Abstract
We analyzed enrichment cultures of ammonia-oxidizing bacteria (AOB) collected from different areas of Salar de Huasco, a high altitude, saline, pH-neutral water body in the Chilean Altiplano. Samples were inoculated into mineral media with 10 mM NH4+ at five different salt concentrations (10, 200, 400, 800 and 1,400 mM NaCl). Low diversity (up to three phylotypes per enrichment) of beta-AOB was detected using 16S rDNA and amoA clone libraries. Growth of beta-AOB was only recorded in a few enrichment cultures and varied according to site or media salinity. In total, five 16S rDNA and amoA phylotypes were found which were related to Nitrosomonas europaea/Nitrosococcus mobilis, N. marina and N. communis clusters. Phylotype 1-16S was 97% similar with N. halophila, previously isolated from Mongolian soda lakes, and phylotypes from amoA sequences were similar with yet uncultured beta-AOB from different biofilms. Sequences related to N. halophila were frequently found at all salinities. Neither gamma-AOB nor ammonia-oxidizing Archaea were recorded in these enrichment cultures.
Keywords: Ammonia-oxidizing bacteria, 16S rDNA, amoA gene, Andean Altiplano, Salinity
Introduction
The Salar de Huasco is an athalassohaline, high altitude (3,800 m) salt-flat with neutral pH, and is located in the Chilean Altiplano. This system exhibits high spatial and temporal variability with water salinities ranging from freshwater to salt-saturated brines (Risacher et al. 1999). Salt-flats located in the Altiplano (locally called “salares”) can have high nutrient concentrations especially at the most saline sites. Within the Salar de Huasco, nutrient concentrations range between 0.08 and 23 mg L−1 for total nitrogen and 0.05 and 2.77 mg L−1 for total phosphorus (Table 1). In altiplanic wetlands, microbial communities are dominated by bacteria instead of Archaea and exhibit a specific pattern according to the type of water body. These salares support an increased diversity relative to local lakes and peatlands locally called “bofedales” (Demergasso et al. 2004; Dorador et al. unpublished).
Table 1.
Temperature, total dissolved salts, dissolved oxygen, total nitrogen, ammonia, nitrate, total phosphorus, phosphate and sulphate concentrations and C:N ratios in water samples of four sites in Salar de Huasco
| Site | Type | Temperature (°C) | Total dissolved salts (g L−1) | C:N | D.O (μM) | Total nitrogen (mg L−1) | N–NH4+ (μM) | N–NO3− (μM) | Total phosphorus (mg L−1) | P–PO43− (μM) | S–SO42− (μM) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| H0 | Stream | 17.2 | 0.47 | 4.33 | 181.26 | 0.08 | 0.28 | 1.22 | 0.05 | 0.58 | 0.27 |
| H1 | Lagoon | 17.9 | 0.39 | 11.78 | 240.63 | 1.38 | 1.33 | 7.74 | 0.08 | 0.92 | 0.32 |
| H4 | Lagoon | 12.3 | 42.77 | 10.55 | 115.63 | 7.75 | 17.89 | ND | 2.77 | 25.28 | 34.92 |
| H6 | Lagoon | 4.9 | 66.99 | 5.82 | ND | 23.38 | 42.44 | ND | 2.16 | 18.69 | 45.00 |
ND not detected
Nitrification is the biological oxidation of the most reduced form of nitrogen (ammonia) to nitrite and nitrate and it plays a central role in the global N-cycle. In freshwater, marine environments and soils, nitrite produced by the ammonia oxidizers is immediately consumed by nitrite oxidizers and thus the nitrite concentration is extremely low in these environments (Bock and Wagner 2006). Under anoxic or oxygen-limited conditions, nitrate and nitrite are used as electron acceptors for anaerobic respiration (if organic matter is available), converted back to ammonia (e.g., by respiratory ammonification) or to dinitrogen (e.g., by denitrification, anammox).
Chemolithoautotrophic ammonia-oxidizing bacteria (AOB) are key components of the nitrogen cycle, and are involved in the aerobic oxidation of ammonia to nitrite, the initial stage of nitrification. Recently, metagenomic studies have revealed that members of the Crenarchaeota within the domain Archaea contain and express genes related to bacterial ammonia monooxygenases (Treusch et al. 2005). Ammonia-oxidizing archaea (AOA) are widespread across oxic to suboxic marine water bodies as well as in soils and sediments (Francis et al. 2005; Wuchter et al. 2006; Leininger et al. 2006). The candidatus Nitrosopumilus maritimus represents the only AOA isolated to date (Könneke et al. 2005).
In the N-limited aquatic systems, AOB populations compete with heterotrophs and benthic algae for reduced nitrogen (Bernhard and Peele 1997; Risgaard-Petersen et al. 2004; Geets et al. 2006). Nitrogen limitation has been reported in Lago Titicaca (Vincent et al. 1984, 1985) and Lago Chungará (Dorador et al. 2003), two lakes located in the tropical Andes, and this phenomenon is likely to occur in other water bodies in the region, including Salar de Huasco. Previous work has demonstrated that nitrification and denitrification rates varied considerably between years in Lago Titicaca (Vincent et al. 1985). Lago Titicaca experiences low levels of oxygen saturation due to the high altitude of the lake, which, in turn favors hypolimnetic anoxia, and thus denitrification.
16S rRNA gene sequence analysis shows that the majority of AOB described to date belong to the Betaproteobacteria including members of Nitrosomonas (including Nitrosococcus mobilis) and Nitrosospira (including Nitrosolobus and Nitrosovibrio). Nitrosococcus oceani and Nitrosococcus halophilus are both members of the Gammaproteobacteria (Purkhold et al. 2000, 2003). AOB have been detected in a variety of environments including soils, marine, estuarine, salt lakes and freshwater systems (e.g., Bothe et al. 2000; Koops et al. 2006). Clone libraries of 16S rDNA in the hypersaline Mono Lake revealed the existence of sequences related to Nitrosomonas europaea and Nitrosomonas eutropha, both shown to exhibit elevated levels of salt tolerance (Ward et al. 2000). In estuarine systems, the dominance of Nitrosospira from marine sites and the prevalence of Nitrosomonas oligotropha and Nitrosomonas sp. Nm143 in freshwater and areas of intermediate salinities have been reported (Bernhard et al. 2005; Freitag et al. 2006). Previous studies have reported reductions in AOB diversity with increased salinity along a salinity gradient in the Plum Island Sound estuary (salinity range 0.5–31.7) and the Schelde estuary (0–28) (Bernhard et al. 2005; Bollmann and Laanbroek 2002). In the present study we used enrichment cultures at different salt concentrations to characterize ammonia oxidizers from different environments of the Salar de Huasco with respect to their salt tolerance.
Materials and methods
Site description and sampling
During May 2006, we collected water samples from four different sites of the Salar de Huasco (20°18′S, 68°50′W) located at 3,800 m altitude in the Chilean Altiplano. The salar exhibits high spatial heterogeneity, represented by a mosaic of streams, bofedales (peatlands), shallow permanent and non-permanent lagoons and salt crusts, with a gradient in salt concentration from north to south. The sampling sites can be described from north to south as follows: (a) H0 is a stream surrounded by abundant macrophytes, e.g., Oxychloe andina (Squeo et al. 2006) and aquatic ferns (mostly Azolla sp.) and is characterized by large amounts of organic matter in the sediments; (b) H1 is a permanent lagoon with low salinity; (c) H4 is a shallow, hypersaline lagoon (approximately 10 cm deep at the time of sampling) with no vegetation; (d) H6 is a anoxic lagoon with fluctuating water levels and high salinity, located in the south of the salar.
Temperature was recorded with a digital Hanna HI thermometer, pH with a Hanna HI 8314 meter, and conductivity with an YSI 33 meter. Total carbon, total nitrogen, nitrate, ammonia, total phosphorus, phosphate and sulfate were analyzed according to Standard Methods (APHA 1999).
Enrichment cultures of ammonia-oxidizing bacteria
Samples of water were collected at four sites (H0, H1, H4 and H6) from the Salar de Huasco. Because of the spatial variability of the sites, several samples were taken from each site (Table 2). On collection they were inoculated into mineral media with 10 mM NH4Cl (Koops et al. 2006) at five different salt concentrations (10, 200, 400, 800 and 1,400 mM NaCl) and pH 8 adjusted with 10% NaHCO3. Because of the high concentration of Li, As and B at the sites (Risacher et al. 2003) LiCl (0.5 mM), NaAsO2 (0.5 mM) and HBO3 (0.2 mM) were added to the culture media. Growth was estimated by measuring nitrite concentration using the Griess–Ilosvay reaction (e.g., Keeney and Nelson 1982) every 2 weeks, and the positive cultures were transferred into fresh media, a total of four times. Incubation temperature was maintained at ∼30°C. Hundred-milliliters Erlenmeyer flasks were filled with 10 ml culture to permit a suitable level of oxygen diffusion into the cultures.
Table 2.
Enrichment cultures of AOB from samples of Salar de Huasco at different salt concentrations
| Site | Sample | NaCl concentration (mM) | ||||
|---|---|---|---|---|---|---|
| 10 | 200 | 400 | 800 | 1,400 | ||
| H0 | H0a | + | + (H0a-200)** | + (H0a-400)** | − | + (H0a-1,400)*, ** |
| H0 | H0b | − | − | − | − | − |
| H1 | H1a | + | + | + (H1a-400)*,** | + (H1a-800)* | + |
| H1 | H1b | + (H1b-10) | + (H1b-200)* | + (H1b-400)* | − | − |
| H4 | H4 | − | − | + (H4-400)* | + | + |
| H6 | H6a | − | + (H6a-200)** | + (H6a-400)*, ** | − | + |
| H6 | H6b | + | − | + (H6b-400)* | + (H6b-800)*, ** | + (H6b-1,400)* |
| H6 | H6c | − | + | + (H6c-400)*, ** | + (H6c-800)*, ** | + |
| H6 | H6d | − | + (H6d-200)** | + (H6d-400)*, ** | + (H6d-800) | + (H6d-1,400) |
The presence (+) or absence (−) of nitrite accumulation, the enrichment used for molecular analysis and the ammonia oxidizers determined from 16S rDNA (*) and amoA (**) gene are indicated
DNA extraction and PCR amplification
DNA from AOB enrichment cultures was extracted with the Ultra Clean Soil DNA Isolation Kit (MoBio Lab., Inc.). Oligonucleotide primers, Eub9-27F and Eub1542R (Stackebrandt and Liesack 1993), were used to PCR-amplify eubacterial 16S rDNA. This PCR product was used as a template to amplify in a nested PCR approach 16S rDNA from ammonia oxidizers of the Betaproteobacteria using NitA and NitB primers as described by Voytek and Ward (1995) and primers NOC1–NOC2 to amplify gamma-AOB (Ward et al. 2000). The bacterial amoA gene was amplified by PCR with primers amoA-1F and amoA-2R according to Rotthauwe et al. (1997) and the archaeal amoA gene with primers CrenAmo1F and CrenAmo1R (Könneke et al. 2005). Each PCR reaction contained 10× PCR-buffer with 2 mM MgCl2 (Roche), 200 mM dNTP mixture (Gibco), 1 pmol of each primer, 2.5 U Taq polymerase (Roche), 10–100 ng template DNA and water to a final volume of 50 μl. PCR was performed using the following conditions for eubacterial primers: initial denaturing step of 5 min at 94°C followed by 34 cycles of denaturing at 94°C for 30 s, annealing at 40°C for 45 s and elongation at 72°C for 1.5 min. Annealing temperatures were 57, 60, 55 and 56°C for NitA–NitB, NOC1–NOC2, bacterial amoA and archaeal amoA primers, respectively. Primers NitA–NitB were selected because they have higher specificity than other primer combinations detecting unknown AOB (e.g., Molina et al. 2007; Kim et al. 2006; Utaker and Nes 1998).
Denaturing gradient gel electrophoresis (DGGE) analysis
DGGE was performed according to Muyzer et al. (1993) with PCR products of eubacterial 16S rDNA generated with the primers P2/P3. The P3 primer contained an additional 40-nucleotide GC-rich sequence (GC clamp) at its 5′ end in order to maintain stable melting behavior during DGGE (Muyzer et al. 1993). PCR conditions were as described above and PCR amplification was carried out using a touchdown protocol as follows: initial denaturing step of 5 min at 94°C, 20 cycles of 30 s at 94°C, 45 s at 65–55°C (decreased by 0.5°C every cycle) and 1.5 min at 72°C and then ten cycles of 30 s at 94°C, 45 s at 55°C and 1.5 min at 72°C. PCR products were applied onto 7.5% polyacrylamide gels containing a linear gradient of 30–60% denaturant where 100% denaturant was defined as 7 M urea and 40% formamide. DGGE was carried out in the BioRad D Gene System (BioRad) at 60°C, 200 V for 6 h. Gels were stained with SYBR Gold nucleic acid gel stain (Molecular Probes). Bands were excised and reamplified for sequencing.
Cloning and 16S rDNA sequencing
Clone libraries of 16S rDNA and amoA were constructed with positive AOB enrichment cultures (Table 2). Beta-AOB 16S rDNA clone libraries were made from all enrichment cultures apart from enrichments H1b-10, H6d-800 and H6d-1,400 where amplifications were unsuccessful. Clone libraries of bacterial amoA gene were made for 11 enrichments (H6b-10, H0a-200, H6d-200, H1a-400, H6c-400, H6a-400, H0a-400, H6d-400, H6c-800, H6b-800, H0a-1,400). Purified amplicons were cloned into pCR-Blunt vector (Invitrogen) according to the manufacturer’s instructions. Twenty-four clones per sample were picked, and the inserts were amplified with M13F/R primers. Cycle sequencing was performed with the BigDye Terminator Cycle Sequencing Kit v3.1 and analyzed on an automated capillary sequencer (model 3100 Gene Analyzer, Applied Biosystems). Sequences were checked for chimeras using Chimera check from the Ribosomal Database Project II (http://www.cme.msu.edu/rdp). Sequences originating from excised DGGE bands and from betaproteobacterial 16S rDNA and amoA clone libraries were analyzed and compared by BLAST search to find similarities with sequences in GenBank.
Phylogenetic analysis
Sequences were aligned using the alignment tool in the ARB package (http://www.arb-home.de). Phylogenetic relationships were calculated using maximum likelihood analysis in the program PhyML (Guindon et al. 2005) with GTR substitution model (generalized time reversible) and 100 bootstrap re-samplings. Trees were edited using MEGA 3 (Kumar et al. 2004). Sequences not included in the ARB database were downloaded from GenBank (e.g., first hit in BLAST) for 16S rDNA and amoA sequences. Similarities of the sequences were analyzed using MEGA 3 (Kumar et al. 2004) and sequences with similarities >99% were considered to represent the same phylotype.
Nucleotide sequence accession numbers
The nucleotide sequences from this study are available in GenBank (http://www.ncbi.nlm.nih.gov) under accession numbers EU116356-EU116365.
Results
Environmental and enrichment culture conditions
Table 1 shows nutrient concentrations and C:N ratios at the four study-sites in Salar de Huasco. Sites H0 and H1 exhibited similar salt, phosphate and sulfate concentrations contrasting with sites H4 and H6 where these concentrations were recorded up to 67, 25 and 45 μM, respectively. Water temperature at the sites ranged between 5 and 18°C. In the enrichment cultures, nitrite production was detected in eight of the nine samples at different salt concentrations (Table 2). Enrichment H1a from site H1 grew in a broad range of salinities (from 10 to 1,400 mM), but enrichment H1b only grew between 10 and 400 mM NaCl. The enrichments from site H4 grew between 400 and 1,400 mM NaCl, while enrichments from H6, H6c and H6d grew at concentrations higher than 200 mM NaCl. There was no apparent relationship between site salinity and growth in the remaining enrichments. Enrichment H0b did not exhibit any growth.
Bacteria in the enrichments
The bacterial composition of the enrichment cultures was analyzed by 16S rDNA PCR-DGGE. Between 7 and 10 bands per sample were found from a total of 171 bands (data not shown), with no apparent relationship between the number and composition of bands and the salt concentration. To identify the members of the communities, both representative (e.g., the most frequent ones) and rare bands were excised and sequenced. The sequences were affiliated to Cytophaga–Flavobacteria–Bacteroidetes (CFB), Gammaproteobacteria and Betaproteobacteria. CFB and Gammaproteobacteria were the most frequent groups found in all the enrichments, and Betaproteobacteria were detected in 16 of 21 enrichments.
AOB identified by 16S rDNA sequences
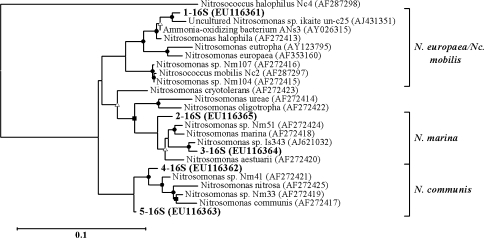
A single phylotype was found in each enrichment. The phylotype 1-16S was found in nine enrichment cultures with a range of salt concentrations (Table 3). It had 98% sequence similarity to an extremely alkali-tolerant ammonia-oxidizing bacterium isolated from composite samples (water and sediment) from Mongolian soda lakes. Phylotype 2-16S was obtained from enrichment H4-400 and had 97% sequence similarity to Nitrosomonas marina. Phylotype 3-16S had 98% sequence similarity to the isolate Nitrosomonas sp. Is343, retrieved from brackish water of the Schelde river in the Netherlands (GenBank information). Phylotype 4-16S was found in three enrichment cultures (H6d-200, H6c-400 and H1b-200) and phylotype 5-16S was recorded from enrichment culture H1b-400. 16S rDNA sequences from both were 96–97% similar to Nitrosomonas sp. Nm41 (Koops et al. 1991). Phylogenetic analysis revealed that the phylotype 1-16S is clustered with N. europaea/Nitrosococcus mobilis, phylotype 2-16S and 3-16S with N. marina and phylotypes 4 and 5 with N. communis (Fig. 1). No amplification was detected using the 16S rDNA primers NOC1 and NOC2 designed to amplify Nitrosococcus oceani, a member of the gamma-AOB (Ward et al. 2000).
Table 3.
Sequence similarity of 16S rDNA from AOB phylotypes with GenBank entries (BLAST search)
| Phylotype | Enrichment culture | Closest relative in BLAST | ||
|---|---|---|---|---|
| Name | Similarity (%) | Environment | ||
| 1-16S | H6b-1,400 | Ammonia-oxidizing bacterium ANs3 (AY026315) | 98 | Surface sediments, mongolian soda lakes |
| H0a-1,400 | Ammonia-oxidizing bacterium ANs3 (AY026315) | 98 | Surface sediments, mongolian soda lakes | |
| H6b-800 | Ammonia-oxidizing bacterium ANs3 (AY026315) | 98 | Surface sediments, mongolian soda lakes | |
| H6d-400 | Ammonia-oxidizing bacterium ANs3 (AY026315) | 98 | Surface sediments, mongolian soda lakes | |
| H6c-800 | Ammonia-oxidizing bacterium ANs3 (AY026315) | 98 | Surface sediments, mongolian soda lakes | |
| H1a-400 | Ammonia-oxidizing bacterium ANs3 (AY026315) | 98 | Surface sediments, mongolian soda lakes | |
| H6b-10 | Ammonia-oxidizing bacterium ANs3 (AY026315) | 98 | Surface sediments, mongolian soda lakes | |
| H6b-400 | Ammonia-oxidizing bacterium ANs3 (AY026315) | 98 | Surface sediments, mongolian soda lakes | |
| H1a-800 | Ammonia-oxidizing bacterium ANs3 (AY026315) | 98 | Surface sediments, mongolian soda lakes | |
| 2-16S | H4-400 | Nitrosomonas marina (AF272418) | 97 | Shell grit, great barrier reef, Australia |
| 3-16S | H6a-200 | Nitrosomonas sp. Is343 (AJ621032) | 98 | Brakish water, River Schelde, Netherlands |
| H6a-400 | Nitrosomonas sp. Is343 (AJ621032) | 98 | Brakish water, River Schelde, Netherlands | |
| 4-16S | H6d-200 | Nitrosomonas sp. Nm41 (AF272421) | 96 | Soil, Leningrad, Russia |
| H6c-400 | Nitrosomonas sp. Nm41 (AF272421) | 96 | Soil, Leningrad, Russia | |
| H1b-200 | Nitrosomonas sp. Nm41 (AF272421) | 96 | Soil, Leningrad, Russia | |
| 5-16S | H1b-400 | Nitrosomonas sp. Nm41 (AF272421) | 97 | Soil, Leningrad, Russia |
Fig. 1.
Phylogenetic tree based on partial betaproteobacterial 16S rDNA sequences (≥800 bp) of AOB enrichment cultures inferred by maximum likelihood analysis. The scale bar represents 10% nucleotide sequence difference. Symbols on the branches indicate bootstrap confidence values as follows: filled circle >80%, filled square 60–80%, open triangle 40–60%. Nitrosococcus halophilus (AF287298) was used as outgroup. Nc. mobilisNitrosococcus mobilis
AOB identified by amoA sequences
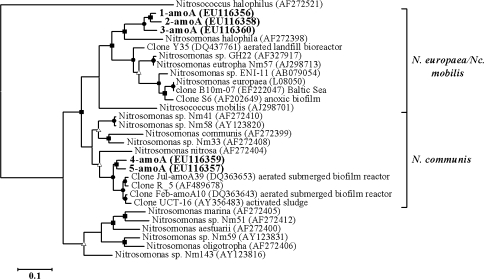
A total of five phylotypes were identified and affiliated to two described clusters of Nitrosomonas: N. europaea/Nitrosococcus mobilis and N. communis (Fig. 2). The topology of the tree was confirmed using amino acid residues (data not shown).
Fig. 2.
Phylogenetic tree based on amoA sequences (≥450 bp) of AOB enrichment cultures inferred by maximum likelihood analysis. Characteristics of the tree as in Fig. 1. Nitrosococcus halophilus (AF272521) was used as outgroup
Phylotype 1-amo, 2-amo and 3-amo were found in ten enrichment cultures, representing a wide range of salt concentrations (from 10 mM to 1,400 mM NaCl) and they had low similarity (>85%) with the best hit in BLAST comparing nucleotide sequences (Table 4). The protein sequence exhibited higher similarity (>95%) with available sequences and the three phylotypes were related to clone S6 retrieved from an anoxic biofilm of a reactor with high anaerobic ammonia oxidation (Schmid et al. 2000). Phylotypes 4-amoA and 5-amoA were found in enrichments with 200 and 400 mM NaCl and they had the same closest protein relative (clone RT_075_01) retrieved from soil (GenBank information) and were related with the N. communis cluster. No amplification was detected using archaeal amoA primers in the enrichment cultures.
Table 4.
Sequence similarity of amoA from AOB phylotypes with GenBank entries (BLAST search)
| Phylotype | Enrichment culture | Closest relative in BLAST | |||||
|---|---|---|---|---|---|---|---|
| Nucleotide | Protein | ||||||
| Name | Similarity (%) | Environment | Name | Similarity (%) | Environment | ||
| 1-amoA | H6a-400 | Clone S6 (AF202649) | 85 | Anoxic biofilm | Clone S6 (AAF22967) | 96 | Anoxic biofilm |
| H1a-400 | Clone S6 (AF202649) | 85 | Anoxic biofilm | Clone S6 (AAF22967) | 96 | Anoxic biofilm | |
| H6c-800 | Clone S6 (AF202649) | 85 | Anoxic biofilm | Clone S6 (AAF22967) | 96 | Anoxic biofilm | |
| H6b-10 | Clone S6 (AF202649) | 85 | Anoxic biofilm | Clone S6 (AAF22967) | 96 | Anoxic biofilm | |
| H6c-400 | Clone S6 (AF202649) | 85 | Anoxic biofilm | Clone S6 (AAF22967) | 96 | Anoxic biofilm | |
| H6d-400 | Clone S6 (AF202649) | 85 | Anoxic biofilm | Clone S6 (AAF22967) | 96 | Anoxic biofilm | |
| H0a-400 | Clone S6 (AF202649) | 85 | Anoxic biofilm | Clone S6 (AAF22967) | 96 | Anoxic biofilm | |
| H6b-800 | Clone S6 (AF202649) | 85 | Anoxic biofilm | Clone S6 (AAF22967) | 96 | Anoxic biofilm | |
| 2-amoA | H1a-400 | Clone Y35 (DQ437761) | 85 | Aerated landfill bioreactor | Clone S6 (AAF22967) | 95 | Anoxic biofilm |
| 3-amoA | H0a-400 | Clone Y35 (DQ437761) | 84 | Aerated landfill bioreactor | Clone S6 (AAF22967) | 96 | Anoxic biofilm |
| H6c-400 | Clone Y35 (DQ437761) | 84 | Aerated landfill bioreactor | Clone S6 (AAF22967) | 96 | Anoxic biofilm | |
| H0a-200 | Clone Y35 (DQ437761) | 84 | Aerated landfill bioreactor | Clone S6 (AAF22967) | 96 | Anoxic biofilm | |
| H0a-1,400 | Clone Y35 (DQ437761) | 84 | Aerated landfill bioreactor | Clone S6 (AAF22967) | 96 | Anoxic biofilm | |
| 4-amoA | H6c-400 | Clone Jul-amoA39 (DQ363653) | 92 | Submerged biofilm reactor | Clone RT-075_01 (ABF20600) | 96 | Soil |
| H6d-200 | Clone Jul-amoA39 (DQ363653) | 92 | Submerged biofilm reactor | Clone RT-075_01 (ABF20600) | 96 | Soil | |
| 5-amoA | H0a-200 | Clone Jul-amoA39 (DQ363653) | 92 | Submerged biofilm reactor | Clone RT-075_01 (ABF20600) | 97 | Soil |
Discussion
High-altitude Altiplanic wetlands are exposed to particularly harsh environmental stresses including aridity, high UV radiation, negative water balance, extreme differences of temperatures between day and night and a wide range of salinity conditions (e.g., Fernández Zenoff et al. 2006; Demergasso et al. 2004; Risacher et al. 2003; Vila and Mühlhauser et al. 1987). The precipitation regime during the austral summer also affects the chemical and physical composition of the basins. Microbial diversity in high altitude wetlands of the Altiplano has been recently studied using cultivation independent techniques (Demergasso et al. 2004; Dorador et al. unpublished) and revealed the predominance of Cytophaga–Flavobacteria–Bacteroidetes and Proteobacteria at a wide range of salinities and sites. CFB and Gammaproteobacteria also were found in the enrichment cultures studied here. Salar de Huasco exhibited considerable variation in salt and nutrient concentrations between the sites (Table 1), and this was reflected in the diversity and composition of microbial communities. Based on C:N ratios, we detected possible N-limitation as defined by Hecky et al. (1993) in some sites of the Salar de Huasco, namely H1 and H4, but not H0 and H6 (Table 1).
Parallel to the establishment of our enrichment cultures, we estimated the presence of AOB, AOA and anammox bacteria in environmental samples of Salar de Huasco using several primer combinations for 16S rDNA of planctomycetes, beta-AOB, gamma-AOB and also amoA for Bacteria and Archaea. We did not detect 16S rDNA sequences of bacterial ammonia oxidizers (data not shown), but found sequences similar (>92%) to Brocadia anammoxidans, a described anammox bacterium (Strous et al. 1999) at the site H4 that frequently is under anoxic conditions, and also with ammonia-oxidizing Archaea at site H0 (Dorador et al. unpublished).
Enrichment cultures of AOB from Salar de Huasco
In the present work, we used enrichment cultures from different sites of Salar de Huasco, which were prepared with salt concentrations ranging from 10 to 1400 mM NaCl. Our results provide a first impression regarding bacterial ammonia oxidizers in this almost unstudied, high-altitude wetland. We revealed that the AOB most commonly found were similar by gene sequences to members of the genus Nitrosomonas (Figs. 1, 2), indicating that this group exhibits good competitive abilities under our enrichment conditions. To detect gamma-AOB in the enrichments we used NOC1–NOC2 primers. This set of primers was developed on the basis of two sequences of Nitrosococcus oceani (Voytek and Ward 1995) but they did not reveal PCR products from several environmental samples (e.g., Ward et al. 2000; Bernhard et al. 2005). Until now, only a few number of gamma-AOB cultures exist, all of them have been isolated from sea water and some sequences have been retrieved also from saline environments like Lake Bonney, a permanently ice-covered lake in Antarctica (Bothe et al. 2000; Ward and O’Mullan 2002; Voytek and Ward 1995).
Enrichment cultures have been used to detect AOB in several environments (e.g., seawater: McCaig et al. 1994; freshwater: Hiorns et al. 1995; calcareous grasslands: Kowalchuk et al. 2000) at several initial ammonia concentrations, from 0.67 mM (Hastings et al. 1998) and up to greater than 100 mM (e.g. McCaig et al. 1994; Bruns et al. 1999). However, enrichments usually result in the isolation of only a fraction of the total diversity of AOB present in a sample (Stephen et al. 1996). Molecular analyses have demonstrated that conclusions from cultivation-based approaches can misrepresent AOB community structure in a sample. Therefore, the present study was not designed to describe the actual community composition of AOB. Both pure and enrichment cultures can provide important clues regarding physiological properties of bacteria and their adaptation to particular conditions of their habitat (e.g., Kowalchuk and Stephen 2001). The use of natural water supplemented with ammonia instead of mineral media positively influenced ammonia oxidation in batch cultures of samples from the Schelde estuary (Bollmann and Laanbroek 2002). We used artificial media to enrich the samples and this might explain the lack of growth in enrichment H0b and the lack of growth in other samples (Table 2). The waters of the Altiplano typically contain high concentrations of specific chemical elements including Li, As, B, Rb and F that are uncommon in other areas (Alonso 1997). For this reason we added Li, As and B to the media. Nevertheless, it is possible that other compounds or trace elements that are necessary for the growth of some AOB were not provided in the media, limiting the recovery of some members of the AOB communities.
Influence of salinity on the growth of AOB in the enrichment cultures
Salinity appears to be an important factor in determining the distribution of AOB in estuarine and river systems (Bernhard et al. 2005; Stehr et al. 1995), with low abundance and low diversity at high salt concentrations. AOB, like all other bacteria, have distinctive ecophysiological preferences, including salt concentration, substrate affinity and habitat (e.g., Geets et al. 2006; Webster et al. 2005; Koops and Pommerening-Röser 2001). A clear correlation of the enriched AOB according to the salinity of site and media was not found. In some samples (e.g., H4) AOB grew only at higher salinities (above 400 mM NaCl), while in sample H1b they only grew at salt concentrations lower than 400 mM NaCl (Table 2). However, this association was not found in all samples and could reflect bias in the composition of the media used for the enrichment cultures (see above). Altogether the range of salt concentrations in which enrichment and growth of AOB from Salar de Husco samples occurred demonstrates that many of these bacteria are adapted to elevated salt concentrations that prevail in the habitats of Salar de Huasco.
AOB identified from enrichment cultures
Analysis of 16S rDNA and the amoA gene revealed that sequences from enrichments belonged to N. europaea/Nitrosococcus mobilis, N. marina and N. communis, and only displayed low levels of similarity with cultured representatives. Most phylotypes were similar to yet uncultured AOB, especially when using amoA as a molecular marker (Table 4). N. marina and N. europaea/ Nitrosococcus mobilis have been described as obligate halophiles or as being halotolerant (e.g., optimal growth of N. marina is between 300 and 400 mM NaCl). Conversely, N. communis has low salt requirements for growth. Phylotype 1-16S was 98% similar to an alkali-tolerant ammonia-oxidizing bacterium representing a subpopulation of N. halophila (Sorokin et al. 2001). This form of N. halophila was highly tolerant to high pH and like the marine N. halophila, had a high salt requirement (Koops et al. 1991). It is likely that phylotype 1-16S and the 1, 2 and 3-amoA represent a new, salt-tolerant species that is closely related to N. halophila.
We also attempted to test for the presence of AOA in the enrichment cultures but our amplification using primers designed for amoA of Nitrosopumilus maritimus failed (Könneke et al. 2005). The medium used to isolate N. maritimus, (the only ammonia-oxidizing Crenarchaeota cultured to-date: Könneke et al. 2005) differed from the culture media used for ammonia oxidizers (inorganic salt medium containing ammonia) with regard to the presence of vitamins and trace elements. Our results suggest that this lack may explain previous failures to isolate ammonia-oxidizing Archaea (Nicol and Schleper 2006).
Acknowledgments
We thank Rodrigo Pardo for help in sampling and Chris Harrod for English corrections. Cristina Dorador was supported by a doctoral fellowship from the Deutscher Akademischer Austausch Dienst (DAAD), Germany. Two anonymous reviewers helped to increase the quality of this manuscript.
References
- Alonso H (1997) Geoquímica de las aguas del Altiplano. Una aproximación. In: González C (ed) El Altiplano: Ciencia y Conciencia en los Andes. Editorial Artegrama, Santiago, pp 105–107
- APHA (1999) Standard methods for the examination of water and wastewater. American Public Health Association, Baltimore
- Bernhard AE, Peele ER (1997) Nitrogen limitation of phytoplankton in a shallow embayment in northern Puget Sound. Estuaries 20:759–769 [DOI]
- Bernhard AE, Donn T, Giblin AE, Stahl DA (2005) Loss of diversity of ammonia-oxidizing bacteria correlates with increasing salinity in an estuary system. Environ Microbiol 7:1289–1297 [DOI] [PubMed]
- Bock E, Wagner M (2006) Oxidation of inorganic nitrogen compounds as an energy source. The prokaryotes. Springer, New York, pp 457–495
- Bollmann A, Laanbroek HJ (2002) Influence of oxygen partial pressure and salinity on the community composition of ammonia-oxidizing bacteria in the Schelde estuary. Aquat Microb Ecol 28:239–247 [DOI]
- Bothe H, Jost G, Schloter M, Ward BB, Witzel K-P (2000) Molecular analysis of ammonia oxidation and denitrification in natural environments. FEMS Microbiol Rev 24:673–690 [DOI] [PubMed]
- Bruns MA, Stephen JR, Kowalchuk GA, Prosser JI, Paul EA (1999) Comparative diversity of ammonia oxidizer 16S rRNA gene sequences in native, tilled and successional soils. Appl Environ Microbiol 65:2994–3000 [DOI] [PMC free article] [PubMed]
- Demergasso C, Casamayor E, Chong G, Galleguillos P, Escudero L, Pedrós-Alió C (2004) Distribution of prokaryotic genetic diversity in athalassohaline lakes of the Atacama Desert, Northern Chile. FEMS Microbiol Ecol 48:57–69 [DOI] [PubMed]
- Dorador C, Pardo R, Vila I (2003) Variaciones temporales de parámetros físicos, químicos y biológicos: el caso del lago Chungará. Rev Chil Hist Nat 76:15–22
- Fernández Zenoff V, Siñeriz F, Farías ME (2006) Diverse responses to UV-B radiation and repair mechanisms of Bacteria isolated from high altitude aquatic environments. Appl Environ Microbiol 72:7857–7863 [DOI] [PMC free article] [PubMed]
- Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB (2005) Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci USA 102:14683–14688 [DOI] [PMC free article] [PubMed]
- Freitag TE, Chang L, Prosser JI (2006) Changes in the community structure and activity of betaproteobacterial ammonia-oxidizing sediment bacteria along a freshwater-marine gradient. Environ Microbiol 8:684–696 [DOI] [PubMed]
- Geets J, Boon N, Verstraete W (2006) Strategies of aerobic ammonia-oxidizing bacteria for coping with nutrient and oxygen fluctuations. FEMS Microbiol Ecol 58:1–13 [DOI] [PubMed]
- Guindon S, Lethiec F, Duroux P, Gascuel O (2005) PHYML Online, a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res 33:W557–W559 [DOI] [PMC free article] [PubMed]
- Hastings RC, Saunders JR, Hall GH, Pickup RW, McCarthy AJ (1998) Application of molecular biological techniques to a seasonal study of ammonia oxidation in a eutrophic freshwater lake. Appl Environ Microbiol 64:3674–3682 [DOI] [PMC free article] [PubMed]
- Hecky RE, Campbell P, Hendzel LL (1993) The stoichiometry of carbon, nitrogen and phosphorus in particulate matter of lakes and oceans. Limnol Oceanogr 38:709–724
- Hiorns WD, Hastings RC, McCarthy AJ, Saunders JR, Pickup RW, Hall H (1995) Amplification of 16S rDNA genes of autotrophic ammonia-oxidizing bacteria demonstrates the ubiquity of nitrosospiras in the environment. Microbiology 141:2793–2800 [DOI] [PubMed]
- Keeney DR, Nelson DW (1982) Nitrogen-inorganic forms. In: Page AL, Miller RH, Keeney DR (eds) Methods of soil analysis, Part 2: chemical and microbiological properties. American Society of Agronomy, Madison
- Kim O-S, Junier P, Imhoff JF, Witzel K-P (2006) Comparative analysis of ammonia-oxidizing bacterial communities in two lakes in North Germany and the Baltic Sea. Arch Hydrobiol 167:335–350 [DOI]
- Koops H-P, Pommerening-Röser A (2001) Distribution and ecophysiology of the nitrifying bacteria emphasizing cultured species. FEMS Microbiol Ecol 37:1–9 [DOI]
- Koops H-P, Böttcher B, Möller UC, Pommerening-Röser A, Stehr G (1991) Classification of eight new species of ammonia-oxidizing bacteria: Nitrosomonas communis sp. nov., Nitrosomonas ureae sp. nov., Nitrosomonas aestuarii sp. nov., Nitrosomonas marina sp. nov., Nitrosomonas nitrosa sp. nov., Nitrosomonas eutropha sp. nov., Nitrosomonas oligotropha sp. nov. and Nitrosomonas halophila sp. nov. J Gen Microbiol 137:1689–1699
- Koops H-P, Purkhold U, Pommerening-Röser A, Timmermann G, Wagner M (2006) The lithotrophic ammonia-oxidizing bacteria. The prokaryotes. Springer, New York, pp 778–811
- Kowalchuk GA, Stephen JR (2001) Ammonia-oxidizing bacteria: a model for molecular microbial ecology. Ann Rev Microbiol 55:485–529 [DOI] [PubMed]
- Kowalchuk GA, Stienstra AW, Heilig GHJ, Stephen JR (2000) Changes in the community structure of ammonia-oxidizing bacteria during secondary succession of calcareous grasslands. Environ Microbiol 2:99–110 [DOI] [PubMed]
- Könneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA (2005) Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543–546 [DOI] [PubMed]
- Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163 [DOI] [PubMed]
- Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW, Prosser JI, Schuster SC, Schleper C (2006) Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442:806–809 [DOI] [PubMed]
- McCaig AE, Embley TM, Prosser JI (1994) Molecular analysis of enrichment cultures of marine ammonia oxidisers. FEMS Microbiol Lett 120:363–368 [DOI] [PubMed]
- Molina V, Ulloa O, Farías L, Urrutia H, Ramírez S, Junier P, Witzel K-P (2007) Ammonia-oxidizing β-proteobacteria from the oxygen minimum zone off Northern Chile. Appl Environ Microbiol 73:3547–3555 [DOI] [PMC free article] [PubMed]
- Muyzer G, de Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700 [DOI] [PMC free article] [PubMed]
- Nicol GW, Schleper C (2006) Ammonia-oxidizing Crenarchaeota: important players in the nitrogen cycle? Trends Microbiol 14:207–212 [DOI] [PubMed]
- Purkhold U, Pommerening-Röser A, Juretschko S, Schmid MC, Koops H-P, Wagner M (2000) Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl Environ Microbiol 66:5368–5382 [DOI] [PMC free article] [PubMed]
- Purkhold U, Wagner M, Timmermann G, Pommerening-Röser A, Koops H-P (2003) 16S rRNA and amoA-based phylogeny of 12 novel betaproteobacterial ammonia-oxidizing isolates: extension of the dataset and proposal of a new lineage within the nitrosomonads. Int J Syst Evol Microbiol 53:1485–1494 [DOI] [PubMed]
- Risacher F, Alonso H, Salazar C (1999) Geoquímica de aguas en cuencas cerradas: I, II y III regiones - Chile. Ministerio de Obras Públicas, Dirección General de Aguas, Santiago, Chile
- Risacher F, Alonso H, Salazar C (2003) The origin of brines and salts in Chilean salars: a hydrochemical review. Earth Sci Rev 63:249–293 [DOI]
- Risgaard-Petersen N, Nicolaisen MH, Revsbech NP, Lomstein BA (2004) Competition between ammonia-oxidizing bacteria and benthic microalgae. Appl Environ Microbiol 70:5528–5537 [DOI] [PMC free article] [PubMed]
- Rotthauwe JH, Witzel K-P, Liesack W (1997) The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol 63:4704–4712 [DOI] [PMC free article] [PubMed]
- Schmid M, Twachtmann U, Klein M, Strous M, Juretschko S, Jetten M, Metzger JW, Schleifer KH, Wagner M (2000) Molecular evidence for genus level diversity of bacteria capable of catalyzing anaerobic ammonium oxidation. Syst Appl Microbiol 23:93–106 [DOI] [PubMed]
- Sorokin D, Tourova TJ, Schmid MC, Wagner M, Koops H-P, Kuenen JG, Jetten M (2001) Isolation and properties of obligately chemolithoautotrophic and extremely alkali-tolerant ammonia-oxidizing bacteria from Mongolian soda lakes. Arch Microbiol 176:170–177 [DOI] [PubMed]
- Squeo F, Warner BR, Aravena D, Espinoza D (2006) Bofedales: high altitude peatlands of the central Andes. Rev Chil Hist Nat 79:245–255 [DOI]
- Stackebrandt E, Liesack W (1993) Nucleic acids and classification. In: Goodfellow M, O’Donnell AG (eds) Handbook of new bacterial systematics. Academic Press, London, pp 152–189
- Stehr G, Böttcher B, Dittberner P, Rath G, Koops H-P (1995) The ammonia-oxidizing nitrifying population of the River Elbe estuary. FEMS Microbiol Ecol 17:177–186 [DOI]
- Stephen JR, McCaig AE, Smith Z, Prosser JI, Embley TM (1996) Molecular diversity of soil and marine 16S rRNA gene sequences related to beta-subgroup ammonia-oxidizing bacteria. Appl Environ Microbiol 62:4147–4154 [DOI] [PMC free article] [PubMed]
- Strous M, Kuenen JG, Jetten MSM (1999) Key physiology of anaerobic ammonium oxidation. Appl Environ Microbiol 65:3248–3250 [DOI] [PMC free article] [PubMed]
- Treusch AH, Leininger S, Kletzin A, Schuster SC, Klenk H-P, Schleper C (2005) Novel genes for nitrite reductase and Amo-related proteins indicate a role of uncultivated mesophilic crenarchaeota in nitrogen cycling. Environ Microbiol 7:1985–1995 [DOI] [PubMed]
- Utaker JB, Nes IF (1998) A qualitative evaluation of the published oligonucleotides specific for the 16S rRNA gene sequences of the ammonia-oxidizing bacteria. Syst Appl Microbiol 21:72–88 [DOI] [PubMed]
- Vila I, Mühlhauser HA (1987) Dinámica de lagos de altura, perspectivas de investigación. Arch Biol Med Exp 20:95–103
- Vincent W, Wurtsbaugh W, Vincent C, Richerson PJ (1984) Seasonal dynamics of nutrients limitation in a tropical high-altitude lake (Lake Titicaca, Perú-Bolivia): application of physiological bioassays. Limnol Oceanogr 29:540–552
- Vincent W, Vincent CL, Downes MT, Richerson PJ (1985) Nitrate cycling in Lake Titicaca (Perú-Bolivia): the effects of high-altitude and tropicality. Freshwater Biol 15:31–42 [DOI]
- Voytek MA, Ward BB (1995) Detection of ammonium-oxidizing bacteria of the beta-subclass of the class Proteobacteria in aquatic samples with the PCR. Appl Environ Microbiol 61:1444–1450 [DOI] [PMC free article] [PubMed]
- Ward BB, O’Mullan GD (2002) Worldwide distribution of Nitrosococcus oceani, a marine ammonia-oxidizing γ-proteobacterium, detected by PCR and sequencing of 16S rRNA and amoA genes. Appl Environ Microbiol 68:4153–4157 [DOI] [PMC free article] [PubMed]
- Ward BB, Martino DP, Diaz MC, Joye SB (2000) Analysis of ammonia-oxidizing bacteria from hypersaline Mono Lake, California, on the basis of 16S rRNA sequences. Appl Environ Microbiol 66:2873–2881 [DOI] [PMC free article] [PubMed]
- Webster G, Embley TM, Freitag TE, Smith Z, Prosser JI (2005) Links between ammonia oxidizer species composition, functional diversity and nitrification kinetics in grassland soils. Environ Microbiol 7:676–684 [DOI] [PubMed]
- Wuchter C, Abbas B, Coolen MJL, Herfort L, van Bleijswijk JPT, Strous M, Teira E, Herndl GJ, Middelburg JJ, Schouten S, Sinninghe Damsté JS (2006) Archaeal nitrification in the ocean. Proc Natl Acad Sci USA 103:12317–12322 [DOI] [PMC free article] [PubMed]