Abstract
Global warming is associated with increasing stress and mortality on temperate seagrass beds, in particular during periods of high sea surface temperatures during summer months, adding to existing anthropogenic impacts, such as eutrophication and habitat destruction. We compare several expressed sequence tag (EST) in the ecologically important seagrass Zostera marina (eelgrass) to elucidate the molecular genetic basis of adaptation to environmental extremes. We compared the tentative unigene (TUG) frequencies of libraries derived from leaf and meristematic tissue from a control situation with two experimentally imposed temperature stress conditions and found that TUG composition is markedly different among these conditions (all P < 0.0001). Under heat stress, we find that 63 TUGs are differentially expressed (d.e.) at 25°C compared with lower, no-stress condition temperatures (4°C and 17°C). Approximately one-third of d.e. eelgrass genes were characteristic for the stress response of the terrestrial plant model Arabidopsis thaliana. The changes in gene expression suggest complex photosynthetic adjustments among light-harvesting complexes, reaction center subunits of photosystem I and II, and components of the dark reaction. Heat shock encoding proteins and reactive oxygen scavengers also were identified, but their overall frequency was too low to perform statistical tests. In all conditions, the most abundant transcript (3–15%) was a putative metallothionein gene with unknown function. We also find evidence that heat stress may translate to enhanced infection by protists. A total of 210 TUGs contain one or more microsatellites as potential candidates for gene-linked genetic markers. Data are publicly available in a user-friendly database at http://www.uni-muenster.de/Evolution/ebb/Services/zostera.
Eletronic Supplementary Material
The online version of this article (doi:10.1007/s10126-007-9065-6) contains supplementary material which is available to authorized users.
Keywords: Gene expression profiling, EST library, Ecological genomics, Temperature stress, Seagrass, Zostera marina
Introduction
Global warming is associated with an increased frequency of environmental extremes, such as heat waves, floods, and droughts (IPCC 2007). Such extremes may be more important for the persistence of local populations than changes in mean conditions (Gaines and Denny 1993). In coastal–habitat-forming species, such as seagrasses or corals, losses caused by summer surface water extremes in temperature have already been reported (Cerrano et al. 2000; Hughes et al. 2003; Reusch et al. 2005). Central questions in marine ecology are thus how organisms physiologically adjust to such stress events, which molecular genetic mechanisms may confer plasticity and tolerance toward extreme conditions, and whether such plasticity itself has a heritable genetic basis and may evolve in the face of global warming (Hofmann et al. 2005; Reusch and Wood 2007).
Ecogenomic techniques are increasingly utilized in the marine realm and hold great promise to address some of above questions (Hofmann et al. 2005; Dupont et al. 2007). Gene transcription profiling, in particular, is one important step toward identifying those genes and metabolic pathways that underlie ecologically important traits, such as stress tolerance (Feder and Mitchell-Olds 2003; Vasemägi and Primmer 2005; Ouborg and Vriezen 2007). In marine systems, transcription profiling has been successful in unravelling the genetic basis of temperature adaptation (Whitehead and Crawford 2006), of calcification in phytoplankton (Fujiwara et al. 2007), and the response of marine plant and animal species to abiotic stresses (Pearson et al. 2001; Jenny et al. 2002; Hashimoto et al. 2004; Kassahn et al. 2007).
The construction of expressed sequence tag (EST) libraries is a convenient entry point for a whole suite of ecogenomic tools (Bouck and Vision 2007). EST libraries enable us to obtain DNA sequence information of expressed genes sufficiently detailed to tentatively characterize the underlying gene function by homology search. Bulk RNA containing messenger (m)RNA is extracted, converted into complementary (c)DNA, and subsequently cloned and sequenced using standard methods. EST libraries are a cost-effective tool to characterize genes important under particular conditions, as well as the starting point for the development of molecular genetic markers, such as gene-linked microsatellites and single nucleotide polymorphisms (SNP). In marine species, gene-linked microsatellites (EST-SSR = simple sequence repeats) were successfully identified, for example, in oyster (Wang and Guo 2007) and shrimp (Pérez et al. 2005).
Any EST library is specific for a certain tissue and experimental condition under which the mRNA was sampled. Conversely, when several EST libraries were obtained under different conditions, the contribution of reads to an identical tentative gene cluster is indicative for its expression strength, provided that libraries were not normalized (Okubo et al. 1992; Bouck and Vision 2007). This way, important inferences on physiologic adjustments to changes in environmental parameters at the level of the transcriptome become possible at least for those genes that are higher expressed in at least one of the experimental conditions to be compared (Kore-eda et al. 2004; Kuo et al. 2004).
We present for the first time several EST libraries of a seagrass, Zostera marina (eelgrass). Z. marina is a habitat-forming, or ecosystem-engineering angiosperm (sensu; Jones et al. 1994), forming dense meadows along sedimentary shorelines, ranging from subarctic to subtropical latitudes. Experimental and observational evidence suggests that local populations from temperate regions experience mortality above a critical summer threshold temperature of 25°C (Williams 2001; Greve et al. 2003; Reusch et al. 2005), and such events are predicted to increase in the next decades (IPCC 2007).
Against this empirical background, our motivation was thus to assess how seagrass physiology is affected by stress events. In an attempt to obtain first data on transcriptional regulation under different temperature conditions, we conducted an analysis of the plants’ transcriptome comparing several nonnormalized EST libraries to identify genes that are critical under temperature stress. Subsequently, the response to temperature stress was compared with transcriptomic changes observable in the plant model Arabidopsis thaliana as a reference. Moreover, we wanted to mine the EST data for gene-linked microsatellites as a valuable resource for gene-linked genetic marker development.
Materials and Methods
Study Species
Zostera marina (eelgrass) is a widespread marine angiosperm or seagrass species exhibiting a mixture of sexual and clonal (vegetative) reproduction (Reusch et al. 1999). The species is monoecious and self-compatible and exhibits true subaqueous pollination (Reusch 2000). Systematically, seagrasses are polyphyletic clades within the monocotyledoneous order Alismatales (Les et al. 1997). Eelgrass populations sampled for constructing the genetic libraries were in the south-western Baltic, a semi-enclosed sea with brackish water. At sampling sites Schilksee and Maasholm (south-western Baltic Sea, Germany), salinities throughout the year range from 12 to 20 g/kg. Samples were collected in 1.6- to 2.5-m depth.
Experimental Conditions
In total, plants were sampled under five different experimental conditions and tissue types. Four of those were the focus of the present study, whereas sequence data from another library (A) solely serve to improve the clustering and homology search (Table 1). Libraries C and D represent plant material under natural conditions, collected from the field in average summer or winter conditions, respectively. In addition, for construction of library E and F, entire plants sampled at the same site during the same time together with material for library D were exposed to increasing heat stress in 60-L aquaria filled with ambient Baltic Sea water. Aquaria were aerated and illuminated for 10 h with approximately 40 μ E x m-2 * s-1 at the water surface. The water temperature was then increased to 17°C in library E during 36 h, and to 25°C within another 24 h in that plants that served as source for library F. Note that, in addition to the temperature stress, we may have elicited additional stress responses, for example to translocation and altered light regime.
Table 1.
Details on EST libraries constructed for eelgrass Zostera marina
| Library | Location | Date | Tissue | Condition | Remark |
|---|---|---|---|---|---|
| A | Maasholm | April 18, 2004 | meristem | 4°C – >6°C | Winter situation* |
| C | Maasholm | Aug 11, 2004 | meristem /leaf | Ambient (23°C) | Summer situation |
| D | Schilksee | Feb 2005 | meristem /leaf | Ambient (4°C) | Winter situation |
| E | Schilksee | Feb 2005 | meristem /leaf | 4°C – >17°C | Mild heat stress |
| F | Schilksee | Feb 2005 | meristem /leaf | 17°C – >25°C | Strong heat stress |
Position Maasholm: 54°41′N 10°00′E ; Schilksee 54°82′N 10°81′E. Both sites are 3 km distant
* Ambient temperature 4°C; plants were kept 1 day in lab before RNA extraction
RNA Extraction
Total RNA was extracted using the RNeasy plant kit (Qiagen, Hilden, Germany). In the case of libraries based on field-sampled material, cleaned tissue was frozen in liquid nitrogen < 20 min after uprooting, while keeping them in ambient water. The RNA quality of an aliquot was checked on EtBr stained agarose gels. RNA concentration was equilibrated among leaf and meristematic tissue in the libraries C to F (Table 1) according to RNA concentration measurements using a NanoDrop spectrophotometer. Each library contained the pooled RNA of four to six genotypes.
Library Construction and DNA Sequencing
All libraries were constructed using the Creator SMART library construction kit (BD Clontech), using the LD PCR based method. Between 22 and 28 PCR cycles were performed before size separation of inserts. The size-selected cDNA (fragments > 800 base pairs) was directionally ligated at the restriction site Sfi1 of the pDNR-lib vector (BD Clontech) and electroporated into E. coli strain DH10B (Invitrogen). Library D shows a low amount of clones carrying an insert. Presence of inserts was controlled via PCR by using M13 primers to exclude empty clones. Dideoxy-termination DNA sequencing was performed on ABI sequencers (ABI 3130XL at MPI for Limnology Plön in case of libraries A, C, D) and ABI 3730XL (libraries E, F, at MPI Molecular Genetics, Berlin) after plasmid preparation using the BigDye 3.1 sequencing chemistry and a forward M13 primer only (GTA AAA CGA CGG CCA GT).
Data Analyses and Bioinformatics
The raw sequence reads were quality-trimmed, poly-A and vector clipped using pregap4 (Staden et al. 2000). Vector flanking sequences surrounding the cloning sites, including the linker used in constructing the library, were included as parameters. Successfully trimmed EST reads were then assembled into tentative gene clusters using CAP3 (Huang and Madan 1999). Two parameters were specified: there was no reverse orientation of sequence reads, and one read of good quality is sufficient to build a consensus at a given position.
Tentative Unigene TUG Annotation
To infer putative functions of the identified tentative unigenes (TUG; >100 nucleotides only), we performed homology searches against three protein databases: SwissProt, Gene Ontology (GO), and Prodom. For the SwissProt search, we used the BLASTX algorithm in conjunction with the database vs. 52.0, with an Expect-value threshold of E ≤ 0.01. For further analysis, the first hit was used if being a plant, otherwise we continued until a plant hit was found. If no higher plant hit was present, we used the other species instead, as our initial material was not axenic and may contain fungal parasites. The AmiGO tool (www.geneontology.org/cgi-bin/amigo/go.cgi) implementing BLASTX searches (Gish and States 1993) was used to identify similarities with proteins of the GO database (www.geneontology.org). The overlap in their annotation of all tentative unigenes was examined using Venn diagrams.
Open Reading Frame Identification and Microsatellite Search
To predict the presence of nontranslated regions of the TUGs, we were interested in identifying the open reading frames (ORFs). ORF prediction was based on BLAST hits to all three databases used: SwissProt (SP), Prodom (PD), and Gene Ontology (GO). We considered the part of the ORF that matched a SP / GO protein or a PD domain as seed. From there, we searched for the beginning and the end of the ORF. The end is defined by the last nucleotide before a STOP codon. As most likely beginning of an ORF, we went in 5′ direction until the last ATG (Methionine) codon before STOP was reached. Alternatively, if no STOP codon was found, the first nucleotide in frame was defined as the beginning. In the latter case, this was most likely to the result of an EST/contig only partly overlapping with an ORF extending 5′. When ORF prediction was solely based on Prodom annotation, a tentative unigene can have more than one domain, which can slightly overlap, and which can be in different frames. We then considered only the open reading frame corresponding to the match with the lowest E-value. Note that 73 TUGs annotated with a reading frame had at least one stop codon within the matching region to a protein or a domain, possibly because of sequencing errors or PCR/reverse transcription artifacts during the preparation of the libraries.
For searching simple sequence repeats or microsatellites, MISA (MIcroSAtellite, http://www.pgrc.ipk-gatersleben.de/misa) PERL script was used. We searched for all possible motifs of a repeat motif length of two to six nucleotides. Minimal length of repetitions for dinucleotide repeats was six, and for repeat units of three to six nucleotides, it was five. Above determination of ORFs was used to predict the position of microsatellite repeats with respect to coding regions.
Comparison of Gene Expression among EST Libraries
We compared global and individual gene expression patterns based on our nonnormalized cDNA libraries using the approaches proposed by Susko and Roger (2004). Our analyses of gene expression differences focussed on two a priori formulated hypotheses, corresponding to experimental conditions under which the libraries were constructed. We were first interested in a comparison of the natural undisturbed gene expression between summer and winter situation, and thus compared EST library C with D. As second comparison, we concentrated on library F as focal experimental condition and compared the composition of gene clusters with libraries D and/or E. If none of the latter nonstress libraries produced a significant outcome, we pooled the frequency of occurrence over D and E.
Binomial and Chi-square tests were used for gene-by-gene comparisons of expression (Susko and Roger 2004). We did not use any adjustment for multiple comparison, for example the cumulative probability of experiment-wise false-positives at less than alpha proposed by Benjamini and Hochberg (1995), and implemented by Susko and Roger (2004). Rather, we see those genes that individually seem to be over- or underexpressed among EST libraries constructed under different conditions as indicative for further studies. As a precaution, we first examined in a global test whether the transcriptome is generally different among conditions. Only if this was true at P ≤ 0.001, we proceeded with single gene tests.
Rocha et al. (2003) proposed that microsatellites are particularly abundant in stress associated genes because they facilitate their rapid expression level evolution as a result of high mutation rates in promoter regions. We were thus interested in comparing the abundance of microsatellites among those genes that were d.e. under heat stress compared with other TUGs. This was done in the comparison of libraries D+E vs. F only, using Chi-square tests.
Comparison with Arabidopsis thaliana
We were interested in similarities among Z. marina and the plant model Arabidopsis thaliana (mouse-ear cress) stress response. Any differentially expressed (d.e.) TUGs from Zostera were matched against the Arabidopsis thaliana protein data set from the MIPS Database website http://mips.gsf.de/proj/plant/jsf/athal/index.jsp) using BLASTX (Expect value threshold E = 1e-04). The global hypergeometric test (Falcon and Gentleman 2007) was performed to detect functional gene ontology categories overrepresented in the resulting set of significant Arabidopsis BLAST hits. Hypergeometric probabilites were computed to assess whether the frequency of BLAST hits associated with a particular GO term was larger than expected. The method ignores the structure of the gene ontology, treats each GO term as independent from all others and returns raw and adjusted P values. Bonferroni correction was used to adjust P values for multiple testing (Boyle et al. 2004). To avoid redundancy, connected GO terms with significant scores were excluded after the analysis.
All d.e. eelgrass genes were then compared to a set of Arabidopsis stress specific proteins, the AtGenExpress (Kilian et al. 2007). This database contains 3095 (of 22,000) genes, which are only expressed in a specific stress condition in at least one of six time points. The genes are unique for a given stress condition and do not overlap with other stresses.
Zostera marina EST Database
Clipped and passed sequence reads were submitted to dbEST within GenBank (accession numbers AM766003–AM773228 and FC822029–FC823189). In addition to deposition of cleared reads in GenBank all processed and assembled data are publicly available in a database at the Institute for Evolution & Biodiversity, accessible through a web interface http://www.uni-muenster.de/Evolution/ebb/Services/zostera). This database is searchable for tentative gene ID (singletons and contigs), gene name and annotation key words, and microsatellites. TUG identifiers (singletons and contigs) used in the present study are identical to those in the database. A local BLAST server also is implemented.
Results
Assembly, ORF Prediction, and Annotation
In the global assembly, i.e. when pooling sequence reads of all 5 libraries, 8573 passed EST reads clustered to 3593 tentative unigenes (TUGs), 2496 of which were singlets and 1097 were contigs, i.e. clusters consisting of 2 or more reads.
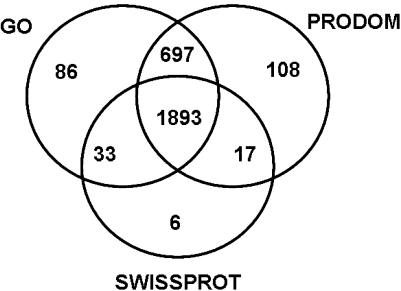
The annotation using the different protein and domain databases yielded largely consistent results. We considered only TUGs that were ≥ 100 nt and hits in a positive reading frame. A core of 1893 TUGs could be annotated with all three databases (Fig. 1), whereas 753 TUGs could not be annotated at all, resulting in total annotated fraction of 79% of all TUGs (2840/3593). Using the Prodom database, a total of 115 TUGs were associated with the key word “transcription,” whereas 35 (0.94%) were specifically designated as transcription factors. The majority of hits to the gene ontology GO database comprised genes of Arabidopsis thaliana (mouse-ear cress; 1857/3593) and Oryza sativa (rice; 853/3593). According to the database comparisons, 761 of all TUGs (singletons or contigs) contained the complete open reading frame, 899 contained no ORF or were not annotated, 1060 and 545 contained portions of the 3′- and 5′ untranslated region (UTR), respectively, whereas 481 genes contained stretches of both UTRs.
Fig. 1.
Venn diagram showing the overlap of significant hits (E-value threshold < 0.0001) of Zostera marina TUGs among different database searched. GO = gene ontology database. Numbers are given for the initial database search before manual editing of some TUGs displaying identical BLASTX hits
After initial annotation, 2 or more TUGs showed the same highly significant BLASTX score for particular SwissProt proteins in 188 cases. In these cases, the assembly into TUGs was manually edited. In 84 cases, we obtained novel “merged” TUGs from 2 or sometimes 3 initial CAP3 TUGs, indicated as “merge” instead of “contig” in the database. As criterion for merging, tentative gene clusters were considered to belong to the same gene if they overlapped < 20% of their read length. Otherwise, they were considered recent duplicates, based on the assumption that clustering in CAP3 is correct in producing different clusters given sufficient sequence overlap. This slightly altered the overall library statistics (total TUGs 3496 including 84 merged TUGs; total annotated TUGs 2743 = 78.5%). The assessment of differential expression among libraries was done with the modified data set.
Microsatellite Identification
In total, we identified 210 genes (6.01%) that contained a total of 223 microsatellite motifs under the specified criteria, i.e. some TUGs contained more than one motif. Among these we found 82 dinucleotide repeats, with the majority being AG/TC and AT-repeats, 113 trinucleotide repeats, and a few tetra-, penta-, and hexanucleotide repeats (Table 2). As hypothesized, trinucleotide repeat motifs are more abundant within the coding region of genes (55) than outside (24, Table 3). In contrast, repeat motifs containing less or more than three nucleotides, resulting in frameshifts when undergoing slip-strand mutations, are primarily found in untranslated regions of a transcript (65), whereas rare in ORFs (16). This difference was statistically significant in a (2× 2 contingeny table, df = 1, Chi-square = 42, P < 0.0001). Consistent with the prediction that microsatellites causing no frame-shift may be more abundant within ORFs, the only detected hexa-nucleotide repeat (AATACC9; unigene ZMD04004) was found within an open reading frame. This TUG had two domains that resembled a zinc-finger domain in PRODOM (PD007661, E = 4e-16). The identity of the gene itself is unclear, it may be a transcription factor, consistent with the zinc-finger domain, or a salt-tolerance like gene (Swiss Prot ID Q9SYM2, Arabidopsis thaliana, E = 4e-20).
Table 2.
Composition and length of microsatellites detected among Zostera marina ESTs
| Motif | No. of repeat units | Total | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | ||
| AC | – | 1 | 1 | 2 | 4 | ||||||
| AG | – | 11 | 5 | 6 | 3 | 2 | 2 | 2 | 1 | 32 | |
| AT | – | 10 | 8 | 3 | 6 | 1 | 1 | 29 | |||
| CT | – | 4 | 2 | 1 | 1 | 1 | 1 | 10 | |||
| GT | – | 5 | 1 | 1 | 7 | ||||||
| AAG | 15 | 6 | 2 | 2 | 1 | 26 | |||||
| AAT | 1 | 1 | |||||||||
| ACC | 1 | 2 | 2 | 1 | 6 | ||||||
| ACG | 1 | 1 | 2 | ||||||||
| AGC | 5 | 5 | |||||||||
| AGG | 5 | 1 | 1 | 7 | |||||||
| ATC | 3 | 1 | 1 | 5 | |||||||
| ATG | 2 | 2 | 4 | ||||||||
| ATT | 6 | 1 | 7 | ||||||||
| CCG | 1 | 1 | |||||||||
| CCT | 3 | 3 | 1 | 7 | |||||||
| CGT | 4 | 4 | |||||||||
| CTG | 6 | 6 | |||||||||
| CTT | 11 | 5 | 3 | 2 | 1 | 22 | |||||
| GGT | 7 | 2 | 9 | ||||||||
| GTT | 1 | 1 | |||||||||
| AAAT | 1 | 1 | 2 | ||||||||
| AATC | 1 | 1 | |||||||||
| AATG | 1 | 1 | |||||||||
| AGAT | 1 | 3 | 1 | 5 | |||||||
| CATC | 2 | 1 | 3 | ||||||||
| ACCT | 1 | 1 | |||||||||
| ATCG | 1 | 1 | |||||||||
| ATGC | 1 | 1 | |||||||||
| AATT | 2 | 2 | |||||||||
| ACTC | 1 | 1 | |||||||||
| ATCT | 1 | 1 | |||||||||
| ATGG | 1 | 1 | |||||||||
| CTGG | 1 | 1 | |||||||||
| GTTT | 1 | 1 | |||||||||
| CTTT | 1 | 1 | 2 | ||||||||
| ATTT | 1 | 1 | |||||||||
| AGAGG | 1 | 1 | |||||||||
| ATTTG | 1 | 1 | |||||||||
| AATACC | 1 | 1 | |||||||||
| Grand total | 223 | ||||||||||
Note that some TUGs contained more than one microsatellite region
Table 3.
Position of microsatellites with respect to putative open reading frames (ORFs)
| Repeat motif | Within ORF | Outside ORF | Mix ORF* | ORF undefined |
|---|---|---|---|---|
| Dinucleotide | 15 | 46 | 1 | 18 |
| Trinucleotide | 54 | 24 | 1 | 31 |
| Tetranucelotide | 0 | 17 | 0 | 7 |
| Pentanucleotide | 0 | 2 | 0 | 0 |
| Hexanucleotide | 1 | 0 | 0 | 0 |
*Mix ORF, microsatellites extend over the predicted border of coding region and UTR
Comparison of Tentative Unigene Frequencies among Libraries
In a global comparison according to Susko and Roger (2004), divergent patterns of gene expression were detected among all pre-planned library contrasts (Table 4). We thus proceeded with a more detailed analysis of single genes that were differentially expressed (d.e.). Given the total number of TUGs in the libraries to be compared, the minimal frequency for detecting d.e. was four reads for the comparison library C vs. library D, and for library D + E vs. F. Accordingly, of the subset of 149 genes where differential expression is detectable, 7 were down- and 19 were up-regulated under winter conditions (library D) versus summer conditions (library C, supplementary Table S1). Qualitatively, many genes of the light reaction, in particular light harvesting proteins (as in TUGs contig 62, 114, merge 29, 71, 188) and reaction subunits themselves (contig 107, 172, 188) are more abundant under summer conditions (Table S1).
Table 4.
Global comparison of EST library composition based on the frequency spectrum of single sequence reads contributing to tentative sequence clusters, according to Susko and Rogers (2000)
| Library comparison | Test statistic (aggregated Chi-square) | P value |
|---|---|---|
| C vs. D | 639.4 | 2.3e-14 |
| D vs. F | 887.4 | 3.5e-06 |
| E vs. F | 166.6 | 1.1e-06 |
| D+E vs. F | 585.1 | 7.5e-09 |
Note that the comparison of libraries C and D lacked the statistical power of the other comparison, as only 1248 passed sequence reads comprise the first library (Table 5). Therefore, despite the very low P value for the global comparison, relatively few individual genes are d.e. In the remainder of this study, we, therefore, focus our discussion on the comparison under experimentally induced stress conditions.
Table 5.
Summary statistics of five eelgrass (Zostera marina) EST libraries
| EST library | Total | |||||
|---|---|---|---|---|---|---|
| A | C | D | E | F | ||
| No. raw EST reads | 390 | 1273 | 2273 | 2688 | 2688 | 9312 |
| No. passed reads | 388 | 1248 | 1883 | 2516 | 2536 | 8573 |
| No. contigs | 49 | 144 | 275 | 342 | 316 | 1097 |
| No. singlets | 265 | 738 | 562 | 1154 | 1240 | 2496 |
| Mean filtered EST length | 446 | 490 | 500 | 450 | 337 | 433 |
| Mean contig length | 576 | 614 | 608 | 616 | 494 | 630 |
| Mean contig depth | 2.5 | 3.5 | 4.8 | 4 | 4.1 | 5.4 |
Pased reads had to be ≥ 100 nucleotides
Our second comparison of libraries concerned the experimental response to heat (and possibly also uprooting and translocation) stress. As for the heat stress response, of 333 TUGs compared among the libraries D + E vs. F, 27 (8%) were up- and 36 (11%) were down-regulated under heat stress (Tables 6 and 7). Among the strongest responses was a 7-fold up-regulation of a putative photosystem I assembly protein (SwissProt Q3BAN1), and a 6-fold increase in a light harvesting, chlorophyll-binding protein (SwissProt P27495). Down-regulations observed were a 15-fold reduction in a chloroplast precursor gene (SwissProt Q6K953), and a 6-fold reduction in a metallothionein-like gene (SwissProt Q40256). Because in several cases, one library contributed no reads to the relevant gene cluster, frequencies could not be estimated, but fold-changes may even be higher in these cases.
Table 6.
Zostera marina TUGs significantly up-regulated in library F (heat stress) with respect to library D (designated DF), E (EF), or both libraries pooled (DE-F)
| Unigene | Contrast | P value | L1 | L2 | Database entry; Gene description – species with best hit |
|---|---|---|---|---|---|
| Contig68 | DE-F/DF/EF | 0.0003 | 33 | 41 | Q41914, low-molecular-weight cysteine-rich protein LCR70 precursor - Arabidopsis thaliana |
| Contig926 | DF | 0.0006 | 19 | 52 | Q7YJV4, chloroplast 30S ribosomal protein S18 - Calycanthus fertilis |
| Contig48 | DF/DE-F/EF | 0 | 0 | 37 | Q9LQI7, probable complex I intermediate-associated protein 30 - Arabidopsis thaliana |
| Contig705 | DF/DE-F/EF | 0.0004 | 6 | 29 | NoHit |
| Contig607 | DF | 0.0003 | 4 | 25 | NoHit |
| Contig787 | DF/DE-F | 0.0007 | 3 | 21 | Q3BAN1, photosystem I assembly protein ycf4 - Phalaenopsis aphrodite subsp. formosana |
| Contig1091 | DF | 0.009 | 4 | 18 | Q05349, auxin-repressed 12.5 kDa protein – Fragaria ananassa |
| Contig126 | DF | 0.0033 | 3 | 18 | NoHit |
| Contig826 | DF | 0.0001 | 1 | 20 | NoHit |
| Contig849 | DF/DE-F/DE | 0.0001 | 0 | 16 | NoHit |
| Merge29 | DE-F/DF | 0.0104 | 5 | 11 | P27521, chlorophyll a-b binding protein 4, chloroplast precursor (LHCI type III CAB-4) — Arabidopsis thaliana |
| Contig65 | DE-F/EF/DF | 0.0035 | 2 | 9 | Q9S7N7, photosystem I reaction center subunit V, chloroplast precursor (PSI-G) — Arabidopsis thaliana |
| Contig736 | DE-F/DF | 0.0035 | 2 | 9 | NoHit |
| Contig592 | DE-F/DF | 0.0084 | 2 | 8 | Q6EW63, ATP synthase subunit alpha (EC 3.6.3.14) - Nymphaea alba |
| Contig606 | DF | 0.0033 | 0 | 10 | Q9MTN0, hypothetical 6.9 kDa protein in psbD-trnT intergenic region (ORF59a) - Oenothera hookeri |
| Contig640 | DE-F/DF | 0.0198 | 2 | 7 | NoHit |
| Contig162 | DF/DE-F | 0.0118 | 0 | 8 | NoHit |
| Contig901 | DF/DE-F | 0.0224 | 0 | 7 | P35017, superoxide dismutase, mitochondrial precursor (EC 1.15.1.1) - Hevea brasiliensis |
| Contig933 | DE-F/DF | 0.0161 | 1 | 6 | P27495, chlorophyll a-b binding protein 40, chloroplast precursor (LHCP) - Nicotiana tabacum |
| Contig945 | DF | 0.0224 | 0 | 7 | NoHit |
| Contig1003 | DE-F | 0.0407 | 1 | 5 | NoHit |
| Contig119 | DF | 0.0425 | 0 | 6 | P07979, lichenase precursor (EC 3.2.1.73) (Endo-beta-1,3-1,4 glucanase) - Nicotiana plumbaginifolia |
| Contig817 | DF/DE-F | 0.0425 | 0 | 6 | GB DQ174248.1, 16S ribosomal RNA, tRNA-Ile - Phaeodactylum tricornutum |
| Contig909 | DE-F/EF/DF | 0.0033 | 0 | 6 | NoHit |
| Merge73 | DF/DE-F | 0.0425 | 0 | 6 | P10708, chlorophyll a-b binding protein 7, chloroplast precursor - Solanum lycopersicum |
| Contig1097 | DE-F | 0.0095 | 0 | 5 | NoHit |
| Merge17 | DE-F | 0.0277 | 0 | 4 | P02889, probable 26S proteasome non-ATPase regulatory subunit 8 - Dictyostelium discoideum (Slime mold) |
The statistical test for each gene was performed by binomial approximation as suggested in Susko and Roger (2004). The P value given is the lowest of all three possible comparisons. Similarly, the number of EST reads L1 and L2 given correspond to the number of reads of the contrast with lowest P value. The gene description starts with a SwissProt entry number if not indicated otherwise. GB Genbank
Table 7.
Zostera marina TUGs significantly down-regulated in library F (heat stress) with respect to library D (designated DF), E (EF), or both libraries pooled (DE-F), in descending order of total expression level
| TUG id | Contrast | P value | L1 | L2 | Database entry, gene description – species of best hit |
|---|---|---|---|---|---|
| Contig479 | DF/DE-F | 0 | 290 | 55 | Q40256, metallothionein-like protein type 3 (MT-3) - Musa acuminata |
| Contig926 | DE-F | 0.0053 | 57 | 52 | Q7YJV4, chloroplast 30S ribosomal protein S18 - Calycanthus fertilis var. ferax |
| Contig341 | DF | 0.0074 | 29 | 13 | P13194, photosystem I reaction center subunit IV, chloroplast precursor (PSI-E) - Hordeum vulgare |
| Contig78 | DE-F/EF | 0.0145 | 30 | 5 | GB ATGSTF8, glutathione S-transferase 8 - Arabidopsis thaliana |
| Contig515 | DF/DE-F | 0.001 | 21 | 5 | TAIR|gene:2201835; photosystem I subunit O - Arabidopsis thaliana |
| Contig292 | DE-F/EF | 0.0225 | 15 | 1 | Q6K953, glutaredoxin-C4, chloroplast precursor (Glutaredoxin-C2 homolog 2) - Oryza sativa |
| Contig3 | EF | 0.0377 | 12 | 3 | Q07764, Protein HVA22 - Hordeum vulgare |
| Contig570 | DE-F | 0.0464 | 13 | 1 | NoHit |
| Contig70 | DF | 0.0135 | 11 | 2 | P46423, glutathione S-transferase (EC 2.5.1.18) (GST class-phi) - Hyoscyamus muticus |
| Contig316 | DF | 0.0037 | 11 | 1 | NoHit |
| Contig326 | DF/DE-F | 0.0244 | 10 | 2 | Q39194, photosystem II reaction center W protein, chloroplast precursor - Arabidopsis thaliana |
| Contig69 | DF | 0.0071 | 10 | 1 | GB AT5G38410.1, ribulose-bisphosphate carboxylase - Arabidopsis thaliana |
| Merge05 | DF | 0.0437 | 9 | 2 | P27347, DNA-binding protein MNB1B (HMG1-like protein) - Zea mays |
| Contig296 | DF | 0.0138 | 9 | 1 | NoHit |
| Contig301 | DF/DE-F | 0.0138 | 9 | 1 | NoHit |
| Contig314 | DF/DE-F | 0.0138 | 9 | 1 | P49107, photosystem I reaction center subunit N, chloroplast precursor (PSI-N) - Arabidopsis thaliana |
| Contig302 | DF/DE-F | 0.0264 | 8 | 1 | P10976, nonspecific lipid-transfer protein precursor (LTP) - Spinacia oleracea |
| Contig365 | DF | 0.0264 | 8 | 1 | NoHit |
| Contig327 | DF/DE-F | 0.0107 | 7 | 0 | P14009, 14 kDa proline-rich protein DC2.15 precursor - Daucus carota |
| Contig373 | DF | 0.0107 | 7 | 0 | NP-849822.1, CAT2 cationic amino acid transporter - Arabidopsis thaliana |
| Contig461 | DF | 0.0107 | 7 | 0 | NP-567911.1, unknown protein - Arabidopsis thaliana |
| Contig631 | EF | 0.0165 | 7 | 0 | P49397, 40S ribosomal protein S3a (CYC07 protein) - Oryza sativa |
| Contig777 | EF | 0.0165 | 7 | 0 | NP-194457.2, quinone reductase family protein - Arabidopsis thaliana |
| Contig89 | DF | 0.0107 | 7 | 0 | Q9M2F1, 40S ribosomal protein S27-2 - Arabidopsis thaliana |
| Contig14 | DF | 0.0226 | 6 | 0 | NoHit |
| Contig158 | DF | 0.0226 | 6 | 0 | NoHit |
| Contig346 | DF | 0.0226 | 6 | 0 | P46077, general regulatory factor 4 , 14-3-3-like protein - Arabidopsis thaliana |
| Contig383 | DF | 0.0226 | 6 | 0 | NoHit |
| Contig388 | DF | 0.0226 | 6 | 0 | P34106, alanine aminotransferase 2 (EC 2.6.1.2) - Panicum miliaceum |
| Contig498 | DF | 0.0226 | 6 | 0 | P29136, metalloendoproteinase 1 precursor (EC 3.4.24) - Glycine max |
| Merge69 | EF | 0.0327 | 6 | 0 | Q687E1, nucleotide pyrophosphatase/phosphodiesterase - Hordeum vulgare |
| Contig275 | DF | 0.0477 | 5 | 0 | P80883, cytochrome b6-f complex subunit 7 - Spinacia oleracea |
| Contig382 | DF | 0.0477 | 5 | 0 | GB AT1G64850, putative calcium ion binding gene - Arabidopsis thaliana |
| Contig404 | DF | 0.0477 | 5 | 0 | O49884, 60S ribosomal protein L30 - Lupinus luteus |
| Contig464 | DF | 0.0477 | 5 | 0 | Q96291, 2-cys peroxiredoxin BAS1, chloroplast precursor (EC 1.11.1.15) - Arabidopsis thaliana |
| Contig474 | DF | 0.0477 | 5 | 0 | GB AT1G42970.1, glyceraldehyde-3-phosphate – Arabidopsis thaliana |
The statistical test for each gene was performed by binomial approximation as suggested in Susko and Roger (2004). The P value given is the lowest of all three possible comparisons. Similarly, the number of EST reads L1 and L2 given correspond to the number of reads of the contrast with lowest P value, which is given first in the column: contrast. The gene description starts with a SwissProt entry number if not indicated otherwise. GB Genbank
Among temperature responsive genes, 7 of 27 (26%) and 5 of 36 (14%) TUGs, respectively, had a role in photosynthesis, predominantly in the light reaction (photosystem I and II). Although under laboratory exposure with higher temperatures, several light harvesting complex proteins were up-regulated (as in contigs 983, 787; merge29 and 73, Table 6), the reaction subunits themselves were down-regulated (as in contig 314, 326 and 341, putative homology to photosystem I and II reaction center subunit genes, Table 7). The dark reaction also was affected. A 10-fold down-regulation upon heat exposure also is observed in the primary gene of photosynthetic carbon fixation, Rubisco (contig69).
Frequency of Microsatellites in Differentially Expressed Genes
When comparing the abundance of microsatellites among differentially expressed genes (comparison D + E vs. F only) with all other TUGs, no significant difference could be detected. Of all up-regulated genes, 5.16% carried a microsatellite, whereas of all down-regulated TUGs, 13.89% carried microsatellites. Both frequencies were not significantly different from the global frequency of microsatellites among all TUGs (6.01%) in Chi-square tests.
Abundance and Diversity of Genes Encoding Stress Proteins
Among 3496 TUGs we found 9 genes encoding for diverse families of heat shock proteins (HSP) that are known to be involved in mediating high temperatures and other stresses (Boston et al. 1996). All of those were genes encoding HSPs of large molecular weight > 60 kDa (Table 8). We also identified one heat shock transcription factor B4 (singleton, ZMD01094, Table 8). Because HSP-genes were too rare to allow frequency-based tests on single genes, we lumped them according to the GO category (biological function) response to heat. Interestingly, we find a higher frequency of HSP genes sensu latu under the summer conditions (library C vs. D, 7 vs. 11 reads; P = 0.035), but no significantly different frequencies among the “winter” and the heat stress libraries, with largely similar contribution of HSPs to the total number of reads [library D (7 reads), E (9), F (13)]. A stress-mediating gene that was significantly up-regulated under heat stress may be involved in scavenging reactive oxygen species, a Mn-superoxide dismutase (contig901, SwissProt P35017, Table 6).
Table 8.
Zostera marina putative HSP (heat shock protein) encoding genes and heat shock transcription factors
| TUG identifier | Symbol | SwissProt ID, gene name | E value | Species with best hit |
|---|---|---|---|---|
| Contig953 | CPN10 | P34893; 10 kDa chaperonin protein | 6.40e-35 | Arabidopsis thaliana |
| ZMC15071 | HSP70-2 | P22954; Heat shock cognate 70 kDa protein | 2.90e-72 | Arabidopsis thaliana |
| Contig325; ZMF06039 | HSC70-1 | P22953; Heat shock cognate 70 kDa protein | 3.00e-67 | Arabidopsis thaliana |
| ZMC10006 | HSP70 | P09189; Heat shock cognate 70 kDa protein | 1.60e-86 | Petunia hybrida |
| Contig49; ZMC02019 | HSP81-1 | P36181; Heat shock cognate protein 80 | 3.40e-100 | Solanum lycopsersicum |
| ZMC13059; ZMF07312 | HSP81-3 | P51818; Heat shock protein 81 | 2.00e-05 | Arabidopsis thaliana |
| Contig19 | HSP81-3 | Q07078; Heat shock protein 81 | 1.80e-92 | Oryza sativa |
| ZME03121 | HSP90 | OJ1540-H01.1; heat shock protein 90 | 1.00e-42 | Oryza sativa |
| ZMD01094 | HSF | Q9C635; Heat stress transcription factor B-4 | 2.00e-07 | Arabidopsis thaliana |
Comparison of Z. marina Differentially Expressed Genes Against Arabidopsis thaliana
Among the 63 TUGs that were differentially expressed (d.e.) in library F (heat stress 25°C) vs. D + E, a BLASTX search with the MIPS Arabidopsis database resulted in 48 significant hits. These were distributed over all five chromosomes in the Arabidopsis genome. There also were three significant hits in the chloroplast genome (ycf4, rps18, atpA). Among those 48 genes, we found a highly significant overrepresentation of several GO categories. There were significantly more photosynthetic genes and those taking part in chromatin binding regulated differentially than expected by chance (GO molecular function, both P < 0.001). In terms of GO category, biological function, photosynthesis was more affected than any other process (P < 0.001). Finally, in terms of cellular components, mainly photosystem I components and light harvesting complexes were d.e., confirming above qualitative finding on composition of significantly up- or down-regulated genes on an individual basis (Tables 6 and 7).
When comparing Z. marina with the ATGenExpress, a stress specific database of Arabidopsis thaliana, 24/63 (38%) d.e. eelgrass genes also were reported to change expression levels in A. thaliana as response to stress (Table 9). The major organ of expression varied and comprised root and shoot. Interestingly, although many were responsive to the same stress type as in our Z. marina data (i.e. heat stress), several of these (10/23 = 43%) are primarily responding to osmotic and salt stress in Arabidopsis. Whether this reflects functional changes of genes in Zostera after adaptation to the marine environment requires further study.
Table 9.
Similarity among eelgrass (Zostera marina) TUGs and Arabidopsis thaliana stress response
| TUG id | Gene description | Accession | E value | Stress type | Organ |
|---|---|---|---|---|---|
| Merge69 | calcineurin-like phosphoesterase family protein | AT1G13900.1 | 1e-020 | cold | root |
| Contig382 | Calcium binding, EF family | AT4G37445.1 | 5e-009 | cold | root |
| Contig119 | Endo-beta-1,3-1,4-glucanase | AT5G20330.1 | 2e-023 | heat | shoot |
| Contig292 | Glutharedoxin | AT3G02000.1 | 3e-012 | heat | root |
| Contig302 | Lipid transfer protein 12 (LTP12) | AT3G51590.1 | 2e-014 | heat | shoot |
| Merge74 | Universal stress A-like protein | AT3G58450.1 | 2e-044 | heat | shoot |
| Contig592 | ATP synthase alpha chain, mitochondrial | AT2G07698.1 | 1e-105 | heat | shoot |
| Contig68 | Cystein-rich protease inhibitor | AT2G02140.1 | 2e-007 | heat | shoot |
| Contig70 | Glutathione S-transferase PM24 | AT1G02940.1 | 5e-046 | heat | shoot |
| Contig777 | Quinone reductase family protein | AT4G36750.1 | 3e-068 | heat | shoot |
| Contig78 | Glutathione S-transferase APIC (EC 2.5.1.18) | AT1G02940.1 | 1e-043 | heat | shoot |
| Contig89 | Metallothionein-like family 15 | AT2G23240.1 | 2e-003 | heat | shoot |
| Merge73 | chlorophyll A-B binding protein CP26 | AT5G51340.1 | 9e-029 | osmotic | root |
| Contig275 | cytochrome B6F complex subunit (PETM) | AT2G26500.1 | 3e-023 | osmotic | root |
| Contig314 | Photosystem I reaction center subunit N | AT5G64040.1 | 6e-049 | osmotic | root |
| Contig341 | Photosystem I reaction center subunit IV | AT2G20260.1 | 8e-030 | osmotic | root |
| Contig474 | Glyceraldehyde-3-phosphate | AT1G42970.1 | 1e-003 | osmotic | root |
| Merge29 | chlorophyll A-B binding protein (LHCP) | AT3G61470.1 | 3e-066 | osmotic | root |
| Contig85 | chlorophyll A-B binding protein (LHCP) | AT1G29930.1 | 1e-105 | osmotic | root |
| Contig901 | Superoxide-dismutase (Fe) | AT5G51100.1 | 1e-024 | osmotic | root |
| Merge05 | High mobility group family (HMG1/2) | AT2G34450.1 | 5e-012 | salt | shoot |
| Contig346 | 14-3-3 protein GF14 lambda (GRF6) | AT5G10450.1 | 2e-060 | salt | root |
| Contig327 | Protease inhibitor/see storage associated | AT4G12500.1 | 2e-020 | wound | shoot |
E-values are given for the BLASTX search of the Z. marina query to the A. thaliana ATGeneExpress database. Accessions, stress type, and tissue refer to A. thaliana
Only approximately half of the Arabidopsis thaliana stress response genes with putative homology to Z. marina TUGs are predominantly expressed in the shoot, whereas the others are characteristic for the root, although the tissue type used for constructing the Z. marina library did not contain root material. Whether this, too, reflects functional dissimilarity driven by the different habitat type or taxonomic affiliation of Arabidopsis (dicot) vs. Zostera (monocot) is unclear.
Further Characteristics of the Heat Stress Response
Under all experimental conditions, the transcriptome of Z. marina is dominated by a gene encoding for a cystein-rich metallothionein-like protein (mt3) comprising between 2.5% and 15% of all transcripts (contig479, SwissProt ID Q40256). Although one reported primary function of such genes is heavy metal homeostasis, in particular copper (Guo et al. 2003), such a dominant frequency suggests that this gene must be responsible for other important functions as well. Note, however, that this putative metallothionein is down-regulated approximately 6-fold under temperature stress. Almost exactly the same down-regulation was observed in winter (library D) compared with average summer conditions (library C, Table S1). Interestingly, in the transcriptome of the Mediterranean seagrass species Posidonia oceanica we also find a similar TUG that is even more abundant (G. Procaccini, personal communication, 2007). Finally, we have probably identified several genes that have a high homology to Dictyostelium, a social amoeba or slime mold (Table 6). One of those genes, encoding a 26S proteasome regulatory subunit, shows a significant up-regulation under heat stress (merge17; SwissProt ID P02889).
Discussion
In this study, we found striking differences in gene expression among experimental conditions in a coastal marine angiosperm, the ecologically important seagrass species Zostera marina (eelgrass). Among the TUGs that consisted of ≥ 4 total sequence reads, we found several individual genes that revealed strikingly different expression patterns when subjected to heat stress, both in terms of up- and down-regulation.
A priori, we would expect the heat stress response among terrestrial and aquatic angiosperms to differ in a number of ways. First, temperature changes are always gradual in the thermally buffered aquatic environment, whereas rapid temperature fluctuations are possible on land. On the other hand, once critical temperatures are reached in the sea, they have to be sustained by the plant for a longer time and cannot be ameliorated by increasing transpiration. This may explain why we identified no small heat shock proteins even under imposed temperature stress in Zostera marina, whereas these genes are a major group of inducible heat shock genes among terrestrial angiosperms (Waters et al. 1996). It also may explain why the response among the HSP encoding genes is relatively weak (≤2-fold induction), suggesting that most HSP genes identified were constitutively expressed and may indicate longer-term acclimation to high temperature. Evidently, the preliminary data obtained in this study need to be verified by quantitative PCR or by macro- or microarray work. The latter is under development in Z. marina.
Notwithstanding, it is notable that in eelgrass, many of the d.e. TUGs have putative homologous in the plant model A. thaliana with its well-characterized stress response associated genes. This demonstrates not only the principal validity of comparative EST-analysis but also is an indication that many components of the stress response may be conserved among the flowering plants. On the other hand, the time to the least common ancestor of the genus Zostera and other families of monocotyledoneous and dicotyledoneous plants is in the order of 100 Mio years (Les et al. 1997). Therefore, a fraction of approximately a fifth of genes that revealed no database hits even when compared against Prodom indicates the phylogenetic distance of the seagrasses relative to well-studied plant genomic model species, such as rice or Arabidopsis. That the majority of data base hits of eelgrass TUG queries revealed A. thaliana as species with lowest E-values probably reflects the abundance of sequences in Genbank, or conversely, the relative paucity of data on monocotyledoneous species. Many groups of monocotyledonous plants are currently poorly represented in transcriptomic/genomic databases (Jackson et al. 2006). Therefore, the TUGs of a seagrass species presented here also may serve as one initial attempt to close this gap within the order of Alismatidae.
Worth mentioning are not only those database hits that are indicative for higher plants but also those indicating a contamination of the EST library. Particularly noteworthy are three genes with high similarity to an amoeba species (Dictyostelium spp.), pointing to possible infection (or symbiotic association) of eelgrass with a protist. It is known that species of slime mold of the genus Labyrinthula are associated with eelgrass and have caused massive die-offs (also dubbed wasting disease) in the 1930s (den Hartog 1970; Muehlstein et al. 1988). It is possible that a related slime mold Dictyostelium yielded the best hit because its genome is much better characterized than those of Labyrinthula species in which only 22 genes are deposited in GenBank (as of September 20, 2007), but this requires further study.
Although some classical stress associated genes are higher expressed under elevated temperatures, such as superoxide-dismutase (SwissProt P35017), most classical heat shock proteins (HSP) had an overall frequency that is too low to allow tests based on single genes (<0.1% of transcripts). Nevertheless, when pooled, a significant 1.8-fold constitutive up-regulation under natural summer vs. winter conditions becomes apparent in libraries prepared from experimentally untreated material (libraries C and D). Such an effect was not detectable under experimentally induced stress, although a tendency of induction was observable in the raw data. Other work in progress (Ransbotyn & Reusch, preliminary data, 2007) revealed using quantitative, real-time PCR assays that two HSP70 genes (contig325, and singleton ZMC10006, SwissProt ID P22953 and P09189) respond to an increase in temperature from 18° to 25°C with moderate up-regulation (2 to 3-fold).
Within our EST libraries we have identified a total of 223 microsatellites or simple sequence repeats in 210 (∼6%) of all TUGs. They may serve as starting points for trait-associated genetic markers (van Tienderen et al. 2002). An equally high frequency of putative microsatellite marker loci associated in direct linkage with genes has now been identified in many other plant species (overview in Li et al. 2004). In Z. marina, PCR-based assays have already been successfully developed for 14 of these candidates, and they were proven to be polymorphic in natural populations (Oetjen and Reusch 2007a, b). As expected, we found a higher fraction of trinucleotide microsatellite repeats within coding regions compared with outside ORFs, because their length variation will not result in frameshifts, whereas the inverse was true for repeat motifs that would result in frameshift mutations.
For constructing the cDNA libraries presented, we used the SMART methodology, which involves a PCR amplification step before cloning to increase full-length representation of transcripts (Herrler 2000). Although this may lead to differential representation of mRNAs, empirical studies have verified that SMART PCR maintains the presentation of transcript abundance (Herrler 2000; Seth et al. 2003). To compare the global gene expression patterns among experimental conditions, tissues or different populations/species, subtractive hybridizations are an alternative to comparing the gene composition of the transcriptome (Diatchenko et al. 1996; for a marine example see Pearson et al. 2001). With sequencing technologies becoming much cheaper in the foreseeable future (Margulies et al. 2005), we predict that global comparison of redundant (i.e. nonnormalized) EST libraries will be a routine tool for gaining first insights into the adjustment of gene regulation as a response to environmental conditions, including stress (this study, Kuo et al. 2004). On the other hand, when sequencing moderate numbers of ESTs, for example, in the range of 1x 104 as in this study, meaningful information is only obtained for the most abundant transcripts. There also will be a negative correlation between global expression strength and the statistical power to detect differential regulation that may bias the outcome of studies (Susko and Roger 2004), in particular when particular gene classes differ nonrandomly in their expression strength under certain conditions. Nevertheless, to obtain a global snapshot of organismal physiology, the quantitatively important physiologic processes are detectable by a moderate-sized EST library.
The metabolic adjustments among the most expressed genes identified in this study suggest a complex stress syndrome in Z. marina subject to adverse conditions. Although light harvesting proteins are up-regulated, we find a strong down-regulation of the photosynthetic active complexes themselves. Several of the major responsive genes as well as stress genes candidates (in particular heat shock proteins and genes involved in scavenging reactive oxygen species) identified may serve as starting points to develop expression profiling techniques (Whitehead and Crawford 2006). The ultimate goal is to obtain a more exhaustive and fine scale picture of plastic and constitutive changes in cellular metabolism associated with short- and long-term adaptation to extreme water temperatures.
Electronic Supplementary Material
Below is the link to the Electronic Supplementary Material.
Zostera marina TUGs differentially regulated among library D (winter situation) and library C (average summer condition) (PDF 87.1 KB).
Acknowledgements
We are grateful to W. Lampert and the Max-Planck-Society for continual support. D. Tautz supported parts of the sequencing. S. Carstensen, I. Dankert and S. Liedtke were indispensable for constructing libraries, plasmid preps and performing sequencing runs. We thank M. Hippler for giving valuable hints for interpreting the data. Funding was partly provided by DFG (Re 1108/7).
Footnotes
Thorsten B. H. Reusch and Amelie S. Veron contributed equally to this work.
Eletronic Supplementary Material
The online version of this article (doi:10.1007/s10126-007-9065-6) contains supplementary material which is available to authorized users.
References
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B l 57:289–300
- Boston RS, Viitanen PV, Vierling E (1996) Molecular chaperones and protein folding in plants. Plant Mol Biol 332:191–222 [DOI] [PubMed]
- Bouck AMY, Vision T (2007) The molecular ecologist’s guide to expressed sequence tags. Mol Ecol 16:907–924 [DOI] [PubMed]
- Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G (2004) GO::TermFinder-open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 20:3710–3715 [DOI] [PMC free article] [PubMed]
- Cerrano C, Bavestrello G, Bianchi CN, Cattaneo-Vietti R, Bava S, Morganti C, Morri C, Picco P, Sara G, Schiaparelli S, Siccardi A, Sponga F (2000) A catastrophic mass-mortality episode of gorgonians and other organisms in the Ligurian Sea (North-western Mediterranean), summer 1999. Ecol Lett 3:284–293 [DOI]
- den Hartog C (1970) The seagrasses of the world. Verhandlingen Koninglijk Nederlandse Akademie Wetenschapen Afdeling Natuurkunde IIl 59:1–275
- Diatchenko L, Lau YFC, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED, Siebert PD (1996) Suppression subtractive hybridization: a method for generating regulated or tissue-specific cDNA probes and libraries. PNAS 93:6025–6030 [DOI] [PMC free article] [PubMed]
- Dupont S, Wilson K, Obst M, Sköld H, Nakano H, Thorndyke MC (2007) Marine ecological genomics: when genomics meets marine ecology. Mar Ecol Prog Ser 332:257–273 [DOI]
- Falcon S, Gentleman R (2007) Using GOstats to test gene lists for GO term association. Bioinformatics 23:257–258 [DOI] [PubMed]
- Feder ME, Mitchell-Olds T (2003) Evolutionary and ecological functional genomics. Nat Rev Genet 4:649–655 [DOI] [PubMed]
- Fujiwara S, Hirokawa Y, Takatsuka Y, Suda K, Asamizu E, Takayanagi T, Shibata D, Tabata S, Tsuzuki M (2007) Gene expression profiling of coccolith-bearing cells and naked cells in haptophyte Pleurochrysis haptonemofera with a cDNA macroarray system. Mar Biotechnol 9:550–560 [DOI] [PubMed]
- Gaines SD, Denny MW (1993) The largest, smallest, highest, lowest, longest, and shortest: extremes in ecology. Ecology 74:1677–1692 [DOI]
- Gish W, States DJ (1993) Identification of protein coding regions by database similarity search. Nat Genet 3:266–272 [DOI] [PubMed]
- Greve TM, Borum J, Pedersen O (2003) Meristematic oxygen variability in eelgrass (Zostera marina). Limnol Oceanogr 48:210–216
- Guo W-J, Bundithya W, Goldsbrough PB (2003) Characterization of the Arabidopsis metallothionein gene family: tissue-specific expression and induction during senescence and in response to copper. New Phytologist 159:369–381 [DOI] [PubMed]
- Hashimoto K, Shibuno T, Murayama-Kayano E, Tanaka H, Kayano T (2004) Isolation and characterization of stress-responsive genes from the scleractinian coral Pocillopora damicornis. Coral Reefs 23:485–491
- Herrler M (2000) Use of SMART-generated cDNA for differential gene expression studies. J Mol Med 78:B23 [PubMed]
- Hofmann GE, Burnaford JL, Fielman KT (2005) Genomics-fueled approaches to current challenges in marine ecology. Trends Ecol Evol 20:305–311 [DOI] [PubMed]
- Huang X, Madan A (1999) CAP3: a seqeunce assembly program. Genome Res 9:868–877 [DOI] [PMC free article] [PubMed]
- Hughes TP, Baird AH, Bellwood DR, Card M, Connolly SR, Folke C, Grosberg R, Hoegh-Guldberg O, Jackson JBC, Kleypas J, Lough JM, Marshall P, Nyström M, Palumbi SR, Pandolfi JM, Rosen B, Roughgarden J (2003) Climate change, human impacts, and the resilience of coal reefs. Science 301:929–933 [DOI] [PubMed]
- IPCC (2007) Climate change 2007: the scientific basis. Intergovernmental Panel of Climate Change, Geneva, Switzerland
- Jackson S, Rounsley S, Purugganan M (2006) Comparative sequencing of plant genomes: choices to make. Plant Cell 18:1100–1104 [DOI] [PMC free article] [PubMed]
- Jenny MJ, Ringwood AH, Lacy ER, Lewitus AJ, Kempton JW, Gross PS, Warr GW, Chapman RW (2002) Potential indicators of stress response identified by expressed sequence tag analysis of hemocytes and embryos from the American oyster, Crassostrea virginica. Mar Biotechnol 4:81–93 [DOI] [PubMed]
- Jones CG, Lawton JH, Chachak M (1994) Organisms as ecosystem engineers. Oikos 69:373–386 [DOI]
- Kassahn KS, Caley MJ, Ward AC, Connolly AR, Stone G, Crozier RH (2007) Heterologous microarray experiments used to identify the early gene response to heat stress in a coral reef fish. Mol Ecol 16:1749–1763 [DOI] [PubMed]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D’Angelo C, Bornberg-Bauer E, Kudla J, Harter K (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50:347–363 [DOI] [PubMed]
- Kore-eda S, Cushman MA, Akselrod I, Bufford D, Fredrickson M, Clark E, Cushman JC (2004) Transcript profiling of salinity stress responses by large-scale expressed sequence tag analysis in Mesembryanthemum crystallinum. Gene 341:83–92 [DOI] [PubMed]
- Kuo J, Chen M-C, Lin C-H, Fang L-S (2004) Comparative gene expression in the symbiotic and aposymbiotic Aiptasia pulchella by expressed sequence tag analysis. Biochem Biophys Res Comm 318:176–186 [DOI] [PubMed]
- Les DH, Cleland MA, Waycott M (1997) Phylogenetic studies in Alismatidae, II: Evolution of marine angiosperms (seagrasses) and hydrophily. System Bot 22:443–463 [DOI]
- Li Y-C, Korol AB, Fahima T, Nevo E (2004) Microsatellites within genes: structure, function, and evolution. Mol Biol Evol 21:991–1007 [DOI] [PubMed]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen Y-J, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer MLI, Jarvie TP, Jirage KB, Kim J-B, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM (2005) Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380 [DOI] [PMC free article] [PubMed]
- Muehlstein LK, Porter D, Short FT (1988) Labyrinthula sp., a marine slime mold producing the symptoms of wasting disease in eelgrass, Zostera marina. Mar Biol 99:465–472 [DOI]
- Oetjen K, Reusch TBH (2007a) Genome scans detect consistent divergent selection among subtidal vs. intertidal populations of the marine angiosperm Zostera marina. Mol Ecol (in press) [DOI] [PubMed]
- Oetjen K, Reusch TBH (2007b) Identification and characterization of 14 polymorphic EST-derived microsatellites in eelgrass (Zostera marina). Mol Ecol Notes 7:777–780 [DOI]
- Okubo K, Hori N, Matoba R, Niiyama T, Fukushima A, Kojima Y, Matsubara K (1992) Large scale cDNA sequencing for analysis of quantitative and qualitative aspects of gene expression. Nat Genet 2:173–179 [DOI] [PubMed]
- Ouborg NJ, Vriezen WH (2007) An ecologist’s guide to ecogenomics. J Ecol 95:8–16 [DOI]
- Pearson G, Serrao E, Cancela ML (2001) Suppression subtractive hybridization for studying gene expression during aerial exposure and dessication in fucoid algae. Eur J Phycol 36:359–366 [DOI]
- Pérez F, Ortiz J, Zhinaula M, Gonzabay C, Calderón J, Volckaert F (2005) Development of EST-SSR markers by data mining in three species of shrimp: Litopenaeus vannamei, Litopenaeus stylirostris, and Trachypenaeus birdy. Mar Biotechnol 7:554–569 [DOI] [PubMed]
- Reusch TBH (2000) Pollination in the marine realm: microsatellites reveal high outcrossing rates and multiple paternity in eelgrass Zostera marina. Heredity 85:459–465 [DOI] [PubMed]
- Reusch TBH, Stam WT, Olsen JL (1999) Size and estimated age of genets in eelgrass Zostera marina L. assessed with microsatellite markers. Mar Biol 133:519–525 [DOI]
- Reusch TBH, Ehlers A, Hämmerli A, Worm B (2005) Ecosystem recovery after climatic extremes enhanced by genotypic diversity. Proc Natl Acad Sci USA 102:2826–2831 [DOI] [PMC free article] [PubMed]
- Reusch TBH, Wood TE (2007) Molecular Ecology of global change. Mol Ecol 16:3973–3992 [DOI] [PubMed]
- Rocha EPC, Matic I, Taddei F (2002) Over-representation of repeats in stress response genes: a strategy to increase versatility under stressful conditions? Nuc Acid Res 30:1886–1894 [DOI] [PMC free article] [PubMed]
- Seth D, Gorrell MD, McGuinness PH, Leo MA, Lieber CS, McCaughan GW, Haber PS (2003) SMART amplification maintains representation of relative gene expression: quantitative validation by real time PCR and application to studies of alcoholic liver disease in primates. J Biochem Biophys Meth 55:53–66 [DOI] [PubMed]
- Staden R, Beal KF, Bonfield JK (2000) The Staden Package. Meth Mol Biol 132:115–130 [DOI] [PubMed]
- Susko E, Roger AJ (2004) Estimating and comparing the rates of gene discovery and expressed sequence tag (EST) frequencies in EST surveys. Bioinformatics 20:2279–2287 [DOI] [PubMed]
- van Tienderen PH, de Haan AA, van der Linden CG, Vosman B (2002) Biodiversity assessment using markers for ecologically important traits. Trends Ecol Evol 17:577–583 [DOI]
- Vasemägi A, Primmer CR (2005) Challenges for identifying functionally important genetic variation: the promise of combining complementary research strategies. Mol Ecol 14:3623–3642 [DOI] [PubMed]
- Wang Y, Guo X (2007) Development and characterization of EST-SSR markers in the eastern oyster Crassostrea virginica. Mar Biotechnol 9:500–511 [DOI] [PubMed]
- Waters ER, Lee GJ, Vierling E (1996) Evolution, structure and function of the small heat shock proteins in plants. J Exp Bot 47:325–338 [DOI]
- Whitehead A, Crawford DL (2006) Neutral and adaptive variation in gene expression. Proc Natl Acad Sci USA 103:5425–5430 [DOI] [PMC free article] [PubMed]
- Williams SL (2001) Reduced genetic diversity in eelgrass transplantations affects both population growth and individual fitness. Ecol Applic 11:1472–1488 [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the Electronic Supplementary Material.
Zostera marina TUGs differentially regulated among library D (winter situation) and library C (average summer condition) (PDF 87.1 KB).