Abstract
We describe the construction of a digital brain atlas composed of data from manually delineated MRI data. A total of 56 structures were labeled in MRI of 40 healthy, normal volunteers. This labeling was performed according to a set of protocols developed for this project. Pairs of raters were assigned to each structure and trained on the protocol for that structure. Each rater pair was tested for concordance on 6 of the 40 brains; once they had achieved reliability standards, they divided the task of delineating the remaining 34 brains. The data were then spatially normalized to well-known templates using 3 popular algorithms: AIR5.2.5’s nonlinear warp (Woods et al., 1998) paired with the ICBM452 Warp 5 atlas (Rex et al., 2003), FSL’s FLIRT (Smith et al., 2004) was paired with its own template, a skull-stripped version of the ICBM152 T1 average; and SPM5’s unified segmentation method (Ashburner and Friston, 2005) was paired with its canonical brain, the whole head ICBM152 T1 average. We thus produced 3 variants of our atlas, where each was constructed from 40 representative samples of a data processing stream that one might use for analysis. For each normalization algorithm, the individual structure delineations were then resampled according to the computed transformations. We next computed averages at each voxel location to estimate the probability of that voxel belonging to each of the 56 structures. Each version of the atlas contains, for every voxel, probability densities for each region, thus providing a resource for automated probabilistic labeling of external data types registered into standard spaces; we also computed average intensity images and tissue density maps based on the three methods and target spaces. These atlases will serve as a resource for diverse applications including meta-analysis of functional and structural imaging data and other bioinformatics applications where display of arbitrary labels in probabilistically defined anatomic space will facilitate both knowledgebase development and visualization of findings from multiple disciplines.
Keywords: probabilistic atlas, brain mapping, neuroanatomy, brain, MRI
1 Introduction
Atlases of human neuroanatomy play important roles in the interpretation of results, in the visualization of information, and in the processing of data. Beginning with the detailed drawings of brain structures produced during the Renaissance by Vesalius, numerous paper atlases comprising collections of neuroanatomical illustrations, photographs, and other imagery have been constructed (see Toga and Mazziotta (2002) for a review). As technology advanced, digital atlases extended these efforts by providing interactive collections of brain data. Most of these brain atlases were based on single subjects or on very limited numbers of individuals. These included the Voxel-Man atlas (Höhne et al., 1992; Tiede et al., 1993; Schiemann et al., 2000), the Karolinska Brain Atlas project (Greitz et al., 1991), the Human Brain Atlas (Roland et al., 1994), the Digital Anatomist project (Eno et al., 1991; Brinkley et al., 1997), the Harvard Brain Atlas (Kikinis et al., 1996), and the Visible Human Project (Spitzer et al., 1996). The focus on individual subjects allowed considerable effort to be spent detailing the neuroanatomy or acquiring multiple types of data from the subjects. However, single subject atlases cannot describe the variability in brain structure that is inherent across the human population. To capture this information, larger numbers of subjects must be examined.
One of the early descriptions of a multisubject atlas was presented by Mazziotta et al. (1995), who proposed the development of a comprehensive probabilistic brain atlas under the banner of the International Consortium for Brain Mapping (ICBM). This project has collected data from 5,300 subjects, including images of the brain using various magnetic resonance imaging (MRI) modalities, genetic material, and demographic information (Mazziotta et al., 2001). Multi-subject studies such as the ICBM project require methods that can bring the image data from different subjects into a common coordinate frame; numerous research efforts have been made to meet these demands (e.g., Bajcsy et al., 1983; Gee et al., 1993; Woods et al., 1993, 1998; Collins et al., 1994; Davatzikos et al., 1996; Wells et al., 1996; Thompson and Toga, 1996; Christensen et al., 1997; Ashburner and Friston, 1997; Ashburner et al., 1999; Fischl et al., 1999; Jenkinson and Smith, 2001; Jenkinson et al., 2002; Johnson and Christensen, 2002; Shen and Davatzikos, 2002; Xue et al., 2006). These techniques extended the method introduced by Talairach and his colleagues (Talairach et al., 1967; Talairach and Tournoux, 1988) for mathematically mapping an individual subject brain to an atlas.
Evans et al. (1993) applied registration methods (Collins et al., 1994) to produce a probabilistic map of MRI data acquired from 305 subjects. The MRI volumes were co-registered using a nine-parameter linear transformation and then averaged at each voxel to produce an average intensity atlas, termed MNI-305. A second atlas was produced by registering a subset of 152 brains to the MNI-305 atlas, again using a 9-parameter linear transformation, to generate the ICBM152 atlas. These templates provide a registration target for various analysis tools, such as SPM (Ashburner et al., 1999) and FSL (Smith et al., 2004), both of which use versions of the ICBM152 atlas as an anatomical reference. ICBM also produced an average intensity atlas from 452 subjects using affine registration, and a second atlas was produced using 5th order polynomial warps that improved cortical definition (Rex et al., 2003). Though affine alignment can bring many subcortical structures into alignment, regions of the brain vary substantially across subjects. Because of this, structures such as cortex are often blurry in average intensity volumetric atlases. This may occur even when non-linear registration methods are applied, as seen in Fig. 1a. Thus, it may be difficult to interpret location within an intensity averaged atlas.
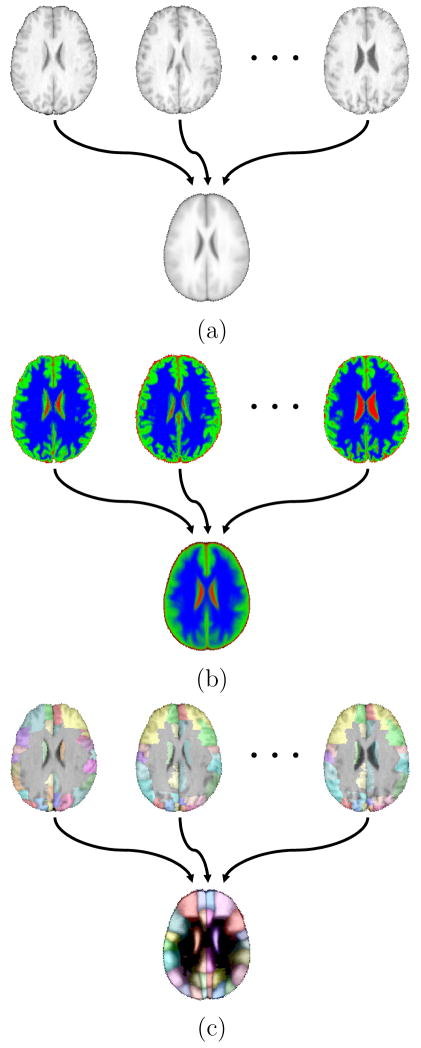
Fig. 1.
Examples of probabilistic atlases. (a) An average intensity atlas computed from co-registered MRI data (b) A tissue-type probability atlas computed from co-registered tissue fraction maps produced from MRI. Each voxel was partitioned into grey matter, white matter, or CSF components in the original volumes (shown here in green, blue, or red, respectively). The values are transformed into the common atlas space, then used to compute the likelihood of each voxel in the atlas space containing the different tissue types. (c) A probabilistic structure atlas computed from co-registered cortical structure delineations. The delineations (see Table 1 for color index) are mapped into the atlas space, then used to create density maps for each structure in the volume. The figure shows a maximum likelihood labeling generated from a probabilistic structure atlas.
One way to address this problem is to create a multi-subject label atlas, in which voxel identifiers from the original subjects are transformed into the atlas space. Statistics of these transformed labels can then be computed at each voxel location to provide probabilistic information about the structures in that atlas. Examples include maps of white matter (WM), grey matter (GM), and cerebrospinal fluid (CSF) (see Fig. 1b) or individual brain structures such as as gyri or nuclei (see Fig. 1c). Probabilistic atlases can also be generated based on deformation maps (e.g., Thompson et al., 2004; Xue et al., 2006). Deformation-map methods analyze the transformations required to bring the subjects into alignment in order to analyze the variation of structures. Measures computed on the deformation processes can then be used to describe the variation seen in the volume or surface models for the studied subjects.
Due to the tremendous burden of manual delineation, as well as concerns of rater reliability, Collins et al. (1999) applied automated labeling techniques to produce a probabilistic mapping of structures within the subject data from composing the ICBM152, thus creating a multi-subject structural atlas. The ICBM project has also produced a series of probabilistic volumes defining the voxel-wise frequency of several structures based upon manual delineations, which included maps of the lobes and of subcortical structures that were manually labeled in the volumetric data and sulcal maps that were generated by sampling curves that were manually traced on cortical surface models 1. Hammers et al. (2003) produced a maximum-probability atlas of the human brain based on a set of 20 manually delineated image volumes. The production of this atlas emphasized labeling structures in the temporal lobe. The data were aligned to the ICBM152 atlas using SPM99 (Ashburner et al., 1999) to produce an atlas that provides a basis for analysis of functional imaging of temporal lobe epilepsy. The collection of labels was later extended by incorporating additional structures and subjects; the augmented collection was used for automated labeling of brain structures (Heckemann et al., 2006). Van Essen (2005) produced a publicly available Population-Average, Landmark- and Surface-based (PALS) atlas from structural volumes of 12 individuals; the atlas can be used for surface-based analysis such as analysis of cortical folding abnormalities (Van Essen et al., 2006). Mega et al. (2005) examined variability in 20 elderly subjects with various mental states (normal cognition, mild cognitive impairment, or Alzheimer’s disease) by constructing an atlas from 68 manually delineated subregions. In that study, the authors also compared the use of 3 registration approaches and concluded that different methods may be appropriate for different areas of the brain. Klein et al. (2005) produced an automated method based on a set of 20 manually labeled brains, each with 36 labels per hemisphere. In their work, each brain serves as an atlas and is registered to serve as atlases; in their labeling method, each atlas brain was registered to the subject brain, and the most frequently occurring label at each voxel was selected to label that voxel in the subject brain. Atlasing is used in numerous other anatomical studies, though it is frequently used as a tool for analyzing the data without the goal of producing a widely distributed reference data set. Atlasing methods have also been applied in multi-subject studies of indvidual substructures of the brain, such as the hippocampus (Csernansky et al., 1998; Styner et al., 2004).
In this paper, we describe the construction of a probabilistic atlas of human brain structures. We produced our atlas from manual delineations of high resolution T1-weighted MRI scans of 40 healthy volunteers. A total of 50 cortical structures, 4 subcortical areas, the brainstem, and the cerebellum were delineated by trained raters following protocols developed during the course of this study. These data were then resampled into common spaces in order to produce estimates of the probability density functions for each structure. We produced 3 versions of our atlas using 3 widely used spatial normalization techniques paired with associated atlases. Each version of the atlas includes probabilistic maps of these structures, probabilistic maps of tissue types, and a volumetric average of the intensity data.
Among its uses, the atlas can serve as a reference template for studies of functional data or neuroanatomy, or as a statistical prior for segmentation algorithms. The delineated data can also provide a basis for the analysis of automated image segmentation methods or as training data for machine learning algorithms. The three versions of the atlas and the protocols used to perform the delineations are all available online.
2 Materials and Methods
2.1 Subjects and data acquisition
Forty volunteers were scanned with MRI at the North Shore - Long Island Jewish Health System (NSLIHS). Inclusion criteria for healthy volunteers included ages 16 to 40 and denial of any history of psychiatric or medical illness as determined by clinical interview. Exclusion criteria for all study participants included serious neurological or endocrine disorder, any medical condition or treatment known to affect the brain, or meeting DSM-IV criteria for mental retardation. The volunteer group was composed of 20 males and 20 females; average age at the time of image acquisition [mean ± S.D.] was 29.2yr±6.4 (min = 19, max = 40). The subjects had an average education level of 2.8±0.8, on a scale from 1 (completed graduate school) to 7 (partial completion of elementary school), where a score of 2 reflects completing college (16 years of education) and a score of 3 reflects completing part of college (12–16 years of education) (Hollingshead and Redlich, 1958; Hollingshead, 1975). The subjects were ethnically diverse, as described by self-selected categories (‘Asian or Pacific Islander’: 4 subjects; ‘Black not of Hispanic Origin’: 7 subjects; ‘Hispanic’: 5 subjects; ‘White not of Hispanic Origin’: 23 subjects; ‘Other’: 1 subject).
High-resolution 3D Spoiled Gradient Echo (SPGR) MRI volumes were acquired on a GE 1.5T system as 124 contiguous 1.5-mm coronal brain slices (TR range 10.0ms–12.5ms; TE range 4.22ms – 4.5ms; FOV = 220mm or 200mm) with in-plane voxel resolution of 0.86mm (38 subjects) or 0.78mm (2 subjects). Data were transmitted to the UCLA Laboratory of Neuro Imaging (LONI) for analysis following IRB approved procedures of both NSLIJHS and UCLA.
2.2 Pre-processing
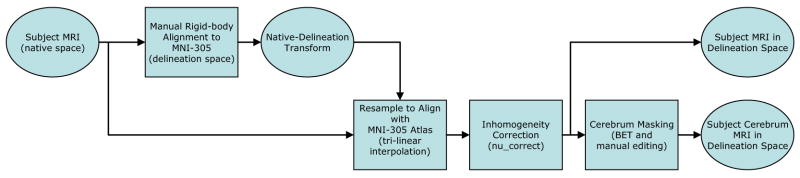
As part of a prior study, the 40 subject MRI volumes were processed to extract the cerebrum and to align the brains rigidly to a canonical space. This processing followed existing protocols used by LONI (see Fig. 2). First, the MRI volumes were aligned to the MNI-305 average brain (181 × 217 × 181 voxels; voxel size 1 × 1 × 1mm3) (Evans et al., 1993) to correct for head tilt and alignment. This step was performed to ensure unbiased anatomical decisions during the delineation process. The alignment was performed with a rigid-body rotation to preserve the native dimensions of the subject. Within each MRI volume, ten standard anatomical landmarks were identified manually in all three planes of section by a trained operator (see (Narr et al., 2002; Sowell et al., 1999) for details on this procedure). The landmarks for the subject were then matched with a set of corresponding point locations defined on the MNI-305 average brain; the landmarks were matched using a least-squares fit to produce a six parameter linear transformation (three-translation, three-rotation rigid-body, no rescaling). This computation was performed using the Register software package 2 (MacDonald et al., 1994). Each image volume was then resampled into a common coordinate system using trilinear interpolation. The resampled image volumes had the same dimensions and resolution as the atlas. Magnetic field inhomegeneities were corrected using a non-parametric non-uniformity normalization method (nu_correct) (Sled et al., 1998). Extra-meningeal tissue in the resampled data was then removed using the Brain Extraction Tool (BET) (Smith, 2002), an automated software program that identified the brain region in the MRI. Errors in the automated segmentation of the brain were corrected manually; the cerebellum and brainstem were also removed manually to produce a cerebrum mask.
Fig. 2.
Image pre-processing sequence. MRI of the brain for each subject were registered to the MNI-305 atlas (181 × 217 × 181 voxels, 1 × 1 × 1mm3 voxel resolution) using a rigid-body transformation. This corrected for head-tilt and alignment to ensure unbiased anatomical decisions during the delineation process. Once in the delineation space, the MRI were corrected for RF inhomogeneity artifacts and processed to identify the cerebrum.
2.3 Delineation of individual subject brains
For each subject MRI volume, a corresponding 3D integer volume was produced that labeled each voxel in the brain (see Fig. 3). Each integer label represents a particular neuroanatomical structure; the set of structures used in this atlas are listed in Table 1. The structures were labeled manually by trained raters following a set of written protocols produced by this project (see online supplemental materials). All delineations were performed using the BrainSuite software package (Shattuck and Leahy, 2002). BrainSuite provided users with the ability to display simultaneous views of three orthogonal planes through the MRI volume, superimposed with a label volume that identifies structures. Additionally, BrainSuite provided synchronized views with 3D surface models of the brain. This allowed raters to view the label information simultaneously in the volumetric space and on the surface model of the brain. The raters edited the volume labels by using the mouse to paint within one of the image slices; the related views updated in real-time with the new label information, providing a 3D context for the delineations.
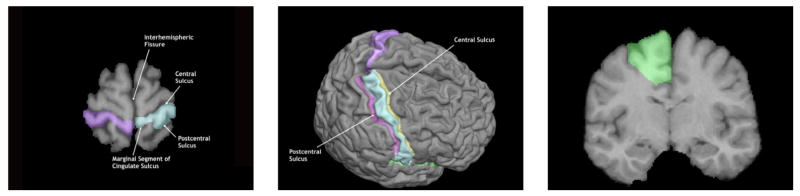
Fig. 3.
Delineation Example figures from the delineation protocol for the post–central gyrus. The left image shows the slice view, while the middle view shows the corresponding surface view. These views are consistent with what the raters see during the delineation process, except for the annotations, which are part of the written protocol. The rightmost figure shows the inclusion of white matter in the delineation of a gyrus. Text and images for all protocols used in this study are available from our website.
Table 1.
Cortical structures in the probabilistic atlas and the corresponding colors used to denote them in this paper. *The hippocampus can be considered to reside in both the temporal lobe and the limbic lobe. ** The fusiform and lingual gyrus can be considered to reside in both the occipital lobe and temporal lobe.

|
The protocols for labeling each structure were defined by two primary raters (M.M. and V.A.) based on neuroanatomical landmarks described in existing atlases, including published references (Duvernoy, 1999; Salamon et al., 2005; Salamon and Huang, 1976) and an online neuroanatomy atlas 3. The protocols were confirmed by a trained neuroanatomist (G.S.) and are available online as a product of this research 4 and are also included as a supplement to this article. The protocols provided detailed written instructions and figures that show both surface renderings and 3D cross-sectional views of reference anatomy. Each protocol also provided a general description of the structure to be delineated and specified a single plane of section (coronal, sagittal, or transaxial) in which that structure should be labeled. Additionally, each protocol defined, in an anatomical sense, where the rater should initiate and complete the labeling. The cortical protocols emphasize inclusion of all grey matter in the cortical structures. Sulci are used as boundaries, and the raters are instructed to follow the internal depths of the sulci. White matter voxels that occurred between the boundaries of sulci and their surrounding grey matter were included in the structure (see Fig. 3); most medial white matter was excluded. To achieve additional refinement of the data into grey matter specific areas, we later combined these results with automated tissue classification methods (see Sec. 2.4 below).
The task of delineating the 40 MRI volumes was divided among 15 human raters. Each structure was assigned to a single pair of raters drawn from the entire group (see Table 3); in the case of bilateral structures, the delineations for the left and right sides of the brain were performed by the same rater pair. Delineation of the structure in the 40 MRI volumes was evenly divided among the two raters assigned to it. At the beginning of the project, six of the subject MRI volumes were selected at random to be used as the training data set for all raters. Raters 1 and 2 (M.M. and V.A.) supervised the training and delineations of the other raters. For each structure, the assigned rater pair was trained by performing delineations of that structure in the training data. Working independently of each other, the two raters delineated the structure in all six training volumes; for bilateral structures, each rater delineated both left and right structures. Once both raters had completed the delineation of the structure in the training data, the total delineated volume of the structure in each volume was computed. These volume measures were then compared using using Intraclass Correlation Coefficient (ICC) methods (Shrout and Fleiss, 1979). We use the Case 1 formula, which is given, for a single pair of raters, by the formula
Table 3.
Rater reliability measures: Shown are the intra-class correlation (ICC) and average Jaccard similarity measures for delineation reliability. For each structure, delineation labels on six image volumes were produced by two raters; multiple pairs of raters were used to delineate the full set of structures. ICC values were computed on measures of structure volume during training to ensure that pairs of raters produced concordant results; raters who did achieve ICC scores of 0.9 or greater were retrained, and Jaccard similarity scores were computed on the same labeled volumes after the training had been completed to assess similarity between the delineated areas.
| ICC | Jaccard | ||||
|---|---|---|---|---|---|
| Rater pair | left | right | left | right | |
| Angular Gyrus | 1,2 | 0.916 | 0.957 | 0.855 | 0.864 |
| Caudate | 1,2 | 0.991 | 0.979 | 0.755 | 0.749 |
| Cingulate Gyrus | 7,2 | 0.935 | 0.921 | 0.699 | 0.639 |
| Cuneus | 9,10 | 0.961 | 0.923 | 0.794 | 0.774 |
| Fusiform Gyrus | 1,2 | 0.987 | 0.943 | 0.820 | 0.804 |
| Gyrus Rectus | 1,2 | 0.917 | 0.922 | 0.966 | 0.980 |
| Hippocampus | 4,5 | 0.933 | 0.982 | 0.796 | 0.787 |
| Inferior Frontal Gyrus | 1,2 | 0.963 | 0.973 | 0.937 | 0.945 |
| Inferior Occipital Gyrus | 1,2 | 0.931 | 0.982 | 0.850 | 0.829 |
| Inferior Temporal Gyrus | 1,2 | 0.928 | 0.924 | 0.785 | 0.788 |
| Insular Cortex | 7, 8 | 0.992 | 0.935 | 0.827 | 0.830 |
| Lateral Orbitofrontal Gyrus | 1,2 | 0.900 | 0.925 | 0.888 | 0.880 |
| Lingual Gyrus | 3,6 | 0.966 | 0.977 | 0.833 | 0.813 |
| Middle Frontal Gyrus | 12,11 | 0.962 | 0.992 | 0.848 | 0.851 |
| Middle Orbitofrontal Gyrus | 1,2 | 0.980 | 0.968 | 0.917 | 0.917 |
| Middle Occipital Gyrus | 1,2 | 0.997 | 0.964 | 0.917 | 0.946 |
| Middle Temporal Gyrus | 1,2 | 0.955 | 0.945 | 0.784 | 0.775 |
| Parrahippocampal Gyrus | 1,2 | 0.943 | 0.927 | 0.755 | 0.773 |
| Postcentral Gyrus | 1,2 | 0.948 | 0.968 | 0.826 | 0.855 |
| Precentral Gyrus | 1,2 | 0.911 | 0.943 | 0.928 | 0.918 |
| Precuneus | 1,2 | 0.944 | 0.970 | 0.816 | 0.739 |
| Putamen | 1,2 | 0.931 | 0.982 | 0.761 | 0.758 |
| Superior Frontal Gyrus | 13,14 | 0.957 | 0.975 | 0.840 | 0.852 |
| Superior Occipital Gyrus | 1,2 | 0.977 | 0.947 | 0.811 | 0.801 |
| Superior Parietal Gyrus | 1,2 | 0.952 | 0.945 | 0.809 | 0.833 |
| Superior Temporal Gyrus | 1,2 | 0.939 | 0.982 | 0.849 | 0.796 |
| Supramarginal Gyrus | 1,2 | 0.943 | 0.969 | 0.697 | 0.686 |
| Cerebellum | 13,14 | 0.926 | 0.903 | ||
| Brainstem | 3,15 | 0.979 | 0.841 | ||
| (1) |
where MSB is the mean squares between the volume measurements of the structure and MSW is the mean squares of the volume measurements within each structure. It assesses the degree to which the variation in the measurements is due to intersubject variation as opposed to the interrater variation. Rater pairs were required to achieve an ICC score above 0.9 on the delineation of a structure in all 6 training images before they were considered to be reliable for that structure. Scores for bilateral structures were computed separately, but raters were required to achieve the score on both sides of the brain. Rater pairs who did not achieve the criteria were retrained; if necessary, the protocol was revised to provide clearer definitions. After retraining, the raters revised their delineations and their results were retested. This process was repeated until raters achieved reliability.
Following training, we also computed the Jaccard similarity metric (Jaccard, 1912) on the sets of delineations. This metric measures the agreement of two sets A and B using the formula
| (2) |
where |A ∩ B| is the size of their intersection and |A ∪ B| is the size of their union. In this context, A and B each represent a set of voxels that were included in the delineation by one of the raters. The metric is zero if the two sets have no common elements and one if the sets are identical. While the ICC method measures the agreement in terms of volume delineated, the Jaccard metric measures the degree of overlap between the two labelings.
Once a rater pair achieved reliability for a structure, the task of delineating that structure in the remaining 34 volumes was divided evenly between the two raters. For each of the 6 subject volumes that were delineated during the training procedure, one of the two label volumes was chosen at random to represent the labeling for that structure in that individual, with 3 labelings being selected from each rater. In some cases, different structures in the same subject volume were delineated in parallel by pairs of raters. These results had to be composited into a single label set for the subject. Conflicting voxel labels from the delineations were identified automatically by computing the differences in the labeled areas of the two data sets. These voxels were examined by a pair of raters, and a single label was selected for each voxel. This merging occurred periodically during the delineation process, and only when the same subset of structures had been labeled in all subjects. This allowing labeling to be performed using partially completed delineations in most cases. The volume labelings were examined for correctness by a trained neuroanatomist (G.S.). The labelings were also corrected for disconnected voxel labels and gaps between structures either by the pairs responsible for those structures or by the two senior raters (M.M. and V.A.) The final result of the manual delineation process was a set of 40 integer volumes, each of which contained 56 contiguously labeled structures in the delineation space of the individual subject.
2.4 Atlas Production
For each structure, we computed spatial density maps after spatial normalization of the delineation and imaging data. Though each brain was previously aligned to the ICBM305 average brain, that alignment did not allow rescaling or stretching. Thus, the individual structures were not well-aligned across the subjects and spatial normalization of each subject’s data to a canonical reference space was required.
Selection of a spatial normalization algorithm and target space is critical in the production of an atlas. Ideally, all variation would be removed by the normalization process, thereby providing an exact labeling of any data that are normalized to the reference space. In practice, registration algorithms will retain some intersubject variation in the spatial location of neuroanatomical structures. Since each registration approach will produce different effects, the individual transformed data sets represent samples of the specific process used. We produced 3 variants of our atlas using widely-used methods matched with appropriate target spaces. The methods and target spaces used were: (1) AIR 5.2.5’s nonlinear warping method 5 (Woods et al., 1998) paired with the ICBM-452 5th order warp atlas 6 (Rex et al., 2003); (2) FSL’s FLIRT algorithm 7 (Smith et al., 2004) paired with its ICBM152 T1 brain atlas; and (3) SPM5’s unified segmentation (Ashburner and Friston, 2005) paired with the ICBM152 T1 whole head atlas used as approach 8 SPM5’s default atlas. Table 2 summarizes the algorithms and registration targets used.
Table 2.
The three variants of the LONI Probabilistic Brain Atlas.
| LPBA40/AIR | LPBA40/FLIRT | LPBA40/SPM5 | |
|---|---|---|---|
| package | AIR | FSL | SPM5 |
| algorithm | align_warp | FLIRT | unified segmentation |
| target | ICBM452 Warp 5 | avg152T1_brain | avg152T1 |
| matrix | 149 × 188 × 148 | 91 × 109 × 91 | 91 × 109 × 91 |
| resolution | 1 × 1 × 1mm3 | 2 × 2 × 2mm3 | 2 × 2 × 2mm3 |
2.4.1 Registration pre-processing
We performed pre-processing on the original MRI data volumes to prepare them for the 3 registration approaches. For SPM5, the whole-head MRI data in their native acquisition space were used in the normalization process. For AIR and FLIRT, we skull-stripped and bias-corrected the MRI data for each subject prior to spatial normalization. For skull-stripping, we generated a whole brain mask in the delineation space by combining the cerebrum mask produced during pre-processing (see Sec. 2.2) with the structural delineation labels. We could have computed a brain mask from the union of the cerebrum mask voxels and the voxels that were delineated as brainstem or cerebellum. However, this mask would not enclose areas such as the 4th ventricle or some portions of white matter in the cerebellum. Since these areas are contained within the atlas target, it is important that we also have them in our subject brains.
We applied a series of mathematical morphological operators to produce a set of whole-brain masks from the delineations and cerebrum masks. First, we produced a mask composed of all voxels labeled as cerebellum and brainstem. We then applied a modified closing operator to this mask, which consisted of a dilation with a structure element, a flood filling of any internal cavities in the resulting mask, and finally an erosion with the same structure element. The structure element used was a digital approximation of a sphere of radius 4 (see (Dogdas et al., 2005) for a detailed description of these operations). We then took the union of this mask with the cerebrum mask, and again applied our modified closing operator to the output mask, this time with a digital sphere of radius 2. We applied this procedure to the cerebrum mask and delineation data of each subject to produce 40 brain masks in the delineation space. We then resampled each of these masks into the native space using the inverse of the affine transform that was used to map the brains into the delineation space. This resampling was performed by applying AIR5.2.5’s reslice command with nearest-neighbor interpolation.
For each subject MRI, we performed bias field correction and tissue classification in its native space. Each MRI volume was first masked using the corresponding resampled whole brain masks, then corrected for RF artifacts using the Bias Field Corrector (BFC) software (Shattuck et al., 2001). The corrected image was then classified into tissue types (GM, WM, and CSF) using the Partial Volume Classifier (PVC) software (Shattuck et al., 2001). We note that the use of BFC here differs from the pre-processing that was performed previously in section 2.2; we selected the combination of BFC and PVC for this stage of the processing because it is a sequence that is in routine use our lab (e.g., Sowell et al. (2004); Apostolova et al. (2007); Chiang et al. (2007)) PVC is a fully automated method that assigns to each voxel a label corresponding to GM, WM, CSF, or partial volume combinations of these; PVC also produces estimates of the tissue fractions at each voxel. We used these estimates to produce fraction volume images for GM, WM, and CSF in each image. Each voxel fraction had a floating point value in the range [0, 1]; these volumes were rescaled to [0, 10000] and stored in 16-bit image volumes to be compatible with the resampling software.
2.4.2 Normalization and averaging
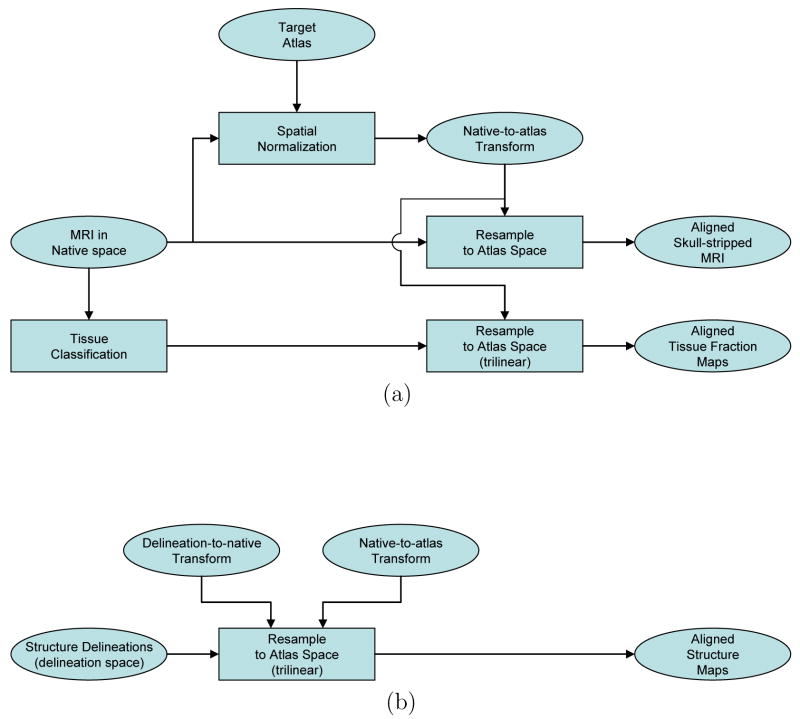
The three variants of our atlas were constructed following a single basic strategy (see Fig. 4). Each MRI volume was first aligned to the atlas target using the selected algorithm to produce a native-to-atlas transform. Then, each of the skull-stripped MRI volumes were transformed to the atlas space and averaged to produce a mean intensity map. Next, the subject tissue fraction maps for GM, WM, and CSF were resampled into the atlas space using trilinear interpolation. These maps were then averaged across the subjects to produce mean tissue fraction maps.
Fig. 4.
These flowcharts show the common approach used in the production of the 3 atlas versions. Once the spatially normalized maps were produced for each subject, they were averaged to produce the average intensity and probability density maps. (a) Normalization of data from native space to the atlas space. The spatial normalization methods and target spaces used are detailed in Table 2. The skull-stripped, RF-corrected MRI were used for the AIR and FLIRT normalization processes; the whole-head MRI were used for the SPM5 normalization. The skull-stripped, RF-corrected MRI were resampled for all 3 versions of the atlas; see the text for details on the resampling parameters used. (b) Normalization of data from delineation space to the atlas space. The delineation labels were separated into individual structure maps and resampled using trilinear interpolation. For the AIR and FLIRT versions of the atlas, this was achieved using a single composited transform; for the SPM5 version, the structure maps were first resampled into the native space, then retransformed into the atlas space. Trilinear interpolation was used in each instance.
We then applied the delineation-to-native and native-to-atlas transforms to resample the subject delineation data into the target atlas space. Integer values in the delineation label volumes represent discrete classes, thus resampling the data by interpolating between values that represent adjacent structure can produce values that are not meaningful. Because of this, nearest-neighbor resampling is often used to resample label or mask data. While this restricts the output label values to those in the original image, it introduces aliasing artifacts at the boundaries of structures. We instead adopted an alternate approach, in which we separated each subject’s label volume into a set of 16-bit volumes for each of its 56 structures. At each voxel site, we represented the presence of a structure label with a large integer value (10000) and the absence of the structure with 0. This allowed us to resample the individual structures using trilinear interpolation, thereby producing partial volume values in the resampled image. We produced a total of 2,040 structure maps, each of which was resampled into the atlas space according to the transforms produced by the automated methods. For each structure, we then averaged across the 40 volumes corresponding to each subject. This produced spatial estimates of the probability densities for the structures in our atlas.
Since most of the structures in our atlas are cortical structures, we also produced a refined set of density maps by combining the partial volume structure maps with the grey matter partial volume map. In the atlas space, we multiplied the density maps for each structure of each individual by the GM map for that individual. We then averaged across the subjects to produce a set of 56 GM-structure density maps.
We computed a maximum likelihood map and a maximum likelihood labeling by identifying, at each voxel, the structure label with the greatest value in the structure density maps. In the case of multiple modes, where multiple structures had maximal likelihood values at a voxel, a label was chosen at random from the modes at that site. We repeated this process using the GM structure maps to produce a GM maximum likelihood map and labeling. The net result of this stage, for each atlas variant, is a set of maps: the average intensity image, the tissue density maps, the structure density maps, and the maximum likelihood maps and labelings. Each map was represented as a 3D integer volume. For a particular atlas variant, all of these data are in spatial register and together compose a single atlas. In the following sections, we detail the key differences in the processing we used to transform the data into the respective atlas spaces for the 3 methods.
2.4.3 LPBA40/AIR
The LPBA40/AIR variant was constructed using AIR5.2.5 (Woods et al., 1998). We computed 5th-order warp transforms to normalize the subject volumes to the ICBM 452 T1 Warp 5 Atlas (Rex et al., 2003), which we denote here as ICBM452W5. The ICBM452W5 target space had a matrix size of 149 × 188 × 148 with 1 ×1 ×1mm3 voxel resolution. We processed each subject data set as follows. First, we applied a crop operation with an 8 voxel pad to the stripped, bias-corrected subject image volume in its native space. This reduced the size of the image volume by trimming empty voxels, which improved the initial alignment to the atlas. We then computed an affine, 12-parameter alignment between the cropped image and the ICBM452W5 target. We next applied AIR’s alignwarp procedure to compute a nonlinear 5th-order polynomial alignment to the atlas, which was initialized with the 12-parameter affine alignment. For both alignment steps, the thresholds were set to 1 since the volumes were masked prior to processing; all other parameters were left as their default values. After correcting for the initial crop, this produced a native-to-ICBM452W5 transform. We combined this with the original delineation-to-native mapping to produce a delineation-to-ICBM452W5 transform. This produced the set of transforms required for the atlas construction. During the resampling process, we applied the native-to-ICBM452W5 transform to the skull-stripped MRI data using AIR’s reslice warp program with 3D sinc interpolation (FWHM=6mm). Resampling of the structure volumes was performed using AIR’s reslice warp program with trilinear interpolation.
2.4.4 LPBA40/FLIRT
The LPBA40/FLIRT variant was constructed using the FSL FLIRT software (Smith et al., 2004) with the skull-stripped version of the ICBM152 average brain (91 × 109 × 91 voxels; 2 × 2 × 2mm3 resolution) that is included with the FSL distribution; we denote that target here as ICBM152brain. The masked, bias corrected brain images for each subject were aligned to the atlas target using the default settings in FLIRT. For each subject, this produced an affine native-to-ICBM152brain transformation; we composited this transform with the affine mapping from the subject’s delineation space to native space, thus producing a delineation-to-ICBM152brain affine transform. Using AIR5.2.5’s reslice program with these transforms, we resampled the subject data into the ICBM152brain space. The skull-stripped bias-corrected MRI brain data were resampled using chirp-Z interpolation; all other structure maps were resampled using trilinear interpolation.
2.4.5 LPBA40/SPM5
We applied the SPM5 unified segmentation approach (Ashburner and Friston, 2005) to each subject’s whole-head MRI in native space. This process simultaneously performed spatial normalization, bias correction, and tissue classification. The registration target for this procedure was the ICBM452 T1 average (91×109 ×91 voxels; 2×2×2mm3 resolution), which is the default registration target used by SPM5. We note here that we could have used the tissue classification volumes or bias-corrected data produced by SPM5 to generate the final atlas products. However, for consistency, we used the same images and tissue fractions for all three variants. Default settings were used consistently, with the exception of the bounding box which was set to [−90 −126 −72; 90 90 108] to provide better coverage of the head. We used SPM5 to resample the native space subject MRI, with spline interpolation. We resampled the tissue fraction data into the ICBM152avg space using SPM5 with trilinear resampling. For the delineation data, we first resampled each structure volume into the native space using AIR5.2.5’s reslice program with trilinear resampling. We then applied SPM5 to resample each of these into the ICBM452avg space, again using trilinear resampling. Using two consecutive resampling steps is not ideal; however, methods for compositing AIR and SPM5 transforms were not available to us.
2.5 Atlas analysis
We performed an initial assessment of the atlas data using measures of volume for each structure. In the resampled structure data, volume measures were computed by summing the voxels in the structure maps and dividing by 10,000, which was the value used to represent a voxel that belonged entirely to a single structure. By doing this, we were able to account for partial volume fractions in our volume measures. We analyzed structures in the data set for asymmetry using the metric:
| (3) |
where VR and VL are the volumes of the right and left structures, respectively. We tested for significance in the asymmetry measures for each bilateral structure using a one sample t-test We also compared the effect of gender on total brain volume by computing a Welch two sample t-test using the sizes of the brain masks divided into groups of males and females. For both types of test, we considered as significant those results for which p < 0.05; we did not correct for multiple comparisons. All statistical comparisons were computed using the statistical software package R v. 2.2.19.
3 Results
3.1 Delineation
The delineation procedures were performed on all 40 subject volumes according to the delineation protocols. The protocol training and reliability testing required between 24 and 40 hours per structure for each rater. The time required for delineation by each rater was approximately one hour per structure per hemisphere. For the entire set of 40 volumes, each structure required on the order of 120 hours of total rater time including training and delineation. Approximately 10 hours of total rater time per volume were required to composite all structure delineations into single volumes and to ensure contiguous labelings.
The similarity measures for the interrater comparisons are detailed in Table 3. All raters achieved the required ICC score of 0.9, though retraining was typically necessary after the initial labeling. We note that we did not record the number of attempts required to achieve reliability, only that the reliability rating was ultimately achieved. The raters showed reasonable Jaccard similarity scores for the training data, with most measures above 0.8. The raters reported that the caudate nucleus, putamen, hippocampus, and lingual gyrus were the most difficult structures to delineate.
3.2 Atlas construction
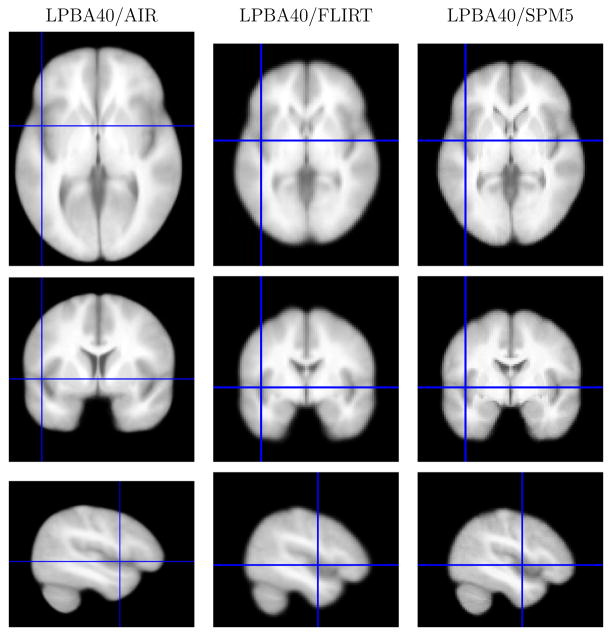
The spatial transforms and resampled data were computed for the three atlas variants as described in the methods section. We note that the LPBA40/FLIRT and LPBA40/SPM5 atlas variants were computed at lower resolution than the LPBA40/AIR version of the atlas, which is evident in the images. Figure 5 shows example slices through the probability density functions for the superior temporal gyrus for each version of the atlas, as well as reference slices from the intensity average data. The GM density maps show improved definition of the structure.
Fig. 5.
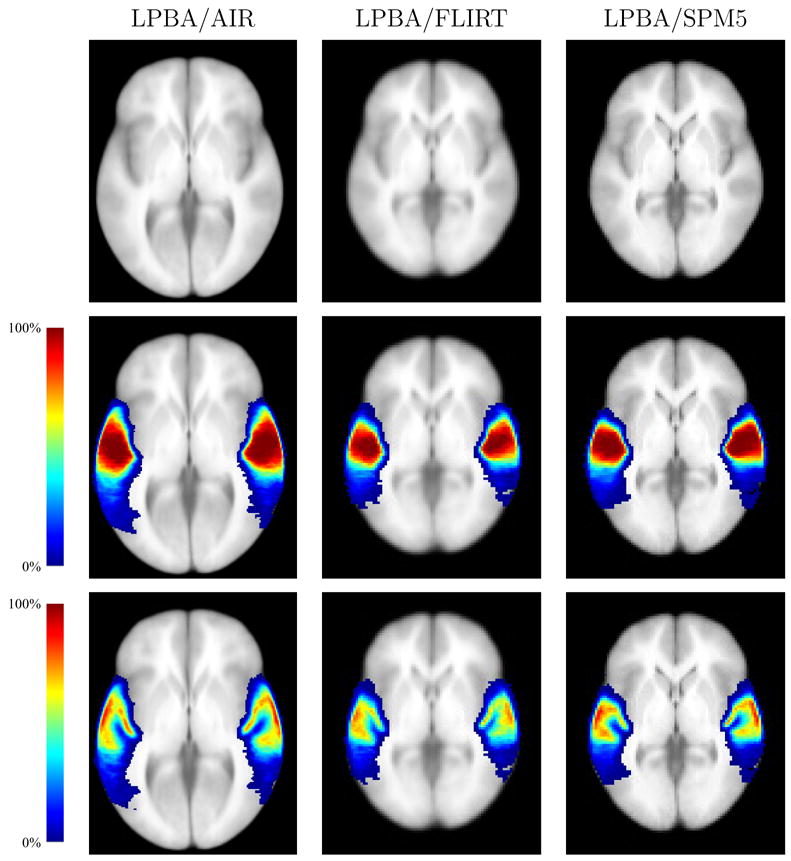
Probability densities for the superior temporal gyrus, computed for each of the three atlas spaces. Each density is superimposed on the average intensity brain image computed for each target space. For the LPBA40/FLIRT and LPBA40/SPM5 images, the slices are taken through the z = 0 plane in SPM5 coordinates for the ICBM152 T1 average; the LPBA40/AIR images were taken from a corresponding plane. All images are shown in neurological convention (anatomical left is displayed on the left) (Top) The average intensity brain image (Middle) The superior temporal gyrus density map (Bottom) The superior temporal gyrus grey matter density map.
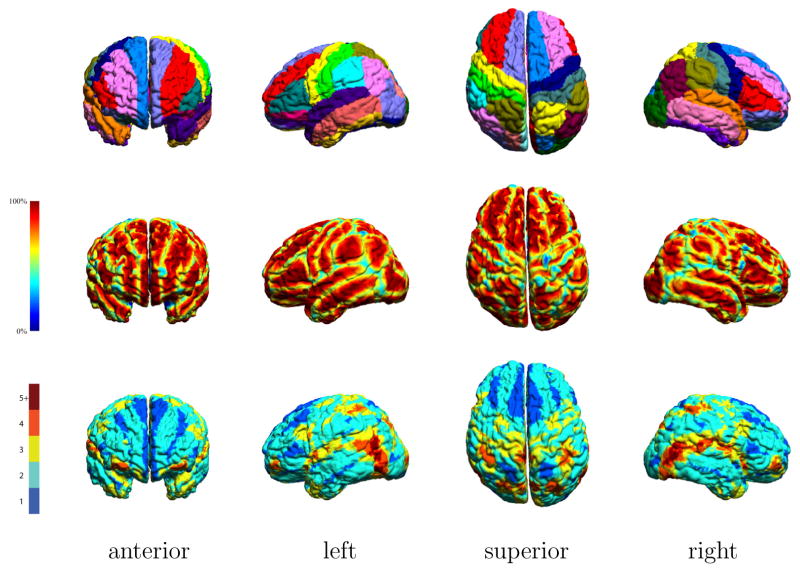
Qualitatively, the maps produced by applying trilinear interpolation separately to each subject’s 56 structures produced smoother maps than we produced using nearest-neighbor interpolation for an earlier version of this atlas. With the nearest-neighbor method, voxels in the resampled atlas space could only be true or false for a give structure, hence the density maps for each structure were quantized to number of subjects, i.e., to 40 levels. The use of trilinear interpolation allowed us to quantize to an arbitrary degree and to use partial volume voxels to reduce the aliasing artifacts. To further illustrate the density functions, we selected a brain at random from the 40 subjects and transformed the LPBA40/AIR density functions back to the native space of that subject using the inverse of the native-to-atlas transform for that subject. Figure 6 shows the mapping of the LPBA40/AIR densities for the middle frontal gyrus, the superior temporal gyrus, and the fusiform gyrus onto a cortical model of the subject.
Fig. 6.
Probability maps for three cortical structures. Probabilistic data from the LPBA40/AIR atlas was mapped back to the space of one of the atlas subjects. The surface model has been colored according to the probability values for middle frontal gyrus, superior temporal gyrus, and fusiform gyrus to indicate the likelihood of one of those structures at a given surface point.
The maximum likelihood maps were computed for each of the three variants. For the LPBA40/AIR variant, multiple modes occurred in 125 voxels for the maximum-likelihood map and 357 voxels for GM maximum-likelihood map. For the LPBA40/FLIRT variant, multiple modes occurred in 29 voxels for the ML map and 51 voxels GM map. For the LPBA40/SPM5 variant, multiple modes occurred in 63 voxels for the ML map and 83 voxels GM map. These numbers are much smaller than those that would have occurred had we used nearest-neighbor interpolation to resample the structure labels, where each density function would be restricted to 40 levels.
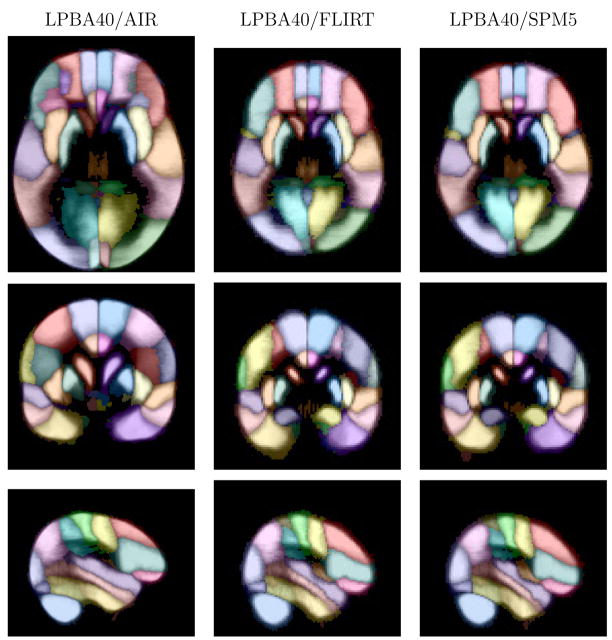
Figure 7 shows orthogonal sections of the intensity average brain images for the three variants of the atlas. Figures 8 and 9 show corresponding slices from the maximum-likelihood maps and the grey matter maximum likelihood maps. The maximum likelihood figures show, at each voxel, the maximum probability value at that voxel colored according to the corresponding most likely structure. The AIR version of the atlas shows the most detail, due in part to the higher resolution at which it was sampled. The FLIRT and SPM5 versions of the atlas are both at 2mm cubic resolution, and the differences between them are more evident in Figs. 7, 8, and 9. The boundaries of the SPM5 atlas appear crisper than those of the FLIRT atlas. This is due to the extra degrees of freedom allowed by the nonlinear warping used by SPM5, which reduces the intersubject variability to a greater degree than the affine transformation used within FLIRT.
Fig. 7.
Average MRI intensity images. On the left are slices from the average brain images produced after spatial alignment with AIR nonlinear warping and the ICBM452 T1 5th order warp atlas. In the middle are similar slices in the average intensity image produced after alignment using FLIRT and its skull-stripped ICBM152 T1 average brain. On the right are the same slices in the images produced using SPM5 to align the subject data to its ICBM152 T1 template. Transaxial and coronal mages are displayed in neurological convention (anatomical left is displayed on the left); the crosshairs indicate the slice positions.
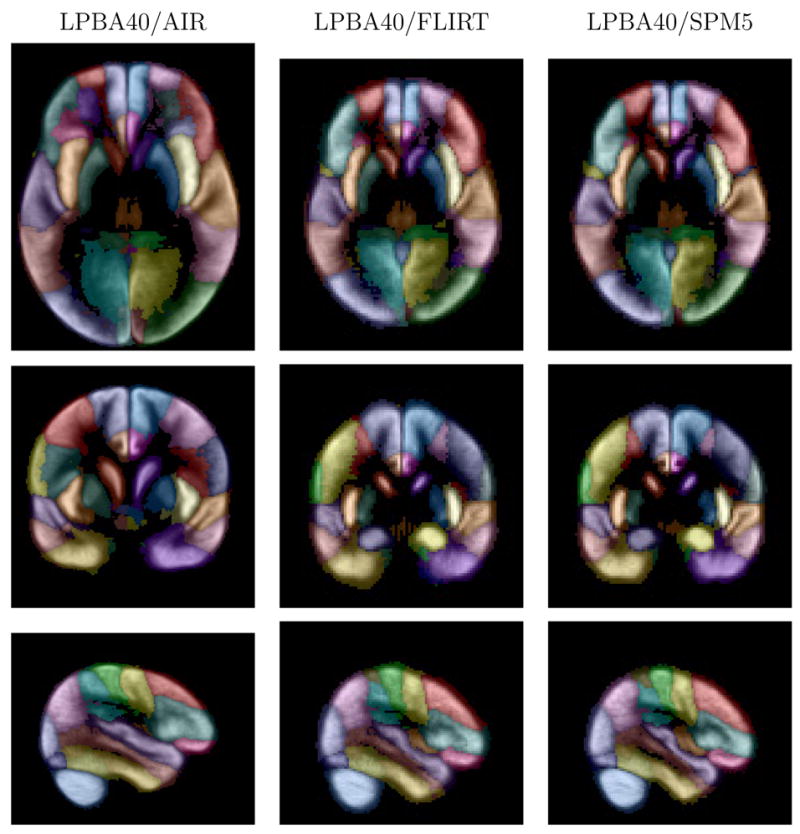
Fig. 8.
Maximum likelihood maps. Shown are images from the maximum likelihood maps for the three versions of the atlas, corresponding to the slices shown in Fig. 7. The intensity represents that maximum probability value at each voxel, computed from the estimated structure probability density maps. The color indicates the most probable structure (see Table 1). Transaxial and coronal images are displayed in neurological convention (anatomical left is displayed on the left)).
Fig. 9.
Maximum likelihood grey matter maps. Shown are images from the maximum likelihood grey matter maps for the three versions of the atlas, corresponding to the slices shown in Fig. 7. The intensity represents that maximum probability value at each voxel, computed from the estimated grey matter structure probability density maps. The color indicates the most probable structure (see Table 1). Transaxial and coronal images are displayed in neurological convention (anatomical left is displayed on the left).
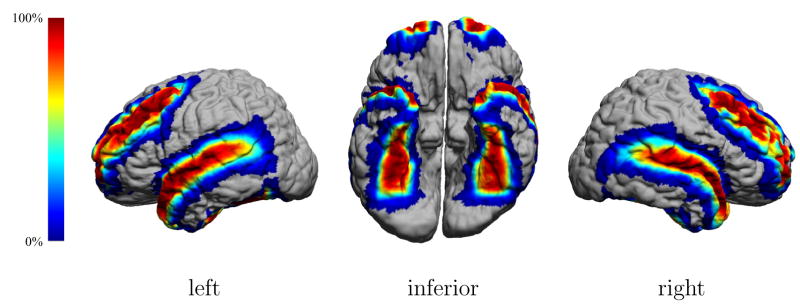
Figure 10 shows the mapping of the LPBA40/AIR maximum likelihood labeling maps to the individual surface model shown in Fig. 6. Also shown are the maximum probability values and the number of structures that have non-zero probability at each vertex of the surface. This figure emphasizes that even with nonlinear warping, most areas of the cortex are not uniquely labeled. In areas where multiple atlas structures are near, such as the posterior portion of the temporal lobe, we see this influence on the number of structures contending for those voxels. Similarly, we see the maximum probabilities decrease, indicating less certainty in the automated labeling. We note here that even with a dataset drawn from the 40 subject volumes used to construct the atlas, the structures in the maximum likelihood labeling will not perfectly match the structures in the individual. This is to be expected, since variation in the spatial alignment of the subject anatomy is what produces the variation in the probability densities in the atlas.
Fig. 10.
The LPBA40/AIR maximimum likelihood atlas data, mapped back to the surface model of an atlas subject. (Top) The surface has been colored according to the most likely structure label (see Table I for key). (Middle) The surface is colored according to the maximum probability value at each surface point. (Bottom) The surface has been colored according to the number of structures with non-zero probabilities at each location on the cortex.
3.3 Volumetric analysis
We computed the total volume for each structure in each subject. The average volumes (mean + S.D.) of each structure are shown in Table 4. The cortical and subcortical structures in the LPBA40/FLIRT and LPBA40/SPM5 versions of the atlas exhibit average expansions in the mean volume measures of 37.7% and 37.8%, respectively, compared to their original sizes in the delineation space. This is due, in part, to the size distortion in the ICBM152 atlas, which is larger than an average brain. For example, the brain mask used in FLIRT for the ICBM152 brain is 2, 066.96 cm3, while the average volume of the brain masks for our population group was 1, 358.25 ± 126.34 cm3. The sizes of these structures in the LPBA40/AIR atlas are more similar to those in the delineation space, undergoing an average increase of 10.3% in their mean structure volume measures. The smaller change in volume is most likely because the ICBM452 target is closer in volume (1, 559.55cm3) to our population group than the the ICBM152 atlas is. We also computed the asymmetry value based on the initial delineation. The hippocampus, for which the right side was larger in 32 of the 40 subjects, had a mean laterality value of 0.0538 with significance (p = 5.40 × 10−5). Other structures showing significant asymmetry in the delineated data were the angular gyrus (0.0765; p = 0.0417), the caudate nucleus (−0.0465; p = 0.00243), the inferior frontal gyrus (0.0615; p = 0.0374); the inferior temporal gyrus (0.0680; p = 0.0192); the insular cortex (−0.0804; p = 3.36×10−5); and the lateral orbitofrontal gyrus (−0.0890; p=0.0291). The total brain volume was larger for males than for females (p = 0.000837), with average sizes (mean+s.d.) of 1422.43±117.70cm3 for males and 1294.05±99.14cm3 for females.
Table 4.
Average volume measures of the delineated structures, computed in the delineation space and after spatial normalization to the three registration targets.
| Structure Volume, in cm^3 (mean + s.d.) | ||||
|---|---|---|---|---|
| structure | delineation space | LPBA40/AIR | LPBA40/FLIRT | LPBA40/SPM5 |
| brainstem | 29.59 ± 3.32 | 31.28 ± 2.28 | 39.57 ± 2.95 | 33.05 ± 1.38 |
| cerebellum | 133.97 ± 17.05 | 160.13 ± 6.23 | 184.37 ± 14.38 | 155.69 ± 6.74 |
| L angular gyrus | 17.44 ± 3.40 | 17.12 ± 4.00 | 24.13 ± 4.86 | 23.86 ± 4.84 |
| R angular gyrus | 18.82 ± 3.65 | 18.36 ± 3.20 | 26.00 ± 4.84 | 25.69 ± 4.37 |
| L caudate | 2.97 ± 0.55 | 3.62 ± 0.63 | 4.10 ± 0.64 | 4.15 ± 0.68 |
| R caudate | 2.86 ± 0.62 | 3.51 ± 0.71 | 3.94 ± 0.75 | 4.05 ± 0.75 |
| L cingulate gyrus | 12.45 ± 2.12 | 19.54 ± 3.92 | 17.28 ± 3.17 | 17.31 ± 3.09 |
| R cingulate gyrus | 12.79 ± 2.30 | 20.15 ± 4.52 | 17.77 ± 3.65 | 17.90 ± 3.55 |
| L cuneus | 6.00 ± 1.51 | 5.97 ± 1.56 | 8.26 ± 1.97 | 8.42 ± 1.94 |
| R cuneus | 6.45 ± 1.69 | 6.73 ± 1.75 | 8.93 ± 2.47 | 9.07 ± 2.29 |
| L fusiform gyrus | 12.25 ± 2.32 | 16.42 ± 2.61 | 16.84 ± 2.70 | 17.04 ± 2.67 |
| R fusiform gyrus | 12.31 ± 2.13 | 16.65 ± 2.75 | 16.98 ± 2.62 | 16.95 ± 2.52 |
| L gyrus rectus | 3.19 ± 0.64 | 3.02 ± 0.71 | 4.41 ± 0.88 | 4.50 ± 0.89 |
| R gyrus rectus | 3.34 ± 0.75 | 3.08 ± 0.67 | 4.58 ± 0.92 | 4.68 ± 0.95 |
| L hippocampus | 3.91 ± 0.52 | 4.44 ± 0.47 | 5.39 ± 0.48 | 5.36 ± 0.54 |
| R hippocampus | 4.12 ± 0.52 | 4.70 ± 0.50 | 5.68 ± 0.58 | 5.64 ± 0.56 |
| L inferior frontal gyrus | 22.26 ± 4.73 | 24.96 ± 5.03 | 30.67 ± 5.97 | 31.52 ± 5.50 |
| R inferior frontal gyrus | 23.76 ± 5.44 | 26.22 ± 5.95 | 32.78 ± 7.23 | 33.23 ± 6.49 |
| L inferior occipital gyrus | 10.78 ± 2.29 | 10.66 ± 2.43 | 14.83 ± 2.81 | 15.43 ± 2.88 |
| R inferior occipital gyrus | 10.79 ± 2.22 | 11.09 ± 2.69 | 14.87 ± 2.74 | 15.43 ± 3.01 |
| L inferior temporal gyrus | 20.62 ± 4.06 | 21.84 ± 3.47 | 28.36 ± 4.66 | 29.04 ± 4.63 |
| R inferior temporal gyrus | 21.97 ± 3.78 | 22.40 ± 3.05 | 30.26 ± 4.09 | 30.18 ± 3.84 |
| L insular cortex | 7.30 ± 1.02 | 9.94 ± 0.90 | 10.03 ± 0.87 | 9.96 ± 0.83 |
| R insular cortex | 6.74 ± 0.89 | 9.20 ± 0.89 | 9.29 ± 1.00 | 9.19 ± 0.91 |
| L lateral orbitofrontal gyrus | 6.31 ± 1.29 | 7.27 ± 1.78 | 8.75 ± 2.03 | 8.89 ± 1.92 |
| R lateral orbitofrontal gyrus | 5.79 ± 1.32 | 6.61 ± 1.82 | 8.00 ± 1.83 | 8.02 ± 1.73 |
| L lingual gyrus | 12.95 ± 2.40 | 16.21 ± 2.75 | 17.88 ± 3.11 | 17.98 ± 3.02 |
| R lingual gyrus | 13.09 ± 2.42 | 16.99 ± 2.85 | 18.05 ± 2.88 | 18.28 ± 2.87 |
| L middle frontal gyrus | 50.96 ± 6.99 | 55.36 ± 6.18 | 70.32 ± 8.08 | 71.95 ± 8.33 |
| R middle frontal gyrus | 51.26 ± 7.10 | 55.02 ± 6.73 | 70.72 ± 8.37 | 71.81 ± 9.07 |
| L middle orbitofrontal gyrus | 9.20 ± 2.00 | 9.75 ± 1.69 | 12.61 ± 2.18 | 12.70 ± 2.28 |
| R middle orbitofrontal gyrus | 9.17 ± 1.89 | 9.68 ± 1.81 | 12.59 ± 2.15 | 12.65 ± 2.23 |
| L middle occipital gyrus | 23.04 ± 5.24 | 19.52 ± 3.87 | 31.61 ± 6.18 | 31.97 ± 5.90 |
| R middle occipital gyrus | 23.79 ± 5.64 | 21.35 ± 5.41 | 32.66 ± 6.79 | 32.97 ± 6.30 |
| L middle temporal gyrus | 25.03 ± 4.50 | 25.83 ± 2.94 | 34.39 ± 4.56 | 36.34 ± 3.94 |
| R middle temporal gyrus | 25.49 ± 4.85 | 25.94 ± 3.45 | 35.05 ± 5.38 | 36.37 ± 4.76 |
| L parahippocampal gyrus | 6.31 ± 1.20 | 7.43 ± 1.23 | 8.70 ± 1.36 | 8.52 ± 1.30 |
| R parahippocampal gyrus | 6.50 ± 1.36 | 7.66 ± 1.42 | 8.92 ± 1.56 | 8.72 ± 1.48 |
| L postcentral gyrus | 22.00 ± 2.70 | 21.99 ± 2.63 | 30.32 ± 2.77 | 29.78 ± 2.62 |
| R postcentral gyrus | 21.35 ± 2.85 | 21.97 ± 2.89 | 29.47 ± 3.25 | 28.63 ± 3.26 |
| L precentral gyrus | 26.44 ± 3.03 | 27.10 ± 2.99 | 36.45 ± 2.83 | 35.82 ± 2.67 |
| R precentral gyrus | 25.88 ± 3.75 | 26.77 ± 3.57 | 35.64 ± 3.64 | 34.62 ± 3.56 |
| L precuneus | 10.68 ± 2.25 | 12.83 ± 2.16 | 14.64 ± 2.25 | 14.06 ± 1.94 |
| R precuneus | 10.59 ± 1.91 | 12.65 ± 2.17 | 14.55 ± 2.10 | 14.02 ± 1.90 |
| L putamen | 4.25 ± 0.52 | 5.17 ± 0.73 | 5.87 ± 0.69 | 5.69 ± 0.68 |
| R putamen | 4.22 ± 0.49 | 5.13 ± 0.61 | 5.83 ± 0.63 | 5.77 ± 0.62 |
| L superior frontal gyrus | 57.67 ± 9.03 | 60.91 ± 6.18 | 79.44 ± 9.53 | 77.26 ± 7.87 |
| R superior frontal gyrus | 57.71 ± 8.66 | 60.74 ± 5.50 | 79.25 ± 7.18 | 76.89 ± 5.89 |
| L superior occipital gyrus | 8.19 ± 2.00 | 7.31 ± 1.79 | 11.29 ± 2.50 | 11.07 ± 2.19 |
| R superior occipital gyrus | 8.70 ± 1.93 | 7.85 ± 1.87 | 12.01 ± 2.47 | 11.81 ± 2.42 |
| L superior parietal gyrus | 26.25 ± 4.30 | 25.05 ± 4.90 | 36.15 ± 4.70 | 34.16 ± 5.02 |
| R superior parietal gyrus | 25.96 ± 4.59 | 24.36 ± 4.50 | 35.63 ± 4.49 | 34.02 ± 4.85 |
| L superior temporal gyrus | 27.66 ± 4.45 | 29.92 ± 3.01 | 38.01 ± 4.06 | 39.22 ± 3.86 |
| R superior temporal gyrus | 26.78 ± 3.28 | 29.10 ± 3.19 | 36.92 ± 3.33 | 37.34 ± 3.51 |
| L supramarginal gyrus | 14.41 ± 3.30 | 13.84 ± 3.00 | 19.77 ± 3.66 | 20.18 ± 3.64 |
| R supramarginal gyrus | 13.95 ± 3.34 | 13.51 ± 2.87 | 19.11 ± 3.72 | 19.02 ± 3.53 |
4 Discussion
In this paper, we have described the production of a probabilistic atlas of human brain structures. The atlas was generated from a set of T1-weighted MRI volumes collected from 40 healthy volunteers. Each MRI volume was delineated by trained raters to identify 56 brain structures. The raters followed carefully written protocols and were tested for interrater reliability on a subset of the data. The atlas data sets produced by this research are being made available publicly via our website, http://www.loni.ucla.edu/Atlases/LPBA40. For each version (LPBA40/AIR, LPBA40/FLIRT, LPBA40/SPM5), we are releasing six product components:
the average intensity brain image
the tissue (GM/WM/CSF) density maps
the estimated probability density maps for each structure
the estimated grey-matter probability density maps for each structure
the maximum probability map and maximum likelihood labeling
the maximum probability map and maximum likelihood labeling for the GM density structure maps
Additionally, we are making the anonymized individual subject data available to investigators. The data that will be available include, for each subject, MRI data, tissue classification data, structure delineation labelings, and the transformations used to map these data between the different spaces used in the production of the versions of the atlas.
The main product of this research – the probabilistic maps of brain structure – can be used as a basis for the analysis of various types of neuroimaging data. The atlases can be used to assign structure probabilities to new images by aligning the images and the atlas. Ideally, the registration methods would remove all variation and match the boundaries of these structures exactly and all anatomical variation would be described by the deformation process itself. In practice, the probabilities in the atlases reflect the variation seen across the subjects in the atlas which was not accounted for by the particular registration methods that were used. Thus, these probabilities can be used to attribute a degree of confidence to a label that was aligned to the atlas space using the same registration technique on data that were similarly processed and acquired. This has direct applications in areas such as functional imaging, where one can align the atlas to a subject data set and associate areas of activation with the probabilistic segmentation.
The probabilistic atlas and associated data can also play several important roles in the development of new algorithms for automated delineation. The atlases provide prior information that can be used by statistical classifiers for automatically labeling features in new images (Collins et al., 1995; Leemput et al., 1999a, b; Heckemann et al., 2006); similarly, the individual labelings and MRI data could also be applied using the multi-atlas approach presented by Klein et al. (2005). Additionally, the manual delineations provide a reference data set to which the results of automated algorithms can be compared. An example the utility of such data sets is the Internet Brain Segmentation Repository 10, which provides delineated reference MRI data specifically for the purpose of analyzing and improving segmentation approaches. While human rater reliability is subject to variability, manual delineation is still the standard for interpretation of imaging data. The validation data produced during the training process for our raters quantifies this variability and can be used to give context to the performance of machine algorithms that automatically label the brain. While the raters approached the ICC metric iteratively during their training, those metrics and the Jaccard similarity metrics can serve as target values for machine algorithms or other raters to achieve. The delineation data may also serve as a training set for automated algorithms that require training. Additional delineations can be performed following the written protocols to obtain additional testing or training data.
While this research shares many features with other multi-subject atlasing efforts, we describe briefly some of its distinguishing features and note that different atlases are likely to be appropriate for different applications. First, few multi-subject atlas studies have been performed where human raters labeled the brain in a comprehensive manner. While Collins et al. (1999) have generated atlases based on delineations of MRI from more subjects (N=152), those results were produced using automated labeling of brain structures due to the resources required for performing manual delineations. Hammers et al. (2003) have also used manual delineation techniques to create a maximum probability labeled brain atlas, but their atlas was constructed from fewer subjects (N=20) and the structures chosen for delineation were concentrated on the temporal lobe. The atlas described by Hammers et al. (2003) was later extended with data from additional subjects and labels for additional structures; these labels emphasize the temporal lobe and the frontal lobe (Heckemann et al., 2006). In contrast, our atlas contains labels for at least 4 sub-structures in each of the frontal, temporal, parietal, and occipital lobes for each hemisphere. The atlases used in the work by Klein et al. (2005) used 20 subjects and 36 cortical areas, with no subdivision of the occipital lobe. The atlas produced by Mega et al. (2005) used 20 subjects and 68 structures, but was composed of data from an aging population, while our atlas is composed of data from a younger population.
A significant component of this work is the public availability of the atlas data via our webset; they currently represent one of the more comprehensive multi-subject structure atlases that is freely available. Through this, we hope this atlas can serve as a resource to the neuroimaging community. In related work, we are developing a validation framework to test the accuracy of segmentation algorithms based on the manual delineations described in this paper. Additionally, we already have identified relations of the delineated structures to corresponding labels in the Foundational Model of Anatomy 11, and several additional atlases, to facilitate bioinformatics applications. For example, we developed a web service that computes associations of the neuroanatomic terms with user-specified search strings (e.g., “working memory”); these results can then be displayed on the probabilistic atlas so that users can visualize and interrogate literature relevant to these associations (seehttp://www.pubbrain.org).
Supplementary Material
Acknowledgments
This work was funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54-RR021813 (PI: Toga). Information on the National Centers for Biomedical Computing can be obtained fromhttp://nihroadmap.nih.gov/bioinformatics. Funding for this project was also provided by P41-RR013642 (PI: Toga), by NIH Roadmap P20-RR020750 (PI: Bilder) and by R01-MH60374 (PI: Bilder).
Footnotes
available online athttp://www.bic.mni.mcgill.ca/software/distribution/
Salamon’s Neuroanatomy and Neurovasculature Web-Atlas Resource, available online athttp://www.radnet.ucla.edu/sections/DINR/index.htm
available online athttp://www.loni.ucla.edu/NCRR/protocols.aspx?id=722
available online athttp://bishopw.loni.ucla.edu
available online athttp://www.loni.ucla.edu/Atlases/ICBM452
available online athttp://www.fmrib.ox.ac.uk/fsl/
available online athttp://www.fil.ion.ucl.ac.uk/spm/software/spm5/
available online athttp://www.r-project.org/
available online athttp://www.cma.mgh.harvard.edu/ibsr/
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Apostolova LG, Lu P, Rogers S, Dutton RA, Hayashi KM, Toga AW, Cummings JL, Thompson PM. 3D mapping of language networks in clinical and pre-clinical Alzheimer’s disease. Brain Lang. 2007 May; doi: 10.1016/j.bandl.2007.03.008. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner J, Andersson JL, Friston KJ. High-dimensional image registration using symmetric priors. NeuroImage. 1999 Jun;9 (6 Pt 1):619–628. doi: 10.1006/nimg.1999.0437. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston K. Multimodal image coregistration and partitioning–a unified framework. NeuroImage. 1997 Oct;6 (3):209–217. doi: 10.1006/nimg.1997.0290. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Unified segmentation. NeuroImage. 2005 Jul;26 (3):839–851. doi: 10.1016/j.neuroimage.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Bajcsy R, Lieberson R, Reivich M. A computerized system for the elastic matching of deformed radiographic images to idealized atlas images. J Comput Assist Tomogr. 1983 Aug;7 (4):618–625. doi: 10.1097/00004728-198308000-00008. [DOI] [PubMed] [Google Scholar]
- Brinkley JF, Bradley SW, Sundsten JW, Rosse C. The digital anatomist information system and its use in the generation and delivery of web-based anatomy atlases. Comput Biomed Res. 1997 Dec;30 (6):472–503. doi: 10.1006/cbmr.1997.1461. [DOI] [PubMed] [Google Scholar]
- Chiang M-C, Reiss AL, Lee AD, Bellugi U, Galaburda AM, Korenberg JR, Mills DL, Toga AW, Thompson PM. 3D pattern of brain abnormalities in Williams syndrome visualized using tensor-based morphometry. NeuroImage. 2007 Apr; doi: 10.1016/j.neuroimage.2005.02.018. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen GE, Joshi SC, Miller MI. Volumetric transformation of brain anatomy. IEEE Trans Med Imaging. 1997 Dec;16 (6):864–877. doi: 10.1109/42.650882. [DOI] [PubMed] [Google Scholar]
- Collins DL, Holmes CJ, Evans TMPAC. Automatic 3-D model-based neuroanatomical segmentation. Human Brain Mapping. 1995;3 (3):190–208. [Google Scholar]
- Collins DL, Neelin P, Peters TM, Evans AC. Automatic 3D intersubject registration of MR volumetric data in standardized Talairach space. J Comput Assist Tomogr. 1994;18 (2):192–205. [PubMed] [Google Scholar]
- Collins DL, Zijdenbos AP, Baarè WFC, Evans AC. In: Kuba A, Samal M, Todd-Pokropek A, editors. ANI-MAL+INSECT: Improved cortical structure segmentation; Information Processing in Medical Imaging: 16th International Conference, IPMI’99, Visegrd; Hungary. June/July 1999; Proceedings. Vol. 1613 of LNCS Springer; Berlin/Heidelberg. 1999. pp. 210–223. [Google Scholar]
- Csernansky JG, Joshi S, Wang L, Haller JW, Gado M, Miller JP, Grenander U, Miller MI. Hippocampal morphometry in schizophrenia by high dimensional brain mapping. Proc Natl Acad Sci U S A. 1998 Sep;95 (19):11406–11411. doi: 10.1073/pnas.95.19.11406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davatzikos C, Prince J, Bryan R. Image registration based on boundary mapping. IEEE Transactions on Medical Imaging. 1996 doi: 10.1109/42.481446. [DOI] [PubMed] [Google Scholar]
- Dogdas B, Shattuck DW, Leahy RM. Segmentation of skull and scalp in 3-D human MRI using mathematical morphology. Hum Brain Mapp. 2005 Dec;26 (4):273–285. doi: 10.1002/hbm.20159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvernoy H. The Human Brain, Surface, Blood Supply, and Three-Dimensional Sectional Anatomy. 2. Springer-Verlag; Vienna: 1999. [Google Scholar]
- Eno K, Sundsten JW, Brinkley JF. A multimedia Anatomy Browser incorporating a knowledge base and 3D images. Proc Annu Symp Comput Appl Med Care; 1991. pp. 727–731. [PMC free article] [PubMed] [Google Scholar]
- Evans AC, Collins DL, Mills SR, Brown ED, Kelly RL, Peters TM. 3D Statistical neuroanatomical models from 305 MRI Volumes. Nuclear Science Symposium and Medical Imaging Conference, 1993., 1993 IEEE Conference Record.1993. [Google Scholar]
- Fischl B, Sereno MI, Tootell RB, Dale AM. High-resolution intersubject averaging and a coordinate system for the cortical surface. Hum Brain Mapp. 1999;8 (4):272–284. doi: 10.1002/(SICI)1097-0193(1999)8:4<272::AID-HBM10>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gee JC, Reivich M, Bajcsy R. Elastically deforming 3D atlas to match anatomical brain images. J Comput Assist Tomogr. 1993;17 (2):225–236. doi: 10.1097/00004728-199303000-00011. [DOI] [PubMed] [Google Scholar]
- Greitz T, Bohm C, Holte S, Eriksson L. A computerized brain atlas: construction, anatomical content, and some applications. J Comput Assist Tomogr. 1991;15 (1):26–38. [PubMed] [Google Scholar]
- Hammers A, Allom R, Koepp MJ, Free SL, Myers R, Lemieux L, Mitchell TN, Brooks DJ, Duncan JS. Three-dimensional maximum probability atlas of the human brain, with particular reference to the temporal lobe. Human Brain Mapping. 2003;19:224–247. doi: 10.1002/hbm.10123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckemann RA, Hajnal JV, Aljabar P, Rueckert D, Hammers A. Automatic anatomical brain mri segmentation combining label propagation and decision fusion. NeuroImage. 2006 Oct;33 (1):115–126. doi: 10.1016/j.neuroimage.2006.05.061. [DOI] [PubMed] [Google Scholar]
- Höhne KH, Bomans M, Riemer M, Schubert R, Tiede U, Lierse W. A volume-based anatomical atlas. IEEE Comput Graph Appl. 1992;12(4):73–78. [Google Scholar]
- Hollingshead . Four Factor Index of Social Status. Yale University; New Haven: 1975. [Google Scholar]
- Hollingshead A, Redlich F. Social Class and Mental Illness. John Wiley; New York: 1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaccard P. The distribution of the flora in the alpine zone. New Phytologist. 1912 Feb;11 (2):37–50. [Google Scholar]
- Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. NeuroImage. 2002 Oct;17 (2):825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- Jenkinson M, Smith S. A global optimisation method for robust affine registration of brain images. Med Image Anal. 2001 Jun;5 (2):143–156. doi: 10.1016/s1361-8415(01)00036-6. [DOI] [PubMed] [Google Scholar]
- Johnson HJ, Christensen GE. Consistent landmark and intensity-based image registration. IEEE Transactions on Medical Imaging. 2002;21 (5):450–461. doi: 10.1109/TMI.2002.1009381. [DOI] [PubMed] [Google Scholar]
- Kikinis R, Gleason PL, Moriarty TM, Moore MR, Alexander E, Stieg PE, Matsumae M, Lorensen WE, Cline HE, Black PM, Jolesz FA. Computer-assisted interactive three-dimensional planning for neurosurgical procedures. Neurosurgery. 1996 Apr;38(4):640–9. discussion 649–51. [PubMed] [Google Scholar]
- Klein A, Mensh B, Ghosh S, Tourville J, Hirsch J. Mindboggle: automated brain labeling with multiple atlases. BMC Medical Imaging. 2005:5. doi: 10.1186/1471-2342-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leemput KV, Maes F, Vandermeulen D, Suetens P. Automated model-based bias field correction of mr images of the brain. IEEE Trans Med Imaging. 1999a Oct;18 (10):885–896. doi: 10.1109/42.811268. [DOI] [PubMed] [Google Scholar]
- Leemput KV, Maes F, Vandermeulen D, Suetens P. Automated model-based tissue classification of mr images of the brain. IEEE Trans Med Imaging. 1999b Oct;18 (10):897–908. doi: 10.1109/42.811270. [DOI] [PubMed] [Google Scholar]
- MacDonald D, Avis D, Evans AC. Multiple surface identification and matching in magnetic resonance images. In: Robb RA, editor. Visualization in Biomedical Computing 1994. Vol. 2359 of Proc. SPIE. 1994. pp. 160–169. [Google Scholar]
- Mazziotta J, Toga A, Evans A, Fox P, Lancaster J, Zilles K, Woods R, Paus T, Simpson G, Pike B, Holmes C, Collins L, Thompson P, MacDonald D, Iacoboni M, Schormann T, Amunts K, Palomero-Gallagher N, Geyer S, Parsons L, Narr K, Kabani N, Goualher GL, Feidler J, Smith K, Boomsma D, Pol HH, Cannon T, Kawashima R, Mazoyer B. A four-dimensional probabilistic atlas of the human brain. J Am Med Inform Assoc. 2001;8 (5):401–430. doi: 10.1136/jamia.2001.0080401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazziotta JC, Toga AW, Evans A, Fox P, Lancaster J. A probabilistic atlas of the human brain: theory and rationale for its development. The International Consortium for Brain Mapping (ICBM) NeuroImage. 1995 Jun;2 (2):89–101. doi: 10.1006/nimg.1995.1012. [DOI] [PubMed] [Google Scholar]
- Mega MS, Dinov ID, Mazziotta JC, Manese M, Thompson PM, Lindshield C, Moussai J, Tran N, Olsen K, Zoumalan CI, Woods RP, Toga AW. Automated brain tissue assessment in the elderly and demented population: construction and validation of a sub-volume probabilistic brain atlas. NeuroImage. 2005 Jul;26 (4):1009–1018. doi: 10.1016/j.neuroimage.2005.03.031. [DOI] [PubMed] [Google Scholar]
- Narr KL, Cannon TD, Woods RP, Thompson PM, Kim S, Asunction D, van Erp TGM, Poutanen VP, Huttunen M, Lnnqvist J, Standerksjld-Nordenstam CG, Kaprio J, Mazziotta JC, Toga AW. Genetic contributions to altered callosal morphology in schizophrenia. J Neurosci. 2002 May;22 (9):3720–3729. doi: 10.1523/JNEUROSCI.22-09-03720.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rex DE, Ma JQ, Toga AW. The loni pipeline processing environment. NeuroImage. 2003 Jul;19 (3):1033–1048. doi: 10.1016/s1053-8119(03)00185-x. [DOI] [PubMed] [Google Scholar]
- Roland PE, Graufelds CJ, Wåhlin J, Ingelman L, Andersson M, Ledberg A, Pedersen J, Åkerman S, Dabringhaus A, Zilles K. Human brain atlas: For high-resolution functional and anatomical mapping. Human Brain Mapping. 1994;1 (3):173–184. doi: 10.1002/hbm.460010303. [DOI] [PubMed] [Google Scholar]
- Salamon G, Huang Y. Radiologic anatomy of the brain Springer Verlag Heidelberg New York 1 vol. 1976. Springer Verlag; New York: 1976. [Google Scholar]
- Salamon N, Sicotte N, Mongkolwat P, Shattuck D, Salamon G. The human cerebral cortex on MRI: value of the coronal plane. Surg Radiol Anat. 2005 Dec;27 (5):431–443. doi: 10.1007/s00276-005-0022-7. [DOI] [PubMed] [Google Scholar]
- Schiemann T, Freudenberg J, Pflesser B, Pommert A, Priesmeyer K, Riemer M, Schubert R, Tiede U, Höhne KH. Exploring the Visible Human using the VOXEL-MAN framework. Comput Med Imaging Graph. 2000;24 (3):127–132. doi: 10.1016/s0895-6111(00)00013-6. [DOI] [PubMed] [Google Scholar]
- Shattuck DW, Leahy RM. BrainSuite: an automated cortical surface identification tool. Medical Image Analysis. 2002;6 (1):129–142. doi: 10.1016/s1361-8415(02)00054-3. [DOI] [PubMed] [Google Scholar]
- Shattuck DW, Sandor-Leahy SR, Schaper KA, Rottenberg DA, Leahy RM. Magnetic resonance image tissue classification using a partial volume model. NeuroImage. 2001;13:856–876. doi: 10.1006/nimg.2000.0730. [DOI] [PubMed] [Google Scholar]
- Shen D, Davatzikos C. HAMMER: hierarchical attribute matching mechanism for elastic registration. IEEE Trans Med Imaging. 2002 Nov;21 (11):1421–1439. doi: 10.1109/TMI.2002.803111. [DOI] [PubMed] [Google Scholar]
- Shrout PE, Fleiss JL. Intraclass correlations: Uses in assessing rater reliability. Psychological Bulletin. 1979;86 (2):420–428. doi: 10.1037//0033-2909.86.2.420. [DOI] [PubMed] [Google Scholar]
- Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imag. 1998 Feb;:17. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- Smith SM. Fast robust automated brain extraction. Hum Brain Mapp. 2002 Nov;17 (3):143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TEJ, JohansenBerg H, Bannister PR, Luca MD, Drobnjak I, Flitney DE, Niazy RK, Saunders J, Vickers J, Zhang Y, Stefano ND, Brady JM, Matthews PM. Advances in functional and structural MR image analysis and implementation as FSL. NeuroImage. 2004;23(Suppl 1):S208–S219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- Sowell ER, Thompson PM, Holmes CJ, Batth R, Jernigan TL, Toga AW. Localizing age-related changes in brain structure between childhood and adolescence using statistical parametric mapping. NeuroImage. 1999 Jun;9 (6 Pt 1):587–597. doi: 10.1006/nimg.1999.0436. [DOI] [PubMed] [Google Scholar]
- Sowell ER, Thompson PM, Leonard CM, Welcome SE, Kan E, Toga AW. Longitudinal mapping of cortical thickness and brain growth in normal children. Journal of Neuroscience. 2004;24 (38):8223–8231. doi: 10.1523/JNEUROSCI.1798-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer V, Ackerman MJ, Scherzinger AL, Whitlock D. The visible human male: a technical report. J Am Med Inform Assoc. 1996;3 (2):118–130. doi: 10.1136/jamia.1996.96236280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styner M, Lieberman JA, Pantazis D, Gerig G. Boundary and medial shape analysis of the hippocampus in schizophrenia. Med Image Anal. 2004 Sep;8 (3):197–203. doi: 10.1016/j.media.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Talairach J, Szikla G, Tournoux P, Prosalentis A, Bordas-Ferrier M, Covello L, Iacob M, Mempel E. Atlas d’anatomie stereotaxique du telencephale. Masson; Paris: 1967. [Google Scholar]
- Talairach J, Tournoux P. Georg Thieme Verlag. Stuttgart; New York: 1988. Co-Planar Stereotactic Atlas of the Human Brain: 3-Dimensional Proportional System: An Approach to Cerebral Imaging. [Google Scholar]
- Thompson PM, Hayashi KM, Sowell ER, Gogtay N, Giedd JN, Rapoport JL, de Zubicaray GI, Janke AL, Rose SE, Semple J, Doddrell DM, Wang YL, van Erp T, Cannon TD, Toga AW. Mapping cortical change in alzheimer’s disease, brain development and schizophrenia. NeuroImage. 2004 Sept;23 (1):S2–S18. doi: 10.1016/j.neuroimage.2004.07.071. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Toga AW. A surface based technique for warping 3-dimensional images of the brain. IEEE Transactions on Medical Imaging. 1996;15 (4):1–16. doi: 10.1109/42.511745. [DOI] [PubMed] [Google Scholar]
- Tiede U, Bomans M, Höhne KH, Pommert A, Riemer M, Schiemann T, Schubert R, Lierse W. A computerized three-dimensional atlas of the human skull and brain. AJNR Am J Neuroradiol. 1993;14(3):551–9. doi: 10.1007/978-1-4899-1701-0_9. discussion 560–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toga AW, Mazziotta JC. Brain Mapping: The Methods. 2. Academic Press; New York: 2002. [Google Scholar]
- Van Essen DC. A Population-Average, Landmark- and Surface-based (PALS) atlas of human cerebral cortex. NeuroImage. 2005 Nov;28 (3):635–662. doi: 10.1016/j.neuroimage.2005.06.058. [DOI] [PubMed] [Google Scholar]
- Van Essen DC, Dierker D, Snyder AZ, Raichle ME, Reiss AL, Korenberg J. Symmetry of cortical folding abnormalities in Williams syndrome revealed by surface-based analyses. J Neurosci. 2006 May;26 (20):5470–5483. doi: 10.1523/JNEUROSCI.4154-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells WM, Viola P, Atsumi H, Nakajima S, Kikinis R. Multi-modal volume registration by maximization of mutual information. Med Image Anal. 1996 Mar;1 (1):35–51. doi: 10.1016/s1361-8415(01)80004-9. [DOI] [PubMed] [Google Scholar]
- Woods RP, Grafton ST, Watson JDG, Sicotte NL, Mazziotta JC. Automated image registration: II. Intersubject validation of linear and nonlinear models. Journal of Computer Assisted Tomography. 1998;22:153–165. doi: 10.1097/00004728-199801000-00028. [DOI] [PubMed] [Google Scholar]
- Woods RP, Mazziotta JC, Cherry SR. MRI-PET registration with automated algorithm. J Comput Assist Tomogr. 1993;17 (4):536–546. doi: 10.1097/00004728-199307000-00004. [DOI] [PubMed] [Google Scholar]
- Xue Z, Shen D, Davatzikos C. Statistical representation of high-dimensional deformation fields with application to statistically constrained 3D warping. Med Image Anal. 2006 Oct;10 (5):740–751. doi: 10.1016/j.media.2006.06.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.