Abstract
The solution conformation of a fully sulfated heparin-derived tetrasaccharide, I, was studied in the presence of a 4-fold excess of Ca2+. Proton–proton and proton–carbon residual dipolar couplings (RDCs) were measured in a neutral aligning medium. The order parameters of two rigid hexosamine rings of I were determined separately using singular value decomposition and ab initio structures of disaccharide fragments of I. The order parameters were very similar implying that a common order tensor can be used to analyze the structure of I. Using one order tensor, RDCs of both hexosamine rings were used as restraints in molecular dynamics simulations. RDCs of the inner iduronic acid were calculated for every point of the molecular dynamics trajectory. The fitting of the calculated RDCs of the two forms of the iduronic acid to the experimental values yielded a population of 1C4 and 2So conformers of iduronic acid that agreed well with the analysis based on proton–proton scalar coupling constants. The glycosidic linkage torsion angles in RDC-restrained molecular dynamics (MD) structures of I are consistent with the interglycosidic three-bond proton–carbon coupling constants. These structures also show that the shape of heparin is not affected dramatically by the conformational flexibility of the iduronic acid ring. This is in line with conclusions of previous studies based on MD simulations and the analysis of 1H-1H NOEs. Our work therefore demonstrates the effectiveness of RDCs in the conformational analysis of glycosaminoglycans.
Keywords: conformation, heparin, iduronic acid, NMR spectroscopy, residual dipolar couplings
Introduction
The essential role of glycosaminoglycan (GAG)–protein interactions in the regulation of physiological processes has been recognized for decades. Nevertheless, the underlying molecular basis of these interactions has only emerged through more recent biophysical investigations (reviewed in Casu and Lindahl (2001); Mulloy and Linhardt (2001); Capila and Linhardt (2002); Hricovíni et al. (2002); Powell et al. (2004); Imberty et al. (2007)). Early insights into the molecular structure of GAGs originated from the studies of heparin, a prominent member of the GAG family, by X-ray diffraction (Nieduszynski et al. 1977) and solution state NMR (Mulloy et al. 1993; Mulloy and Forster 2000).
The repeating disaccharide unit of many GAGs, including heparin, heparan sulfate (HS), and dermatan sulfate (DS), contains iduronic acid (IdoA), a hexopyranose which is known to adopt several conformations (4C1, 1C4, and 2So). This flexibility of IdoA has attracted considerable attention (Ferro et al. 1990; Forster and Mulloy 1993; Ernst et al. 1998; Hricovíni 2006; Pol-Fachin and Verli 2008) as it can potentially affect how GAGs interact with proteins. Indeed, significant experimental evidence points to the importance of IdoA for protein GAG interactions. To mention a few more recent examples, the lack of IdoA in HS derived from Hsepi−/− mouse affected its interactions with grow factors and cell signaling (Jia et al. 2009). Binding and activity of less-sulfated species interacting with the hepatocyte growth factor/scatter factor are greatly enhanced by the presence of IdoA in the GAG sequence with otherwise comparable overall sulfation levels (Catlow et al. 2008). IdoA plays a crucial role in achieving the appropriate 3D structure of the heparin-pentasaccharide (Hricovíni et al. 2001) and heparin-octasaccharide when interacting with antithrombin (Guerrini et al. 2006) or heparin-tetrasaccharides interacting with the basic fibroblast growth factor (Guglieri et al. 2008).
X-ray structures of protein–GAG complexes show the IdoA residue in either 1C4 or 2So conformation. Interestingly, the two internal IdoA residues of a heparin hexasaccharide bound to the basic fibroblast growth factor exist in different conformations (Faham et al. 1996). Furthermore, solution state NMR studies of a heparin-like hexasaccharide in a complex with the acidic fibroblast growth factor (Canales et al. 2005) revealed that IdoA2S is present in the bound state in an equilibrium of 1C4 and 2So forms.
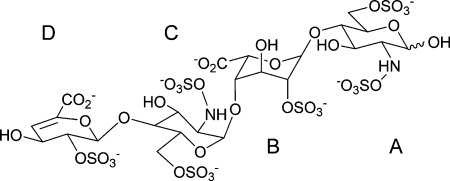
The question that then arises is why are the binding properties of IdoA so favorable? Is it that the flexibility of IdoA affects the geometry of glycosidic linkages or is it the larger conformational space available to sulfate groups that enhances its binding properties? The current evidence, based mostly on the analysis of NOE data of heparin and heparin oligosaccharides, indicates that the chair-skew boat equilibrium of IdoA has only a limited effect on the glycosidic linkage conformation of heparin (Ferro et al. 1986; Mulloy et al. 1993; Mikhailov et al. 1997; de Paz et al. 2001; Zhang et al. 2008). It should be pointed out that although NOEs are invaluable in biomolecular structure determination their two potential drawbacks are: (i) NOEs only report on the local structure and (ii) their interpretation in flexible systems is notoriously difficult (Neuhaus and Williamson 2000). On the other hand, residual dipolar coupling constants (RDCs) provide global structural information and thus have the potential to characterize the overall shape of molecules (Tjandra and Bax 1997). We have therefore decided to investigate the shape of heparin by using this recently introduced NMR methodology. In order to make our investigation tractable, we have limited the size of the studied heparin fragment to a tetrasaccharide. Our sample was prepared by the enzymatic cleavage of heparin, which resulted in the nonreducing terminal IdoA being converted to an unsaturated uronic acid. We refer to this compound as heparin ΔU-tetrasaccharide, I (Figure 1).
Fig. 1.
Fully sulfated heparin ΔU-tetrasaccharide, I, obtained through enzymatic cleavage of heparin. The monosaccharide rings are labeled A–D from the reducing to the nonreducing end of the molecule.
The activity of heparin can be modulated by calcium ions (Rabenstein et al. 1995). Similarly, the structurally related heparan sulfate interacts with extracellular proteins and in several cases these interactions involve cations such as Ca2+ and Mg2+ (Chevalier et al. 2004). We have therefore conducted our studies in a 4-fold molar excess of Ca2+ with regard to I. The presence of Ca2+ has been shown not to affect the overall shape of a heparin hexasaccharide (Angulo et al. 2000; de Paz et al. 2001; Chevalier et al. 2004); however, it is known to affect the equilibrium of 1C4 and 2So forms of the IdoA (Ferro et al. 1990; Chevalier et al. 2004). As a beneficial side effect for NMR studies, by influencing the kinetics of the chair-skew boat equilibrium of IdoA, the presence of Ca2+ coincidently sharpens NMR spectral lines (de Paz et al. 2001).
Results
Internal dynamics of rings A and D
The internal dynamics of the constituent monosaccharide rings of ΔU-tetrasaccharide I was analyzed using vicinal proton–proton coupling constants. The coupling constants (Table I) were determined using intensity-based methods (Pham et al. 2002, 2004; Jin et al. 2007) discussed later in the paper. As confirmed by the large 3J23, 3J34, and 3J45 coupling constants, the two glucosamine rings (A and C) are stable in the 4C1 conformation. On the other hand, the internal iduronic acid ring (B) and the nonreducing terminal unsaturated uronic acid ring (D) undergo conformational averaging. The internal iduronic acid can in principle adopt three different conformations, 4C1, 1C4, and 2So, but only 1C4 and 2So were found previously in fully sulfated species (Mulloy and Forster 2000). Based on an idealized geometry of 1C4 and 2So forms, the theoretical 3JHH coupling constants were calculated using the recently derived Karplus equation parameterized for sulfated GAGs (Hricovíni and Bízik 2007; Hricovíni et al. 2007). The population of each conformer was calculated by fitting the calculated coupling constants to the experimental data (Table I) yielding 76% of 1C4 and 24% 2So forms for the sulfated IdoA residue. This is in line with 79% of 1C4 observed in heparin (Ferro et al. 1990) or 70% of 1C4 in heparin hexasaccharide (Chevalier et al. 2004), both in the presence of Ca2+. A similar analysis performed for the D ring using Karplus equations (Haasnoot et al. 1980) incorporating the effects of substituents yielded 78% and 22% of 1H2 and 2H1 conformers, respectively.
Table I.
3JHH coupling constants (Hz) of heparin-derived fully sulfated ΔU-tetrasaccharide, I
| Population of | |||||
|---|---|---|---|---|---|
| Ring/3JHH | H1-H2 | H2-H3 | H3-H4 | H4-H5 | conformersd |
| Aa | 3.5 | 10.2 | 8.8 | 9.8 | – |
| Ca | 3.7 | 10.6 | 9.0 | 10.1 | – |
| Ba | 2.4 | 4.8 | 3.4 | 2.5 | – |
| (1C4)b | 1.7 | 3.0 | 3.1 | 2.3 | 76% |
| (2S0)b | 4.8 | 10.5 | 3.4 | 2.5 | 24% |
| Bd | 2.4 | 4.8 | 3.4 | 2.7 | (rmsd = 0.12 Hz) |
| Da | 3.4 | 2.6 | 4.7 | – | – |
| (1H2)c | 3.1 | 0.2 | 6.0 | – | 78% |
| (2H1)c | 8.3 | 10.2 | 1.7 | – | 22% |
| Dd | 4.2 | 2.4 | 5.0 | – | (rmsd = 0.57 Hz) |
aExperimental values.
bCalculated using the Karplus curve according to Hricovíni and Bízik (2007).
cCalculated using the Karplus curve according to Haasnoot et al. (1980).
dObtained by least square analysis of the theoretical and experimental values.
Unrestrained molecular dynamics simulations of I
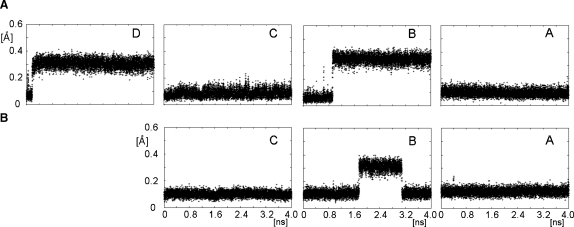
Amber (Case et al. 2002) force field solution structures of I were generated by free molecular dynamics using the protocols described in the Material and methods. Various starting conformations for rings B and D were tried, while for rings A and C, the conformation was always set to 4C1. As expected, the latter conformation did not change during MD simulations. Ring B showed transitions between 1C4 and 2So forms, while the 4C1 conformer was not observed. Ring D quickly converted from the 1H2 form to the 2H1 form. This transition was faster when the B ring was in the 2So form; when simulations were started with the D ring in the 2H1 conformation, it never changed to the 1H2 form. A 4 ns nonrestrained MD of I is illustrated in Figure 2).
Fig. 2.
The evolution of rmsd of ring atoms of monosaccharides D–A from their mean structures during a 4 ns (A) unrestrained MD (B) restrained MD.
Almost exclusive appearance of the 2H1 conformer for ring D during the unrestrained molecular dynamics contradicts the results of our analysis of experimental proton–proton coupling constants, which indicate that the 1H2 form dominates (78%). This discrepancy could be explained by inaccurate parameterization of the AMBER force field for this unsaturated ring or inaccurate parameterization of the Karplus curves used to analyze the coupling constants. The latter is unlikely as the experimental 3J23 = 2.65 Hz is too small to be consistent with a large diaxial 3J23 (10.2 Hz) coupling constant of the 2H1 conformer. Inadequate performance of the AMBER force field for Δ4,5-uronic acids has been noted previously (Adeyeye et al. 2003). As MD simulations are an integral part of our methodology, the discrepancy between the experimental data and simulations had to be taken into account. We have therefore concluded that proceeding with the analysis of the D ring would add little value to our work and decided to concentrate on the trisaccharide fragment CBA of I. This is in line with the main focus of our investigation on the conformation of native heparin/HS and the effects of conformational flexibility of the internal iduronic acid on the overall shape of the molecule. Ring D, a modified terminal iduronic acid, is a consequence of preparation of oligosaccharides by enzymatic cleavage and does not have a role in the biology of heparin/HS.
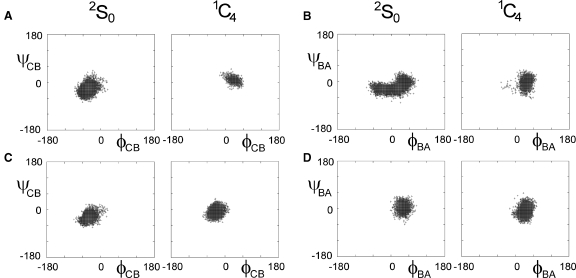
Our MD simulations show that the dynamics of the BA and CB glycosidic linkages is affected by the state of ring B as illustrated in Figure 3 using Φ/Ψ maps. These maps show the behavior of two dihedral angles, ΦH (H1 − C1 − O4′ − C4′) and ΨH (C1 − O4′ − C4′ − H4′) describing the conformation of glycosidic linkages. Considering the BA linkage first, a population of conformers exists that has very similar dihedral angles irrespective of the state of ring B. However, there are other conformers that differ by 60° in the ΦBA angle between the 2So and 1C4 forms of the B ring. Characterizing the CB linkage, the ΦCB and ΨCB angles differ by 90° and 40°, respectively, depending on the state of the B ring. The geometry of the glycosidic linkages has a major influence on the conformation of carbohydrates and their overall shape. Therefore, the results of unrestrained MD indicate that the conformation of ring B could affect the overall shape of heparin/HS polysaccharide.
Fig. 3.
Φ/Ψ maps of CB (A and C) and BA (B and D) glycosidic linkages of I during a 4 ns MD. (A) and (B) show unrestrained and (C) and (D) restrained MD simulations, respectively. Conformation of ring B is shown at the top. Ring D was always in 2H1 conformation.
In the next stage, two types of experimental parameters, which have not been used previously in the conformational analysis of heparin, are employed to assess the effects of the conformational flexibility of ring B on the overall shape of I: residual dipolar coupling constants (RDCs) and the interglycosidic long-range proton-carbon coupling constants (nJCH). The former are used as restraints in MD simulations, while the latter are used to cross validate the results of these simulations.
Measurement, validation, and analysis of residual dipolar coupling constants
Provided that a sufficient amount of compound is available, the most easily accessible RDCs in carbohydrates are those between the directly attached protons and carbons (1DCH). However, in rigid hexopyranose rings, the majority of equatorial positions are occupied by OR groups, while all axial CH orientations point in the same direction, i.e., are spatially degenerated. In α-d-GlcNS hexopyranose rings of I, only two independent orientations are sampled by the CH vectors – the axial and one equatorial, along the C1αH1α bond. When a limited number of RDCs are available, an independent determination of the order tensor is required (Zweckstetter and Bax 2000; Landersjo et al. 2000; Almond et al. 2001; Lycknert et al. 2001; Almond and Axelsen 2002; Azurmendi and Bush 2002). This approach therefore relies on an initial model describing the overall shape of the molecule. A self-consistent analysis of RDCs via the Saupe order matrix (Saupe 1968; Losonczi et al. 1999) on the other hand only requires structures of rigid fragments to be known. However, this approach requires ≥5 RDCs necessitating RDCs between remote nuclei, such as nDHH or nDCH, to be measured. Due to the dependence of RDCs on the gyromagnetic ratios of the coupled nuclei, accurate measurement of nDCH coupling constants requires stronger alignment. Such alignment can easily induce higher order effects in proton spectra of carbohydrates, which typically show little chemical shift dispersion. This can compromise the measurement of all types of RDCs. On the other hand, nDHH coupling constants are larger, which allows the use of a very weak alignment. We have therefore adapted this approach and supplemented 1DCH with nDHH coupling constants.
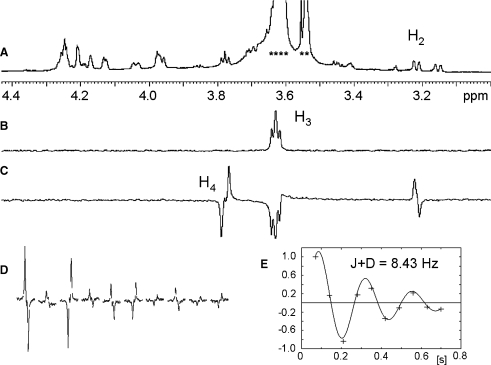
We have recently developed a suite of NMR experiments for the accurate measurement of small proton–proton RDCs from highly overlapped proton spectra and weakly aligned samples (Jin L et al. 2007). The reader is referred to the original references for their full description. In short, these methods are based on the selective 1D directed-COSY experiment (Pham et al. 2002, 2004) and provide coupling constants through fitting of signal intensities measured in a series of experiments. Their key feature is the ability to excite selectively a single proton resonance from severely overlapping spectra via the use of double-selective HOHAHA (Konrat et al. 1991) or gradient-enhanced, chemical shift-selective filters (Robinson et al. 2004) as illustrated in Figure 4.
Fig. 4.
Determination of proton–proton RDCs of I. An example of the measurement of the H3, H4 J+D splitting of ring C. (A) 1D CPMG spectrum of aligned sample of I. Residual signal from the medium is labeled with asterisks. (B) 1D double selective HOHAHA spectrum of I with a selective transfer of magnetisation from H2 to H3 of ring C. (C) variable time J-modulated 1D double selective HOHAHA-COSY spectrum of I with a selective transfer of magnetization from H2 to H3 and a selective COSY transfer to H4 using a COSY evolution interval of 205 ms. (D) Variation of the H4 signal intensity as a functional of the COSY evolution delay and (E) corresponding fitting yielding the J+D splitting.
Table II summarizes experimental 1DHH and 1DCH coupling constants of I measured in neutral aligning media (Ruckert and Otting 2000). The latter were determined from the F2 dimension of 1H-coupled nonrefocused 1H-13C HSQC spectra. The measured RDCs were validated using the rigid monosaccharide rings A and C. For this purpose, ab initio disaccharide structures, IdoA2S-GlcNS,6S-OMe and GlcNS,6S-IdoA2S-Ome representing BA and CB fragments of I, were used. Five 1DCH and four 3DHH coupling constants for either the A or C ring were used to calculate separately the order tensors via the singular value decomposition method (Losonczi et al. 1999). The theoretical RDCs were then back calculated using REDCAT (Valafar and Prestegard 2004). These calculations were performed for both 1C4 and 2So conformations of the B ring BA and CB disaccharides. A comparison of the theoretical and experimental RDCs (Table III) showed a very good agreement indicating a high accuracy of the measured coupling constants. For both BA and CB disaccharides, a better agreement between the theoretical and experimental RDCs was obtained when the iduronic ring was in the 1C4 conformation. In the case of the CB disaccharide, this agreement was much better. From now on, we will therefore focus on the disaccharides with this conformation of ring B, although the conclusions presented below were the same when the 2So conformers were analyzed.
Table II.
Scalar and residual dipolar coupling constants of ΔU-tetrasaccharide I
| One-bond | Three-bond | |||||||
|---|---|---|---|---|---|---|---|---|
| 1H-13 C couplings | J (Hz) | J + D (Hz) | RDC (Hz) | 1H-1H couplings | J (Hz) | J + D (Hz) | RDC (Hz) | |
| Ring | C1H1 | 172.2 | 168.8 | −3.4 | H1H2 | 3.53 | 2.71 | −0.82 |
| A | C2H3 | 139.1 | 144.9 | 5.8 | H2H3 | 10.25 | 11.13 | 0.88 |
| C3H3 | 148.0 | 154.1 | 6.1 | H3H4 | 8.79 | 9.75 | 0.96 | |
| C4H4 | 147.2 | 152.6 | 5.4 | H4H5 | 9.85 | 10.75 | 0.90 | |
| C5H5 | 146.9 | 153.1 | 6.2 | – | – | – | – | |
| B | C1H1 | 174.1 | 178.9 | 4.8 | H1H2 | 2.42 | 3.97 | 1.55 |
| C2H3 | 151.4 | 152.5 | 1.1 | H2H3 | 4.76 | 3.53 | −1.22 | |
| C3H3 | 151.5 | 148.9 | −2.6 | H3H4 | 3.41 | 3.30 | −0.11 | |
| C4H4 | 148.8 | 152.8 | 4.0 | – | – | – | – | |
| C5H5 | 146.1 | 145.7 | −0.4 | – | – | – | – | |
| C | C1H1 | 172.6 | 177.7 | 5.1 | H1H2 | 3.67 | 2.46 | −1.21 |
| C2H3 | 138.7 | 135.6 | −3.1 | H2H3 | 10.59 | 11.14 | 0.55 | |
| C3H3 | 147.4 | 144.4 | −3.0 | H3H4 | 9.04 | 8.43 | −0.61 | |
| C4H4 | 147.2 | 142.4 | −4.8 | H4H5 | 10.16 | 8.80 | −1.36 | |
| C5H5 | 147.0 | 143.4 | −3.6 | – | – | – | – | |
Table III.
Experimental and back-calculated RDCs of rings A and C of I analyzed using separately ab initio structures of IdoA2S-GlcNS,6S-OMe (ring A) and GlcNS,6S-IdoA2S-OMe (ring C)
| 4C1–1C4 | 4C1–2SO | 4C1 –1C4 | 4C1 – 2SO | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| RDC (Hz) | Expt. Ring A | Calc. | Diff. | Calc. | Diff. | Expt. Ring C | Calc. | Diff. | Calc. | Diff. |
| C1H1 | −3.4 | −3.40 | 0.00 | −3.41 | 0.01 | 5.1 | 5.11 | −0.01 | 5.14 | −0.04 |
| C2H2 | 5.8 | 5.89 | −0.09 | 5.89 | −0.09 | −3.1 | −2.98 | −0.12 | −2.65 | −0.45 |
| C3H3 | 6.1 | 6.07 | 0.03 | 6.07 | 0.03 | −3.0 | −3.09 | 0.09 | −4.10 | 1.10 |
| C4H4 | 5.4 | 5.65 | −0.25 | 5.59 | −0.19 | −4.8 | −4.88 | 0.08 | −4.39 | −0.41 |
| C5H5 | 6.2 | 5.94 | 0.26 | 6.04 | 0.16 | −3.6 | −3.57 | −0.03 | −3.22 | −0.38 |
| H1H2 | −0.82 | −0.82 | 0.00 | −0.77 | −0.05 | −1.21 | −1.22 | 0.01 | −1.10 | −0.11 |
| H2H3 | 0.88 | 0.70 | 0.18 | 0.74 | 0.14 | 0.55 | 0.46 | 0.09 | 0.03 | 0.52 |
| H3H4 | 0.96 | 0.86 | 0.10 | 0.76 | 0.20 | −0.61 | −0.48 | −0.13 | −0.32 | −0.29 |
| H4H5 | 0.90 | 0.91 | −0.01 | 0.43 | 0.47 | −1.36 | −1.23 | −0.13 | −1.06 | −0.30 |
| rmsd | 0.14 | 0.20 | 0.09 | 0.49 | ||||||
Inspection of the experimental 1DCH coupling constants of rings C and A showed their ratio to be close to 2:−1. This suggested that the orientation of CH vectors of the two rings is approximately parallel and perpendicular with regard to the z-axis of the principal axis frame (PAF) of the respective order tensor. This was confirmed through the analysis of the PAF systems of rings A and C. Four axial CH bonds of ring A in the disaccharide BA were found to be oriented at angles 88° ± 6° and 7.5° ± 3° relative to Szz- and Syy-axes of the order tensor, while for ring C in the disaccharide CB these were at 31° ± 5° and 81° ± 5°. An implication of these observations is that the CH bonds of rings A and C in I are approximately perpendicular to each other. This statement assumes that a single-order tensor can be used to characterize the alignment of I. In order to test this assumption, the Eigen values of the order matrix of ring A in the BA disaccharide (1.33 × 10−4, −1.32 × 10−4, −4.15 × 10−7) and those of ring C in the CB disaccharide (−1.64 × 10−4, 1.61 × 10−4, 3.75 × 10−6) were compared using the 1C4 conformers or ring B. Almost identical absolute values of Szz and Syy in both cases (note that these components therefore occasionally swap) indicate that both order tensors can be characterized as oblate. The alignment of ring A is 19% weaker, which could be attributed to the fact that this is the terminal, reducing monosaccharide of I. In order to detect the presence of conformational averaging, Tian et al. (2001) have introduced the generalized degree of order (GDO). A large difference between GDOs of two rigid fragments of a molecule indicates conformational flexibility. In our study, the GDOs of rings A and C are 1.53 × 10−4 and 1.88 × 10−4 for 1C4 conformers of ring B. Even closer agreement (GDO = 1.47 × 10−4 and 1.42 × 10−4) was observed for 2So conformation of the B ring. A similar difference in GDO of around 20% has been interpreted as originating from a limited relative movement of two monosaccharide rings in a trimannoside (Tian et al. 2001). Adopting this assumption, we postulated that the relative orientation of rings A and C is not affected significantly by the presence of the conformational flexibility of the B ring, and we performed restrained MD simulations of I using RDCs of both rings simultaneously as restraints.
RDC-restrained molecular dynamics simulations of I
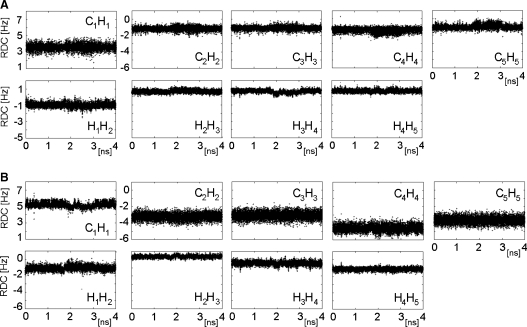
RDCs of two rigid monosaccharide rings, A and C, are used simultaneously to define the overall order tensor of I. This analysis differs from the one presented above, where RDCs were analyzed separately for each of the two rings using static structures. Here, they are used as experimental restraints during the AMBER molecular dynamics while the order tensor parameters are allowed to float. RDCs of ring B are not included as restraints; they are merely calculated for every point of the MD trajectory and their values are compared with experimental data. During the RDC-restrained MD, and similarly to the unrestrained MD, rings A and C were stable in their 4C1 conformations, while ring B showed transitions between the 1C4 and 2So forms. The effect of RDCs on the MD simulation was assessed by calculating rmsds of ring atoms of monosaccharides A, B, and C. The magnitude of rmsd fluctuations (Figure 2B) was identical to those observed in the unrestrained MD simulations (Figure 2A) indicating that the inclusion of RDCs did not destabilize the structures. RDCs of ring A showed smaller rmsds (5% for 1DCH and 19% for 3DHH) than those of ring C (10% for 1DCH and 24% for 3DHH) (Figure 5); in either case, the rmsds did not depend on the state of the B ring.
Fig. 5.
Evolution of calculated RDCs during the restrained MD simulation (A) ring A and (B) ring C.
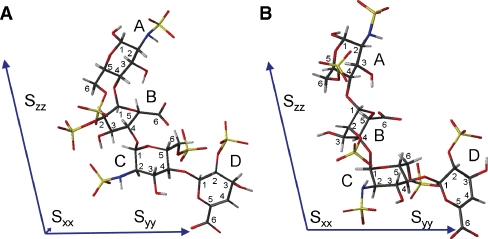
In order to analyze the order tensor parameters, the structures were divided into two groups depending on the conformation of ring B and their diagonalized order tensors were inspected. The average Saupe matrix of the 1C4 forms was found to have principal axis components: Szz = 1.44e-04, Syy = 1.36e-04, Sxx = −0.70e-05, and (GDO = 1.62e-04), while the 2So form gave Szz = 1.58e-04, Syy = 1.43e-04, and Sxx = −1.44e-05 (GDO = 1.74e-04). The two largest components of these tensors, Szz and Syy, differ by less than 10%, which caused their occasional swapping, seen in particular for the B ring in the 1C4 conformer. Importantly, there is <10% difference between the Szz (Syy) values of the two groups differing in the conformations of the B ring; their GDOs are also very similar (<7% difference). In addition, the orientation of the principal axes of the two tensors was very similar. Figure 6 shows that Szz is parallel to the long axis of the trisaccharide fragment CBA of I. The large Syy component arises due to the presence of bulky sulfate groups and the fact that the D ring is positioned at an acute angle to the Szz-axis. The smallest component, Sxx, is at least an order of magnitude smaller than the other two. Although there is 100% difference between the two forms of the B ring, this can easily be a result of small changes in the average orientation of some sulfate groups.
Fig. 6.
Orientation of the PAF of the order tensor relative to the structure of I: (A) ring B in 2So and (B) ring B in 1C4 conformation. The relative length of the principal axis corresponds to the sizes of the principal order parameters.
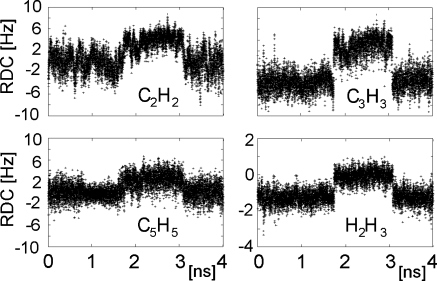
Eight RDCs of ring B, which were not used as structural restraints during MD, were then back calculated using MD structures. Four of them were practically identical between the two sets of molecules differing in the conformation of the B ring; the MD trajectories of the remaining RDCs are shown in Figure 7. Our MD simulations are too short to allow for a reliable calculation of the average theoretical RDCs by using weighted averages of the two forms. Instead, least-square analysis was performed using the average theoretical RDCs of the individual conformers (Table IV). This yielded the best fit with the experimental data for 82% of the 1C4 and 18% of the 2So forms, which is remarkably similar to the results of the analysis of the proton–proton scalar couplings of ring B (76% and 24%, respectively).
Fig. 7.
Back-calculated RDCs of ring B during the restrained MD simulations. Only the RDCs that were different between the two forms of ring B are shown.
Table IV.
Population analysis of the 1C4 and 2So forms of B ring using the back-calculated average RDCs
| RDC (Hz) | C2H2 | C3H3 | C5H5 | H2H3 | Population |
|---|---|---|---|---|---|
| 1C4 | −0.20 | −3.72 | −0.01 | −1.27 | 82% |
| 2SO | 3.80 | 2.78 | 2.38 | −0.07 | 18% |
| Expt. | 1.1 | −2.6 | −0.4 | −1.22 | rmsd = 0.51 |
| Theory | 0.52 | −2.55 | 0.42 | −1.05 |
Focusing on rings A and C in structures with the best agreement between the experimental and calculated RDCs, the average angles between the corresponding CH vectors of individual rings of 118° ± 7° and 110° ± 9° were found for 1C4 and 2So conformations of ring B, respectively. These results are in line with approximately perpendicular orientations of both rings as implied by the analysis of RDCs using the disaccharide structures discussed above.
Any conformational changes of the trisaccharide fragment CBA during the restrained MD simulations will affect the dihedral angles along CB and BA glycosidic linkages (Figure 3C and D). Table V shows that the differences between the average values of Φ and Ψ angles corresponding to two forms of ring B are less than 15° – a value comparable to the natural fluctuations of these parameters. This also illustrates that the conformational space sampled during MD simulations is smaller than observed for the unrestrained MD dynamics (cf. Figure 3A and B) and also implies that the overall shape of I is not affected significantly by the conformational change of ring B.
Table V.
Dihedral angles across the glycosidic linkages of rings C–B–A
| Dihedral angles (°) | ΦCB | ΨCB | ΦBA | ΨBA |
|---|---|---|---|---|
| 3JCOCH baseda | −28 | −19 | 35 | ± 23 |
| Retrained MD, 1C4(B)b | −33 ± 12 | −10 ± 13 | 34 ± 11 | −7 ± 16 |
| Retrained MD, 2So(B)b | −38 ± 4 | −25 ± 14 | 41 ± 11 | 5 ± 14 |
| Heparinc, 1C4(B)b | −39 | −32 | 41 | 14 |
| Heparinc, 2So(B)b | −9 | −41 | 61 | 16 |
| Solution structure of Id | −43 | −42 | 45 | 15 |
| Crystal structuree of I | −18 | −4 | 57 | 25 |
Verification of RDC-restrained MD structures using interglycosidic 3JCH coupling constants of I
Interglycosidic dihedral angles can be conveniently probed by three bond proton–carbon coupling constants. The corresponding Karplus curve, relating J couplings to dihedral angles, has been parameterized for glycosidic linkages of carbohydrates (Tvaroška et al. 1989). The dihedral angles that satisfy experimental J couplings are not unique due to the 4-fold degeneracy of the Karplus curve. However, relatively large measured coupling constants (Table V) immediately indicate that the average absolute dihedral angles they report on are small (quadrants I and IV) or large (quadrants II and III) angles. The latter possibility could be eliminated as there is no experimental evidence for the existence of anticonformer for either Φ or Ψ dihedral angle in heparin structures. Taking into account NOESY data (data not shown) and the MD simulation, three out of the four angles can therefore be interpreted unambiguously, leaving ΨBA ambiguous between the first and the fourth quadrant. Table V shows the dihedral angles derived from the experimental 3JCH coupling constants; their values are within ± 15° of the average dihedral angles obtained for both tetrasaccharides containing the B ring in either 1C4 or 2So conformation. Assuming a 4:1 ratio of the 1C4 and 2So conformers, the average values of 3JCH coupling constants were calculated using the average dihedral angles of each form obtained in restrained MD simulations (Table VI). The rmsd between the measured and calculated values was 0.49 Hz indicating a good agreement between dihedral angles in RDC-restrained MD structures and J-based dihedral angles.
Table VI.
3JCOCH coupling constants (Hz) across the glycosidic linkages CB and BA
| 3JCH (Hz) | J(CB4HC1) | J(CC1HB4) | J(CA4HB1) | J(CB1HA4) | rmsdc |
|---|---|---|---|---|---|
| Experimental | 4.4 | 5.0 | 3.8 | 4.8 | – |
| Retrained MD, 1C4(B)a | 4.0 | 5.4 | 3.9 | 5.6 | – |
| Retrained MD, 2So(B)a | 3.6 | 4.7 | 3.3 | 5.5 | – |
| Retrained MD, Javeb | 3.9 | 5.3 | 3.8 | 5.6 | 0.49 |
| Heparinc, 1C4(B)a | 3.5 | 4.1 | 3.3 | 5.3 | – |
| Heparinc, 2So(B)a | 5.5 | 3.3 | 1.6 | 5.2 | – |
| Heparinc, Javeb | 3.9 | 3.9 | 2.9 | 5.3 | 0.79 |
| Solution structure of Id | 5.0 | 5.6 | 1.9 | 4.6 | 1.05 |
| Crystal structure of Ie | 3.1 | 3.2 | 2.9 | 5.2 | 1.21 |
Discussion
The order tensor defining the direction and the strength of the alignment is required for the interpretation of RDCs. If structures of the studied compound are available, the order tensor can be obtained using a steric model (Almond et al. 2001) or mass distribution (Lycknert et al 2001; Azurmendi and Bush 2002). Starting from NOE-restrained structure, the latter approach was recently used to analyze the conformation of a dermatan sulfate tetrasaccharide (Silipo et al. 2008). Linearly averaged RDCs calculated for every frame of MD simulations were compared with the experimental values and showed a good agreement.
Alternatively, the tensor parameters can be obtained by analyzing RDCs without the need for an initial molecular model. This requires more RDCs to be measured, typically five to ten for any rigid part of the molecule. In our work, we have supplemented the standard 1DCH by 3DHH coupling constants; the latter type was obtained by 1D intensity-based methods. Altogether 18 RDCs, 9 per each ring were used. They sampled 12 independent orientations: two and four described by 1DCH and 3DHH couplings per ring, respectively. We have analyzed these RDCs using the singular value decomposition (Losonczi et al. 1999) method in two different ways. Firstly, by focusing separately on the rigid A and C rings of I and using disaccharides related to BA and CB fragments of I. A major conclusion of this analysis was a relative agreement between the GDO parameters of the order tensors of rings A and C that allowed us to suggest that one order tensor can be used to describe the alignment of I. Based on this premise, we were able to show that the relative orientation of rings A and C is such that their CH bonds are approximately perpendicular.
One observation made during this analysis deserves a brief discussion. According to the analysis of 3JHH coupling constants, the B ring of I exists mainly (76%) in 1C4 conformation. The analysis of RDCs of rings A and C using disaccharides BA and CB showed better agreement with the experimental data when ring B was in the 1C4 conformation. It is therefore tempting to suggest that RDCs of rigid rings A or C reflect the conformational state of the B ring. However, this might be an over interpretation of our data. First, we are using methylated disaccharide structures to interpret RDCs of monosaccharide rings of I, i.e., we are not taking into account possible effects of the third carbohydrate ring. Second, the structures of the BA and CB disaccharides were calculated in an isolated state and, as we have shown previously (Pham et al. 2004), RDCs are extremely sensitive to small changes in dihedral and bond angles and the different environments could therefore play a role. Nevertheless, a proposition that the type or a conformation of the neighboring ring can affect the monosaccharide ring geometry which could be probed by RDCs is intriguing and warrants further investigation using simpler systems. This suggestion is supported by theoretical data indicating that influence of the IdoA2S residue on the geometry of neighboring GlcNS residues could be important (Hricovíni et al. 2007).
We performed restrained MD simulations of I using RDCs of rings A and C simultaneously as restraints. The resulting structures were then used to back calculate RDCs for the flexible B ring in both of its conformations. The relative amounts of 1C4 and 2So forms deduced from the back-calculated RDCs agreed well with the results of the analysis of the proton–proton scalar couplings of ring B.
The experimental RDCs reflect the dynamics of glycosidic linkages, internal dynamics of rings B and D, as well as any possible accompanying changes of the order tensor. They therefore mirror a highly complex system. Due to the long time scale of NMR measurements, we can be confident that the sampling of these RDCs was comprehensive. While the fluctuations of the glycosidic linkages are fast, the B ring is only slowly interconverting between its two forms, 1C4 and 2So, and on a picosecond time scale exists in a particular conformation. This means that average RDCs were used here to refine immediate conformations of I during MD. Meaningful results are therefore only obtained if the order tensor is largely independent of the complex dynamics experienced by the ΔU-tetrasaccharide. This assumption was based on the analysis of two disaccharide fragments showing similar GDOs for both conformations of the B ring.
In order to verify this assumption, albeit in an indirect way, we need another experimental parameter. This was provided by interglycosidic 3JCH coupling constants, measured for the first time on a heparin oligosaccharide. Their relatively large values (Table VI) immediately indicated that the corresponding dihedral angles are small, which is in agreement with the results of our restrained MD simulations. We used a 4:1 ratio of 1C4 and 2So conformers of ring B, as determined by the analysis of proton–proton scalar coupling constants, and calculated the 3JCH coupling constants based on RDC-restrained MD structures. The obtained rmsd between the theoretical and experimental coupling constants of 0.49 Hz indicated a good agreement with the experimental data.
As stated in the Introduction, the structure of heparin and heparin oligosaccharides has been investigated in the past by standard NMR techniques. When making a comparison with our studies, it should be kept in mind that none of the cited work has carried out conformational analysis in the presence of Ca2+, which could be a source of discrepancies. ΔU-tetrasaccharide I was studied previously using NOESY spectroscopy and the authors (Mikhailov et al. 1996) have reported the interglycosidic dihedral angles of their final structure. The 3JCH coupling constants for this structure showed rmsd of 1.05 Hz to our experimental values (Table VI). This larger rmsd is a direct consequence of more acute dihedral angles (Table V) found by the previous study based on 1H-1H NOESY data only.
Our solution structures of I can also be compared with the heparin structure (Mulloy et al. 1993). The reported NOE- and MD-based structure of a dodecasaccharide shows angles between the CH vectors of pairs of consecutive glucosamine rings of 111° ± 9.0 and 86° ± 5.9 for 1C4 and 2So conformation of IdoA2S, respectively. For 1C4 conformation, this agrees very well with our RDC-based solution structure (118°) while a 25° difference was found for the 2So conformation of ring B (111°). This could be due to much shorter fragment approximating the terminal tetrasaccharide of heparin used in our study. We can also compare interglycosidic dihedral angles of both structures (Table V). Here, all the dihedral angles are within one rmsd of our restrained MD structures, with an exception of ΨCB glycosidic linkage. The theoretical 3JCH coupling constants, calculated as weighted averages of ring B 1C4 and 2So conformers using the heparin structure (Mulloy et al. 1993), show rmsd of 0.79 Hz from our experimental values, i.e., larger than for our RDC-restrained MD structures (rmsd of 0.49 Hz). Again, this difference could be attributed to the end effects of I that are diluted in heparin dodecasaccharide. Nevertheless, the observed values indicate that heparin helix recently also found through NOE and MD investigation of 13C-labeled heparin (Zhang et al. 2008) is already starting to form in the fully sulfated heparin ΔU-tetrasaccharide I.
Table V also includes the dihedral angles reported for an X-ray structure of I, containing the iduronic acid in 1C4 conformation, in complex with the basic fibroblast growth factor (Faham et al. 1996). While a good agreement was obtained for the dihedral angles along the CB glycosidic linkage, differences of up to 30° were observed for the BA linkage (Table V). This is reflected in a large rmsd (1.21 Hz) for the corresponding 3JCH coupling constants reported in Table VI. This comparison should be taken with caution as average values reflecting the presence of two forms in solution are compared here with only one conformer (1C4) of I observed in the solid state, which in addition is in complex with protein.
As 3JCH coupling constants were not used during the structure refinement, they represent an experimental parameter that may be used to verify our structure and therefore justify the assumptions that underline our analysis of RDCs. However, we would like to emphasize that this reasoning is not strictly consequential. All we can conclude from this analysis is that the structures obtained using RDC-restrained MD simulations reproduce well the experimental inter-glycosidic 3JCH coupling constants.
Although showing some differences in interglycosidic dihedral angles to our structure of I that were verified by inter-glycosidic 3JCH coupling constants, previous investigations of heparin (Mulloy et al. 1993) and its oligosaccharides (Ferro et al. 1986; Mikhailov et al. 1996, 1997; de Paz et al. 2001) using 1H-1H NOESY spectroscopy and force field calculations concluded that the flexibility of the IdoA has only a limited effect on the overall shape of heparin. Our conclusions based on the analysis of additional NMR parameters, inter-glycosidic 3JCH coupling constants and RDCs in particular, support these findings. The nature of RDCs makes this NMR parameter particularly suitable for the study of mutual orientation of remote molecular fragments providing superior information to NOE data. This is the main contribution of this work. From the structural biology point of view, enhancement of the protein binding properties of IdoA containing GAGs acts through more flexible orientation of sulfate groups, rather than significantly affecting the conformation of glycosidic linkages. At the same time however, the overall shape of the heparin molecules is not changing significantly, as shown by similar order parameters irrespective of the conformation of the internal IdoA.
Conclusion
We have investigated the structure of a fully sulfated, heparin-derived ΔU-tetrasaccharide using NMR spectroscopy and MD simulations. Our analysis of RDCs showed that the inter-conversion of the iduronic acid between its 1C4 and 2So forms has only modest effect on the overall shape of the molecule. This conclusion is born out of our observation that the average RDCs of the two rigid carbohydrate rings surrounding the flexible iduronic acid were equally well reproduced using similar order tensors calculated for the two IdoA conformations. Following RDC-restrained MD simulations obtained using a single order tensor produced the final structures of I. The ratios of 1C4 and 2So forms of the iduronic acid derived using 3JHH and RDC were practically identical. The RDC-restrained structures were validated by the interglycosidic 3JCH coupling constants. Experimentally determined interglycosidic dihedral angles of the terminal trisaccharide fragment indicate that the heparin helix already starts forming in this minimal structure. Thus, by using RDCs and interglycosidic coupling constants, the two parameters previously not applied to the study of heparin, our investigation confirmed the limited effect of the flexibility of IdoA on the overall shape of heparin molecule. Our work therefore demonstrates the effectiveness of the methods for the measurement and analysis of RDCs in the conformational analysis of glycosaminoglycans.
Material and methods
Preparation of fully heparin-derived ΔU-tetrasaccharide I
The heparin ΔU-tetrasaccharide (Figure 1) was prepared by partial enzymatic digestion of low-molecular-weight heparin (Innohep; Leo Laboratories, Princes Risborough, UK, with heparinase I; Grampian Enzymes, Orkney, UK). The digest was resolved into its constituent oligosaccharide size fractions by gel-filtration chromatography on a Bio-Gel P10 (Bio-Rad Laboratories, Hemel Hempstead, UK) column (2.5 × 120 cm) eluted with 0.25 M NH4HCO3 at a flow rate of 10 mL/h. The elution profile was monitored by UV absorption at 232 nm, specific to the unsaturated nonreducing ends of the oligosaccharides introduced by the lyase action of the enzyme. The tetrasaccharide fractions were pooled, freeze-dried twice, desalted on PD-10 columns (Amersham Biosciences Ltd, Chalfont St. Giles, UK) and then dried. The tetrasaccharide fractions were resolved into their constituent species by strong anion-exchange HPLC chromatography on an IonPac AS17 column (0.4 × 25 cm; Dionex, Camberley, UK). After a brief wash with water (pH 3.5), the column was eluted with a 60 mL linear gradient of 0–1.0 M NaCl, pH 3.5, at a flow rate of 1 mL/min with in line monitoring of UV absorption at 232 nm. ΔU-tetrasaccharide eluted as the last fraction; its structure was identified as described previously (Jin et al. 2005).
Preparation of isotropic and aligned samples for NMR
Five hundred fifty micrograms of I was dissolved in 0.5 mL deuterium oxide (D2O >99.96%) and lyophilized to remove exchangeable protons. The powder was then redissolved in 320 μL D2O, which contained CaCl2 corresponding to a 4-fold molar access with regard to I. The pH was adjusted to 7.5 by adding NaOH dissolved in D2O. A 5 mm Shigemi tube with or without plunger was used for isotropic and aligned samples, respectively. The C12E5/hexanol medium (Ruckert and Otting 2000) with r = 0.9 and wt% = 3.7% was used for the aligned sample of I. The composition of the mixture was 18.6 μL C12E5, 6.2 μL hexanol, and 430 μL D2O. The splitting of the deuterium signal was 22.5 Hz at 25°C.
Measurements of 1DCH and 3DHH coupling constants
NMR experiments were performed on a 600 or 800 MHz Bruker Advance spectrometer equipped with a 5 mm cryoprobe with inverse geometry and z-gradients. RDCs were calculated as a difference between the splittings observed in the isotropic and aligned samples determined using identical NMR experiments acquired under the same conditions. Nonrefocused 2D 1H-13C HSQC spectra were acquired without decoupling and refocusing in the directly detected dimension. Acquisition times of both isotropic and aligned samples in t1 and t2 were 63.6 and 341 ms, respectively; the total experimental times were 62.5 and 45.5 h, respectively. SPFGSE-COSY, CSSF-COSY, and double-selective HOHAHA-COSY spectra, acquired using the parameters described elsewhere (Jin et al. 2007) were used for the measurement of 1H-1H scalar and dipolar coupling constants. The spectra of the isotropic sample were normally acquired using 80 scans per increment, while those of the aligned sample were recorded using 200 scans. Twenty to thirty millisecond Gaussian pulses were used to selectively invert proton resonances. CSSF-COSY spectra were typically acquired using eight 10 ms CSSF increments; 30–45 ms Gaussian pulses were used during the CSSF and also during the COSY part.
DFT calculations of disaccharide fragments
The geometry of heparin disaccharides CB (GlcNS,6S-IdoA2S-OMe) and BA (IdoA2S-GlcNS,6S-OMe) has been optimized (JAGUAR 1998) using density functional theory (DFT) with Lee–Young–Parr (B3LYP) (Lee et al. 1988) correlation functional and the 6-311++G** basis set. Two conformers (1C4 and 2So) were considered for the IdoA2S residue with the starting structures obtained previously (Hricovíni 2006). The GlcN,6S residue was in the 4C1 conformation. Geometry optimizations were obtained with the gradient optimization routine, and the convergence criteria were set to 1.10−6.
Free and restrained MD simulations of I using AMBER
ΔU-tetrasaccharide I was built using the atom types contained in the recently parameterized force field (Huige and Altona 1995; Jin et al. 2005). Partial atom charges were calculated with the RESP (Restrained ElectroStatic Potential fit) procedure using Gaussian98 with the 6-31G* basis set. The single point charge calculation was carried out with the convergence criteria of 1.00D-02 and 1.00D-04 for the maximal and RMSDs change of the density matrix elements, respectively. The net charge was set to −8. Firstly, a simulated annealing procedure was used to generate low potential energy structures of I. The calculation was run in vacuum without any solvent or periodic box. In the initial energy minimization period, the starting conformation was optimized with a 3 ps steepest descent step followed by a gradient method of 0.5 ns duration. The optimized structure was subjected to 30 ps of molecular dynamics at 800 K with constraint on the bonds involving H-atoms (SHAKE). The system was then cooled to 0 K during a period of 10 ps using 10 cooling steps of 80 K in order to freeze out one conformation. This geometry was then optimized by minimizing the energy in the same way as the initial energy minimization was carried out. The first candidate conformation was obtained in this way and used as the input for the next round of the simulated annealing.
The structure with the lowest potential energy from gas phase simulation was dissolved in a water box for explicit solution periodical calculation. Before adding the water molecules, eight sodium ions were added randomly to neutralize the whole unit. A cubic 40.675 Å3 water box with a spacing of 8 Å containing 1938 triangulated 3-point water molecules was generated. The water box was first energy minimized with a tight restraint on the ΔU-tetrasaccharide I atoms to relax the water molecules. The whole system was energy minimized without any restraints. The temperature of the system was then increased from 0 K to 300 K in 2 ps and stabilized at 300 K for 20 ps at a constant volume with constraint on the bonds involving H-atoms, while the ΔU-tetrasaccharide molecule was restrained weakly. Another 0.1 ns equilibration molecular dynamic at 300 K and a constant pressure (1 atm) was simulated without any restraint before the MD production calculations. Free and RDC-restrained molecular dynamics were performed at constant pressure (1 atm) and temperature of 300 K for 4 ns. RDCs of both glucosamine rings (A and C) of I were used as experimental restraints.
Funding
The Wellcome Trust (078780/Z/05/Z to DU); ORS scheme and University of Edinburgh (to L.J.); CRUK Programme Grant (to M.L. and J.A.D.); and VEGA (2/0108/08 to M.H.).
Conflict of interest statement
None declared.
Glossary
Abbreviations
- AMBER
assisted model building with energy refinement
- COSY
correlated spectroscopy
- CSSF
chemical shift selective filter
- DS
dermatan sulfate
- GDO
generalized degree of order
- GlcNS
2-deoxy-2-sulfamido-α-d-glucopyranosyl
- HOHAHA
homonuclear Hartmann–Hahn spectroscopy
- HS
heparan sulfate
- HSQC
heteronuclear single quantum coherence
- IdoA
iduronic acid
- MD
molecular dynamics
- RDC
residual dipolar coupling
- REDCAT
residual dipolar coupling analysis tool
- SPFGSE
single pulse field gradient spin echo
References
- Adeyeye J, Azurmendi HF, Stroop CJM, Sozhamannan S, Williams AL, Adetumbi AM, Johnson JA, Bush CA. Conformation of the hexasaccharide repeating subunit from the Vibrio cholerae O139 capsular polysaccharide. Biochemistry. 2003;42:3979–3988. doi: 10.1021/bi026700t. [DOI] [PubMed] [Google Scholar]
- Almond A, Axelsen JB. Physical interpretation of residual dipolar couplings in neutral aligned media. J Am Chem Soc. 2002;124:9986–9987. doi: 10.1021/ja026876i. [DOI] [PubMed] [Google Scholar]
- Almond A, Bunkenborg J, Franch T, Gotfredsen CH, Duus JO. Comparison of aqueous molecular dynamics with NMR relaxation and residual dipolar couplings favors internal motion in a mannose oligosaccharide. J Am Chem Soc. 2001;123:4792–4802. doi: 10.1021/ja0025696. [DOI] [PubMed] [Google Scholar]
- Angulo J, De Paz JL, Nieto PM, Martin-Lomas M. Interaction of heparin with Ca2+: A model study with a synthetic heparin-like hexasaccharide. Isr J Chem. 2000;40:289–299. [Google Scholar]
- Azurmendi HF, Bush CA. Tracking alignment from the moment of inertia tensor (TRAMITE) of biomolecules in neutral dilute liquid crystal solutions. J Am Chem Soc. 2002;124:2426–2427. doi: 10.1021/ja017524z. [DOI] [PubMed] [Google Scholar]
- Canales A, Angulo J, Ojeda R, Bruix M, Fayos R, Lozano R, Gimenez-Gallego G, Martin-Lomas M, Nieto PM, Jimenez-Barbero J. Conformational flexibility of a synthetic glycosylaminoglycan bound to a fibroblast growth factor. FGF-1 recognizes both the C-1(4) and S-2(0) conformations of a bioactive heparin-like hexasaccharide. J Am Chem Soc. 2005;127:5778–5779. doi: 10.1021/ja043363y. [DOI] [PubMed] [Google Scholar]
- Capila I, Linhardt RJ. Heparin–protein interactions. Angew Chem Int Ed. 2002;41:391–412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Case DA, Pearlman DA, Caldwell JW, Cheatham TE, III, Wang J, Ross WS, Simmerling CL, Darden TA, Merz KM, Stanton RV, et al. AMBER 7. San Francisco: University of California; 2002. [Google Scholar]
- Casu B, Lindahl U. Structure and biological interactions of heparin and heparan sulfate. Adv Carbohydr Chem Biochem. 2001;57:159–206. doi: 10.1016/s0065-2318(01)57017-1. [DOI] [PubMed] [Google Scholar]
- Catlow KR, Deakin JA, Wei Z, Delehedde M, Fernig DG, Gherardi E, Gallagher JT, Pavao MSG, Lyon M. Interactions of hepatocyte growth factor/scatter factor with various glycosaminoglycans reveal an important interplay between the presence of iduronate and sulfate density. J Biol Chem. 2008;283:5235–5248. doi: 10.1074/jbc.M706589200. [DOI] [PubMed] [Google Scholar]
- Chevalier F, Lucas R, Angulo J, Martin-Lomas M, Nieto PM. The heparin-Ca2+ interaction: The influence of the O-sulfation pattern on binding. Carbohydr Res. 2004;339:975–983. doi: 10.1016/j.carres.2003.12.023. [DOI] [PubMed] [Google Scholar]
- de Paz JL, Angulo J, Lassaletta JM, Nieto PM, Redondo-Horcajo M, Lozano RM, Gimenez-Gallego G, Martin-Lomas M. The activation of fibroblast growth factors by heparin: Synthesis, structure, and biological activity of heparin-like oligosaccharides. Chembiochem. 2001;2:673–685. doi: 10.1002/1439-7633(20010903)2:9<673::AID-CBIC673>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Ernst S, Venkataraman G, Sasisekharan V, Langer R, Cooney CL, Sasisekharan R. Pyranose ring flexibility. Mapping of physical data for iduronate in continuous conformational space. J Am Chem Soc. 1998;120:2099–2107. [Google Scholar]
- Faham S, Hileman RE, Fromm JR, Linhardt RJ, Rees DC. Heparin structure and interactions with basic fibroblast growth factor. Science. 1996;271:1116–1120. doi: 10.1126/science.271.5252.1116. [DOI] [PubMed] [Google Scholar]
- Ferro DR, Provasoli A, Ragazzi M, Casu B, Torri G, Bossennec V, Perly B, Sinay P, Petitou M, Choay J. Conformer populations of l-iduronic acid residues in glycosaminoglycan sequences. Carbohydr Res. 1990;195:157–167. doi: 10.1016/0008-6215(90)84164-p. [DOI] [PubMed] [Google Scholar]
- Ferro DR, Provasoli A, Ragazzi M, Torri G, Casu B, Gatti G, Jacquinet JC, Sinay P, Petitou M, Choay J. Evidence for conformational equilibrium of the sulfated l-iduronate residue in heparin and in synthetic heparin monosaccharides and oligosaccharides – NMR and force-field studies. J Am Chem Soc. 1986;108:6773–6778. [Google Scholar]
- Forster MJ, Mulloy B. Molecular-dynamics study of iduronate ring conformation. Biopolymers. 1993;33:575–588. [Google Scholar]
- Guerrini M, Guglieri S, Beccati D, Torri G, Viskov C, Mourier P. Conformational transitions induced in heparin octasaccharides by binding with antithrombin III. Biochem J. 2006;399:191–198. doi: 10.1042/BJ20060656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrini M, Hricovíni M, Torri G. Interaction of heparins with fibroblast growth factors: Conformational aspects. Curr Pharm Des. 2007;13:2045–2056. doi: 10.2174/138161207781039733. [DOI] [PubMed] [Google Scholar]
- Guglieri S, Hricovíni M, Raman R, Polito L, Torri G, Casu B, Sasisekharan R, Guerrini M. Minimum FGF2 binding structural requirements of heparin and heparan sulfate oligosaccharides as determined by NMR spectroscopy. Biochemistry. 2008;47:13862–13869. doi: 10.1021/bi801007p. [DOI] [PubMed] [Google Scholar]
- Haasnoot CAG, Deleeuw F, Altona C. The relationship between proton–proton NMR coupling-constants and substituent electronegativities: 1. An empirical generalization of the Karplus equation. Tetrahedron. 1980;36:2783–2792. [Google Scholar]
- Hricovíni M. B3LYP/6–311++G** study of structure and spin–spin coupling constant in methyl 2-O-sulfo-alpha-l-iduronate. Carbohydr Res. 2006;341:2575–2580. doi: 10.1016/j.carres.2006.07.010. [DOI] [PubMed] [Google Scholar]
- Hricovíni M, Bízik F. Relationship between structure and three-bond proton–proton coupling constants in glycosaminoglycans. Carbohydr Res. 2007;342:779–783. doi: 10.1016/j.carres.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Hricovíni M, Guerrini M, Bisio A, Torri G, Petitou M, Casu B. Conformation of heparin pentasaccharide bound to antithrombin III. Biochem J. 2001;359:265–272. doi: 10.1042/0264-6021:3590265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hricovíni M, Nieto PM, Torri G. NMR of sulfated oligo- and polysaccharides. In: Jimenéz-Barbero J, Peters T, editors. NMR Spectroscopy of Glycoconjugates. Weinham: Wiley-VCH; 2002. pp. 325–334. [Google Scholar]
- Hricovíni M, Scholtzova E, Bízik F. B3LYP/6-31 1++G** study of structure and spin-spin coupling constant in heparin disaccharide. Carbohydr Res. 2007;342:1350–1356. doi: 10.1016/j.carres.2007.03.020. [DOI] [PubMed] [Google Scholar]
- Huige CJM, Altona C. Force-field parameters for sulfates and sulfamates based on ab-initio calculations – Extensions of amber and CHARMM fields. J Comput Chem. 1995;16:56–79. [Google Scholar]
- Imberty A, Lortat-Jacob H, Perez S. Structural view of glycosaminoglycan–protein interactions. Carbohydr Res. 2007;342:430–439. doi: 10.1016/j.carres.2006.12.019. [DOI] [PubMed] [Google Scholar]
- JAGUAR 3.5. Schrodinger, Inc., Portland, 1998.
- Jia J, Maccarana M, Zhang X, Bespalov M, Lindahl U, Li JP. Lack of l-iduronic acid in heparan sulfate affects interaction with growth factors and cell signaling. J Biol Chem. 2009;284:15942–15950. doi: 10.1074/jbc.M809577200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Barran PE, Deakin JA, Lyon M, Uhrín D. Conformation of glycosaminoglycans by ion mobility mass spectrometry and molecular modelling. Phys Chem Chem Phys. 2005;7:3464–3471. doi: 10.1039/b508644b. [DOI] [PubMed] [Google Scholar]
- Jin L, Pham TN, Uhrín D. Measurement of H-1-H-1 residual dipolar coupling constants for structural studies of medium size molecules. Chemphyschem. 2007;8:1228–1235. doi: 10.1002/cphc.200700071. [DOI] [PubMed] [Google Scholar]
- Konrat R, Burghardt I, Bodenhausen G. Coherence transfer in nuclear-magnetic-resonance by selective homonuclear Hartmann–Hahn correlation spectroscopy. J Am Chem Soc. 1991;113:9135–9140. [Google Scholar]
- Landersjo C, Hoog C, Maliniak A, Widmalm G. NMR investigation of a tetrasaccharide using residual dipolar couplings in dilute liquid crystalline media: Effect of the environment. J Phys Chem. 2000;B 104:5618–5624. [Google Scholar]
- Lee CT, Yang WT, Parr RG. Development of the Colle–Salvetti correlation-energy formula into a functional of the electron-density. Phys Rev. 1988;B 37:785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- Losonczi JA, Andrec M, Fischer MWF, Prestegard JH. Order matrix analysis of residual dipolar couplings using singular value decomposition. J Magn Reson. 1999;138:334–342. doi: 10.1006/jmre.1999.1754. [DOI] [PubMed] [Google Scholar]
- Lycknert K, Maliniak A, Widmalm G. Analysis of oligosaccharide conformation by NMR spectroscopy utilizing H-1,H-1 and H-1,C-13 residual dipolar couplings in a dilute liquid crystalline phase. J Phys Chem. 2001;A 105:5119–5122. [Google Scholar]
- Mikhailov D, Linhardt RJ, Mayo KH. NMR solution conformation of heparin-derived hexasaccharide. Biochem J. 1997;328:51–61. doi: 10.1042/bj3280051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhailov D, Mayo KH, Vlahov IR, Toida T, Pervin A, Linhardt RJ. NMR solution conformation of heparin-derived tetrasaccharide. Biochem J. 1996;318:93–102. doi: 10.1042/bj3180093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulloy B, Forster MJ. Conformation and dynamics of heparin and heparan sulfate. Glycobiology. 2000;10:1147–1156. doi: 10.1093/glycob/10.11.1147. [DOI] [PubMed] [Google Scholar]
- Mulloy B, Forster MJ, Jones C, Davies DB. NMR and molecular-modeling studies of the solution conformation of heparin. Biochem J. 1993;293:849–858. doi: 10.1042/bj2930849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulloy B, Linhardt RJ. Order out of complexity – Protein structures that interact with heparin. Curr Opin Struct Biol. 2001;11:623–628. doi: 10.1016/s0959-440x(00)00257-8. [DOI] [PubMed] [Google Scholar]
- Neuhaus D, Williamson MP. The Nuclear Overhauser Effect in Structural and Conformational Analysis. 2nd ed. New York: Willey-VCH; 2000. [Google Scholar]
- Nieduszynski IA, Gardner KH, Atkins EDT. Cellulose Chemistry and Technology. ACS Symp. Ser. 2nd ed. Vol. 48. Washington (DC): American Chemical Society; 1977. pp. 73–80. [Google Scholar]
- Pham TN, Hinchley SL, Rankin DWH, Liptaj T, Uhrín D. Determination of sugar structures in solution from residual dipolar coupling constants: Methodology and application to methyl beta-d-xylopyranoside. J Am Chem Soc. 2004;126:13100–13110. doi: 10.1021/ja047242+. [DOI] [PubMed] [Google Scholar]
- Pham TN, Liptaj T, Barlow PN, Uhrín D. J-Modulated 1D directed COSY for precise measurement of proton–proton residual dipolar coupling constants of oligosaccharides. Magn Reson Chem. 2002;40:729–732. [Google Scholar]
- Pol-Fachin L, Verli H. Depiction of the forces participating in the 2-O-sulfo-alpha-l-iduronic acid conformational preference in heparin sequences in aqueous solutions. Carbohydr Res. 2008;343:1435–1445. doi: 10.1016/j.carres.2008.04.016. [DOI] [PubMed] [Google Scholar]
- Powell AK, Yates EA, Fernig DG, Turnbull JE. Interactions of heparin/heparan sulfate with proteins: Appraisal of structural factors and epxerimental approaches. Glycobiology. 2004;14:17R–30R. doi: 10.1093/glycob/cwh051. [DOI] [PubMed] [Google Scholar]
- Rabenstein DL, Robert JM, Peng J. Multinuclear magnetic resonance studies of the interaction of inorganic cations with heparin. Carbohydr Res. 1995;278:239–256. doi: 10.1016/0008-6215(95)00263-4. [DOI] [PubMed] [Google Scholar]
- Robinson PT, Pham TN, Uhrín D. In phase selective excitation of overlapping multiplets by gradient-enhanced chemical shift selective filters. J Magn Reson. 2004;170:97–103. doi: 10.1016/j.jmr.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Ruckert M, Otting G. Alignment of biological macromolecules in novel nonionic liquid crystalline media for NMR experiments. J Am Chem Soc. 2000;122:7793–7797. [Google Scholar]
- Saupe A. Recent results in field of liquid crystals. Angew Chem Int Ed. 1968;7:97–118. [Google Scholar]
- Silipo A, Zhang ZQ, Canada FJ, Molinaro A, Linhardt RJ, Jimenez-Barbero J. Conformational analysis of a dermatan sulfate-derived tetrasaccharide by NMR, molecular modeling, and residual dipolar couplings. Chembiochem. 2008;9:240–252. doi: 10.1002/cbic.200700400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjandra N, Bax A. Direct measurement of distances and angles in biomolecules by NMR in a dilute liquid crystalline medium. Science. 1997;278:1111–1114. doi: 10.1126/science.278.5340.1111. [DOI] [PubMed] [Google Scholar]
- Tian F, Al-Hashimi HM, Craighead JL, Prestegard JH. Conformational analysis of a flexible oligosaccharide using residual dipolar couplings. J Am Chem Soc. 2001;123:485–492. doi: 10.1021/ja002900l. [DOI] [PubMed] [Google Scholar]
- Tvaroška I, Hricovíni M, Petráková E. An attempt to derive a new Karplus-type equation of vicinal proton carbon coupling-constants for C-O-C-H segments of bonded atoms. Carbohydr Res. 1989;189:359–362. [Google Scholar]
- Valafar H, Prestegard JH. REDCAT: A residual dipolar coupling analysis tool. J Magn Reson. 2004;167:228–241. doi: 10.1016/j.jmr.2003.12.012. [DOI] [PubMed] [Google Scholar]
- Zhang ZQ, McCallum SA, Xie J, Nieto L, Corzana F, Jimenez-Barbero J, Chen M, Liu J, Linhardtt RJ. Solution structures of chemoenzymatically synthesized heparin and its precursors. J Am Chem Soc. 2008;130:12998–13007. doi: 10.1021/ja8026345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweckstetter M, Bax A. Prediction of sterically induced alignment in a dilute liquid crystalline phase: Aid to protein structure determination by NMR. J Am Chem Soc. 2000;122:3791–3792. [Google Scholar]