Abstract
Screening of glycan arrays represents a powerful, high-throughput approach to defining oligosaccharide ligands for glycan-binding receptors, commonly referred to as lectins. Correlating results from such arrays with structural analysis of receptor–ligand complexes provide one way to validate the arrays. Using examples drawn from the family of proteins that contain C-type carbohydrate-recognition domains, this review illustrates how information from the arrays reflects the way that selectivity and affinity for glycan ligands is achieved. A range of binding profiles is observed, from very restricted binding to a small set of structurally similar ligands to binding of broad classes of ligands with related terminal sugars and even to failure to bind any of the glycans on an array. These outcomes provide insights into the importance of multiple factors in defining the selectivity of these receptors, including the presence of conformationally defined units in some oligosaccharide ligands, local and extended interactions between glycans and the surfaces of receptors, and steric factors that exclude binding of some ligands.
Keywords: glycan array, glycan-binding receptor, lectin, modeling, structure
Introduction
Glycan-binding receptors are involved in biological processes that require recognition of selected sets of oligosaccharide ligands. Receptor-mediated glycan-binding events underlie selective adhesion between mammalian cells, sorting and trafficking of soluble glycoproteins, and detection of microbial pathogens based on their surface glycosylation (Taylor and Drickamer 2006; Varki et al. 2009). Glycan arrays are increasingly being used as a means of characterizing the glycan-binding specificity of these receptors (Feizi et al. 2003; Paulson et al. 2006).
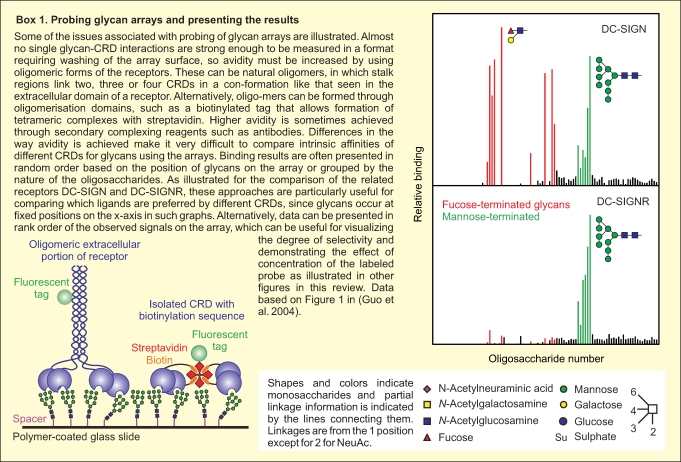
There are important practical issues that have been addressed in the construction and probing of glycan arrays (Box 1). Multiple different platforms have been described for the display of glycans using a range of immobilization strategies. These include both noncovalent adsorption, such as direct immobilization on nitrocellulose (Wang et al. 2002), binding of lipid-conjugated glycans to nitrocellulose surfaces (Fukui et al. 2002), and binding of biotinylated ligands to streptavidin-coated surfaces (Guo et al. 2004), and various types of covalent linkages to surfaces through the reducing ends of oligosaccharides (Blixt et al. 2004; Manimala et al. 2006; Karamanska et al. 2008). The nature of the surface and the spacer between the glycan and the surface can affect the interaction with glycan-binding proteins, often in unpredictable ways. Data shown here were obtained with glycans covalently attached to polymer-coated glass surfaces through spacers of different lengths in a format developed by the Consortium for Functional Glycomics (Blixt et al. 2004). Arrays are generally probed with soluble lectins or soluble fragments of membrane receptors that have been fluorescently labeled. Because of the extensive washing steps needed to remove unbound receptors, binding to arrays is generally enhanced through avidity resulting from multivalent binding of naturally oligomeric receptor fragments or clusters of carbohydrate-binding domains generated by oligomerization motifs such as immunoglobulin Fc domains or biotinylation tags that can be clustered on streptavidin tetramers (Paulson et al. 2006; Powlesland et al. 2006).
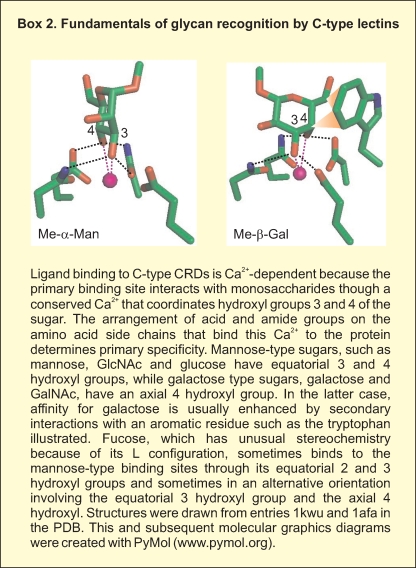
Although it is clear that glycan arrays are powerful tools for highly parallel analysis of the interaction of receptors with a broad spectrum of potential ligands, an understanding of the significance of the positive and negative results obtained is still being developed. Multiple approaches have been employed for array validation, most commonly involving solid-phase binding assays employing labeling strategies to detect bound receptors (van Vliet et al. 2005), quantitative binding competition assays (Guo et al. 2004), isothermal titration calorimetry (Gregg et al. 2008; Neu et al. 2008), or surface plasmon resonance (Bochner et al. 2005; van Liempt et al. 2006). The focus of this review is on structural features of the binding proteins and the glycan ligands that explain some of the behaviors observed on the arrays. Examples are drawn from the family of glycan-binding proteins that contain C-type carbohydrate-recognition domains (CRDs) (Box 2). Many of these domains have been tested on the array created by the Consortium for Functional Glycomics (CFG), generating one of the largest collections of reasonably comparable datasets available. Importantly, both the glycan array binding data and coordinates for the crystal structures of receptor–glycan complexes are publicly available through the CFG web site (www.functionalglycomics.org) and the Protein Data Bank (DPB) (www.rcsb.org).
Materials and methods
Receptors that display highly selective ligand binding
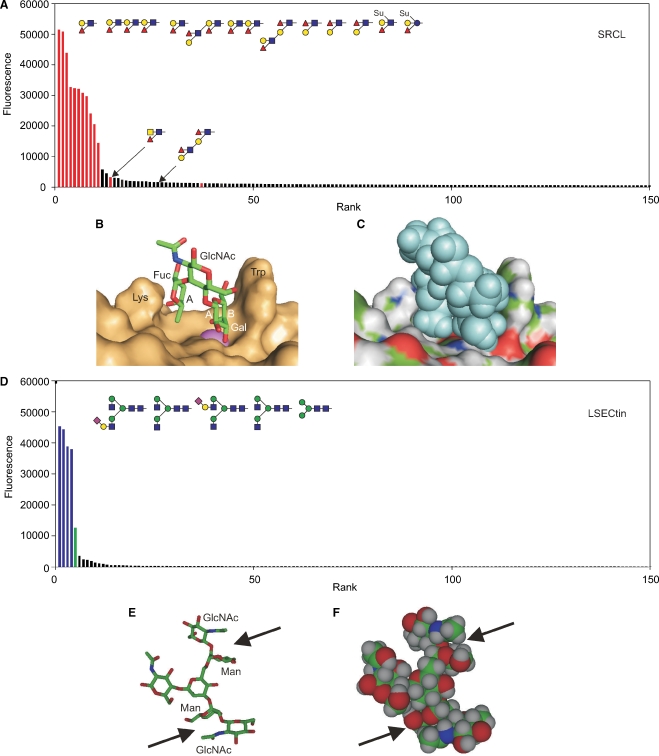
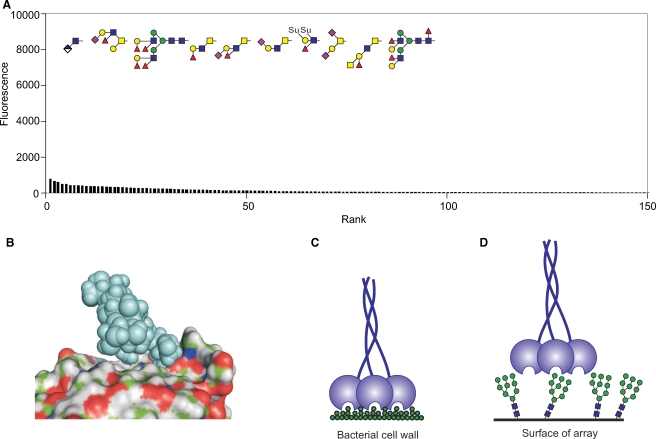
The most readily interpreted results on the glycan array are those that reflect binding to a very limited set of glycans that clearly share a structural motif. The endothelial scavenger receptor C-type lectin illustrates this type of situation since probing of the array even at relatively high protein concentrations reveals selective binding to only a few glycans, all of which contain the Lewisx or Lewisa trisaccharide motifs (Figure 1A) (Coombs et al. 2005; Feinberg, Taylor, et al. 2007). Plotting the data in rank order of the signal strength observed on the array reveals a sharp drop-off in binding, with relatively few structures that fall in a twilight zone between strong signals and background. In addition to the presence of the Lewisx and Lewisa motifs in the group of positive ligands, it is also important to note the absence of these motifs in most of the glycans that do not bind to the receptor.
Fig. 1.
Ligand binding by the scavenger receptor C-type lectin (SRCL) and LSECtin. (A) Glycan array data for the mouse receptor (200 μg/mL) plotted in rank order of ligand binding. The 10 oligosaccharides giving the highest signals are highlighted in red, along with other structurally similar glycans that are ranked lower. (B) Structure of Lewisx trisaccharide bound to SRCL. The surface of the protein is shown, with Ca2+ highlighted in violet. The A face of fucose and the A and B faces of galactose are labeled. These designations are based on the order of the ring carbons, which is clockwise when the sugar is viewed from the A face. (C) Structure of the complex in (B) with surface colored by underlying atom type and ligand presented as space-filling spheres. (D) Results for glycan arrays probed with labeled human LSECtin CRD-streptavidin tetramers (4.5 μg/mL). The structures of the five oligosaccharides giving the highest signals are shown. Bars are coded based on terminal sugars in the glycans: green for mannose and blue for GlcNAc. (E and F) Structure of a portion of a bi-antennary glycan terminating in GlcNAc-β1-2Man disaccharides showing packing of the N-acetyl group of GlcNAc on top of mannose. The figure was created from CFG primscreen_GLYCAN_v3_72_02172005 and CFG primscreen_1043 and structures 2OX9 and 1SLA in the PDB.
The Lewisx trisaccharide is a commonly recognized structural motif in ligands for glycan-binding receptors. In addition to the scavenger receptor C-type lectin, DC-SIGN (discussed below) and the selectins bind to ligands that contain this unit (Vestweber and Blanks 1999; van Die et al. 2002; Guo et al. 2004; Rosen 2004). An important feature of these ligands is that the three sugar residues that make up the Lewisx epitope are consistently in the same conformation in glycans in solution and in crystals as well as in glycans bound to receptors, as analyzed by NMR and crystallography (Miller et al. 1992; Perez et al. 1996; Somers et al. 2000; Feinberg, Taylor, et al. 2007). This structure results from van der Waals packing of the fucose and galactose residues against each other, forming a rigid and stable structure (Figure 1B). In the complex with the scavenger receptor C-type lectin, the 3- and 4-hyroxyl groups of the galactose residue of the Lewisx ligand form coordination bonds with a bound calcium ion, an arrangement that is characteristic of the C-type lectins (Box 2). A further feature of the Lewisx conformation is that the relatively nonpolar B face of galactose remains exposed, allowing it to pack with the side chain of tryptophan and other aromatic amino acids (Figure 1B). In addition to these interactions with the galactose residue, the fucose residue also makes contact with other residues projecting from the protein surface as can be seen in a surface representation of the bound complex (Figure 1C).
A key consequence of the preformed, rigid conformation of the glycan ligand is that there is little or no conformational entropy penalty associated with the binding interaction. Crystal structures of C-type CRDs in the absence of glycan ligands reveal that the binding site is also preformed, so that the only change needed to accommodate the glycan ligand is release of water molecules that occupy the coordination positions that will be replaced by the hydroxyl groups of the sugar. Thus, the enthalpy gain associated with the secondary contacts with the surface of the protein is not negated by an extensive entropy penalty, resulting in an overall more favorable free energy of the interaction that is reflected in a 70-fold enhanced affinity for Lewisx trisaccharide compared to galactose, as measured in binding competition assays (Coombs et al. 2005; Feinberg, Taylor, et al. 2007).
Although many oligosaccharides display conformational heterogeneity, the Lewisa and Lewisx trisaccharides are not the only examples of preferred local conformations that create preformed structural features that can interact with receptors with a minimal entropy penalty associated with glycan immobilization. In the case of another endothelial cell receptor, LSECtin, glycan array screening at high receptor concentration reveals binding to multiple groups of ligands with terminal fucose and GlcNAc residues, but at reduced concentrations much more selective binding is observed to glycans bearing terminal GlcNAcβ1-2Man disaccharides (Figure 1D) (Powlesland et al. 2008). Analysis of the conformation of this disaccharide in multiple crystal structures reveals that in many cases it assumes a common preferred conformation in which the N-acetyl group of GlcNAc is positioned above the mannose residue, making van der Waals interactions with the B face of the hexose (Figure 1E and F) (Petrescu et al. 1999). This is one of two conformations of the disaccharide that have been observed, but the fact that it appears to be a preferred if not necessarily completely stable, preformed conformation would still reduce the entropy penalty associated with binding to a receptor. Although potential interactions with the binding site in LSECtin have been modeled, these remain to be examined experimentally.
In summary, glycan array results indicating highly selective binding to a group of glycans can reflect the presence of a preformed conformational feature in the ligands, which provides an extended surface for local interactions with the receptor surface at minimal entropy cost associated with glycan immobilization.
Receptors that bind to broad classes of glycan ligands
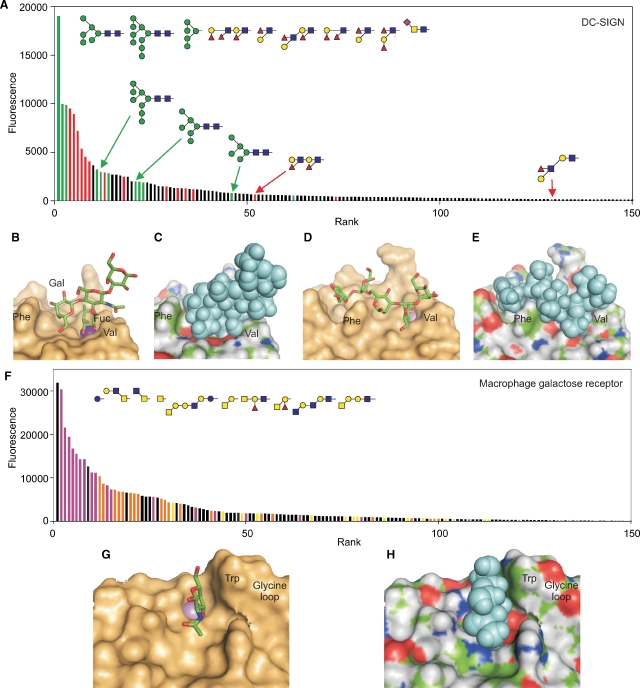
One of the first receptors tested against glycan arrays was DC-SIGN, which appears to have a dual role in the interaction of dendritic cells with pathogen surfaces as well as with T cells (Geijtenbeek et al. 2000; Engering et al. 2002). The glycan array analysis reveals binding to two classes of ligands: high mannose oligosaccharides and Lewis-type structures (Figure 2A), while structural analysis suggests that the mechanisms underlying interactions with the two classes of ligands differ significantly (Guo et al. 2004). The Lewisx trisaccharide sits in the binding site as a rigid unit, as in the case of the scavenger receptor C-type lectin, but the placement of the fucose rather than the galactose residue in the primary binding site results in a more limited set of interactions (Figure 2B and C). Notably, the prominent phenylalanine residue on the surface of the CRD is not positioned sufficiently close to the oligosaccharide to allow packing interactions with the galactose residue. Nevertheless, the limited contacts observed are probably sufficient to generate specificity because of the rigid structure of the oligosaccharide ligand.
Fig. 2.
Binding of multiple classes of ligands to DC-SIGN and the macrophage galactose receptor. (A) Data for screening of a recent version of the Consortium for Functional Glycomics glycan array with human DC-SIGN (200 μg/mL). Results are presented in rank order of binding, with bars for mannose-terminated glycans colored green and bars for glycans containing the Lewisa or Lewisx epitopes highlighted in red. The 10 oligosaccharides giving the strongest signals are shown at the top and additional related glycans are indicated below. (B and C) Structure of the CRD from DC-SIGN in complex with a tetrasaccharide containing the Lewisx epitope. (D and E) Structure of the CRD with Man5 modeled into the binding site based on the crystal structure with a GlcNAc2Man3 oligosaccharide. (F) Binding data for the human macrophage galactose receptor (4.5 μg/mL) on the CFG printed glycan array. The results are similar to previous studies with the human and rat receptors on earlier streptavidin-based arrays (Coombs et al. 2005; van Vliet et al. 2005). The data are color-coded to indicate glycans bearing GalNAc with free 3 and 4 hydroxyl groups in purple and galactose with free 3 and 4 hydroxyl groups in orange, except galactose residues in α linkage or adjacent to fucose residues, which are in yellow. In spite of the preference for exposed GalNAc and galactose residues, binding of other glycans indicated in black also occurs, including the simple sugar glucose which gives the highest signal. (G and H) Model of the binding site in the macrophage galactose receptor with a bound GalNAc residue, based on the structure of the galactose-binding mutant of mannose-binding protein that was created by insertion of key binding site residues from the galactose-binding receptor. These residues include the marked tryptophan residue and the glycine-rich loop positioned just to the right of this residue, which serves to hold it in the position for packing against galactose or GalNAc residues. For the model, additional surface residues were substituted into the structure using Insight software. Data are from CFG primscreen_2010, and the model structures were created starting from entries 1sl5, 1k9i, and 1afb in the PDB.
In contrast, binding to the more flexible high mannose ligands involves a different energetic balance. Because high mannose oligosaccharides are known to assume multiple conformations (Petrescu et al. 1999), no one of which would exactly match the conformation needed to dock into the binding site, the entropy cost associated with freezing out one of these structures would be substantial. The structure of a ligand with five mannose residues in the binding site can be modeled on the available crystal structure of a mimic containing the core trimannose structure capped with two GlcNAc residues (Figure 2D and E) (Feinberg, Castelli, et al. 2007). The model reveals that by snaking though a groove on the surface of the CRD and wrapping around the surface phenylalanine residue, the oligosaccharide can make many favorable interactions. Thus, binding of this class of ligand must involve a significant trade-off between entropy costs for immobilization of the ligand and additional favorable contacts in a much more extended binding site. The binding of DC-SIGN to high mannose oligosaccharides thus provides a second paradigm for how relatively narrow specificity for a class of ligands can be achieved.
Other receptors display binding to a still broader spectrum of ligands on the glycan array, which structurally appears to correlate with binding largely to terminal residues in glycans. For example, ranking of ligands for the macrophage galactose receptor detected on the glycan array reveals preferential binding to glycans bearing GalNAc residues that are either unsubstituted or substituted only on the 6-position (Figure 2F), with simple GalNAc being one of the most prominent ligands. Although binding to galactose-terminated structures is also detected, these ligands generally give weaker signals. These results are consistent with other profiling methods that indicate a strong preference for GalNAc over galactose (van Vliet et al. 2005). Structural and mutagenesis studies indicate that GalNAc and galactose bind in the primary binding site on the CRD, ligated to the binding site calcium ion and packing against a tryptophan residue for increased affinity and specificity, with further selectivity for GalNAc resulting from favorable contacts with the 2-acetamido group (Figure 2G and H) (Iobst and Drickamer 1996; Kolatkar et al. 1998).
The overall trend in the types of glycans bound to the macrophage galactose receptor is evident from the array data, and all of the ligands with high signals can be accounted for by interactions of a terminal sugar with the primary binding site. The fact that there are no other obvious common features of the most prominent ligands is consistent with the suggestion that specificity derives from the interaction with the terminal sugar. However, many glycans that feature apparently appropriate terminal residues do not rank amongst the best ligands, and there are no obvious rules describing which ligands bind and which do not. Two factors are generally correlated with lower ranking in the binding: linkage in α- rather than β-configuration and linkage adjacent to a fucose residue. These observations illustrate the importance of a further mechanism that has been invoked to explain binding specificity, which is exclusion of some ligands by steric hindrance. Surface features of the CRD near to the binding site may block binding of some ligands, particularly those with fixed geometry such as the Lewisx structure, rather than enhance binding through favorable interactions. In such cases, affinity must be generated largely though the interactions with the terminal monosaccharides, which makes the packing interaction of the galactose with the tryptophan residue particularly important. Selectivity then results more from exclusion of some classes of ligands. A similar principle of exclusion probably explains why DC-SIGN binds poorly to some fucose-containing ligands and well to others, particularly given the position of the surface phenylalanine residue near to the primary binding site.
Comparing the glycan array profiles for the scavenger receptor C-type lectin and LSECtin (Figure 1A and D) with those for DC-SIGN and the macrophage galactose receptor (Figure 2A and F) reveals differences in the shapes and color distributions of the profiles. The first set of receptors, in which the binding sites make multiple favorable interactions with a relatively rigid structural motif, give strong signals with a few structurally related glycans, after which there is a sharp drop-off in signal, reflecting the fact that binding is dependent on the presence of the specific binding epitope. In contrast, the profiles for the second set of receptors are more extended, with a continuous decline in signal and no strict segregation of structurally distinct ligands. Although binding to these receptors reflects a nucleating interaction with a mannose-type or a galactose-type monosaccharide, the affinity for oligosaccharides results from a complex balance between entropy costs of immobilizing larger oligosaccharides, with resulting favorable interactions, and steric exclusion.
Receptors with low selectivity binding sites
At the most extreme end of the glycan array results are receptors that are known to bind sugar ligands but which interact poorly with all of the oligosaccharides on the array. Serum mannose-binding protein represents such a case since very little binding is evident even with highly fluorescent probe at high receptor concentrations (Figure 3A). There are no obvious common features to the glycans which give the highest signals, and the pattern is not reproduced in multiple screens of the array. The binding site in this protein is open, so that there are very few constraints on what binds beyond the key hydroxyl groups that coordinate with the binding site calcium ion (Weis et al. 1992; Ng et al. 2002). However, as a result of this arrangement, there are very few possibilities for further favorable interactions to enhance affinity as in the case of DC-SIGN (Figure 3B). The binding of mannose, GlcNAc, and fucose in the primary binding site is not enhanced by packing with an adjacent tryptophan residue as seen for the galactose- and GalNAc-binding receptors. Also, binding involves only a limited portion of the terminal residue of oligosaccharide ligands because other portions of the glycans are not in direct contact with the surface of the protein, which contrasts sharply with the arrangement in DC-SIGN.
Fig. 3.
Mechanisms of mannose-binding protein interaction with ligands. (A) Screening of the glycan array with the trimeric terminal fragment of rat serum mannose-binding protein at a 250 μg/mL concentration. The oligosaccharides giving the 10 highest signals are illustrated at the top. Scale of the y-axis is expanded compared to other figures. (B) Structure of a Man6 ligand complexed with the CRD of mannose-binding protein. (C and D) Diagrams illustrating the potential effect of glycan spacing on the avidity with which mannose-binding protein trimers bind to dense clusters of sugars on bacterial or fungal surfaces compared to their interaction with glycans on the array. The figure is based on data from CFG primscreen_1618 and structure 1kx1 in the PDB.
The inherently weak and low specificity of the interaction of mannose-binding protein with terminal sugars means that biologically significant binding can only occur through multivalent interactions. Although multivalency plays a role in increasing avidity of interactions of many glycan-binding receptors, for the mannose-binding protein the geometry of the placement of multiple binding sites facing a single direction is a determining feature for broad recognition of glycan-rich surfaces without requiring specific complex oligosaccharide structures (Figure 3C) (Weis and Drickamer 1994). The wide spacing between the binding sites in mannose-binding protein trimer and their rigid arrangement means that although they are able to engage with repetitive sugar structures on surfaces of bacteria and fungi, they probably cannot adapt well to the spacing of glycans on the arrays (Figure 3D). Thus, the lack of binding to the glycan array reflects an alternative structural mechanism of optimizing interactions with sugars, in this case with minimal local selectivity so that spatial distribution becomes the dominant factor in determining specificity.
Conclusions and perspective
The description of some of the factors that lead to specificity and affinity in glycan–receptor interactions presented in this review illustrate how the power of both glycan arrays and structural analysis is enhanced by examining the results of the two approaches in combination. The fact that the interpretations are largely consistent with each other provides validation for the use of the arrays to define binding specificity of receptors. The array allows screening of far more ligands than could be undertaken in a structural analysis. Viewing the limited set of structures of receptor–ligand complexes in the light of information about the relative binding signals obtained with additional ligands makes it possible to suggest how the specificity is achieved. Combining such information may provide a realistic and reliable basis for predicting which ligands will bind to particular receptors as well as for modeling of how they bind. It may also provide a foundation for prediction and modeling of glycan ligand binding to other receptors for which structural information is not available.
Funding
The Wellcome Trust (075565) and the National Institute of General Medical Sciences (GM62116 to the Consortium for Functional Glycomics).
Supplementary Material
Acknowledgments
We thank Eliot Ward and Adrián Quintero-Martinez for providing the human macrophage galactose receptor for glycan array screening and David Smith of the Consortium for Functional Glycomics for the array screening experiments. Funding to pay the Open Access publication charges for this article was provided by the Wellcome Trust.
Conflict of interest statement
None declared.
Abbreviations
- CFG
Consortium for Functional Glycomics
- CRD
carbohydrate-recognition domain
- PDB
Protein Data Bank
References
- Blixt O, Head S, Mondala T, Scanlan C, Huflejt ME, Alvarez R, Bryan MC, Fazio F, Calarese D, Stevens J, et al. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc Natl Acad Sci USA. 2004;101:17033–17038. doi: 10.1073/pnas.0407902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochner BS, Alvarez RA, Mehta P, Bovin NV, Blixt O, White JR, Schnaar RL. Glycan array screening reveals a candidate ligand for siglec-8. J Biol Chem. 2005;280:4307–4312. doi: 10.1074/jbc.M412378200. [DOI] [PubMed] [Google Scholar]
- Coombs PJ, Graham SA, Drickamer K, Taylor ME. Selective binding of the scavenger receptor C-type lectin to Lewisx trisaccharide and related glycan ligands. J Biol Chem. 2005;280:22993–22999. doi: 10.1074/jbc.M504197200. [DOI] [PubMed] [Google Scholar]
- Engering A, Geijtenbeek TBH, van Vliet SJ, Wijers M, van Liempt E, Demaurex N, Lanzacecchia A, Fransen J, Figdor CG, Piguet V, et al. The dendritic cell-specific adhesion receptor DC-SIGN internalizes antigen for presentation to T cells. J Immunol. 2002;168:2118–2126. doi: 10.4049/jimmunol.168.5.2118. [DOI] [PubMed] [Google Scholar]
- Feinberg H, Castelli R, Drickamer K, Seeberger PH, Weis WI. Multiple modes of binding enhance the affinity of DC-SIGN for high-mannose N-linked glycans found on viral glycoproteins. J Biol Chem. 2007;282:4202–4209. doi: 10.1074/jbc.M609689200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg H, Mitchell DA, Drickamer K, Weis WI. Structural basis for selective recognition of oligosaccharides by DC-SIGN and DC-SIGNR. Science. 2001;294:2163–2166. doi: 10.1126/science.1066371. [DOI] [PubMed] [Google Scholar]
- Feinberg H, Taylor ME, Weis WI. Scavenger receptor C-type lectin binds to the leukocyte cell surface glycan Lewis x by a novel mechanism. J Biol Chem. 2007;282:17250–17258. doi: 10.1074/jbc.M701624200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feizi T, Fazio F, Chai W, Wong CH. Carbohydrate microarrays—A new set of technologies at the frontiers of glycomics. Curr Opin Struct Biol. 2003;13:637–645. doi: 10.1016/j.sbi.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Fukui S, Feizi T, Galustian C, Lawson AM, Chai W. Oligosaccharide microarrays for high-throughput detection and specificity assignments of carbohydrate–protein interactions. Nat Biotechnol. 2002;20:1011–1017. doi: 10.1038/nbt735. [DOI] [PubMed] [Google Scholar]
- Geijtenbeek TBH, Torensma R, van Vliet SJ, van Duijnhoven GCF, Adema GJ, van Kooyk Y, Figdor CG. Identification of DC-SIGN, a novel dendritic cell-specific ICAM-3 receptor that supports primary immune responses. Cell. 2000;100:575–585. doi: 10.1016/s0092-8674(00)80693-5. [DOI] [PubMed] [Google Scholar]
- Gregg KJ, Finn R, Abbott DW, Boraston AB. Divergent modes of glycan recognition by a new family of carbohydrate-binding modules. J Biol Chem. 2008;283:12604–12613. doi: 10.1074/jbc.M709865200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Feinberg H, Conroy E, Mitchell DA, Alvarez R, Blixt O, Taylor ME, Weis WI, Drickamer K. Structural basis for distinct ligand-binding and targeting properties of the receptors DC-SIGN and DC-SIGNR. Nat Struct Mol Biol. 2004;11:591–598. doi: 10.1038/nsmb784. [DOI] [PubMed] [Google Scholar]
- Iobst ST, Drickamer K. Selective sugar binding to the carbohydrate-recognition domains of rat hepatic and macrophage asialoglycoprotein receptors. J Biol Chem. 1996;271:6686–6693. doi: 10.1074/jbc.271.12.6686. [DOI] [PubMed] [Google Scholar]
- Karamanska R, Clarke J, Blixt O, MacRae JI, Zhang JQ, Crocker PR, Laurent N, Wright A, Flitsch SL, Russell DA, et al. Surface plasmon resonance imaging for real-time, label-free analysis of protein interactions with carbohydrate microarrays. Glycoconjugate J. 2008;25:69–74. doi: 10.1007/s10719-007-9047-y. [DOI] [PubMed] [Google Scholar]
- Kolatkar AR, Leung AK, Isecke R, Brossmer R, Drickamer K, Weis WI. Mechanism of N-acetylgalactosamine binding to a C-type animal lectin carbohydrate-recognition domain. J Biol Chem. 1998;273:19502–19508. doi: 10.1074/jbc.273.31.19502. [DOI] [PubMed] [Google Scholar]
- Manimala JC, Roach TA, Li Z, Gildersleeve JC. High-throughput carbohydrate microarray analysis of 24 lectins. Angew Chem Int Ed. 2006;45:3607–3610. doi: 10.1002/anie.200600591. [DOI] [PubMed] [Google Scholar]
- Miller KE, Mukhopadhyay C, Cagas P, Bush CA. Solution structure of the Lewis x oligosaccharide determined by NMR spectroscopy and molecular dynamics simulations. Biochemistry. 1992;31:6703–6709. doi: 10.1021/bi00144a009. [DOI] [PubMed] [Google Scholar]
- Neu U, Woellner K, Gauglitz G, Stehle T. Structural basis of GM1 ganglioside recognition by simian virus 40. Proc Natl Acad Sci USA. 2008;105:5219–5224. doi: 10.1073/pnas.0710301105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng KK-S, Kolatkar AP, Park-Snyder S, Feinberg H, Clark DA, Drickamer K, Weis WI. Orientation of bound ligands in mannose-binding proteins: Implications of multivalent ligand recognition. J Biol Chem. 2002;277:16088–16095. doi: 10.1074/jbc.M200493200. [DOI] [PubMed] [Google Scholar]
- Paulson JC, Blixt O, Collins BE. Sweet spots in functional glycomics. Nat Chem Biol. 2006;2:238–248. doi: 10.1038/nchembio785. [DOI] [PubMed] [Google Scholar]
- Perez S, Mouhous-Riou N, Nifant'ev NE, Tsvetkov YE, Bachet B, Imberty A. Crystal and molecular structure of a histo-blood group antigen involved in cell adhesion: The Lewis x trisaccharide. Glycobiology. 1996;6:537–542. doi: 10.1093/glycob/6.5.537. [DOI] [PubMed] [Google Scholar]
- Petrescu AJ, Petrescu SM, Dwek RA, Wormald MR. A statistical analysis of N- and O-glycan linkage conformations from crystallographic data. Glycobiology. 1999;9:343–352. doi: 10.1093/glycob/9.4.343. [DOI] [PubMed] [Google Scholar]
- Powlesland AS, Fisch T, Taylor ME, Smith DF, Tissot B, Dell A, Pöhlmann S, Drickamer K. A novel mechanism for LSECtin binding to Ebola virus surface glycoprotein through truncated glycans. J Biol Chem. 2008;283:593–602. doi: 10.1074/jbc.M706292200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powlesland AS, Ward EM, Sadhu SK, Guo Y, Taylor ME, Drickamer K. Novel mouse homologs of human DC-SIGN: Widely divergent biochemical properties of the complete set of mouse DC-SIGN-related proteins. J Biol Chem. 2006;281:20440–20449. doi: 10.1074/jbc.M601925200. [DOI] [PubMed] [Google Scholar]
- Rosen SD. Ligands for L-selectin: Homing, inflammation, and beyond. Annu Rev Immunol. 2004;22:129–156. doi: 10.1146/annurev.immunol.21.090501.080131. [DOI] [PubMed] [Google Scholar]
- Somers WS, Tang J, Shaw GD, Camphausent RT. Insights into the molecular basis of leukocyte tethering and rolling revealed by structures of P- and E-selectin bound to SleX and PSGL-1. Cell. 2000;103:467–479. doi: 10.1016/s0092-8674(00)00138-0. [DOI] [PubMed] [Google Scholar]
- Taylor ME, Drickamer K. Introduction to Glycobiology. Oxford: Oxford University Press; 2006. 2nd ed. [Google Scholar]
- van Die I, van Vliet SJ, Schiphorst WECM, Bank CMC, Appelmelk B, Nyame AK, Cummings RD, Geijtenbeek TBH, van Kooyk Y. The dendritic cell specific C-type lectin DC-SIGN recognizes Lewis x, a major glycan epitope of Schistosoma mansoni egg antigen. Glycobiology. 2002;12:641–642. doi: 10.1093/glycob/cwg052. [DOI] [PubMed] [Google Scholar]
- van Liempt E, Bank CMC, Mehta P, Garcia-Vallejo JJ, Kawar ZS, Geyere R, Alvarez RA, Cummings RD, van Kooyk Y, van Die I. Specificity of DC-SIGN for mannose- and fucose-containing glycans. FEBS Lett. 2006;580:6123–6131. doi: 10.1016/j.febslet.2006.10.009. [DOI] [PubMed] [Google Scholar]
- van Vliet SJ, van Liempt E, Saeland E, Aarnoudse CA, Appelmelk B, Irimura T, Geijtenbeek TB, Blixt O, Alvarez R, van Die I, et al. Carbohydrate profiling reveals a distinctive role for the C-type lectin MGL in the recognition of helminth parasites and tumor antigens by dendritic cells. Int Immunol. 2005;17:661–669. doi: 10.1093/intimm/dxh246. [DOI] [PubMed] [Google Scholar]
- Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME. Essentials of Glycobiology. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor, Laboratory Press; 2009. Chapters 26-33. [PubMed] [Google Scholar]
- Vestweber D, Blanks JE. Mechanisms that regulate the function of selectins and their ligands. Physiol Rev. 1999;79:181–213. doi: 10.1152/physrev.1999.79.1.181. [DOI] [PubMed] [Google Scholar]
- Wang D, Liu S, Trummer BJ, Deng C, Wang A. Carbohydrate microarrays for the recognition of cross-reactive molecular markers of microbes and host cells. Nat Biotechnol. 2002;20:275–281. doi: 10.1038/nbt0302-275. [DOI] [PubMed] [Google Scholar]
- Weis WI, Drickamer K. Trimeric structure of a C-type mannose-binding protein. Structure. 1994;2:1227–1240. doi: 10.1016/S0969-2126(94)00124-3. [DOI] [PubMed] [Google Scholar]
- Weis WI, Drickamer K, Hendrickson WA. Structure of a C-type mannose-binding protein complexed with an oligosaccharide. Nature. 1992;360:127–134. doi: 10.1038/360127a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.