Abstract
Arabinogalactan (AG) and lipoarabinomannan (LAM) are the two major cell wall (lipo)polysaccharides of mycobacteria. They share arabinan chains made of linear segments of α-1,5-linked d-Araf residues with some α-1,3-branching, the biosynthesis of which offers opportunities for new chemotherapeutics. In search of the missing arabinofuranosyltransferases (AraTs) responsible for the formation of the arabinan domains of AG and LAM in Mycobacterium tuberculosis, we identified Rv0236c (AftD) as a putative membrane-associated polyprenyl-dependent glycosyltransferase. AftD is 1400 amino acid-long, making it the largest predicted glycosyltransferase of its class in the M. tuberculosis genome. Assays using cell-free extracts from recombinant Mycobacterium smegmatis and Corynebacterium glutamicum strains expressing different levels of aftD indicated that this gene encodes a functional AraT with α-1,3-branching activity on linear α-1,5-linked neoglycolipid acceptors in vitro. The disruption of aftD in M. smegmatis resulted in cell death and a decrease in its activity caused defects in cell division, reduced growth, alteration of colonial morphology, and accumulation of trehalose dimycolates in the cell envelope. Overexpression of aftD in M. smegmatis, in contrast, induced the accumulation of two arabinosylated compounds with carbohydrate backbones reminiscent of that of LAM and a degree of arabinosylation dependent on aftD expression levels. Altogether, our results thus indicate that AftD is an essential AraT involved in the synthesis of the arabinan domain of major mycobacterial cell envelope (lipo)polysaccharides.
Keywords: arabinogalactan, arabinosyltransferase, lipoarabinomannan, Mycobacterium, tuberculosis
Introduction
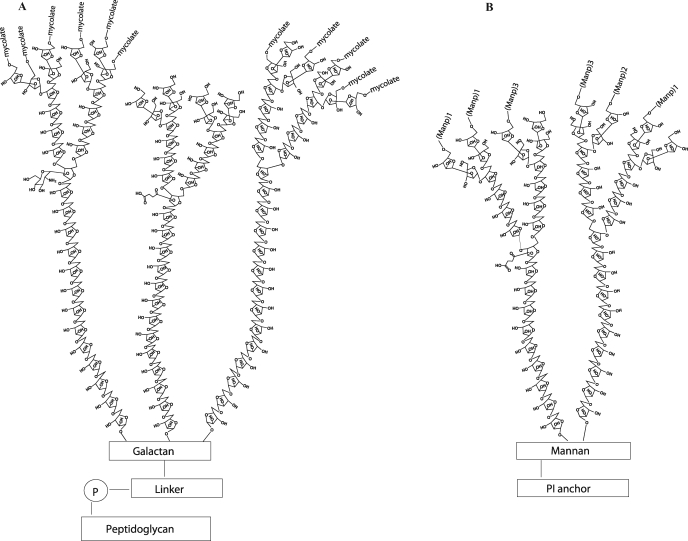
The biology of Mycobacterium tuberculosis (M. tb) and other mycobacteria is dominated by their characteristic cell envelope, based on carbohydrates of unique nature. A covalently linked complex consisting of long-chain α-branched, β-hydroxylated fatty acids (the mycolic acids), the heteropolysaccharide arabinogalactan (AG), and peptidoglycan constitute the core of the cell wall (Crick and Brennan 2008). This structure is intercalated with abundant quantities of various lipids and glycolipids, among which lipoarabinomannan (LAM), which confer upon pathogenic mycobacteria important biological activities in the course of infection. Intense research efforts for more than 30 years have led to detailed models of the structures of AG and LAM (see for a review, Berg et al. 2007; Gilleron et al. 2008; Kaur et al. 2009). The most recent model of AG indicates that it contains on average 125 glycosyl residues in total distributed between a galactan domain made of 30 Galf residues, three arabinan domains each containing 31 Araf residues, and a specific linker unit made of a rhamnosyl residue attached to a N-acetylglucosaminosyl-1-phosphate residue (Bhamidi et al. 2008). The characteristic nonreducing termini of the arabinan domain of AG consist of an Ara6 motif, Arafβ1→2Arafα1→5(Arafβ1→2Arafα1→3)-Arafα1→5Arafα1→, where both the terminal β-Araf and the penultimate 2-α-Araf serve as the anchoring points for the mycolic acids. The inner core of the arabinan domain is essentially made of stretches of α-1,5-linked Araf residues with a critically positioned α-3,5-branch site (Figure 1). The major LAM glycoforms contain about 110 glycosyl residues (approximately 60 Araf and 50 Manp units) and consist of a d-arabinan chain attached to a linear α-1,6-linked d-mannan backbone frequently branched with single α-1,2-linked mannoses, with a phosphatidyl-myo-inositol anchor that intercalates in the inner and outer membranes of the cell envelope (Gilleron et al. 2008; Pitarque et al. 2008). The d-arabinan portion of LAM is very similar to that of AG in that the same linkages of Araf units are found and both structures share an Ara18 motif extending from the α-3,5-Araf interior residues (Figure 1) (Shi et al. 2006; Bhamidi et al. 2008). However, the d-arabinan structure of LAM has been found to be more variable than that of AG in terms of the length of this particular motif (Ara18–Ara22) (Shi et al. 2006). Other distinctive features of the d-arabinan of LAM are found in its nonreducing termini which, in addition to the branched Ara6 motif found in AG, may consist of linear Ara4. Although considerable progress has been made over the last 6 years in deciphering the biosynthetic pathways leading to the biogenesis of these complex molecules (reviewed in Berg et al. (2007), Gilleron et al. (2008), Kaur et al. (2009)), the enzymes involved in the elongation and/or assembling of the d-arabinan structures of AG and LAM remain essentially unknown. Likewise, the fundamentals of how the different domains of LAM and AG are assembled, if on a lipid carrier, growing stepwise from the reducing toward the nonreducing end through the sequential addition of glycosyl residues or assemble through the polymerization of building blocks are at present not known. Arabinofuranosyltransferases (AraT) characterized to date include AftA, involved in the transfer of the very first Araf residue to the galactan domain of AG (Alderwick et al. 2006), the terminal β-1,2-capping AraT AftB (Seidel et al. 2007), AftC involved in the internal α-1,3-branching of AG (Birch et al. 2008), the EmbA and EmbB proteins involved in the formation of the Ara6 motif of AG (Escuyer et al. 2001; Khasnobis et al. 2006), and EmbC, required for the elongation of the arabinan domain of LAM (Zhang et al. 2003). Although elongating α-1,5 AraT activities – some of which are apparently unrelated to the Emb proteins – have been detected in cell-free assays using mycobacterial cell wall preparations and synthetic arabinan acceptors (Lee et al. 1997, 1998; Zhang et al. 2007), the identity of the underlying enzymes remains to be determined. Likewise, it is at present not known whether AftC is the only AraT committed to the α-1,3-branching of the inner core and nonreducing termini of the arabinan domain of AG and if the same enzyme also acts in the branching of LAM. With polyprenol-monophosphoryl-β-d-arabinose (C35/C50-P-Araf; DPA) being the only known Araf donor in mycobacteria, it is expected that the arabinosylation of AG and LAM is catalyzed by membrane-associated polyprenyl-dependent glycosyltransferases (GTs) on the periplasmic side of the plasma membrane. We and others have recently reported on the potential existence of 17 such enzymes in M. tb H37Rv, the implication of 12 of which in the glycosylation of various proteins, glycolipids, or polysaccharides has now been established (see for a review, Berg et al. 2007; Kaur et al. 2009). In the context of our continuing efforts to decipher the biosynthetic pathways of phosphatidylinositol mannosides, AG and LAM in M. tb, we have undertaken to characterize the function of the putative polyprenyl-dependent GT, Rv0236c. Although Rv0236c clearly encodes a membrane-associated protein carrying the proposed GT-C motif of polyprenyl-dependent GTs, it differs from other GT-Cs in its membrane topology and location of the GT-C motif (Berg et al. 2007). With 1400 amino acids compared to 430–670 on average for other GT-Cs (with the exception of the Emb proteins which contain about 1090 amino acids), Rv0236c is also the largest predicted GT-C protein of the M. tb genome. These deviating features raised doubts as to the proper classification of this protein as a GT. We report here, the characterization of Rv0236c as an essential AraT required for mycobacterial growth. In line with the nomenclature of other mycobacterial AraTs, we have termed this enzyme AftD.
Fig. 1.
Structures of the arabinan domains of arabinogalactan (A) and lipoarabinomannan (B). See text for details.
Results
AftD is an essential gene of M. smegmatis mc2155
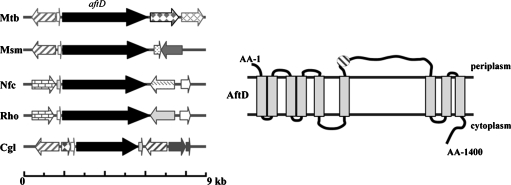
With the early steps of the arabinosylation of AG and LAM taking place on the periplasmic side of the plasma membrane (see for a review, Berg et al. 2007; Kaur et al. 2009), most if not all of the arabinosylation of these heteropolysaccharides are expected to be catalyzed by GTs dependent on DPA as the d-Araf donor. Our bioinformatics studies recently identified Rv0236c (AftD) as a putative polyprenyl-phosphate-linked sugar-dependent GT of the GT-C superfamily (Berg et al. 2007). This putative enzyme is predicted to contain 1400 amino acids and nine transmembrane domains making it the largest of its class. It also differs from other GT-Cs in its membrane topology and in that its GT-C signature motif (D474E475-19AA-P495P496), although predicted to reside in a periplasmic loop, is not located at the beginning of the protein (Figure 2). The aftD gene has close orthologs in all mycobacterial species whose genomes have been sequenced to date as well as in Rhodococcus and Nocardia. An ortholog of aftD also exists in Corynebacterium glutamicum (NClg2757), although in this species the encoded protein is significantly shorter than its mycobacterial, rhodococcal, and nocardial counterparts (1043 amino acids) and shares only 37% identity (50% similarity) with AftD from M. tb on a 767-amino-acid overlap.
Fig. 2.
Comparison of the aftD locus within Corynebacterianeae (A) and topology of AftD (B). Mtb, Mycobacterium tuberculosis; Msm, M. smegmatis; Nfc, Nocardia farcinica; Rho, Rhodococcus sp. RHA1_1; Cgl, Corynebacterium glutamicum ATCC 13032. The genomic regions flanking aftD are not well conserved among mycobacteria and other Corynebacterianeae (orthologous genes are highlighted accordingly). AftD is a hydrophobic protein predicted to span the membrane nine times. The hatched circle indicates the position of the predicted GT-C motif of the enzyme (DE-X19-PP) (Berg et al. 2007).
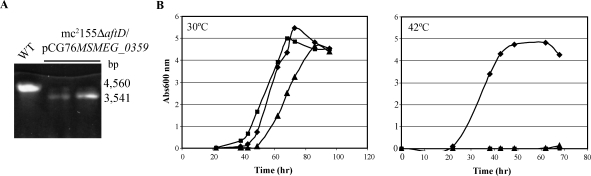
To investigate the possible involvement of aftD in the biosynthesis of LAM and/or AG, we attempted to disrupt the ortholog of this gene in the fast-growing nonpathogenic species, M. smegmatis, by allelic replacement. MSMEG_0359 encodes a 1414-amino-acid protein which is about 70% identical (80% similar) to the M. tb enzyme. Care was taken in constructing the disrupted copy of the MSMEG_0359 gene to delete the entire GT-C motif. Essentially the same strategy with the temperature-sensitive-sacB plasmid pPR27 was used to knock-out MSMEG_0359 as was used earlier to disrupt the phosphatidylinositol synthase gene in the same species (Jackson et al. 2000). Briefly, clones having undergone a single crossover at the MSMEG_0359 locus were first selected upon plating of mc2155/pPR27MSMEG_0359KX transformants on LB-Kan plates at 42°C and their genotype was confirmed by PCR. Two single crossover recombinants (SCO1 and SCO2) were then selected, grown in LB-Kan broth, and plated onto sucrose containing plates at 30°C or 37°C to select for allelic exchange mutants. No knock-out mutants could be isolated following this approach strongly suggesting that MSMEG_0359 was essential for growth regardless of the temperature. To confirm this assumption, we next constructed a conditional (temperature-sensitive) mutant of M. smegmatis. To this end, a temperature-sensitive rescue plasmid carrying a WT copy of the MSMEG_0359 gene, pCG76MSMEG_0359, was introduced in one of the single-crossover recombinants, and the resulting merodiploids were plated onto LB-Kan-sucrose plates at 30°C. Candidate conditional mutants were then obtained in which allelic replacement at the chromosomal MSMEG_0359 locus was confirmed by PCR (Figure 3A). The conditional mutants grew normally at 30°C in liquid broth or on plates, a temperature at which pCG76MSMEG_0359 replicates, but lost viability at 42°C where the rescue plasmid is lost (Figure 3B). Results thus indicated that MSMEG_0359 was essential for the growth of M. smegmatis under the experimental conditions used. Importantly, allelic exchange mutants were also obtained when pVVRv0236c was used as the rescue plasmid, indicating that the aftD gene from M. tb H37Rv displays the same function as its M. smegmatis counterpart in whole cells (data not shown).
Fig. 3.
aftD is an essential gene of M. smegmatis mc2155. (A) Evidence for allelic replacement at the aftD locus of M. smegmatis in the presence of a rescue copy of this gene expressed from an episomal plasmid. Allelic exchange mutants were rescued with the aftD gene from M. smegmatis expressed from pCG76MSMEG_0359. Allelic replacement was confirmed by PCR using primers smg0236c.3 and smg0236c.6 (see Material and methods). The WT 4560-bp amplification signal is replaced by a 3541-bp fragment in the mutants due to the 2219-bp AgeI deletion in the aftD gene and insertion of a 1.2 kb-kanamycin resistance cassette. (B) Growth characteristics of WT mc2155 (diamonds) and two independent mc2155ΔMSMEG_0359/pCG76MSMEG_0359 conditional mutants (squares and triangles) in the LB-Kan-Tween80 medium at 30°C (where the rescue plasmid replicates) and 42°C (where the rescue plasmid is lost).
Effect of overexpressing aftD in M. smegmatis
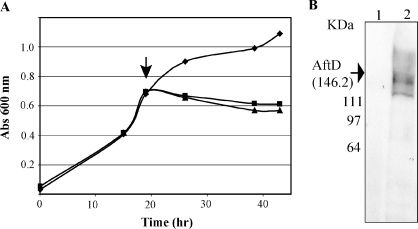
Attempts to stably produce the AftD enzyme from M. tb H37Rv in M. smegmatis mc2155 from the constitutive expression system pVV16 (Korduláková et al. 2002) proved unsuccessful. Transformants failed to grow at 37°C and only yielded colonies at 30°C after 8 days, i.e., 3 days later than M. smegmatis transformants carrying the empty vector. Although a His6-tagged recombinant protein of the expected size (∼146 KDa) could be detected by Western blot in some of the transformants (consistent with the ability of pVVRv0236c to rescue M. smegmatis MSMEG_0359 allelic exchange mutants), production was extremely unstable. Because these results suggested that the overexpression of aftD was toxic to M. smegmatis, we thus resorted to the inducible acetamidase-based expression system, pJAM2 (Triccas et al. 1998). A recombinant His6-tagged AftD was efficiently produced in mc2155/pJAMRv0236c upon induction of the expression of the aftD gene with acetamide (Figure 4A and B). However, induction of mc2155/ pJAMRv0236c cultures at 30°C or 37°C immediately caused the cells to cease growing, confirming the toxic effect of overexpressing this gene (Figure 4A).
Fig. 4.
Production of a recombinant form of AftD in M. smegmatis mc2155 using the inducible pJAM2 system. (A) Growth characteristics of mc2155/pJAM2 (diamonds) and two independent mc2155/pJAMRv0236c clones (squares and triangles) in the MM63 medium before and after induction with acetamide (indicated by an arrow). (B) Production of a recombinant C-ter His6-tagged form of AftD from M. tb 8-h post-induction with acetamide was detected by Western blot using a monoclonal anti-His tag antibody in the mc2155/pJAMRv0236c strain (lane 2) but not in the control strain, mc2155/pJAM2 (lane 1).
The control and overexpressing strains were then compared for their lipoglycan and AG contents. Analyses of the monosaccharide composition and structures of the AG from mc2155/pJAM2 and mc2155/pJAMRv0236c upon induction with acetamide for 13 h did not reveal any significant quantitative or qualitative differences between the two strains. The AG of the overexpressor displayed an Ara:Gal ratio of about 3:1, comparable to that of the control strain mc2155/pJAM2, and the expected glycosyl linkage profile normally found in wild-type M. smegmatis.
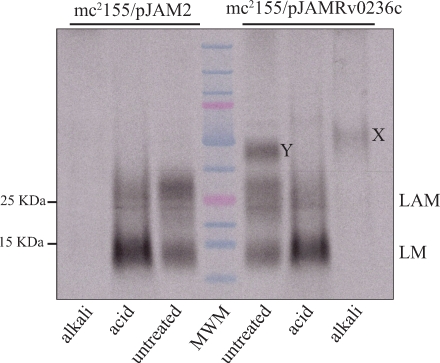
The two acetamide-induced strains were further labeled with [U-14C]glucose and their lipoglycans analyzed by SDS–PAGE followed by autoradiography. This analysis clearly and reproducibly revealed the accumulation of two co-migrating glycoconjugates (compounds X, only detectable upon alkali-treatment, and compound Y), migrating above LAM in the mc2155/pJAMRv0236c cells which were barely or not visible in the control strain (Figure 5).
Fig. 5.
Effect of overexpressing aftD on the lipoglycan content of M. smegmatis. Autoradiogram of the lipoglycans extracted from identical amounts of mc2155/pJAM2 and mc2155/pJAMRv0236c cells labeled with [U-14C]glucose and separated on a Tricine SDS–PAGE gel. The positions of products X and Y are indicated. Samples were either untreated or submitted to mild-acid (acid) or mild-alkali (alkali) treatments. MWM, molecular weight marker.
To gain insight into the structures of LAM and of compounds X and Y produced by the control and overexpressing strains, these products were purified from nonradiolabeled cells and analyzed for their monosaccharide composition. The LAM from mc2155/pJAMRv0236c showed an Ara/Man ratio of 0.9:1 identical to LAM from the control strain. Compound Y, which was consistently only detectable in the overexpressing strain, exhibited an Ara/Man ratio of 1.15:1. Owing to its monosaccharide composition, sensitivity to mild-alkali treatment, and chromatographic behavior suggestive of a higher apparent molecular weight than LAM (Figure 5), we concluded that Y most likely corresponds to a new form of LAM with a larger arabinan domain.
Compound X was about 3-fold more abundant in the overexpressing strain than in the control. Monosaccharide analyses showed that while compounds X produced by the two strains contained Ara and Man, the Ara/Man ratio of 0.25:1 measured for the control strain increased to 0.4:1 in the aftD overexpressor. Methylation analyses further revealed that the larger arabinan domain exhibited by compound X from the overexpressor relative to the control was apparently attributable to an increase of all types of Araf linkages (Table I). Interestingly, the glycosidic linkages found in compound X were the same as those found in LAM (Table I) (Kaur et al. 2007). However, compared to LAM, compound X displayed dramatically reduced proportion of Araf units and showed no detectable alkyl chains. Radiolabeling of the acetamide-induced control and overexpressing strains with myo-[2-3H]Inositol showed that X does not contain inositol (data not shown), further suggesting that, unlike LAM, it is devoid of a phosphatidyl-myo-inositol anchor. More analyses are in progress to characterize the complete structure of product X.
Table I.
Methylation analysis data of compound X from M. smegmatis mc2155/pJAM2 and mc2155/pJAMRv0236c strains. Molar ratio values are normalized relative to 2,6-Manp units
| Abbreviated name | ||
|---|---|---|
| of the glycosyl residue | mc2155/pJAM2 | mc2155/pJAMRv0236c |
| Mol ratio | ||
| t-Araf | 0.01 | 0.02 |
| 2-Araf | 0.14 | 0.25 |
| 5-Araf | 0.81 | 1.49 |
| 3,5-Araf | 0.28 | 0.47 |
| t-Manp | 1.23 | 1.38 |
| 6- Manp | 0.84 | 0.93 |
| 2,6-Manp | 1.0 | 1.0 |
Overall, the overexpression of aftD in whole M. smegmatis cells thus resulted in the accumulation and/or increased arabinosylation of two LAM-like compounds.
Characterization of AftD as a functional arabinofuranosyltransferase
Enzymatic assays were next designed to compare the ability of mc2155/pJAM2 and mc2155/pJAMRv0236c cell-free extracts to transfer Manp from GDP-[14C]Manp, and Araf from DP[14C]A (formed in situ from p[14C]Rpp) (Scherman et al. 1996; Zhang et al. 2007) onto endogenous substrates and synthetic mannopyranosyl- and arabinofuranosyl-based glycoconjugates. No effect of overexpressing aftD was detected on the transfer of Manp from GDP-[14C]Man onto endogenous substrates (PIM or polyprenyl-phospho-mannoses) or a synthetic linear Man5 octylthiomethyl α-d-Manp-(1→6)-α-d-Manp-(1→6)-α-d-Manp-(1→6)-α-d-Manp-(1→6)-α-d-Manp acceptor (Holemann et al. 2006).
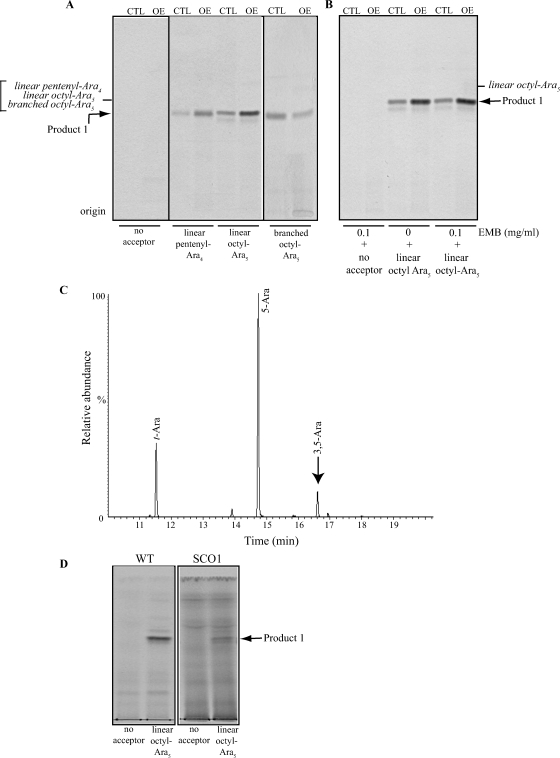
Four different arabinosyl acceptors were tested in the arabinofuranosyl transferase assay including a linear Ara3 pentenyl α-d-Araf-(1→5)-α-d-Araf-(1→5)-α-d-Araf, a linear Ara4 pentenyl α-d-Araf-(1→5)-α-d-Araf-(1→5)-α-d-Araf-(1→5)-α-d-Araf, a linear Ara5 octyl α-d-Araf-(1→5)-α-d-Araf-(1→5)-α-d-Araf-(1→5)-α-d-Araf-(1→5)-α-d-Araf, and a branched Ara5 octyl (α-d-Araf )2-(1→3,5)-α-d-Araf-(1→5)-α-d-Araf-(1→5)-α-d-Araf. TLC analysis of the products formed in the various reactions showed evidence of single [14C]Araf transfer onto each of the acceptors tested and a clear stimulatory effect of overexpressing aftD on the transfers onto the linear ones (Figure 6A). Measurement of the radioactivity incorporated in the 1-butanol phase of the reactions indicated that overexpression of aftD increased 1.7-fold the transfer of [14C]Araf onto linear Ara3 (data not shown), 2.1-fold the transfer onto linear Ara4, and 2.5-fold the transfer onto linear Ara5. The greatest effect of overexpression was thus observed with the linear Ara5 acceptor suggestive of a preference of AftD for linear Ara5 over shorter linear arabinofuranosyl or branched Ara5 acceptors. The addition of ethambutol to the assays using linear Ara5 as the acceptor substrate had no effect on [14C]Araf transfer indicating that the AraT activity of AftD, unlike that of the Emb proteins, is insensitive to this drug (Figure 6B). Importantly, no stimulation of arabinosyl transfer was detected when cell-free extracts from M. smegmatis strains overexpressing unrelated GTs such as the α-1,4-glucosyltransferase GlgA (Sambou et al. 2008) and the α-1,2-mannosyltransferases PimE (Morita et al. 2006) were used (data not shown), ruling out an unspecific effect of expressing toxic GT genes on Araf transfer in vitro.
Fig. 6.
Arabinofuranosyltransferase assays using synthetic arabinosyl acceptors. (A) TLC autoradiographs of the products of the reactions using mc2155/pJAM2 (CTL) and mc2155/pJAMRv0236c (OE) cell-free extracts as enzyme sources, p[14C]Rpp as the donor substrate and different synthetic arabinofuranosyl acceptors. The lower minor product formed in the reaction utilizing a linear Ara5 acceptor is likely to result from the activity of a β-1,2 AraT, consistent with previous studies using mycobacterial extracts and short linear Araf acceptors (Birch et al. 2008). The synthesis of this product is not affected by the overexpression of aftD. (B) The same assays using linear Ara5 acceptor were run in the presence of 0.1 mg/mL ethambutol (EMB). Samples were prepared and analyzed as described under Material and methods. Twenty percent of each reaction was loaded onto the TLC plate. The products of the reactions were identified by co-migration with synthetic arabinofuranosyl standards (italicized). The presence of different aglycon moieties (pentenyl or octyl) on the synthetic acceptors accounts for the similar Rf of the radiolabeled Ara5 and Ara6 products on the TLC plate. (C) GC/MS analysis of the Ara6 product of the reaction. The per-O-methylated purified product was hydrolyzed with 2 M TFA, reduced, per-O-acetylated, and analyzed as described under Material and methods. (D) Assays using WT mc2155 (WT) and SCO1 cell-free extracts, p[14C]Rpp as the donor substrate and linear synthetic Ara5 as the acceptor substrate.
To establish the nature of the product whose synthesis was increased in the assays using mc2155/pJAMRv0236c extracts and linear Ara5 as the acceptor substrate (i.e., product 1), a nonradioactive assay was up-scaled and the product of the reaction, purified by preparative TLC, was submitted to MALDI-TOF/MS and linkage analyses. The mass of the purified product as determined by MALDI-TOF/MS (m/z = 1127.57) was consistent with that of octyl-Ara6. GC/MS analysis of the partially per-O-methylated, per-O-acetylated alditol acetate derivative of the product further revealed the occurrence of an α-(1→3)-linked Araf residue (Figure 6C). We conclude that the product of the reaction corresponds to the branched Ara6 glycoconjugate (α-d-Araf )2-(1→3,5)-α-d-Araf-(1→5)-α-d-Araf-(1→5)-α-d-Araf-(1→5)-α-d-Araf-octyl. Altogether, the results of our assays thus indicate that AftD displays branching α-1,3 AraT activity on linear α-(1→5)-linked Araf acceptors in vitro.
Further supporting this conclusion, the expression of MSMEG_0359 (the M. smegmatis ortholog of aftD) in Corynebacterium glutamicum conferred upon cell-free extracts from this bacterium the ability to transfer Araf from DP[14C]A onto the linear Ara5 acceptor, an activity which was not detectable in extracts from wild-type C. glutamicum (Supplementary Material 1, see lane denoted by an asterisk).
Not unexpectedly in light of the reported difficulty of expressing polytopic membrane GTs in Escherichia coli (Morita et al. 2006), attempts to produce AftD under an active form in E. coli BL21-AI, BL21(DE3)pLysS and C43(DE3) using different expression systems (pBAD/mycHis, pET29), growth conditions, and induction protocols at 16 and 37°C were unsuccessful, precluding the functional characterization of the enzyme in a microorganism devoid of endogenous AraT activity.
Effect of decreasing aftD expression on colonial morphology and growth
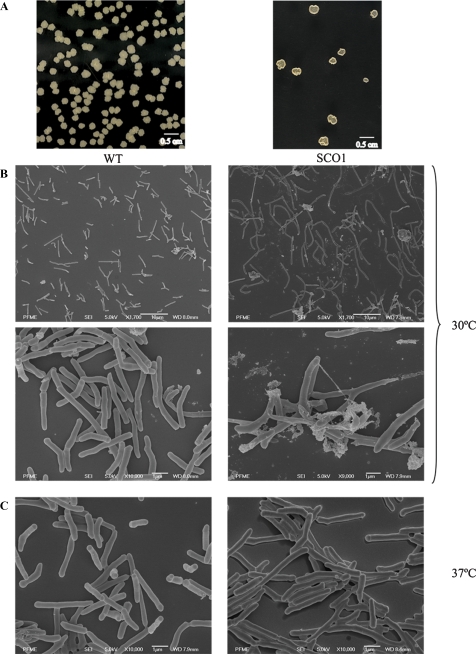
In constructing the aftD mutant of M. smegmatis, it was noted that the single crossover strains isolated at the first selection step displayed a dramatically altered colonial morphology and a 4-day delay in growth at 30°C on LB-Km plates (Figure 7A). Single crossover strains also suffered a 3-day growth delay and a tendency to form filaments in liquid broth at 30°C. However, they showed a WT morphology and growth rate when cultured on agar or liquid broth at 37°C (or higher temperatures) (data not shown). Scanning electron microscopy analyses indicated that the single crossover strains formed longer bacilli (10.2 ± 3.2 μm) than the WT parent (4.2 ± 1.7 μm) when grown at 30°C, some of them with distorted shapes, suggestive of alterations in cell division (Figure 7B). Consistent with macroscopic observations, no significant differences were found between WT and recombinant strains grown at 37°C (Figure 7C).
Fig. 7.
Morphology of the aftD single crossover strain of M. smegmatis. (A) Colony morphology of WT M. smegmatis mc2155 and the aftD single crossover strain SCO1 on LB agar plates at 30°C. (B and C) Scanning electron micrographs of the same strains cultured in 7H9-ADC-Tween 80 broth at 30 (B) or 37°C (C).
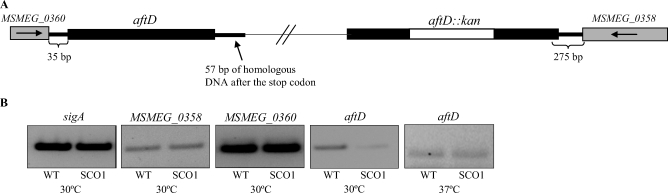
Although single crossover strains carry both a WT and a disrupted copy of the aftD gene in their chromosome, the very short length of homologous DNA (57 bp) flanking the 3′-end of the gene in our knock-out construct, pPR27MSMEG_0359KX, and the orientation with which single crossover events had occurred in all of the clones analyzed (all single homologous recombination events had taken place downstream of the kan cassette) (Figure 8A) suggested that their altered growth might have been due to defects in the expression of aftD. The relatively short length of homologous DNA downstream of the stop codon of gene aftD in these strains might indeed have affected the stability of its transcript with perhaps more pronounced effects at 30°C than at 37°C. Alternatively, altered growth could have been due to polar effects of the insertion of the pPR27MSMEG_0359KX plasmid affecting the expression of adjacent genes. To distinguish between these two hypotheses, RT-PCR experiments were undertaken on the aftD, MSMEG_0358, and MSMEG_0360 genes. sigA served as a control in the experiment. Results showed a clear reduction in the expression of aftD in the single crossover strain relative to WT mc2155 in the culture batches grown at 30°C (Figure 8B). Both strains expressed aftD at similar levels at 37°C and the expression of MSMEG_0358 and MSMEG_0360 appeared relatively unaffected in the single crossover recombinants at both temperatures. The growth defects of the single crossover strain at 30°C thus seemed to result from the decreased expression of aftD. Consistent with the RT-PCR results, arabinosyl transfer onto the linear Ara5 acceptor was reduced in the single crossover strain SCO1 grown at 30°C relative to the control (Figure 6D).
Fig. 8.
RT-PCR analysis of the expression of aftD and adjacent genes in M. smegmatis mc2155 and the aftD single crossover strain SCO1. (A) Schematic representation of the aftD genomic region in the single crossover strain SCO1. The thin line symbolizes the body of the integrated plasmid, pPR27MSMEG_0359KX, and is not represented to scale. The lengths of the intergenic regions are indicated. (B) RT-PCR analysis of the sigA, MSMEG_0358, aftD, and MSMEG_0360 transcripts in WT M. smegmatis mc2155 and the aftD single crossover strain SCO1. One quarter of the RT-PCR reactions were analyzed on 1% agarose gels.
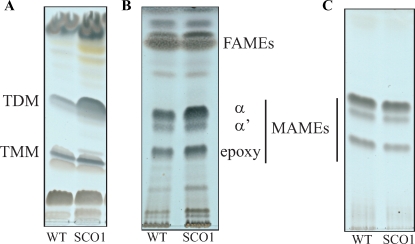
To determine whether the cell division defects of the single crossover strains correlated with alterations in the biogenesis of their cell wall, we next compared the lipid, lipoglycan, mycolate, and AG contents of the WT and SCO1 strains grown at 30°C. Equivalent starting amounts of biomass was used for each strain. Analyses of the monosaccharide composition and glycosidic linkages of AG failed to reveal any significant qualitative or quantitative differences between the two strains (Supplementary Material 2). Likewise, both strains produced comparable quantities of LM and LAM that migrated similarly on a Tricine gel (data not shown). The relative proportions and amounts of α-, α′-, and epoxy-mycolic acid methyl esters (MAMEs) esterifying the AG of strain SCO1 were also indistinguishable from those of the WT strain (Figure 9C). In fact, the only noticeable difference between the two strains was at the level of extractable lipids; approximately 2- to 3-fold more trehalose dimycolates (TDM) were found in strain SCO1 than in the WT parent, mc2155 (Figure 9A). Consistent with this finding, more MAMEs were recovered from the extractable lipids of strain SCO1 compared to WT mc2155 (Figure 9B). No differences in the TMM content were observed (Figure 9A). Assuming that AftD participates in the synthesis of the arabinan domain of AG in addition to LAM and LAM-like compounds as suggested by our data, it is possible that the accumulation of TDM in the single crossover strain results from the unbalanced production rate of mycolates relative to AG. In the absence of sufficient attachment sites at the nonreducing terminal Ara6 motif of AG, mycolic acids may be channeled into TDM.
Fig. 9.
Analysis of the extractable lipids and cell wall-bound mycolates from M. smegmatis mc2155 and the aftD single crossover strain SCO1. (A) Equal amounts of total cellular lipids from WT M. smegmatis mc2155 and the aftD single crossover strain SCO1 were analyzed by TLC developed in the solvent system chloroform/methanol/water (20:4:0.5). (B) Fatty acid methyl esters (FAMEs) and mycolic acid methyl esters (MAMEs) prepared from the same amount of WT and SCO1 extractable lipids and (C) cell wall-bound MAMEs prepared from the same amount of WT and SCO1 cells were analyzed by TLC developed thrice in the solvent system n-hexane/ethyl acetate (95:5). TLC plates were revealed by spraying with cupric sulfate (10% in a 8% phosphoric acid solution) and heating. Analyses reveal an increase in the MAME content of the extractable lipids from strain SCO1 (B) consistent with the greater quantities of TDM recovered from this strain (A).
Discussion
The envelopes of Mycobacterium spp. and related Actinomycetales are the source of unique carbohydrates. Among these, AG was recognized as the major cell wall polysaccharide of mycobacteria as early as the 1950s. AG shares with LAM arabinan chains made of linear segments of α-1,5-linked Araf residues with some α-1,3-branching. Given the confinement of Araf to the prokaryotic world and the important physiological and structural roles played by AG and LAM in the biology of the tubercle bacillus (Berg et al. 2007; Kaur et al. 2009), the enzymes leading to the synthesis and assembling of the arabinan domains of these polysaccharides have long been suggested to present opportunities for new chemotherapeutics (Wolucka 2008). In support of this assumption, the mode of action of ethambutol, a first-line drug in the treatment of TB, involves inhibition of the synthesis of the d-arabinans (Mikušová et al. 1995). More recently, two classes of compounds capable of inhibiting the formation of DPA from decaprenyl-phosphoribose were shown to be potent inhibitors of M. tb grown intracellularly and in vivo, thereby validating the heteromeric decaprenyl-phosphoribose 2′-epimerase DprE1 (Rv3790c)/DprE2 (Rv3791c) as a valuable antimycobacterial drug target (Makarov et al. 2009; Christophe et al., unpublished).
In search of the missing AraTs responsible for the polymerization of the arabinan domains of AG and LAM, we identified Rv0236c (AftD) as a putative candidate. AftD encodes a membrane-associated protein carrying the proposed GT-C motif of polyprenyl-dependent GTs known to catalyze glycosyl transfer on the periplasmic face of the plasma membrane (Berg et al. 2007). AftD, which contains 1400 amino acids, is the largest predicted GT-C protein of the M. tb genome. Assays using extracts from recombinant M. smegmatis and C. glutamicum strains expressing aftD at different levels all indicated that this gene encodes a functional AraT with α-1,3-branching activity on a linear α-1,5-linked Ara5 neoglycolipid acceptor in vitro. The activity of this enzyme is ethambutol-insensitive and essential for the in vitro growth of M. smegmatis mc2155. A decrease in activity dramatically impairs cell division, colonial morphology, and growth and results in the accumulation of TDM in the cells. Conversely, overexpression of this gene in M. smegmatis leads to the accumulation of two arabinosylated glycoconjugates with carbohydrate structures reminiscent of that of LAM and an overall increase of all types of Araf linkages in the case of product X. The addition of new branching points to the arabinan domain – and thus number of side chains – potentially catalyzed by AftD might account for this observed increase in all types of Araf linkages. Altogether, our results suggest that AftD is a novel AraT involved in the synthesis of the arabinan domains of LAM and AG. That AftD exclusively contributes to the formation of the arabinan domain of LAM is unlikely given that a M. smegmatis embC knock-out mutant deficient in LAM synthesis was found to be viable (Zhang et al. 2003).
The fact that no structural differences were found between the AGs of M. smegmatis mc2155, mc2155/pJAMRv0236c, and SCO1 suggests that the activity of this enzyme in whole cells is tightly regulated and that any decrease in expression impacts the synthesis of AG as a whole, with immediate consequences on cell division and mycolic acids accumulation under the form of TDM. Actually, in view of the significantly larger size of AftD compared to other GT-Cs, it is tempting to speculate that this protein may display additional functions in the biogenesis of AG and LAM – e.g., controlling the length of the various interior or exterior segments of the arabinan polymer, acting as a scaffold for a multi-enzyme machinery involved in arabinosylation and/or cell division – beyond the addition of Araf residues from DPA. Similar functions have been proposed for the Emb proteins of M. tb based on the presence of conserved proline motifs reminiscent of those found in the Wzz-type O-antigen chain-length regulators of Gram-negative bacteria (Berg et al. 2005; Shi et al. 2006). Clearly, further studies are required to investigate these hypotheses.
Another AraT, AftC, has recently been reported to display like AftD α-1,3-branching AraT activity on a synthetic linear α-1,5-linked Ara5 acceptor in vitro (Birch et al. 2008). The analysis of aftC knock-out mutants of M. smegmatis and C. glutamicum revealed that this enzyme is responsible for the α-1,3-branching of the inner core of the arabinan domain of AG and that it is apparently the only enzyme engaged in this function. Contrary to aftD, aftC is not an essential gene of M. smegmatis. If AftD is in whole cells an α-1,3-branching AraT as the results of our cell-free assays suggest, it thus most likely participates in the α-1,3-branching of the nonreducing termini of the arabinan domain of LAM and AG. The essentiality of this enzyme would then arise from its putative other regulatory functions in the assembling of the arabinan domain. Alternatively, one cannot exclude that AftD displays in vivo another activity than that detected in vitro using synthetic acceptors, acting for instance as an α-1,5-elongating AraT. Unfortunately, attempts to verify this hypothesis by generating a C. glutamicum knock-out mutant of aftD (NClg2757) proved inconclusive. The NClg2757 knock-out strain of C. glutamicum produced AG and lipoglycans that were structurally undistinguishable from those of its WT parent (data not shown). Different functions of the corynebacterial and mycobacterial orthologs most likely account for this result. Indeed, NClg2757 is significantly shorter than its mycobacterial counterparts (1043 instead of 1400 amino acids) and shares only 37% identity (50% similarity) with AftD from M. tb on a 767-amino-acid overlap. A similar divergence between the functions of two orthologous GTs of mycobacterial and corynebacterial sources has previously been noted in the case of NCgl1505 (MptB) and Rv1459c (Mishra et al. 2008).
The identification of AftD as a key component of the biosynthetic machinery involved in the synthesis of LAM and presumably AG advances our knowledge of the arabinosylation of the mycobacterial cell wall and provides the first direct evidence of what seems to be an intricate link between AG synthesis and cell division.
Material and methods
Bacterial strains and growth conditions
M. smegmatis mc2155 was grown in the Middlebrook 7H9 medium (Difco) supplemented with ADC and 0.05% Tween 80, in Luria Bertani (LB) broth (pH 7.5) (Bactotryptone, 10 g/L, BactoTM yeast extract, 5 g/L, NaCl, 5 g/L) (Becton Dickinson, Sparks, MD) or in minimal MM63 medium with 0.2% succinate and 0.025% tyloxapol. E. coli XL1-blue, the strain used for cloning experiments, was propagated in LB broth. Corynebacterium glutamicum was propagated in Brain Heart Infusion medium (BHI; pH 7.4) (Becton Dickinson, Sparks, MD). Where indicated, ampicillin (Amp), kanamycin (Kan), hygromycin (Hyg), and streptomycin (Str) were added to final concentrations of 100 μg/mL, 20 μg/mL, 50 μg/mL, and 20 μg/mL, respectively. When required, 10% sucrose was added to the solid medium.
Construction of the M. smegmatis and C. glutamicum aftD mutants
A two-step procedure employing the counterselectable marker sacB (Jackson et al. 2001) was used to achieve allelic replacement at the MSMEG_0359 locus of M. smegmatis mc2155 and NClg2757 locus of C. glutamicum. Briefly, the M. smegmatis MSMEG_0359 gene and flanking regions was PCR-amplified from M. smegmatis mc2155 genomic DNA using the primers smg0236c.KOS (5′-gggtctagatcgtcgtaccctctgcggccag-3′) and smg0236c.KOAS (5′-gggtctagagatcaccgtaagtactgcacc-3′), and a disrupted allele, MSMEG_0359::kan, was obtained by replacing 2219 bp of the coding sequence of this gene flanked by two AgeI restriction sites by the kanamycin resistance cassette from pUC4K (Amersham Pharmacia Biotech, Pittsburgh, PA). MSMEG_0359::kan was then cloned into the NotI-cut and blunt-ended pPR27-xylE (Pelicic et al. 1997) to obtain pPR27MSMEG_0359KX, the construct used for allelic replacement. pCG76MSMEG_0359, the plasmid used to rescue the M. smegmatis MSMEG_0359 conditional mutant was obtained by cloning the 4500 bp PCR fragment described above into the XbaI-cut and blunt-ended pCG76 (Guilhot et al. 1994). Allelic replacement at the MSMEG_0359 locus was confirmed by PCR using primers smg0236c.3 (5′-cgccgtgttaggttggccggg-3′) and smg0236c.6 (5′-cctcgatgccgccgaaacagc-3′) located just outside the smg0236c.KOS and smg0236c.KOAS primers used to generate the knock-out construct.
The C. glutamicum NClg2757 gene and flanking regions was PCR-amplified from C. glutamicum ATCC 13032 Kitasato genomic DNA using the primers NCgl2757KOfw (5′-gccactagtacgtcatggcatacgaaactg-3′) and NCgl2757KOrev (5′-cggcgcggccgccagccgtttgtggtggat-3′). A disrupted allele of NClg2757 was obtained by substituting 1340 bp of the coding sequence of this gene flanked by two AgeI restriction sites by the kanamycin resistance cassette from pUC4K (Amersham Pharmacia Biotech, Pittsburgh, PA). NCgl2757::kan was then cloned into the SpeI and NotI-cut pJQ200-xylE, yielding pJQNClg2757KX to obtain pJQ200NCgl2757KX, the construct used for allelic replacement. Allelic replacement at the NCgl2757 locus was confirmed by PCR using primers NCgl2757fw (5′-gagcgccatcgttgatgctgct-3′) and NCgl2757rev (5′-ctgttggtgttcatcgtgacg-3′). pJKS1MSMEG_0359, the plasmid used to complement the C. glutamicum NClg2757 mutant was constructed by cloning the MSMEG_0359 gene from M. smegmatis, PCR-amplified as described above, into the PstI-cut and blunt-ended E. coli/Corynebacterium shuttle plasmid, pJKS1 (a kind gift from Drs. Haller and Holmes, UCHSC, Denver, CO).
Overexpression of AftD in M. smegmatis
The entire coding sequence of Rv0236c was PCR amplified from M. tb H37Rv genomic DNA using the primers Rv0236.3 (5′-gggtctagagtggcgccgttgtctcgcaaatgg-3′) and Rv0236c.4 (5′-gggtctagattgcatgcggtcttgaccacgag-3′) and cloned into the XbaI restriction site of the expression vector pJAM2 (Triccas et al. 1998). The resulting plasmid, pJAMRv0236c, allows the inducible expression of aftD under control of the acetamidase promoter. Production of AftD in mc2155/pJAMRv0236c cells grown at 37°C in MM63 broth was induced as described (Triccas et al. 1998) during log phase with 0.2% acetamide for 12 h. The entire coding sequence of aftD was also PCR amplified from M. tb H37Rv genomic DNA using primers Rv0236.1 (5′-gggccggcatatggcgccgttgtctcgcaaatgg-3′) and Rv0236c.2 (5′-gggaagcttttgcatgcggtcttgaccacgag-3′) and cloned into the NdeI and HindIII restriction sites of the expression vector pVV16 (Korduláková et al. 2002). The resulting plasmid, pVVRv0236c, allows the constitutive expression of aftD under control of the hsp60 promoter. The recombinant proteins produced with both systems carry an hexa-histidine at their carboxyl terminus allowing their detection by immunoblotting with the monoclonal Penta-His antibody from Qiagen (Valencia, CA).
Whole cell radiolabeling experiments
Radiolabeling of whole M. smegmatis cells with [U-14C]glucose (0.5 μCi/mL; specific activity, 310 Ci/mol, MP Biomedicals Inc., Solon, OH) and myo-[2-3H]Inositol (10 μCi/mL; specific activity, 20.0 Ci/mmol, Perkin Elmer, Waltham, MA) was performed in MM63 broth at 37°C, for 12 h with shaking. Radiochemicals were added 1 h after the induction of the cells with acetamide.
Preparation and analysis of lipids and lipoglycans
Lipoglycans from cold or radiolabeled bacterial cultures were prepared as described previously (Kaur et al. 2007). For structural characterization requiring large amounts of products, WT mc2155, mc2155/pJAM2, mc2155/pJAMRv0236c, and SC01 cells were harvested from 24 L of MM63 medium and lyophilized. Lipoglycan fractions and AG were prepared from the dried cells as described previously (Pitarque et al. 2005; Bhamidi et al. 2008). Lipoglycans were analyzed by SDS–PAGE on commercial 10–20% gradient Tricine SDS-polyacrylamide gels (Invitrogen, Carlsbad, CA). Blotting to the nitrocellulose membrane was performed at 50 V for 1 h. Radiolabeled lipoglycans were visualized by exposure of nitrocellulose membranes to Kodak X-Omat AR films at −80°C. Mild acid hydrolysis of [U-14C]glucose-labeled lipoglycans was conducted on samples resuspended in 20 μL of 1-propanol with 40 μL 0.02 M HCl at 60°C for 30 min. Mild alkali hydrolysis was performed on samples resuspended in 50 μL of 60% CH3OH with 50 μL 0.2M NaOH in CH3OH at 37°C for 60 min. After incubation, reactions were cooled on ice, dried under a stream of nitrogen and washed three times with methanol. The products of the reactions were analyzed by SDS–PAGE as described above.
Total lipids were extracted from bacterial cells as previously described (Stadthagen et al. 2005), and TMM and TDM analyzed by TLC on aluminum-backed silica gel 60-precoated plates F254 (E. Merck, Darmstadt, Germany) using chloroform/methanol/water (20:4:0.5, by volume) as the eluent. Mycolic acid methyl esters (MAMEs) from delipidated cells were prepared as described earlier (Stadthagen et al. 2005) and analyzed by TLC using n-hexane/ethyl acetate (95:5; three developments) as the eluent. TLC plates were revealed by spraying with cupric sulfate (10% in a 8% phosphoric acid solution) and heating.
Analytical procedures
Determination of glycosyl linkage patterns, preparation of alditol acetates and analyses of samples by MALDI-TOF/TOF were performed as described by Kaur et al. (2007). LAM and compounds X and Y were purified from two independent cold cultures by extraction from DOC-PAGE gels using a procedure that will be described in details elsewhere. Their monosaccharide composition was determined by capillary electrophoresis monitored by laser-induced fluorescence after total acid hydrolysis (TFA 2 M, 2 h at 110°C) and APTS derivatization (Nigou et al. 2000). The amount of X for a fixed amount (2 mg) of the total lipoglycan fraction deposited on the gel was determined for its accurate quantification by capillary electrophoresis (Nigou et al. 2000).
Preparation of enzymatically active membranes and cell envelope fraction (P60) from M. smegmatis
M. smegmatis mc2155/pJAM2 and mc2155/pJAMRv0236c cultures were grown and induced as described above. The M. smegmatis aftD single crossover strain SCO1 was grown in MM63 broth at 30°C. Enzymatically active membranes and cell envelopes (P60) (cell wall plus membranes) were prepared essentially as described by Zhang et al. (2007) in buffer A (50 mM MOPS buffer, pH 7.9, containing 5 mM 2-mercaptoethanol and 10 mM MgCl2). The cell envelope and membrane fractions were resuspended in buffer A to final protein concentrations of 10– 15 mg/mL and 15–20 mg/mL, respectively.
Arabinofuranosyltransferase assays
Reaction mixtures for the AraT assays with synthetic arabinosyl acceptors contained buffer A, 62.5 μM ATP, 1.8 μM p[14C]Rpp (200,000 dpm; specific activity 300 mCi/mmol), arabinosyl acceptors (300 μM) (Zhang et al. 2007) (see below), membranes (0.5 mg), and P60 (0.3 mg) in a total volume of 200 μL. The reaction mixtures were incubated at 37°C for 2 h and terminated by the addition of 200 μL of ethanol. Reaction mixtures were then centrifuged at 14,000 × g, the supernatants were loaded onto pre-packed strong anion exchange (SAX) columns (Thermo, Waltham, MA), and the columns were washed with 2 mL of water. Eluates were evaporated to dryness and partitioned between water-saturated 1-butanol and water (1:1, by volume). The 1-butanol fractions were measured for radioactivity incorporation by liquid scintillation counting prior to analysis by TLC in the solvent system chloroform/methanol/1 M ammonium acetate/ammonium hydroxide:water (180:140:9:9:23; by volume). In some assays, membranes and P60 were pre-incubated with 0–100 μg/mL ethambutol for 30 min at 37°C, prior to the addition of the acceptor and p[14C]Rpp and further incubation for another 1.5 h. The values reported are averages of duplicate reactions from representative experiments. Radioactive p[14C]Rpp was generated from uniformly labeled [d-14C]glucose (American Radiolabeled Chemicals Inc., St. Louis, MO) as described (Scherman et al. 1996).
For generating sufficient amount of cold product for structural analyses, assays were run using nonradiolabeled commercial pRpp (Sigma, St. Louis, MO). This substrate was first further purified by Dionex HPAEC. The fractions found to co-elute with radioactive p[14C]Rpp were pooled and stored in −20°C. The reaction mixtures contained buffer A, 62.5 μM ATP, 2.0 mM pRpp, linear Ara5 acceptor (1.0 mM), membranes (0.6 mg), and P60 (0.4 mg) in a total volume of 160 μL (Zhang et al. 2007). Subsequent procedures were exactly the same as for the radioactive reactions. The product of the reaction eluted from the SAX column was further purified by preparative TLC run in the same solvent system as above and eluted thrice from the silica with 1 mL of Milli-Q water. The pooled water phases were then washed once with 2 mL of 1-butanol and the 1-butanol extract was dried, methylated and/or alditol acetylated, and subjected to MALDI-TOF/MS and linkage analyses.
d-Araf-based glycoconjugates
The synthesis of the branched octyl (α-d-Araf )2-(1→3,5)-α-d-Araf-(1→5)-α-d-Araf-(1→5)-α-d-Araf Ara5 acceptor was described previously (Zhang et al. 2007). The linear Ara5 octyl α-d-Araf-(1→5)-α-d-Araf-(1→5)-α-d-Araf-(1→5)-α-d-Araf-(1→5)-α-d-Araf acceptor was synthesized using the thioglycoside glycosylation method (Zhang et al. 2007) and its structure confirmed by 1H, 13C NMR, and HRMS. This synthetic work will be published elsewhere. The linear Ara3 pentenyl α-d-Araf-(1→5)-α-d-Araf-(1→5)-α-d-Araf and Ara4 pentenyl α-d-Araf-(1→5)-α-d-Araf-(1→5)-α-d-Araf-(1→5)-α-d-Araf acceptors were provided to us by Dr. P. Seeberger (from the Swiss Federal Institute of Technology, Zürich, Switzerland).
Scanning electron microscopy
Scanning electron microscopy analyses were performed on the mc2155 wild-type and aftD single crossover (SCO1) strains grown in 7H9-ADC-Tween 80 medium at 37°C and 30°C. Bacterial samples were prepared as described previously (Rousseau et al. 2003) and examined with a JEOL JSM 6700F scanning electron microscope operating at 4.0 kV using a SEI detector.
RNA extraction and RT-PCR
Cultures of M. smegmatis mc2155 wild-type and aftD single crossover (SCO1) strains were grown in MM63 broth at 37°C or 30°C to OD600 0.6. Bacteria resuspended in 1 mL Trizol solution (Invitrogen, Carlsbad, CA) were broken with mini glass beads using the FastPrep®-24 apparatus and Lysing Matrix B from MP Biomedicals (Solon, OH). RNA was extracted with 200 μL chloroform/isoamylalcohol (24/1; Sigma, St. Louis, MO) for 3 min at RT. After centrifugation, the aqueous phase was transferred to a tube containing 500 μL 2-propanol. Total RNA was allowed to precipitate overnight at −20°C and the resulting centrifugation pellet was washed twice with 1 mL 80% DEPC-treated ethanol, dried, and finally resuspended in DEPC-treated water. Contaminating DNA was removed by digestion with DNase I (NEB, Ipswich, MA) followed by extraction with chloroform/isoamylalcohol. RNA was precipitated with DEPC-treated NaCl and ethanol for 4 h at −80°C. After centrifugation and washing, the RNA pellet was resuspended in DEPC-treated water and the concentration of RNA in each sample determined. The absence of DNA contamination in the RNA preparations was confirmed using Taq polymerase (Roche, Basel, Switzerland) and the same sets of primers as those used for RT-PCR (Table II). RT-PCR was performed using the C. therm. polymerase one-step RT-PCR system (Roche, Basel, Switzerland) with 0.7 μg total RNA. Fifteen pmol antisense primers specific for the MSMEG_0358, aftD, and MSMEG_0360 genes of M. smegmatis were used for reverse transcription (Table II). PCR was performed with specific sets of primers for the same genes (Table II). sigA was used as an internal standard. One quarter of the RT-PCR reactions were analyzed on 1% agarose gels. RT-PCR experiments were performed on the RNA extracted from two independent bacterial cultures.
Table II.
Primers used for RT-PCR experiments
| Gene | Forward primer (5′-3′) | Reverse primer (5′-3′) | Product (bp) |
|---|---|---|---|
| SigA | CATCTCGCTGGACCAGACC | TAGTCGCGCAGCACCTGC | 317 |
| MSMEG_0358 | CCTACGTCCAGATCTTCTGCG | CTGCGTGAGGCGCAGCGCAAC | 340 |
| MSMEG_0359 | GCACTGGACTCGGTGCAACGC | GGTCATCGCGTCGCTGTCGAC | 360 |
| MSMEG_0360 | GTGAACCGGTTCGTCGTACC | CTAAGAACGGTCGCCGTACT | 173 |
Supplementary Data
Supplementary data for this article is available online at http://glycob.oxfordjournals.org/.
Funding
The National Institute of Allergy and Infectious Diseases/National Institutes of Health (AI064798 and AI018357), and the Slovak Research and Development Agency (APVV-0499-07).
Acknowledgments
We gratefully acknowledge A. Amin (Colorado State University) for the preparation of p[14C]Rpp, Dr. A. Dasgupta (Institut Pasteur, Paris) for his help with plasmid constructs, and Dr. P. Seeberger (Swiss Federal Institute of Technology, Zürich, Switzerland) for providing us with the synthetic Ara3, Ara4, and Man5 linear acceptors.
Conflict of interest statement
None declared.
Abbreviations
- Araf
arabinofuranosyl
- AraT
arabinofuranosyltransferase
- GC
gas chromatography
- LAM
lipoarabinomannan
- MALDI-TOF
matrix-assisted laser desorption-ionization time-of-flight
- Manp
mannopyranosyl
- TDM
trehalose dimycolates
- TLC
thin-layer chromatography
References
- Alderwick LJ, Seidel M, Sahm H, Besra GS, Eggeling L. Identification of a novel arabinosyl transferase (AftA) involved in cell wall arabinan biosynthesis in Mycobacterium tuberculosis. J Biol Chem. 2006;281:15653–15661. doi: 10.1074/jbc.M600045200. [DOI] [PubMed] [Google Scholar]
- Berg S, Kaur D, Jackson M, Brennan PJ. The glycosyltransferases of Mycobacterium tuberculosis- roles in the synthesis of arabinogalactan, lipoarabinomannan, and other glycoconjugates. Glycobiology. 2007;17:35R–56R. doi: 10.1093/glycob/cwm010. [DOI] [PubMed] [Google Scholar]
- Berg S, Starbuck J, Torrelles JB, Vissa VD, Crick DC, Chatterjee C, Brennan PJ. Roles of the conserved proline and glycosyltransferase motifs of EmbC in biosynthesis of lipoarabinomannan. J Biol Chem. 2005;280:5651–5663. doi: 10.1074/jbc.M411418200. [DOI] [PubMed] [Google Scholar]
- Bhamidi S, Scherman MS, Rithner CD, Prenni JE, Chatterjee D, Khoo K-H, McNeil MR. The identification and location of succinyl residues and the characterization of the interior arabinan region allows for a model of the complete primary structure of Mycobacterium tuberculosis mycolyl arabinogalactan. J Biol Chem. 2008;283:12992–13000. doi: 10.1074/jbc.M800222200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch HL, Alderwick LJ, Bhatt A, Rittmann D, Krumbach K, Singh A, Bai Y, Lowary TL, Eggeling L, Besra GS. Biosynthesis of mycobacterial arabinogalactan: Identification of a novel α(1→3) arabinofuranosyltransferase. Mol Microbiol. 2008;69:1191–1206. doi: 10.1111/j.1365-2958.2008.06354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick DC, Brennan PJ. Biosynthesis of the arabinogalactan-peptidoglycan complex of Mycobacterium tuberculosis. In: Daffé M, Reyrat J-M, editors. The Mycobacterial Cell Envelope. Washington (DC): ASM Press; 2008. pp. 25–40. [Google Scholar]
- Escuyer VE, Lety M-A, Torrelles JB, Khoo K-H, Tang J-B, Rithner CD, Frehel C, McNeil MR, Brennan PJ, Chatterjee C. The role of the embA and embB gene products in the biosynthesis of the terminal hexaarabinofuranosyl motif of Mycobacterium smegmatis arabinogalactan. J Biol Chem. 2001;276:48854–48862. doi: 10.1074/jbc.M102272200. [DOI] [PubMed] [Google Scholar]
- Gilleron M, Jackson M, Nigou J, Puzo G. Structure, activities and biosynthesis of the phosphatidyl-myo-inositol-based lipoglycans. In: Daffé M, Reyrat J-M, editors. The Mycobacterial Cell Envelope. Washington, (DC): ASM Press; 2008. pp. 75–105. [Google Scholar]
- Guilhot C, Otal I, Van Rompaey I, Martin C, Gicquel B. Efficient transposition in mycobacteria: Construction of Mycobacterium smegmatis insertional mutant libraries. J Bacteriol. 1994;176:535–539. doi: 10.1128/jb.176.2.535-539.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holemann A, Stocker BL, Seeberger PH. Synthesis of a core arabinomannan oligosaccharide of Mycobacterium tuberculosis. J Org Chem. 2006;71:8071–8088. doi: 10.1021/jo061233x. [DOI] [PubMed] [Google Scholar]
- Jackson M, Camacho LR, Gicquel B, Guilhot C. Gene replacement and transposon delivery using the negative selection marker sacB. In: Parish T, Stocker NG, editors. Mycobacterium Tuberculosis Protocols. Vol. 54. Totowa (NJ): Humana Press; 2001. pp. 59–75. [DOI] [PubMed] [Google Scholar]
- Jackson M, Crick DC, Brennan PJ. Phosphatidylinositol is an essential phospholipid of mycobacteria. J Biol Chem. 2000;275:30092–30099. doi: 10.1074/jbc.M004658200. [DOI] [PubMed] [Google Scholar]
- Kaur D, Guerin M, Skovierova H, Brennan PJ, Jackson M. Biogenesis of the cell wall and other glycoconjugates of Mycobacterium tuberculosis. Adv Appl Microbiol. 2009;69 doi: 10.1016/S0065-2164(09)69002-X. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur D, McNeil MR, Khoo K-H, Chatterjee D, Crick DC, Jackson M, Brennan PJ. New insights into the biosynthesis of mycobacterial lipomannan arising from deletion of a conserved gene. J Biol Chem. 2007;282:27133–27140. doi: 10.1074/jbc.M703389200. [DOI] [PubMed] [Google Scholar]
- Khasnobis S, Zhang J, Angala SK, Amin AG, McNeil MR, Crick DC, Chatterjee D. Characterization of a specific arabinosyltransferase activity involved in mycobacterial arabinan synthesis. Chem Biol. 2006;13:787–795. doi: 10.1016/j.chembiol.2006.05.016. [DOI] [PubMed] [Google Scholar]
- Korduláková J, Gilleron M, Mikušová K, Puzo G, Brennan PJ, Gicquel B, Jackson M. Definition of the first mannosylation step in phosphatidylinositol synthesis: PimA is essential for growth of mycobacteria. J Biol Chem. 2002;277:31335–31344. doi: 10.1074/jbc.M204060200. [DOI] [PubMed] [Google Scholar]
- Lee RE, Brennan PJ, Besra GS. Mycobacterial arabinan biosynthesis: The use of synthetic arabinoside acceptors in the development of an arabinosyl transfer assay. Glycobiology. 1997;7:1121–1128. doi: 10.1093/glycob/7.8.1121. [DOI] [PubMed] [Google Scholar]
- Lee RE, Brennan PJ, Besra GS. Synthesis of β-d-arabinofuranosyl-1-monophosphoryl polyprenols: Examination of their function as mycobacterial arabinosyl transferase donors. Bioorg Med Chem Lett. 1998;8:951–954. doi: 10.1016/s0960-894x(98)00147-4. [DOI] [PubMed] [Google Scholar]
- Makarov V, Manina G, Mikušová K, Mollmann U, Ryabova O, Saint-Joanis B, Dhar N, Pasca MR, Buroni S, Lucarelli AP, et al. Benzothiazinones kill Mycobacterium tuberculosis by blocking arabinan synthesis. Science. 2009;324:801–804. doi: 10.1126/science.1171583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikušová K, Slayden RA, Besra GS, Brennan PJ. Biogenesis of the mycobacterial cell wall and the site of action of ethambutol. Antimicrob Agents Chemother. 1995;39:2484–2489. doi: 10.1128/aac.39.11.2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra AK, Alderwick LJ, Rittmann D, Wang C, Bhatt A, Jacobs WR, Jr, Takayama K, Eggeling L, Besra GS. Identification of a novel alpha(1→6) mannopyranosyltransferase MptB from Corynebacterium glutamicum by deletion of a conserved gene, NCgl1505, affords a lipomannan- and lipoarabinomannan-deficient mutant. Mol Microbiol. 2008;68:1595–1613. doi: 10.1111/j.1365-2958.2008.06265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita YS, Sena CCB, Waller RF, Kurokawa K, Sernee MF, Nakatani F, Haites RE, Billman-Jacobe H, McConville MJ, Maeda Y, et al. PimE is a polyprenol-phosphate-mannose-dependent mannosyltransferase that transfers the fifth mannose of phosphatidylinositol mannoside in mycobacteria. J Biol Chem. 2006;281:25143–25155. doi: 10.1074/jbc.M604214200. [DOI] [PubMed] [Google Scholar]
- Nigou J, Vercellone A, Puzo G. New structural insights into the molecular deciphering of mycobacterial lipoglycan binding to C-type lectins: Lipoarabinomannan glycoform characterization and quantification by capillary electrophoresis at the subnanomole level. J Mol Biol. 2000;299:1353–1362. doi: 10.1006/jmbi.2000.3821. [DOI] [PubMed] [Google Scholar]
- Pelicic V, Jackson M, Reyrat JM, Jacobs WR, Jr, Gicquel B, Guilhot C. Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 1997;94:10955–10960. doi: 10.1073/pnas.94.20.10955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitarque S, Herrmann J-L, Duteyrat J-L, Jackson M, Stewart GR, Lecointe F, Payré B, Schwartz O, Young DB, Marchal G, et al. Deciphering the molecular bases of Mycobacterium tuberculosis binding to DC-SIGN reveals an underestimated complexity. Biochem J. 2005;392:615–624. doi: 10.1042/BJ20050709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitarque S, Larrouy-Maumus G, Payré B, Jackson M, Puzo G, Nigou J. The immunomodulatory lipoglycans, lipoarabinomannan and lipomannan, are exposed at the mycobacterial cell surface. Tuberculosis. 2008;88:560–565. doi: 10.1016/j.tube.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau C, Neyrolles O, Bordat Y, Giroux S, Sirakova TD, Prevost M-C, Kolattukudy PE, Gicquel B, Jackson M. Deficiency in mycolipenate- and mycosanoate-derived acyltrehaloses enhances early interactions of Mycobacterium tuberculosis with host cells. Cell Microbiol. 2003;5:405–415. doi: 10.1046/j.1462-5822.2003.00289.x. [DOI] [PubMed] [Google Scholar]
- Sambou T, Dinadayala P, Stadthagen G, Barilone N, Bordat Y, Constant P, Levillain F, Neyrolles O, Gicquel B, Lemassu A, et al. Capsular glucan and intracellular glycogen of Mycobacterium tuberculosis: Biosynthesis and impact on the persistence in mice. Mol Microbiol. 2008;70:762–774. doi: 10.1111/j.1365-2958.2008.06445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherman MS, Kalbe-Bournonville L, Bush D, Xin Y, Deng L, McNeil MR. Polyprenylphosphate-pentoses in mycobacteria are synthesized from 5-phosphoribose pyrophosphate. J Biol Chem. 1996;271:29652–29658. doi: 10.1074/jbc.271.47.29652. [DOI] [PubMed] [Google Scholar]
- Seidel M, Alderwick LJ, Birch HL, Sahm H, Eggeling L, Besra GS. Identification of a novel arabinofuranosyltransferase AftB involved in a terminal step of cell wall arabinan biosynthesis in Corynebacterianeae, such as Corynebacterium glutamicum and Mycobacterium tuberculosis. J Biol Chem. 2007;282:14729–14740. doi: 10.1074/jbc.M700271200. [DOI] [PubMed] [Google Scholar]
- Shi L, Berg S, Lee A, Spencer JS, Zhang J, Vissa V, McNeil MR, Khoo K-H, Chatterjee C. The carboxy terminus of EmbC from Mycobacterium smegmatis mediates chain length extension of the arabinan in lipoarabinomannan. J Biol Chem. 2006;281:19512–19526. doi: 10.1074/jbc.M513846200. [DOI] [PubMed] [Google Scholar]
- Stadthagen G, Korduláková J, Griffin R, Constant P, Bottova I, Barilone N, Gicquel B, Daffé M, Jackson M. p-hydroxybenzoic acid synthesis in Mycobacterium tuberculosis. J Biol Chem. 2005;280:40699–40706. doi: 10.1074/jbc.M508332200. [DOI] [PubMed] [Google Scholar]
- Triccas JA, Parish T, Britton WJ, Gicquel B. An inducible expression system permitting the efficient purification of a recombinant antigen from Mycobacterium smegmatis. FEMS Microbiol Lett. 1998;167:151–156. doi: 10.1111/j.1574-6968.1998.tb13221.x. [DOI] [PubMed] [Google Scholar]
- Wolucka BA. Biosynthesis of d-arabinose in mycobacteria—a novel bacterial pathway with implications for antimycobacterial therapy. FEBS J. 2008;275:2691–2711. doi: 10.1111/j.1742-4658.2008.06395.x. [DOI] [PubMed] [Google Scholar]
- Zhang J, Khoo K-H, Wu S-W, Chatterjee C. Characterization of a distinct arabinofuranosyltransferase in Mycobacterium smegmatis. J Am Chem Soc. 2007;129:9650–9662. doi: 10.1021/ja070330k. [DOI] [PubMed] [Google Scholar]
- Zhang N, Torrelles JB, McNeil MR, Escuyer VE, Khoo K-H, Brennan PJ, Chatterjee D. The Emb proteins of mycobacteria direct arabinosylation of lipoarabinomannan and arabinogalactan via an N-terminal recognition region and a C-terminal synthetic region. Mol Microbiol. 2003;50:69–76. doi: 10.1046/j.1365-2958.2003.03681.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.