2006. Technical Report No 2006/04. Available from: http://mcs.open.ac.uk/nlg/publications/TR2006_04.pdf. [Google Scholar]
- 118.Green N. Representing normative arguments in genetic counseling. Proceedings of the AAAI-2006 Spring Symposium on Argumentation for Consumers of Healthcare; 2006 Mar 27–29; Stanford, California. 2006. [Google Scholar]
- 119.Smith R. What clinical information do doctors need? BMJ. 1996;313:1062–1068. doi: 10.1136/bmj.313.7064.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Demner-Fushman D, Lin J. Answering Clinical Questions with Knowledge-Based and Statistical Techniques. Computational Linguistics. 2007;33(1):63–103. [Google Scholar]
- 121.Jacquemart P, Zweigenbaum P. Towards a medical question-answering system: a feasibility study. Stud Health Technol Inform. 2003;95:463–8. [PubMed] [Google Scholar]
- 122.Niu Y, Zhu X, Hirst G. Using outcome polarity in sentence extraction for medical question-answering. AMIA Annu Symp Proc. 2006:599–603. [PMC free article] [PubMed] [Google Scholar]
- 123.Lee M, Cimino J, Zhu HR, Sable C, Shanker V, Ely J, Yu H. Beyond information retrieval--medical question answering. AMIA Annu Symp Proc. 2006:469–73. [PMC free article] [PubMed] [Google Scholar]
- 124.Pennebaker JW, Mehl MR, Niederhoffer KG. Psychological aspects of natural language. use: our words, our selves. Annu Rev Psychol. 2003;54:547–77. doi: 10.1146/annurev.psych.54.101601.145041. [DOI] [PubMed] [Google Scholar]
- 125.Pennebaker JW, Mayne TJ, Francis ME. Linguistic predictors of adaptive bereavement. J Pers Soc Psychol. 1997 Apr;72(4):863–71. doi: 10.1037//0022-3514.72.4.863. [DOI] [PubMed] [Google Scholar]
- 126.Owen JE, Giese-Davis J, Cordova M, Kronenwetter C, Golant M, Spiegel D. Self-report and linguistic indicators of emotional expression in narratives as predictors of adjustment to cancer. J Behav Med. 2006 Aug;29(4):335–45. doi: 10.1007/s10865-006-9061-8. Epub 2006 Jul 15. [DOI] [PubMed] [Google Scholar]
- 127.Lyons EJ, Mehl MR, Pennebaker JW. Pro-anorexics and recovering anorexics differ in their linguistic Internet self-presentation. J Psychosom Res. 2006 Mar;60(3):253–6. doi: 10.1016/j.jpsychores.2005.07.017. [DOI] [PubMed] [Google Scholar]
- 128.Stirman SW, Pennebaker JW. Word use in the poetry of suicidal and nonsuicidal poets. Psychosom Med. 2001 Jul–Aug;63(4):517–22. doi: 10.1097/00006842-200107000-00001. [DOI] [PubMed] [Google Scholar]
- 129.Pestian J, Matykiewicz P, Grupp-Phelan J, Lavanier SA, Combs J, Kowatch R. Using Natural Language Processing to Classify Suicide Notes. Proceedings of the Workshop on Current Trends in Biomedical Natural Language Processing (BioNLP’08); 2008 Jun 19; Columbus, Ohio. pp. 96–7. [DOI] [PubMed] [Google Scholar]
- 130.Roark B, Mitchell M, Hollingshead K. Syntactic complexity measures for detecting Mild Cognitive Impairment. ACL’07 workshop on Biological, translational, and clinical language processing (BioNLP’07); Prague, Czech Republic. 2007. pp. 1–8. [Google Scholar]
- 131.Butler SF, Venuti SW, Benoit C, Beaulaurier RL, Houle B, Katz N. Internet surveillance: content analysis and monitoring of product-specific internet prescription opioid abuse-related postings. Clin J Pain. 2007 Sep;23(7):619–28. doi: 10.1097/AJP.0b013e318125c5cf. [DOI] [PubMed] [Google Scholar]
- 132.Malouf R, Davidson B, Sherman A. Mining web texts for brand associations. Proceedings of the AAAI-2006 Spring Symposium on Computational Approaches to Analyzing Weblogs; 2006 Mar 27–29; Stanford, California. 2006. pp. 125–6. [Google Scholar]