Abstract
In plants, the C-repeat binding factors (Cbfs) are believed to regulate low-temperature (LT) tolerance. However, most functional studies of Cbfs have focused on characterizing expression after an LT shock and have not quantified differences associated with variable temperature induction or the rate of response to LT treatment. In the Triticeae, rye (Secale cereale L.) is one of the most LT-tolerant species, and is an excellent model to study and compare Cbf LT induction and expression profiles. Here, we report the isolation of rye Cbf genes (ScCbfs) and compare their expression levels in spring- and winter-habit rye cultivars and their orthologs in two winter-habit wheat (Triticum aestivum L.) and barley (Hordeum vulgare L.) cultivars. Eleven ScCbfs were isolated spanning all four major phylogenetic groups. Nine of the ScCbfs mapped to 5RL and one to chromosome 2R. Cbf expression levels were variable, with stronger expression in winter- versus spring-habit rye cultivars but no clear relationship with cultivar differences in LT, down-stream cold-regulated gene expression and Cbf expression were detected. Some Cbfs were expressed only at warmer acclimation temperatures in all three species and their expression was repressed at the end of an 8-h dark period at warmer temperatures, which may reflect a temperature-dependent, light-regulated diurnal response. Our work indicates that Cbf expression is regulated by complex genotype by time by induction–temperature interactions, emphasizing that sample timing, induction–temperature and light-related factors must receive greater consideration in future studies involving functional characterization of LT-induced genes in cereals.
Electronic supplementary material
The online version of this article (doi:10.1007/s00438-009-0451-9) contains supplementary material, which is available to authorized users.
Keywords: Secale cereale, Rye, Cold acclimation, Cbf, Triticeae, Induction temperature
Introduction
In plants, low-temperature (LT) tolerance is induced upon exposure to low, non-freezing temperatures. This phenomenon, known as cold acclimation, is a cumulative process that is activated once temperatures fall below a critical threshold temperature (Fowler 2008). Once acclimation starts, the differences in genetic potentials are quickly magnified such that genotypes with warmer acclimation induction temperatures respond more rapidly to LT treatment. The cold acclimation process is complex, but involves the induction of cold-regulated (Cor) genes that code for the cryoprotective proteins necessary for protection against LT stress.
It has been established that an important component of cold acclimation is the C-repeat binding factor (Cbf)-induced cold response pathway (Thomashow 2001). The Cbfs are transcriptional activators that belong to the AP2/EREBP transcription factor family and are known to induce Cor transcription by binding to a C-repeat/dehydration-responsive element (CRT/DRE) in the regulatory regions of those genes (Yamaguchi-Shinozaki and Shinozaki 1994). All reported Cbfs lack introns and possess an AP2 DNA-binding domain which is required for Cor gene activation (Jaglo et al. 2001). In all Cbfs, the AP2 DNA-binding domain is flanked by two signature motifs (CMIII-3-PKK/RPAGRxKFxETRHP and CMIII-1-DSAWR). Cbf expression is induced by one or more abiotic stresses (cold, drought and salt) (Xue 2003; Skinner et al. 2005), and in the case of cold, their induction may be regulated in part by an inducer of Cbf expression (ICE) gene (Guo et al. 2002; Chinnusamy et al. 2003).
Large Cbf families are present in cereals and are subdivided into four phylogenetic groups, each with two or more sub-groups (Badawi et al. 2007; Skinner et al. 2005; Miller et al. 2006). In the Triticeae, which includes economically important cereals such as wheat (Triticum spp.), barley (Hordeum vulgare L.) and rye (Secale cereale L.), several Cbfs have been characterized, including 20 from barley (Skinner et al. 2005), 13 from T. monococcum (Miller et al. 2006) and 37 from common wheat (T. aestivum L.) (Badawi et al. 2007). The Cbf subgroups IVb and IVc appear to be unique within the Triticeae tribe (Badawi et al. 2007). Current information indicates that the Cbf genes cluster on the homeologous group 5 chromosomes of the Triticeae, and coincide with quantitative trait loci (QTL) for LT tolerance (Båga et al. 2007; Francia et al. 2004; Tondelli et al. 2006, Miller et al. 2006; Vágújfalvi et al. 2003). However, it is still not clear which Cbf gene(s) most influence LT tolerance or whether they have an additive effect. Recently, Knox et al. (2008) identified a deletion in the AP2 domain of T. monococcumCbf12 that prevented the coded protein from binding to the CRT/DRE motif of Cor genes, and this molecular variation was linked to phenotypic differences in frost survival.
The Triticeae form a homogeneous genetic system and comparative genetic studies suggest that the key components of LT tolerance are conserved among members. However, there is large variation in the ability to survive freezing temperatures, with winter-habit rye cultivars having greater LT tolerance compared to winter common wheat and barley cultivars (Fowler and Carles 1979). Although the genetic factors conferring the superior LT tolerance of rye are not known, acclimation is induced at a higher temperature (Fowler 2008) and the rate of response to cold is more rapid than in wheat and barley (Fowler et al. 1996). This suggests that rye has a superior cold responsive pathway that could be due in part to more rapid induction of one or more Cbf genes at higher acclimation temperatures. However, few rye Cbfs have been sequenced and characterized (Jaglo et al. 2001). In addition most functional studies of Cbfs have characterized Cbf expression after a single LT treatment at a constant acclimation temperature and differences associated with variable temperature induction or the rate of response to LT treatment have not been quantified. To better understand the role of this gene family in determining LT tolerance in the Triticeae, we initiated a study to identify and characterize Cbf genes from the cold hardy rye cultivar “Puma” and to compare Cbf gene expression in rye, wheat and barley cultivars in response to differing acclimation times and induction temperatures.
Materials and methods
Plant material, growth conditions and low-temperature tolerance measurements
For these studies, two winter-habit rye cultivars “Puma” and “Sangaste”, “Gazelle” spring rye, “Norstar” winter-habit wheat and “Kold” winter-habit barley were used. For gene expression studies, plants were grown hydroponically. Imbibed seeds were held in the dark for 2 days at 4°C and then incubated at 25°C for 1 day. Actively germinating seeds were transferred, embryos down, into white light-blocking plastic trays with holes backed by a 1.6-mm mesh screen. Trays were incubated at 25°C for 2 days to allow for further root growth. The seedlings were then placed in hydroponics tanks filled with continuously aerated one-half strength modified Hoagland’s solution in a controlled environment chamber at 20°C with a 16 h light and a photosynthetically active radiation (PAR) of 250 μmol m−2 s−1. Once seedlings had developed two to three fully expanded leaves and visible crowns, trays were transferred to LT-acclimation chambers set at 6, 10, 15 and 18°C (as measured at the crown level) for 2, 4, 8 and 48 h and 16 h light at a PAR of 230 μmol m−2 s−1. The experiment was replicated three times in time and space.
The procedure outlined by Fowler (2008) was used to determine the LT50 of each cultivar acclimated at 6°C for 0, 2 and 49 days. Threshold induction temperatures, defined as the warmest temperature at which plants achieved an LT50 of −3°C after 2 days acclimation, were also determined for each cultivar using the method outlined by Fowler (2008). Briefly, plants were grown hydroponically as outlined above. At each sampling date, five crowns per cultivar were frozen for each of five freezing temperatures previously established for each cultivar (Fowler 2008). The crowns were placed in aluminum weighing cans, covered in moist sand and then loaded into a programmable freezer that was held at −3°C for 12 h. After 12 h, they were cooled at a rate of 2°C h−1 down to −17°C and then cooled at a rate of 8°C h−1 until the established temperatures in each treatment were reached. The crowns were thawed overnight at 3°C and then transplanted into 52 × 26 × 6 cm black plastic trays (Kord Products, Bramalea, ON, Canada) containing “Sunshine” artificial soil medium for re-growth at 20°C with a 16 h day and 8 h night. Plant recovery was rated (alive vs. dead) after 3 weeks and the LT50 was calculated for each treatment within each replicate. The experiment was repeated three times in time and space.
Isolation of Scvrn-1 sequence, Cbf sequences and their 3′ UTR regions
Primers (Supplementary Table Ia, Sigma-Genosys Canada, Oakville, ON, Canada) designed based on wheat TaVRT-1 (AY280870, Danyluk et al. 2003) were used to amplify a 699 bp fragment of its rye ortholog from Puma rye cDNA. First-strand cDNA was synthesised from 5 μg of RNA (extracted from leaf samples of plants cold-acclimated for 70 days at 6°C) using 0.5 μg of poly T primer, and 200 U of M-MLV Reverse Transcriptase (Invitrogen Canada Inc., Burlington, ON, Canada). Subsequently 2 μl of the synthesised cDNA was amplified in a 25 μl PCR reaction volume containing 0.15 μM of each primer, 1 U of Taq DNA polymerase (Invitrogen Canada Inc., Burlington, ON, Canada), 0.2 mM dNTP, 1.5 mM MgCl2 and 5% DMSO. Amplification conditions were as follows: 95°C for 5 min, 35 cycles of 95°C (1 min), the established annealing temperature (1 min) and 72°C (1 min), followed by an extension step at 72°C (15 min). The annealing temperatures for the primers used are listed in supplementary Table Ia. Amplification products were separated at 150 V for 1 h on a 1% (w/v) agarose gel, fragments of the expected size were gel-purified and cloned using pCR®4-TOPO® vector cloning kit (Invitrogen Canada Inc., Burlington, ON, Canada). Five positive clones were identified using colony-PCR and sequenced using DNA sequencing services at Plant Biotechnology Institute-National Research Council, Saskatoon, Saskatchewan, Canada.
Nine primer pairs, designed based on wheat Cbf orthologous sequences, were used to clone 11 rye ScCbfs (Table 2) from Puma. Cbf fragments were amplified from 100 ng of genomic DNA using the PCR conditions indicated above. The primers used and their annealing temperatures and expected fragment lengths are listed (Supplementary Table Ia). All primer sets amplified the complete coding sequences, except those used to amplify ScCbfIVa. Amplification products were separated at 150 V for 1 h on a 1% agarose gel and fragments of the expected size were cloned and sequenced. The 3′ UTR sequence of ScCbfIIId-12 was cloned with a 3′ RACE approach as follows: cDNA was synthesised from 5 μg of RNA (Puma rye, 4 h treated at 6°C) as indicated in the previous section but using 0.5 μg of T17-ADAPTER primer. Subsequently 2 μl of the synthesised cDNA was amplified in a reaction containing a forward gene-specific primer and the ADAPTER reverse primer. The 3′ UTR of the remaining Cbfs were amplified as described above except using reverse primers designed on the 3′ UTR of their wheat orthologs. Partial 3′ UTR sequences of ScCbfIVa-2A and ScCbfIVa-2B were cloned directly from gDNA. Amplification products were run on a 1% agarose gel and fragments of the expected size were cloned and sequenced. Primers, annealing temperatures and expected fragment sizes are listed in Supplementary Table Ib. Sequence alignments, in silico translations, and protein isoelectric point and molecular weight calculations were performed using programs within VectorNTI Advance 10 (Invitrogen Canada Inc., Burlington, ON, Canada. MEGA software version 4 (Tamura et al. 2007) and ClustalW alignment of the nucleotide sequences used to coding for the three conserved domains of Cbfs were used to generate a phylogenetic dendrogram.
Table 2.
Nomenclature and characteristics of rye Cbf genes cloned from the cultivar Puma rye
| Gene name | Genbank accession number | Group | Chromosome location | Sequence length (bp) | Protein length (aa) | pI | MW (kDa) | Best BLAST | Identity (%) |
|---|---|---|---|---|---|---|---|---|---|
| ScCbfIa-11 | EU194240 | Ia | 2R | 614 | 204 | 5.60 | 23.8 | TaCbfIa11 | 92.9 |
| ScCbfII-5 | EU194241 | II | – | 641 | 213 | 6.06 | 23.8 | TaCbfII-5.2 | 86.0 |
| ScCbfIIIa-6 | EU194242 | IIIa | 5RL | 689 | 229 | 4.92 | 25.9 | TaCbfIIIa-D6 | 91.2 |
| ScCbfIIIc-10 | EU194243 | IIIc | 5RL | 666 | 221 | 4.80 | 24.8 | TmCbf10 | 92.4 |
| ScCbfIIIc-3A | EU194244 | IIIc | 5RL | 686 | 228 | 4.57 | 25.7 | TaCbfIIIc-D3 | 94.6 |
| ScCbfIIIc-3B | EU194245 | IIIc | 5RL | 695 | 231 | 4.61 | 25.8 | TaCbfIIIc-D3 | 94.0 |
| ScCbfIIId-12 | EU194246 | IIId | 5RL | 698 | 232 | 5.70 | 26.2 | TaCbfIIId-B12 | 93.7 |
| ScCbfIIId-15 | EU194247 | IIId | 5RL | 662 | 220 | 5.11 | 24.8 | TaCbfIIId-A15 | 93.4 |
| ScCbfIIId-19 | EU194248 | IIId | 5RL | 665 | 221 | 5.13 | 25.2 | TaCbfIIId-D19 | 94.4 |
| ScCbfIVa-2Aa | EU194249 | IVa | 5RL | 605 | >193 | (5.76) | (22.0) | TaCbfIVa-A2 | 94.7 |
| ScCbfIVa-2Ba | EU194250 | IVa | 5RL | 651 | >193 | (5.98) | (22.0) | TaCbfIVa-A2 | 94.3 |
| ScCbfIVb-20b | AF370728 | IVc | 5RL | 850 | 212 | 8.53 | 23.3 | TaCbfIVb-A20 | 91.4 |
| ScCbfIVd-9Ab | AF370729 | IVd | 5RL | 1040 | 268 | 9.47 | 28.9 | TaCbfIVd-B9 | 90.9 |
| ScCbfIVd-9Bb | AF370730 | IVd | 5RL | 1145 | 270 | 9.00 | 29.2 | TaCbfIVd-B9 | 92.8 |
aCloned sequences were truncated at their 5′ ends (lacked the first 78 bp based on their wheat ortholog). Partial protein sequences were used to calculate pI and MW values (in brackets)
bPreviously reported by Jaglo et al. (2001)
All ScCbfsequences isolated in this study were named based on the nomenclature proposed by Badawi et al. (2007). For example, ScCbfIIIc-10 indicates a rye (Sc) Cbf sequence belonging to group/subgroup IIIc that shares the highest homology with wheat ortholog Cbf10. For clarity, we have re-designated the three previously reported ScCbfs (Jaglo et al. 2001) using the same nomenclature such that AF370728, AF370729 and AF370730 are now designated as ScCbfIVb-20, ScCbfIVd-9A and ScCbfIVd-9B, respectively. All sequences presented here have been deposited in the DDBJ/EMBL/GenBank databases and the accession numbers are reported.
Chromosomal localization of Scvrn-1 and ScCbf genes
The chromosomal localization of the isolated ScCbfs were performed using nine Chinese Spring wheat × Imperial rye [CS/IMP-1R, -2R, -3R, -4R, -5R, -5RL (long arm only), -5RS (short arm only), -6R, -7R] disomic addition lines, kindly provided by Dr. J.P. Gustafson (USDA, ARS, University of Missouri). Rye chromosome-specific microsatellite markers (RMS markers, Chebotar et al. 2003; WRM, Bolibok et al. 2006; REMS and GWM, Khlestkina et al. 2004; and SCM, Saal and Wricke 1999) were used to confirm the presence of each single rye chromosome in each wheat/rye addition line -1R (RMS10, SCM9), -2R (SCM75), -3R (RMS28), -4R (WRM216), -5R (SCM138, SCM268), -5RL (RMS115, REMS1205, REMS1218, REMS1237, REMS1264; GWM1059, SCM109, SCM120), -6R (GWM1103, SCM180) and -7R (SCM86) as previously described (Silkova et al.2006). Cleaved amplified polymorphic markers (CAPS) were used to map many of the ScCbfs. The amplification products for ScCbfIIIc-3A/B, ScCbfIIId-15 and ScCbfIVa-2A/B were digested with restriction endonucleases BsrI, XhoI and RsaI, respectively, which cut only within wheat fragments. Amplified fragments of ScCbfIIId-12 and ScCbfIIId-19 were digested with MboI, while the ScCbfII-5 product was digested with TseI. Amplification products for the remaining Cbfs (ScCbfIa-11, ScCbfIIIa-6, ScCbfIIIc-10, ScCbfIVb-20, ScCbfIVd-9A and ScCbfIVd-9B) and Scvrn-1 did not require restriction because the primers were rye specific. Chinese spring and Puma gDNA were used as controls. Primers, restriction enzymes, annealing temperatures, fragment sizes and restriction patterns after digestion are listed (Supplementary Table Ic).
Expression analysis of Cbf and Cor genes
Whole plant samples, excluding the roots, were collected before LT treatment (0 h = control sample) and at five time points (0, 2, 4, 8 and 48 h) after induction of acclimation at 6, 10, 15 and 18°C. The 0 and 48 h tissue samples were collected within 15 min after dawn at the start of a 16 h day. Samples were immediately frozen in liquid nitrogen and stored at −80°C until required for molecular analyses. For each treatment, total RNA was extracted from 100 mg of tissue using TRIZOL® reagent (Invitrogen) following the manufacturer’s instructions. First-strand cDNA synthesis was performed on 5 μg of total RNA, as described above, but a poly T primer was used instead. Semi-quantitative RT-PCRs were performed on cDNA samples using gene-specific primers (sequences, annealing temperatures and expected fragment sizes are listed in Supplementary Table Id). A rye actin fragment was amplified from 1 μl cDNA (Puma rye, 4 h treated at 6°C) using primers designed from a wheat actin sequence (AB181991). A 120 bp amplification product was cloned and sequenced (submitted as GenBank accession number EU525892). The sequenced fragment was 98.3% identical to the AB181991 and was designated as Scactin. All samples were normalized to this gene. Expression analyses were performed by amplifying 1 μl of cDNA for 18–34 cycles, using gene-specific primers and PCR conditions described above except that 5% DMSO (v/v) was used for Cbf amplification. Primer annealing temperatures are listed in Supplementary Table Id.
Results
Low-temperature tolerance
There were significant differences (P < 0.05) in temperatures at which plants began to acclimate (Table 1). At 6°C, all cultivars acclimated rapidly and large differences in LT tolerance were obvious by 2 days. Gazelle spring rye reached maximum LT tolerance after 2 days of acclimation at 6°C, whereas winter-habit genotypes continued to acclimate, with the greatest differences in LT tolerance observed after 49 days. The winter rye cultivar Puma began acclimating at 17°C, compared to 15°C for Sangaste and 14.5°C for Norstar winter wheat. Acclimation in Kold and Gazelle was induced at approximately 10°C (Table 1).
Table 1.
Threshold induction temperature and LT50 (°C) after 0, 2 and 49 days acclimation at 6°C for Kold winter barley, Norstar winter wheat, Puma winter rye, Sangaste winter rye and Gazelle spring rye
| Cultivar | Threshold induction temperature (°C) | Acclimation time (days) | ||
|---|---|---|---|---|
| 0 | 2 | 49 | ||
| Kold | 10.5b | −2.0a | −5.3a | −11.7b |
| Gazelle | 10.0b | −2.0a | −6.3a | −5.0a |
| Sangaste | 15.0c | −2.0a | −7.7ab | −16.5c |
| Norstar | 14.5c | −2.7a | −9.3b | −21.7d |
| Puma | 17.0d | −3.3a | −13.3c | −25.0e |
Within columns, means followed by the same letter are not different as determined by Duncan’s new multiple range test (P < 0.05)
Rye Cbf gene isolation
Prior to conducting expression studies, 11 ScCbf genes were isolated and their characteristics are summarized in Table 2. The isolated 3′ UTR of ScCbf sequences ranged from 23 bp (ScCbfIVa-2A) to 172 bp (ScCbfIa-11) in length (see supplementary information for 3′ UTR sequences) and showed a sequence similarity of 50–100% with their wheat orthologs. The translated proteins ranged from 204 to 232 amino acids in length and shared conserved motifs typical of other Cbfs (Fig. 1), including the AP2 DNA-binding domain flanked by CMIII-3 (PKK/RPAGRxKFxETRHP) and CMIII-1 (DSAWR, Jaglo et al. 2001) and the LWSY sequence near the C-terminal (Dobouzet et al. 2003). The majority of isolated rye sequences from groups I to III coded for proteins with a predicted acidic character, with pI values ranging from 4.57 to 6.06 (Table 2). In contrast, the group IV ScCbfs possessed a predicted basic character, with pI values ranging from 8.53 to 9.47. ScCbfIVa-2A and 2B could not be evaluated for this feature because the complete CDS’ were not isolated.
Fig. 1.
Alignment of the predicted amino acid sequences of the Cbf sequences cloned from the rye cultivar Puma. The Genbank accession numbers of the sequences used in this analysis are listed in Supplementary Table I.e and rye Cbf Genbank accession numbers are reported in Table 2. Designations are as follows: AP2/EREBP domain (over line), signature sequences PKK/RPAGRxKFxETRHP (open circles) and DSAWR (filled circles), conserved C-terminal region LWSY (asterisks)
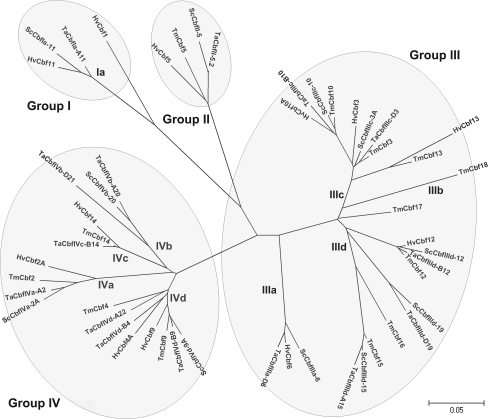
The nucleotide sequences coding for the three conserved domains of the ScCbf genes were aligned with reported hexaploid wheat (Ta, n = 15), barley (Hv, n = 12), diploid wheat (Tm, n = 13) to develop a phylogenetic tree. All of the ScCbfs clustered into one of four phylogenetic groups (Fig. 2). Seven group III ScCbf genes were identified and were 91.2–94.6% identical to their wheat orthologs (Table 2). ScCbfIIIc-3A and ScCbfIIIc-3B were 97.3% identical and likely represent gene duplications when a species cut-off of 95% identity is used to separate homeologous copies and recently duplicated genes. Group IV Cbfs ScCbfIVd-9A and ScCbfIVd-9B had identical protein sequences and likely also represent gene duplications. Most ScCbfs showed greater similarity to their wheat orthologs (Table 2) except for ScCbfIIIc-10 which showed a higher identity with a T. monococcumCbf gene (94.0%). A ScCbf was not found from subgroup IVc, and no rye orthologs to HvCbf1 and HvCbf13 were isolated.
Fig. 2.
Phylogenetic relationship of PoaceaeCbfs isolated from rye (Sc), wheat (Ta), diploid wheat (Tm) and barley (Hv). The nucleotide sequences corresponding to the conserved AP2 domain and the flanking CMIII-3 and CMIII-1 sequences were used to construct the dendrogram. Cbfs belonging to the same group are contoured, while subgroups are indicated near the branch
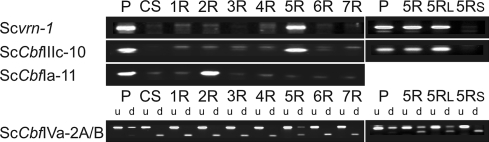
ScCbf genes and Scvrn-1 map on long arm of chromosome 5R
The Chinese Spring/Imperial wheat/rye addition lines (CS/IMP 1R-7R, 5RL and 5RS) were used to determine the chromosomal location of the ScCbfs (Table 2). The vernalization gene Scvrn-1 has been mapped to 5R in rye (Plaschke et al. 1993) and was isolated to confirm the chromosome 5 substitutions. The 699 bp partial cDNA sequence of Scvrn-1 (EU525891) was 97% identical to TaVRT-1 (AY280870, Danyluk et al.2003). Rye-specific primers were designed for Scvrn-1 and localized to 5RL (Fig. 3), confirming the CS/IMP 5R addition. All group III (seven sequences) and IV (five sequences) ScCbfs also mapped to 5RL (data not shown), whereas ScCbfIa-11 mapped to 2R (Fig. 3). It was not possible to assign a chromosomal localization to ScCbfII-5 using the addition lines as the CAPS marker was not polymorphic between Chinese spring (wheat parent) and Imperial (rye parent).
Fig. 3.
Chromosomal localization of Scvrn-1 and three ScCbfs using Chinese Spring (CS) wheat × imperial rye addition lines (1R-7R, 5RL and 5RS). Scvrn-1 is known to map to 5R in rye and was used as a control sequence. The undigested (u) and digested (d) fragments for the CAPS marker used to map ScCbf1Va-2A/B are presented
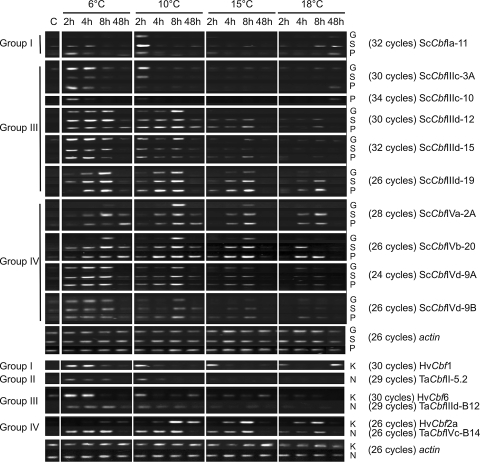
Expression levels of Cbfs genes during cold acclimation
The expression profiles of ten ScCbf genes were evaluated in three rye cultivars at five time points (0, 2, 4, 8 and 48 h) after induction of acclimation at 6, 10, 15 and 18°C (Fig. 4). The expressions of ScCbfIVa-2B and ScCbfIIIc-3B were excluded because primers could not be designed to distinguish them from ScCbfIVa-2A and ScCbfIIIc-3A, respectively. ScCbfII-5 and ScCbfIIIa-6 were tested but their expression levels were below visible detection in all three rye genotypes, regardless of temperature (data not shown).
Fig. 4.
Expression profile of Cbf genes in rye cultivars Gazelle (G), Sangaste (S), Puma(P) and Norstar (N) winter wheat and Kold (K) winter barley in response to varying acclimation temperatures (6, 10, 15 and 18°C) and sampling times [before treatment (C) and after 2, 4, 8 and 48 h]. Actin was used as the reference gene for all cultivars
Comparison of ScCbf expression in Gazelle spring rye with the two winter-habit rye cultivars showed comparable expression of ScCbfIIId-12, ScCbfIIId-19 and ScCbfIVd-9B among the three genotypes at 6 and 10°C, while at higher temperatures the expression level was greater in the winter-habit cultivars (Fig. 4). In contrast, delayed induction of ScCbfIVa-2A and ScCbfIVb-20 was observed in Gazelle, with maximum expression only after 8 h of cold treatment. The expression of ScCbfIIId-15 and ScCbfIVd-9A generally increased as temperature decreased and was similar in all three rye cultivars. ScCbfIIIc-10 was detectable at low levels only in Puma and after 2 h at 6°C (data not shown for Sangaste and Gazelle). The expression of ScCbfIVb-20 was pronounced after 8 h at 18°C in Puma compared to no detectable expression in the less hardy cultivar Sangaste. ScCbfIIId-19, ScCbfIVa-2A and ScCbfIVb-20 were the only genes that were expressed at high levels at 18°C, but only at 4 and 8 h after treatment and only in the winter cultivars. In the winter-habit rye cultivars, most of the group III and group IV ScCbfs showed strong initial LT induction that peaked by 8 h and then decreased at 48 h, which was the end of an 8-h dark period.
A subset of Cbfs belonging to groups I, II, III and IV were evaluated in winter wheat (Norstar) and barley (Kold) cultivars (Fig. 4) for comparison with rye to determine if variability in expression could be associated with variation in LT induction among species. Interactions among Cbf genes, species and induction temperature were evident and the results are summarized in Table 3. In barley, HvCbf1 showed a stronger expression level than ScCbfIa-11 after exposure at 6°C, while at 10°C the expression of Sangaste was comparable to that of Kold. TaCbfII-5.2 was detectable in Norstar (Fig. 4), while expression of its ortholog ScCbfII-5 was not detectable in any of the rye cultivars. In winter wheat and rye cultivars, the group III Cbfs showed prolonged expression until 8 h, while in barley HvCbf6 transcript levels started to decrease after 4 h (6°C) or 2 h (10°C). TaCbfIIId-B12 and HvCbf6 showed weaker expression than the Group III ScCbfs that were characterized. The expression levels of ScCbfIVa-2A, TaCbfIVc-B14 and HvCbf2A were comparable in the winter species, while the spring cultivar Gazelle rye showed delayed induction and reduced expression (Fig. 4).
Table 3.
Comparison of Cbf expression levels among different rye cultivar and different species (wheat, barley, rye)
| Rye Cbfs | Comparison among rye cultivars | Comparison among species | Wheat/barley Cbfs | |
|---|---|---|---|---|
| Group I | ScCbfIa-11 | Higher expression in winter cv | Higher expression in barley | HvCbf1 |
| Group II | ScCbfII-5 | Not detectable | Detectable only in wheat | TaCbfII-5.2 |
| Group III | ScCbfIIIa-6 | Not detectable | Higher expression in rye | TaCbfIIId-B12/HvCbf6 |
| ScCbfIIIc-3A | Higher expression in spring Gazelle | |||
| ScCbfIIIc-10 | Detectable only in winter Puma | |||
| ScCbfIIId-12 | Comparable expression level at 6–10°C, higher expression level at 15–18°C | |||
| ScCbfIIId-15 | Comparable expression level | |||
| ScCbfIIId-19 | Comparable expression level at 6–10°C, higher expression level at 15–18°C | |||
| Group IV | ScCbfIVa-2A | Higher expression in winter cv | Comparable | TaCbfIVc-B14/HvCbf2a |
| ScCbfIVb-20 | Higher expression in winter Puma | |||
| ScCbfIVd-9A | Comparable expression level | |||
| ScCbfIVd-9B | Comparable expression level at 6–10°C, higher expression level at 15–18°C |
The Genbank accession numbers of genes presented in the table are listed in Table 2 (rye) and Supplementary table I.e (wheat/barley)
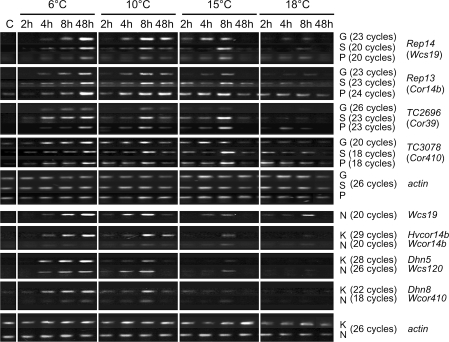
Induction of Cor genes during cold acclimation
Expression levels of Cor genes were profiled in rye, wheat and barley cultivars to compare the cold induction response of genes downstream from the Cbfs (Fig. 5). Under our test conditions, Cor genes were up-regulated slightly after 4 h of LT exposure and expression levels were maintained until 48 h at 6°C, but gradually decreased by 48 h, particularly at 15 and 18°C (Fig. 5). Rye Cor gene Rep14 (AF491840; Ndong et al. 2002) is orthologous to wheat wcs19 (Chauvin et al. 1993) and showed similar induction levels among spring and winter rye cultivars. However, higher expression was observed in Norstar, especially at 18°C. Expression of the rye Cor gene Rep13 (AF491839; Ndong et al. 2002), which is orthologs to wheat Wcor14b (Tsvetanov et al. 2000) and barley Hvcor14b (Dal Bosco et al. 2003), was higher in Puma at all acclimation temperatures while expression levels were similar in Gazelle and Kold (Fig. 5). The two reported rye ESTs, TC2696 and TC3078, were annotated, respectively, as wheat Cor39 and wheat/barley Cor410/Dhn5 homologues (Close et al. 1995; Danyluk et al. 1994) and expression of these was induced at higher acclimation temperatures in the winter cultivars Puma and Sangaste compared to Gazelle spring rye. Wheat Wcs120 (M93342; Houde et al. 1992) and barley Dhn8 (AF043093) expression was similar in Norstar and Kold.
Fig. 5.
Expression profile of Cor genes in rye cultivars Gazelle (G), Sangaste (S), Puma(P) and Norstar (N) winter wheat and Kold (K) winter barley in response to varying acclimation temperatures (6, 10, 15 and 18°C) and sampling times [before treatment (C) and after 2, 4, 8 and 48 h]. The Genbank accession numbers of genes used in this analysis are: Rep14 (AF491840), Rep13 (AF491839), Cor39hom (TC2696), Cor410hom (TC3078), Wcs19 (L13437), Hvcor14b (AJ512944), Wcor14b (AF207546), Dhn5 (M95810), Wcs120 (M93342), Dhn8 (AF181458), Wcor410 (L29152). Samples were normalized with wheat actin-specific primers (AB181991)
Discussion
Cbf gene isolation and chromosomal location
Plant cold acclimation is a complex process and has been studied extensively in Arabidopsis and in commercially important cereal species such as wheat and barley. The Cbf transcription factors play a central role in plant response to cold stress, and in the present work, 11 Cbf genes were cloned and characterized from the LT-tolerant rye cultivar Puma and their expression levels were compared in rye cultivars with different growth habits, acclimation thresholds and LT-tolerance genetic potentials (Table 1). The majority of ScCbfs were mapped to 5RL, which is consistent with reports for wheat and barley where clusters of Cbf genes have been localized to the LT-tolerance QTL on the group 5 chromosomes. ScCbf1a-11 was localized to 2R and is orthologous to TaCbf1a-11 which has been mapped to 2AL in common wheat (Badawi et al. 2007). We were unable to identify a chromosomal location for ScCbfII-5, probably because the single nucleotide polymorphism used to generate the TseI CAPS marker for mapping was specific to the Norstar sequence (EF028752, EF028753, EF028754) and not present in the sequence of Chinese spring, the wheat parent of the Imperial line substitution series. However, the T. monococcum and barley orthologs have been mapped on 7Am and 7HS, respectively (Miller et al. 2006; Skinner et al. 2005).
Alignment of the coding sequence for the AP2 and Cbf signature-conserved domains allowed us to assign ScCbfs to different phylogenetic groups (Fig. 2). Groups I and II contain a reduced number of sequences compared to other Cbf groups. Group I is the most ancient monocot Cbf group and it shares the highest homology with dicotyledonous Cbfs (Badawi et al. 2007). Despite repeated attempts, we could not identify a rye ortholog of HvCbf1. Perhaps this gene does not exist in rye as it also appears to be absent in common wheat and T. monococcum. The absence of this Cbf in wheat and rye suggests that it does not play a critical role in determining LT tolerance in the Triticeae. Seven out of 11 of the rye Cbfs sequences were assigned to Cbf group III (Fig. 2). However, no genes belonging to subgroup IIIb were found in rye, which is also the case in wheat and barley. To date, the only subgroup IIIb Cbf reported in Pooideae is TmCbf18 identified in T. monococcum. The remaining five ScCbfs genes were assigned to group IV, but we did not identify a rye orthologs of TaCbfIVb-D21, TaCbfIVc-B14 and TaCbIVd-A22, so continued effort is warranted to determine if these are present in rye. In addition, orthologs of barley and T. monococcumCbf13, Cbf16 and Cbf17 have yet to be identified in rye and common wheat.
Low-temperature induction and expression of Cbfs and Cor genes during cold acclimation
Rye is very responsive to temperatures in the acclimation range, generating rapid increases in LT acclimation and high levels of expression of LT-associated genes (Fowler et al. 1996). In this study, Puma winter rye displayed the best LT tolerance followed by Norstar winter wheat, Sangaste winter rye and Kold winter barley (Table 1), which is consistent with earlier reports (Fowler and Carles 1979). Once acclimation started, the differences in LT-tolerance genetic potentials were quickly magnified such that cultivars with acclimation induced at warmer temperatures were the most cold hardy. While the most rapid changes in freezing tolerance occurred during the initial stages of acclimation, the largest differences among cultivars were observed after 49 days of acclimation. These observations are consistent with our hypothesis that cold-sensing mechanisms and responses in the early stages of acclimation play a critical role in determining the genetic potential for LT tolerance (Fowler 2008). Full expression of cold hardiness genes only occurs in the vegetative stage (Limin and Fowler 2006) and the inability of Gazelle spring rye to continue accumulating LT tolerance after an early burst is consistent with the theory that the delayed vegetative/reproductive transition allows winter-habit genotypes to be more responsive to extended periods of acclimation (Fowler et al. 1999).
We compared the expression of several rye, wheat and barley Cbfs to assess their rate of response to varying acclimation temperatures. We found that the group I CbfScCbfIa-11 had a very low-induction level after cold exposure and we were unable to detect mRNA accumulation for group II ScCbfII-5 under our test conditions. This was not surprising because Cbfs are induced not only by cold, but also by drought and osmotic stress (Haake et al. 2002; Dobouzet et al. 2003). Skinner et al. (2005) found that HvCbf1 was the only Cbf strongly induced after drought exposure in barley, and in this study, its expression was induced following 2 h at lower acclimation temperatures, but only after 48 h at 18°C. In general, the expression patterns of most of group III and IV Cbfs were more closely associated with the threshold induction temperatures than the other groups (Table 1; Fig. 4). In particular, the group III Cbfs where induced at higher temperatures and had prolonged expression at 6°C in the most cold tolerant winter rye cultivars. However, this was not true of all group III subgroups because Cbf subgroups IIIa and IIIc were either not expressed (IIIa-6) or had low-expression levels at warmer temperatures (IIIc-3A, IIIc-10) in rye. TaCbfIIId-B12 was also expressed at very low levels at all temperatures (Fig. 4), despite high expression of LT tolerance in Norstar (Table 1). However, expression of TaCbfIVc-B14 was high at 6 and 10°C in Norstar, with reduced expression at 18°C, which reflects the acclimation induction temperature of that cultivar. These observed differences in expression levels among the Cbf genes could be due to different upstream regulatory factors that monitor temperature, or to mutations within a common regulatory factor controlling gene response.
Once the threshold induction temperature for cereals is reached, there is an inverse relationship between rate of LT acclimation and exposure temperature (Fowler et al. 1999). There are also genotypic differences in the threshold induction temperature (Fowler 2008) with the result that the warmer the induction temperature, the sooner LT tolerance starts to accumulate. If Cbfs are involved in the regulation of plant response to LT, their expression should also be delayed in cultivars with colder induction temperatures and the level of expression should be higher in hardier genotypes. However, we found that differences in Cbf expression did not necessarily correspond to the acclimation response of the cultivars considered in this study. After 2 days acclimation at 6°C, the LT50 of Gazelle and Sangaste were similar and significantly warmer than Puma (Table 1), but the threshold responses of the Cbfs were similar for Sangaste and Puma and generally much warmer than Gazelle (Fig. 4). Furthermore, ScCbf genes belonging to the same subgroups showed similar expression in the two winter rye cultivars evaluated despite the large difference in expression of LT tolerance at all three acclimation temperatures (Table 1). These results suggest that Cbf expression is being dictated to a greater extent by the winter/spring growth habit than by genetic differences in cold hardiness potential.
Interestingly, for those rye Cbfs expressed at higher induction temperatures (15 and 18°C), expression was strongly repressed at 48 h, just as plants came out of the dark. This was also true for rye ScCbfIVd-9A and TaCbfIVc-B14, where expression at 6°C was repressed at 48 h. These results suggest a diurnal fluctuation in Cbf expression in grasses, particularly at higher induction temperatures. These observations are consistent with results in Arabidopsis, where endogenous circadian rhythms have been reported to regulate Cbf transcripts at higher temperatures (Edwards et al. 2006). In contrast, expression of several Cbfs at 48 h was noted at 6 and 10°C, suggesting that their turnover rate is temperature-dependent or induction of these Cbfs at low temperatures is not gated by the diurnal response, as is the case in Arabidopsis (Fowler et al. 2005).
Although there is a rapidly expanding pool of information on Cbf sequencing, mapping and expression analyses in cereals, only TmCbf-12 and TmCbf-14 have been correlated with variation in LT tolerance in T. monococcum (Knox et al. 2008). In this study the expression patterns of these Cbfs were variable, with complex genotype by time by induction–temperature interactions (Fig. 4). In barley, the CbfIIId-12 ortholog (HvCbf6) was highly expressed at 2 and 4 h after treatment at 6°C, with variable expression as temperature increased. In contrast, except for the 48 h sampling time, the rye ortholog was strongly expressed at 15°C in winter-habit cultivars. In Norstar, TaCbfIIId-B12 was expressed after 2, 4 and 8 h exposure to 6°C and decreased dramatically at 15°C (Fig. 5). Wcs120 expression in wheat is highly correlated with LT50 (Limin et al. 1997) and Wcs120 expression more closely followed CbfIIId-B12 than CbfIVc-14 expression. Puma began to acclimate at temperatures as high as 17°C, and only the Cor14b ortholog was expressed in Puma at temperatures greater than 15°C (Fig. 5). However, accumulation of the Cor14b ortholog in Puma was not correlated to ScCbfIIId-12 or CbfIVc-14 expression. These observations show that Cbf gene(s) have different temperature thresholds and that the interaction of several different Cbf gene(s) and/or regulatory factors may determine Cor gene expression and the superior LT tolerance in rye compared to wheat and barley.
Conclusions
We analyzed the expression of Cbfs in rye, the most cold tolerant cereal species, and found LT acclimation induction of winter and spring rye cultivars was most associated with the expression levels of Cbf sequences belonging to groups III and IV. However, none of the Cbfs considered adequately explained the differences in response to LT temperature tolerance observed in the cultivars examined, and Cbf expression was dictated to a greater extent by winter/spring growth habit. Comparison of Cbf and Cor gene expressions among wheat, barley and rye revealed complex threshold induction temperature by time by LT-induced gene interactions indicating that expression of Cbf and Cor genes is staged at different temperatures and times during the day, making it difficult to determine cause-and-effect relationships. Our results also suggest that Cbf expression in cereals is regulated not only by induction temperature, but also by diurnal rhythm, particularly at higher temperatures. To date, most Cbfs expression studies in cereals have focused on sampling at a single time point, usually after treatment with a single LT shock. These methods do not adequately address the variable time/temperature and rate of response that we show to be critical in determining LT tolerance (Table 1). Our results indicate that sample timing, induction temperature and light-related factors must be considered in future studies involving functional characterization of LT-induced genes in cereals.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table Ia: List of primers used to clone Scvrn-1 and ScCbfs (XLS 47 kb)
Acknowledgments
Appreciation is expressed to our technical staff for their assistance in growing and collecting plant material, conducting LT-tolerance evaluations (Garcia Schellhorn, Twyla Krug and Kendra Smith) and cloning/chromosomal localization studies (Akiko Tomita). Kathleen Ross (USDA-ARS; Columbia, Missouri) provided the Chinese Spring/Imperial wheat/rye addition lines. Seed of Sangaste rye was provided by Külli Annamaa, Jõgeva Plant Breeding Institute, Estonia. Financial support for this research was provided by Genome Prairie/Genome Canada (C. J. P. and D. B. F.), and Saskatchewan Agriculture, Food and Rural Revitalization (C. J. P.). C. C. was supported by a Ph.D. internship from Genome Prairie and funding from the Centre for Genomic Research, Italy (L. C.) and Ducks Unlimited Canada (D. B. F.).
Abbreviations
- PAR
Photosynthetically active radiation
- LT
Low temperature
- Cbfs
C-repeat/dehydration-responsive binding factors
- Cor
(Cold regulated) genes
- ICE1
Inducer of Cbf expression 1
- VRN
Vernalization
- Fr
Frost resistance
- LT50 value
Lethal temperature for 50% of the plants
- CDS
Complete coding sequence
Footnotes
Nucleotide sequence data reported are available in the DDBJ/EMBL/GenBank databases under the following accession numbers: EU194240 (ScCbfIa-11), EU194241 (ScCbfII-5), EU194242 (ScCbfIIIa-6), EU194243 (ScCbfIIIc-10), EU194244 (ScCbfIIIc-3A), EU194245 (ScCbfIIIc-3B), EU194246 (ScCbfIIId-12), EU194247 (ScCbfIIId-15), EU194248 (ScCbfIIId-19), EU194249 (ScCbfIVa-2A), EU194250 (ScCbfIVa-2B), EU525891 (ScVrn-1), EU525892 (ScActin).
Electronic supplementary material
The online version of this article (doi:10.1007/s00438-009-0451-9) contains supplementary material, which is available to authorized users.
Contributor Information
Chiara Campoli, Email: campoli@mpiz-koeln.mpg.de.
Curtis J. Pozniak, Phone: +1-306-9662361, FAX: +1-306-9665015, Email: curtis.pozniak@usask.ca
References
- Badawi M, Danyluk J, Boucho B, Houde M, Sarhan F (2007) The CBF gene family in hexaploid wheat and its relationship to the phylogenetic complex of cereal CBFs. Mol Gen Genomics 277:533–554 [DOI] [PMC free article] [PubMed]
- Båga M, Chodaparambil SV, Limin AE, Pecar M, Fowler DB, Chibbar RN (2007) Identification of quantitative trait loci and associated candidate genes for low-temperature tolerance in cold-hardy winter wheat. Funct Integr Genomics 7:53–68 [DOI] [PubMed]
- Bolibok H, Rakoczy-Trojanowska M, Wyrzykowska M, Radecka M, Orczyk W (2006) Identification of microsatellite markers in the rye genome. Cell Mol Biol Lett 11:291–298 [DOI] [PMC free article] [PubMed]
- Chauvin LP, Houde M, Sarhan F (1993) A leaf-specific gene stimulated by light during wheat acclimation to low temperature. Plant Mol Biol 23:255–265 [DOI] [PubMed]
- Chebotar S, Roder MS, Korzun V, Saal B, Weber WE, Borner A (2003) Molecular studies on genetic integrity of open-pollinating species rye (Secale cereale L.) after long-term genebank maintenance. Theor Appl Genet 107:1469–1476 [DOI] [PubMed]
- Chinnusamy V, Ohta M, Kanrar S, Lee B-H, Hong X, Agarwal M, Zhu J-K (2003) ICE1: a regulator of cold induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev 17:1043–1054 [DOI] [PMC free article] [PubMed]
- Close TJ, Meyer NC, Radik J (1995) Nucleotide sequence of a gene encoding a 58.5-Kilodalton barley dehydrin that lacks a serine tract. Plant Physiol 107:289–290 [DOI] [PMC free article] [PubMed]
- Dal Bosco C, Busconi M, Govoni C, Baldi P, Stanca AM, Crosatti C, Bassi R, Cattivelli L (2003) cor Gene expression in barley mutants affected in chloroplast development and photosynthetic electron transport. Plant Physiol 131(2):793–802 [DOI] [PMC free article] [PubMed]
- Danyluk J, Houde M, Rassart E, Sarhan F (1994) Differential expression of a gene encoding an acidic dehydrin in chilling sensitive and freezing tolerant gramineae species. FEBS Lett 344:20–24 [DOI] [PubMed]
- Danyluk J, Kane NA, Breton G, Limin AE, Fowler DB, Sarhan F (2003) TaVRT-1, a putative transcription factor associated with vegetative to reproductive transition in cereals. Plant Physiol 132:1849–1860 [DOI] [PMC free article] [PubMed]
- Dobouzet JG, Sakuma Y, Ito Y, Kasuga M, Dobouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Os-DREB genes in rice, Oriza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J 33:751–763 [DOI] [PubMed]
- Edwards KD, Anderson PE, Hall A, Salathia NS, Locke JCW, Lynn JR, Straume M, Smith JQ, Millar AJ (2006) FLOWERING LOCUS C mediated natural variation in the high-temperature response of the Arabidopsis circadian clock. Plant Cell 18:639–650 [DOI] [PMC free article] [PubMed]
- Fowler DB (2008) Cold acclimation threshold induction temperatures in cereals. Crop Sci 48:1147–1154 [DOI]
- Fowler DB, Carles RJ (1979) Growth, development, and cold tolerance of fall-acclimated cereal grains. Crop Sci 19:915–922
- Fowler DB, Chauvin LP, Limin AE, Sarhan F (1996) The regulatory role of vernalization in the expression of low temperature-induced genes in wheat and rye. Theor Appl Genet 93:554–559 [DOI] [PubMed]
- Fowler DB, Limin AE, Ritchie JT (1999) Low-temperature tolerance in cereals: model and genetic interpretation. Crop Sci 39:626–633
- Fowler SG, Cook D, Thomashow MF (2005) Low temperature induction of Arabidopsis CBF1, 2, and 3 is gated by the circadian clock. Plant Physiol 137:961–968 [DOI] [PMC free article] [PubMed]
- Francia E, Rizza F, Cattivelli L, Stanca AM, Galiba G, Toth B, Hayes PM, Skinner JS, Pecchioni N (2004) Two loci on chromosome 5H determine low-temperature tolerance in a ‘Nure’(winter) × ‘Tremois’ (spring) barley map. Theor Appl Genet 108:670–680 [DOI] [PubMed]
- Guo Y, Xiong L, Ishitani M, Zhu J-K (2002) An Arabidopsis mutation in translation elongation factor 2 causes superinduction of CBF/DREB1 transcription facto genes but blocks the induction of their downstream targets under low temperature. Proc Natl Acad Sci USA 99:7786–7791 [DOI] [PMC free article] [PubMed]
- Haake V, Cook D, Riechmann JL, Pineda O, Thomashow MF, Zhang JZ (2002) Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol 130:639–648 [DOI] [PMC free article] [PubMed]
- Houde M, Danyluk J, Laliberte JF, Rassart E, Dhindsa RS, Sarhan F (1992) Cloning, characterization, and expression of a cDNA encoding a 50-Kilodalton protein specifically induced by cold acclimation in wheat. Plant Physiol 99:1381–1387 [DOI] [PMC free article] [PubMed]
- Jaglo KR, Kleff S, Amundsen KL, Zhang X, Haake V, Zhang JZ, Deits T, Thomashow MF (2001) Components of the Arabidopsis C-repeat/dehydration-responsive element binding factor cold-response pathway are conserved in Brassica napus and other plant species. Plant Physiol 127:910–917 [DOI] [PMC free article] [PubMed]
- Khlestkina EK, Myint-Than MH, Pestsova EG, Roder MS, Malyshev SV, Korzun V, Corner A (2004) Mapping of 99 new microsatellites-derived loci in rye (Secale cereale L.) including 39 expressed sequence tags. Theor Appl Genet 109:725–732 [DOI] [PubMed]
- Knox AK, Li CX, Vágújfalvi A, Galiba G, Stockinger EJ, Dubcovsky J (2008) Identification of candidate CBF genes for the frost tolerance locus Fr-A(m)2. Plant Mol Biol 67:257–270 [DOI] [PubMed]
- Limin AE, Fowler DB (2006) Low-temperature tolerance genetic potential in wheat (Triticum aestivum L.): response to photoperiod, vernalization, and plant development. Planta 224:139–146 [DOI] [PubMed]
- Limin AE, Danyluk J, Chauvin LP, Fowler DB, Sarhan F (1997) Chromosome mapping of low-temperature induced Wcs120 family genes and regulation of cold-tolerance expression in wheat. Mol Gen Genet 253:720–727 [DOI] [PubMed]
- Miller AK, Galiba G, Dubcovsky J (2006) A cluster of 11 CBF transcription factors is located at the frost tolerance locus Fr-Am2 in Triticum monococcum. Mol Gen Genomics 275:193–203 [DOI] [PubMed]
- Ndong C, Danyluk J, Wilson KE, Pocock T, Huner NP, Sarhan F (2002) Cold-regulated cereal chloroplast late embryogenesis abundant-like proteins. Molecular characterization and functional analyses. Plant Physiol 129(3):1368–1381 [DOI] [PMC free article] [PubMed]
- Plaschke J, Börner A, Xie DX, Koebner RMD, Schlegel R, Gale MD (1993) RFLP mapping of genes affecting plant eight and growth habit in rye. Theor Appl Genet 85:1049–1054 [DOI] [PubMed]
- Saal B, Wricke G (1999) Development of simple sequence repeat markers in rye (Secale cereale L.). Genome 42:964–972 [DOI] [PubMed]
- Silkova OG, Dobrovolskaya OB, Dubovets NI, Adonina IG, Kravtsova LA, Roeder MS, Salina EA, Shchapova AI, Shumny VK (2006) Production of wheat-rye substitution lines and identification of chromosome composition of karyotypes using C-banding, GISH, and SSR markers. Russ J Genet 42:645–653 [DOI] [PubMed]
- Skinner JS, Zitzewitz J, Szucs P, Marquez-Cedillo L, Filichkin T, Amundsen K, Stockinger EJ, Thomashow MF, Chen THH, Hayes PM (2005) Structural, functional, and phylogenetic characterization of a large CBF gene family in barley. Plant Mol Biol 59:533–551 [DOI] [PubMed]
- Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599 [DOI] [PubMed]
- Thomashow MF (2001) So what's new in the field of plant cold acclimation? Lots! Plant Physiol 125:89–93 [DOI] [PMC free article] [PubMed]
- Tondelli A, Francia E, Barabaschi D, Aprile A, Skinner JS, Stockinger EJ, Stanca AM, Pecchioni N (2006) Mapping regulatory genes as candidates for cold and drought stress tolerance in barley. Theor Appl Genet 112:445–454 [DOI] [PubMed]
- Tsvetanov S, Ohno R, Tsuda K, Takumi S, Mori N, Atanassov A, Nakamura C (2000) A cold-responsive wheat (Triticum aestivum L.) gene wcor14 identified in a winter-hardy cultivar ‘Mironovska 808’. Genes Genet Syst 75(1):49–57 [DOI] [PubMed]
- Vágújfalvi A, Galiba G, Cattivelli L, Dubcovsky J (2003) The cold-regulated transcriptional activator Cbf3 is linked to the frost-tolerance locus Fr-A2 on wheat chromosome 5A. Mol Gen Genomics 269:60–67 [DOI] [PMC free article] [PubMed]
- Xue GP (2003) The DNA binding activity of an AP2 transcriptional activator HvCBF2 involved in regulation of low-temperature responsive genes in barley is modulated by temperature. Plant J 33:373–383 [DOI] [PubMed]
- Yamaguchi-Shinozaki K, Shinozaki K (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low temperature, or high salt stress. Plant Cell 6:251–264 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the electronic supplementary material.
Table Ia: List of primers used to clone Scvrn-1 and ScCbfs (XLS 47 kb)