Abstract
The budding yeast CenH3 histone variant Cse4 localizes to centromeric nucleosomes and is required for kinetochore assembly and chromosome segregation. The exact composition of centromeric Cse4–containing nucleosomes is a subject of debate. ChIP-chip experiments and high resolution quantitative PCR confirm that there is a single Cse4 nucleosome at each centromere, and additional regions of the genome contain Cse4 nucleosomes at low levels. Using unbiased biochemical, cell biological, and genetic approaches we have tested the composition of Cse4-containing nucleosomes. Using micrococcal nuclease-treated chromatin, we find that Cse4 is associated with the histones H2A, H2B, and H4, but not H3 or the non-histone protein Scm3. Overexpression of Cse4 rescues the lethality of a scm3 deletion, indicating Scm3 is not essential for the formation of functional centromeric chromatin. Additionally, octameric Cse4 nucleosomes can be reconstituted in vitro. The Cse4-Cse4 interaction domain appears to be essential and interaction occurs in vivo in the centromeric nucleosome. Taken together, our experimental evidence supports the model that the Cse4-nucleosome is an octamer, containing two copies each of Cse4, H2A, H2B, and H4.
Keywords: Cse4/CENP-A, centromere, yeast, Scm3, chromatin-immunoprecipitation, nucleosome
Introduction
One of the most critical tasks for a dividing cell is to make sure that the two new cells each have an accurate and complete copy of the genome. In all eukaryotes, each chromosome contains a specialized DNA sequence that helps to ensure proper segregation, known as the centromere. At each centromere a large proteinaceous structure called the kinetochore is formed. Microtubules attach to the kinetochore and pull sister chromatids to opposite spindle poles. Two types of centromeres have been identified: point centromeres and regional centromeres. Regional centromeres are typically found in higher eukaryotes, and are composed of numerous copies of repetitive DNA sequences (Waye and Willard, 1986). In humans, some centromeres can be as long as a megabase (Cleveland et al., 2003). S. cerevisiae and other hemiascomycetous fungi contain point centromeres. In these organisms the centromere sequence is ~125 base pairs (bp) and is comprised of three DNA elements: CDEI, CDEII, and CDEIII (Fitzgerald-Hayes et al., 1982; Hegemann and Fleig, 1993).
Although their DNA sequences are highly variable between species, all eukaryotic centromeres are thought to be epigenetically marked by the presence of a centromere specific histone variant (CenH3). This variant, known as Cse4 in budding yeast and more generally as CENP-A, is essential for the formation of a kinetochore and for proper chromosome segregation (Henikoff and Dalal, 2005; Meluh and Koshland, 1997). Structurally, CENP-A homologs have two major domains: an evolutionarily conserved histone fold domain (HFD) and a divergent amino-terminal domain. The HFD of CENP-A homologs has a high degree of amino acid identity to histone H3, while the amino-terminal portion of the protein can vary greatly between species. The function of the CenH3 nucleosome is highly conserved, as demonstrated by the functional complementation of RNAi-depleted CENP-A in human cells by yeast Cse4 (Wieland et al., 2004).
In S. cerevisiae, Cse4 localizes in vivo to a single nucleosome directly at CDE I-II-III, and is thought to replace histone H3 at this site (Furuyama and Biggins, 2007; Meluh and Koshland, 1997). In organisms with regional centromeres, CenH3 is found in multiple nucleosomes at or near the centromeric repeats. In human cells, CENP-A localizes to the large arrays of α-satellite DNA, interspersed among canonical histone H3-containing nucleosomes (Blower et al., 2002). In the case of human (CENP-A) or Drosophila melanogaster (CID) CenH3 octamers assembled in vitro, it has been shown that CENP-A and CID can substitute for histone H3 (Furuyama et al., 2006; Yoda et al., 2000).
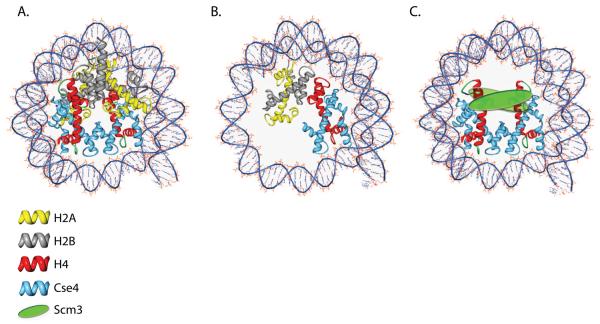
Although it is generally accepted that Cse4 is assembled into nucleosomes at the budding yeast centromere, the overall composition of this nucleosome is a topic of debate. At present, there are three models for the composition of the budding yeast centromeric nucleosome. The first model for the centromeric nucleosome is that Cse4 simply replaces H3 in an octameric nucleosome which contains Cse4, H2A, H2B, and H4 (Figure 1A). Given the crystal structure of a canonical nucleosome (Luger et al., 1997), this model is based in part on the strong sequence identity between the HFD of the CENP-A homologues and that of H3. In addition, octameric nucleosomes containing human or Drosophila CenH3 can be reconstituted in vitro (Furuyama et al., 2006; Yoda et al., 2000). Another more provocative model suggested for centromeric nucleosomes is that they contain a single molecule of each CenH3, H2A, H2B, and H4, which forms a tetrameric structure called a “hemisome” (Figure 1B) (Dalal et al., 2007a). This hemisomal complex was purified from interphase Drosophila S2 cells by crosslinking and immunoprecipitation (IP) of CID. Hemisomes are predicted to wrap <120bp of DNA, and when analyzed by atomic force microscopy appear to be half the height of canonical bulk nucleosomes (Dalal et al., 2007b).
Figure 1.
Three models of the budding yeast centromeric nucleosome. The cartoon schematic represents each of the proposed centromeric nucleosomes wrapped by DNA. The DNA is represented by the blue lines. Actual nucleosome/DNA contact points are unknown for all three models. A. The octamer model proposes that Cse4 is found in a canonical-like octameric configuration. B. The hemisome model predicts that the centromeric nucleosomes consist of a monomer of each Cse4, H2A, H2B, and H4. The hemisome is predicted to wrap less DNA than an octameric or hexameric nucleosome (~120bp). C. The hexamer model proposes that the centromeric nucleosome is comprised of a tetramer of Cse4-H4 and two molecules of Scm3.
The final model for centromeric nucleosome composition involves the newly identified budding yeast kinetochore protein Scm3. Scm3 was initially identified in budding yeast as a high copy suppressor of a Cse4-HFD mutant (Chen et al., 2000), and has since been shown to be an essential kinetochore protein required for proper localization of the CenH3 histone variant to the centromere in both budding and fission yeast (Camahort et al., 2007; Mizuguchi et al., 2007; Pidoux et al., 2009; Stoler et al., 2007; Williams et al., 2009). SCM3 homologs are found in fungi with both point and regional centromeres (Aravind et al., 2007). Scm3 has been shown to facilitate the exclusion of histones H2A and H2B from preassembled Cse4-containing octamers in vitro. Additionally, chromatin immunoprecipitation studies (ChIP) suggest that histones H2A and H2B are absent from the centromeric nucleosome in vivo in budding yeast (Mizuguchi et al., 2007). Based on these findings it has been proposed that along with Cse4 and H4, Scm3 forms a unique hexameric nucleosome specifically at centromeres (Figure 1C). Unlike canonical nucleosomes which contain two heterodimers of the histones H2A/H2B complexed with a tetramer of the histones H3/H4 (Luger et al., 1997), this specialized centromeric nucleosome is predicted to contain a tetramer of Cse4-H4 and two copies of Scm3 (Mizuguchi et al., 2007). For a more in-depth review of non-canonical nucleosome structures please see (Zlatanova et al., 2009).
Using unbiased and multi-faceted approaches, we sought to gain a better understanding of the content and structure of Cse4-containing chromatin. We have mapped the genomic locations of Cse4 in budding yeast using chromatin immunoprecipitation followed by hybridization to DNA microarrays (ChIP-chip) and high resolution quantitative PCR (qPCR). As expected, we find evidence for localization of Cse4 to a single centromeric nucleosome at every centromere, and also at low levels at select non-centromeric sites. Solubilization and immunoprecipitation of Cse4-containing mononucleosomes indicate that the Cse4 nucleosome contains histones H2A, H2B, and H4, but not Scm3 or H3. Additionally, we find that overexpression of Cse4 can rescue a scm3Δ strain, indicating that a specialized Scm3-containing nucleosome is not essential for centromere function. Cse4-containing octamers can be assembled in vitro that contain histones H2A, H2B, and H4. These octamers can wrap DNA to form nucleosomes. Moreover, comprehensive mutagenesis of Cse4 indicates that Cse4-Cse4 interactions are essential for its function. Finally, we find that Cse4-Cse4 interaction occurs in vivo in the context of the centromeric nucleosome. Taken together, our experimental data is consistent with the model that Cse4-containing nucleosomes have a structure similar to canonical octameric nucleosomes.
Results
Cse4 localization at centromeres and non-centromeric locations
Cse4 has been shown to localize to centromeres in budding yeast by both ChIP and microscopy (Chen et al., 2000; Collins et al., 2004; Meluh et al., 1998). In the case of Cse4 fused to GFP, the only signal reported corresponds to centromeres (Pearson et al., 2004). In the ChIP studies, the presence of Cse4 was only tested at a centromere or a select few other non-centromeric loci (Collins et al., 2004; Meluh et al., 1998). In order to take an unbiased approach and discover all the locations of Cse4 in the genome, we utilized formaldehyde crosslinking and immunoprecipitation followed by hybridization to DNA microarrays. We refer to ChIP performed with crosslinked chromatin as XChIP. The microarrays we used contain approximately 13,000 PCR fragments corresponding to each open reading frame and each intergenic region in the yeast genome.
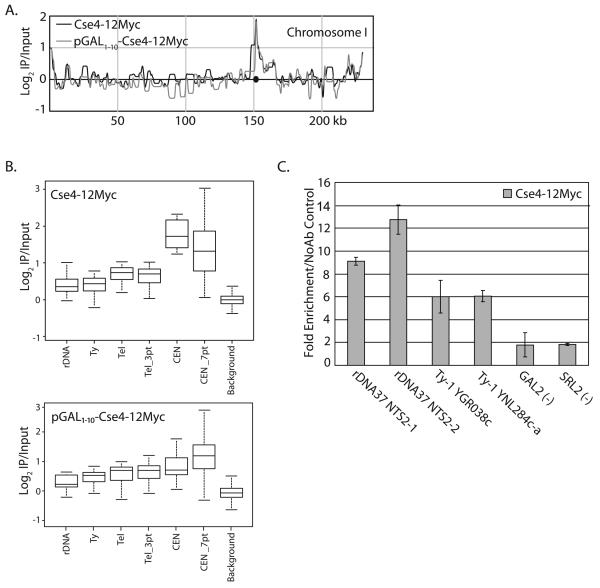
We analyzed the location of Cse4 in two different strains: 1) a strain in which the endogenous Cse4 is tagged with 12Myc epitopes (Cse4-12Myc), and 2) a strain that contains both the endogenous Cse4 (untagged) and an ectopic copy of Cse4-12Myc which is inserted into the URA3 locus and expressed from the GAL1-10 promoter (pGAL1-10-Cse4-12Myc). DNA recovered from XChIPs and “no antibody” controls was tested for enrichment of CEN3 sequence by semi-quantitative PCR prior to hybridization to microarrays. When antibody was omitted from the XChIP, we did not detect centromere sequences by semi-quantitative PCR (data not shown). In all cases, the strongest signal detected by microarray on each chromosome was at the centromere (Figure 2A). However, a few repetitive regions consistently showed a low level of Cse4 enrichment, including Ty transposable elements, telomeres, and the rDNA repeats (Figure 2B, Supplementary Table 3-4). Stringent statistical analysis of the microarray data was performed to confirm enrichment at these non-centromeric sites (see Materials and Methods). To ensure that the non-centromeric localization of Cse4 was not due to the addition of the Myc epitope tag, we also performed the same XChIP-chip using a HA epitope tagged Cse4 (Cse4-3HA). We find localization of Cse4 to the same non-centromeric sites in the strain expression Cse4-3HA (Supplementary Table 5). Notably, overexpression of Cse4 does not appear to cause a large increase in non-centromeric localization, as has been shown for overexpression of CENP-A and CID (Ahmad and Henikoff, 2002; Van Hooser et al., 2001).
Figure 2.
Genome-wide localization of Cse4. Cultures expressing Cse4-12Myc (SBY617) or pGAL1-10-Cse4-12Myc (SBY1071) were grown to mid-log phase and arrested in metaphase with nocodazole. XChIP-chip and qPCR was performed using two independent biological samples and the results were averaged. A. PeakFinder display (Glynn et al., 2004) of the pattern of Cse4-12Myc and pGAL1-10-Cse4-12Myc on Chromosome 1. The centromere is indicated by the circle. B) Box plot representing Cse4-12Myc and pGAL1-10-Cse4-12Myc localization at all centromeres (CEN), centromeres along with 3 array spots on both sides (CEN_7pt), the sequence at the rDNA locus, Ty elements, the telomeres, 3 array spots proximal to all telomeres (Tel_3Pt) and all other array spots (Background). The span of each box represents a range where 95% of the array spots fall. The line in the box represents the median of each sample. The whiskers represent the 1.5 interquartile range. C. XChIP/qPCR was carried out for several non-centromeric regions. Cse4-12Myc ChIP/total chromatin signal was divided by the no antibody/total chromatin control for each region to obtain the fold-enrichment. Error bars represent +/− the average deviation of biological replicates.
In order to verify the low levels of Cse4 at non-centromeric locations, we performed qPCR utilizing primer pairs representing NTS2 (rDNA), and two different Ty-1 transposable elements. As a negative control, we used primers which amplified regions of two genes where no detectable Cse4 signal was observed by DNA microarray analysis (GAL2, SRL2). All ChIP intensities are shown relative to the signal from total chromatin, so we have controlled for the fact that these sequences may be repeated in the genome. We found that sequences from NTS2 and Ty elements were significantly enriched relative to a control XChIP performed without antibody, and relative to the negative control regions (GAL2, SRL2) (Figure 2C). We were unable to verify the low level of Cse4 enrichment we observed at telomeres by qPCR.
High resolution mapping of centromeric chromatin
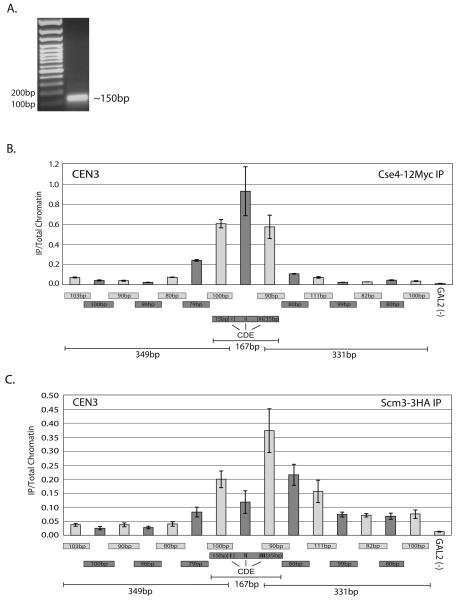
In order to define centromeric chromatin, we utilized a high resolution XChIP/qPCR approach. To do this we isolated chromatin from a yeast strain expressing Cse4-12Myc. The culture was arrested with nocodazole, and crosslinked with formaldehyde. We then treated the chromatin with micrococcal nuclease (MNase) to generate mononucleosomes (Figure 3A). Using αMyc antibody, we immunoprecipitated DNA, and used this DNA as a template for qPCR. To achieve the highest resolution possible we designed tiled qPCR primers for ~0.4 kb flanking CDE I-II-III of CEN3.
Figure 3.
Cse4 and Scm3 location at high resolution in vivo. XChIP/qPCR analysis was performed on MNase treated chromatin from strain RC82 using overlapping primers that span ~ 400 bp across the centromere on Chromosome 3. GAL2 is a gene located on the arm of Chromosome XII and is a negative control for Cse4 localization. A. MNase-treated chromatin was visualized with ethidium bromide on an agarose gel. B. Quantitative PCR results of the Cse4-12Myc XChIP for the region surrounding CEN3. Only three primer pairs contained sequences from CDEI-II-III, as indicated. The size of each primer pair product is indicated below its respective bar on the histogram. Without antibody, the XChIP/qPCR signal was <10% of the total signal; this has been subtracted from the values presented. The signal from each XChIP has been divided by the signal obtained with total chromatin. Error bars represent +/− the average deviation of biological replicates. C. Quantitative PCR results of the Scm3-3HA XChIP for the region surrounding CEN3. Primers and error bars are the same as in 3B. The Scm3-3HA localization pattern differs from the Cse4-12Myc localization pattern, exhibiting the strongest XChIP signal at the primer which amplifies CDE III and the sequence directly downstream of the centromeric DNA elements.
This primer set generated products that averaged ~95 bp and overlapped by ~50-60 bp. Of the 16 primer sets utilized, we found that the three PCR primer pairs which directly included sequences of CDE I-II-III had the highest signal for Cse4 (Figure 3B). Similar results were observed for CEN1 (Supplementary Figure 1). Based on the observed DNA fragments amplified by these primers and the absence of signal directly adjacent, we narrowed down the possible location of Cse4 to <200 bps of DNA, which includes the centromeric DNA elements. Assuming that a Cse4-containing nucleosome is wrapped around a similar amount of DNA as a canonical H3-containing nucleosome, this finding is consistent with the observation that Cse4 is confined to a single centromeric nucleosome (Furuyama and Biggins, 2007).
To elucidate the exact centromeric location of Scm3, we performed the same high resolution XChIP/qPCR using a strain expressing HA epitope tagged Scm3 (Scm3-3HA, Figure 3C). Interestingly, this experiment revealed that the localization pattern of Scm3-3HA differs from that of Cse4-12Myc. Scm3-3HA localized to the centromere with a bias towards the CDE III end of the centromeric sequence. This result is very similar to what has been observed for the inner kinetochore proteins Ndc10 and Mif2 (Cohen et al., 2008; Espelin et al., 1997). Scm3 has been shown to bind to Ndc10 (Camahort et al., 2007).
Next, using antibodies against histones H2A, H2B, H3, and H4, and epitope-tagged 3FLAG-H2A, we attempted high resolution XChIP/qPCR. We found that we could not reproducibly XChIP any core histones to the centromere in MNase-treated chromatin (data not shown). Using sheared chromatin as a substrate for XChIP, we also found that we could not detect any of the core histones directly at CEN3, although we could readily detect Cse4-12Myc and Scm3. We observed signal for all core histones at nucleosomes directly flanking the centromere (~750 bp away) (Supplementary Figure 2), indicating our methods are working in principle. Based on our inability to obtain signal for any of the core histones at the centromere, including H4, we conclude that XChIP is not a useful tool to elucidate the complete composition of the centromeric nucleosome. We suspect that the formaldehyde may crosslink other kinetochore proteins to the centromeric nucleosome, and physically block the histone epitopes located in the underlying chromatin from recognition by the antibody.
Cse4 co-immunoprecipitates with H2A, H2B, and H4 in MNase-solubilized chromatin
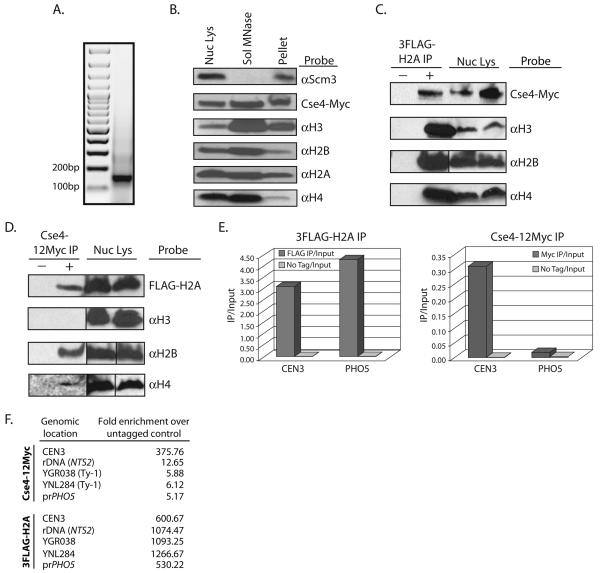
Using sub-cellular fractionation and salt challenge experiments, we have previously shown that Scm3 and Cse4 strongly associate with chromatin (Camahort et al., 2007). However, these experiments did not directly address whether these proteins were actual components of nucleosomes or just intimately associated with them. To test this, we performed chromatin fractionation followed by co-immunoprecipitation (co-IP) and Western blot analysis. Yeast nuclei were isolated and the soluble nuclear proteins were separated from the chromatin fraction. The chromatin fraction was then treated with MNase and the solubilized chromatin was recovered. We refer to this fraction as MNase-solubilized chromatin. To ensure that the majority of the MNase-solubilized chromatin was mononucleosome in nature, DNA was isolated from the MNase-solubilized fraction and subjected to ethidium bromide staining (Figure 4A). Western blotting was performed to detect proteins in the nuclear lysate, the MNase-solubilized chromatin and the insoluble chromatin fraction. Using antibodies against epitope-tagged 3FLAG-H2A and Cse4-12Myc, and endogenous H2B, H3, H4, and Scm3 we could detect all of the above proteins in the total nuclear lysate (Figure 4B). Interestingly, Cse4, H2A, H2B, H3, and H4 were all detected in the MNase-solubilized fraction but Scm3 was not (Figure 4B). Scm3 remained insoluble following MNase treatment. This result is identical to that from a recent study in fission yeast which reported that Schizosaccharomyces pombe Scm3 (Scm3sp) remained in the insoluble pellet after bulk chromatin was treated with MNase (Pidoux et al., 2009). The simplest interpretation of this result is that Scm3 is not a core component of the centromeric nucleosome, although we cannot rule out the possibility that a Scm3-containing nucleosome species exists that cannot be solubilized by MNase.
Figure 4.
Co-Immunoprecipitation of histones from MNase-solubilized chromatin. Nuclear lysates were made from a strain expressing both 3FLAG-H2A and Cse4-12Myc (RC188). The chromatin fraction was pelleted and treated with MNase. Western blots were probed with each antibody listed. For each antibody, the lanes are from a single exposure of the same gel. Vertical lines indicate that intervening lanes were cropped. A. DNA was isolated from the MNase solubilized chromatin and was visualized with ethidium bromide on an agarose gel to ensure a majority of mononucleosomes. B. Total nuclear lysate, the MNase-solubilized chromatin fraction, and the insoluble chromatin pellet after MNase treatment were loaded for Western blot analysis. C. Co-immunoprecipitation was performed using the MNase-solubilized chromatin from strain RC188. αFLAG-conjugated beads were used to pulldown 3FLAG-H2A, and the pulldown was probed using the antibodies listed. The negative control pulldown was performed using chromatin from a strain lacking 3FLAG-H2A (SBY617). D. αMyc-conjugated beads were used to pulldown Cse4-12Myc from chromatin made from RC188, and the pulldown was probed using the antibodies listed. The negative control pulldown was performed using chromatin from a strain lacking Cse4-12Myc (RC177). E. DNA was isolated from both the αFLAG and αMyc pulldowns (NChIP). qPCR was then performed to look for enrichment of DNA at either CEN3 or at the site of a well-positioned canonical nucleosome at the PHO5 promoter. Cse4 is significantly enriched at CEN3 when compared to the PHO5 promoter. Levels of 3FLAG-H2A are similar at both sites. F. qPCR signal from immunoprecipitated Cse4-12Myc and 3FLAG-H2A compared to an untagged control strain at CEN3, the rDNA, Ty elements and the PHO5 promoter. Fold enrichment was calculated by dividing IP/input ratios from the untagged control by the IP/input ratios of the epitope-tagged samples.
Next we performed co-IP using the MNase-solubilized chromatin. Pulldowns were performed using beads conjugated to αFLAG or αMyc antibodies. The immunoprecipitations were probed for all other core histones. Immunoprecipitation of 3FLAG-H2A pulled down Cse4-12Myc, H3, H2B, and H4 (Figure 4C). When we immunoprecipitated Cse4-12Myc, we were able to detect an interaction with H2A, H2B, and H4, but not H3 by Western blot (Figure 4D). The lack of detectable histone H3 signal confirms the lack of any significant contamination of oligonucleosomes in the MNase-solubilized chromatin. To further ensure that a positive interaction between Cse4 and histone H2A could be detected from purified mononucleosomes, we isolated MNase-solubilized chromatin from the above strain (Cse412Myc, 3FLAG-H2A), and subjected the soluble nucleosomes to gel filtration chromatography to isolate fractions which contain mononucleosomes (Supplementary Figure 3A). When we used this mononucleosome pool as a substrate for co-IP, we detected and an interaction between Cse4-12Myc and 3FLAG-H2A (Supplementary Figure 3B).
To alleviate the concern that the MNase-solubilized chromatin fraction did not contain centromeric nucleosomes, we performed qPCR analysis using DNA prepared from both the 3FLAG-H2A and the Cse4-12Myc immunoprecipitated samples. This experiment is in essence a modified ChIP procedure, without formaldehyde crosslinking (NChIP). Using primers that amplify either CEN3, or the region of a positioned canonical nucleosome at the PHO5 promoter (Almer et al., 1986), we observed that the 3FLAG-H2A immunoprecipitation was enriched for sequences from both sites, while the Cse4-12Myc immunoprecipitation was significantly enriched only for CEN3 DNA (Figure 4E). We also performed qPCR to look for enrichment of DNA from either the rDNA or Ty elements. We found only a slight enrichment for rDNA (2-fold over control) and no enrichment for Ty elements in the DNA that immunoprecipitated with Cse4-12Myc, an indication that the majority of nucleosomes in the Western blot analysis were centromeric in origin (Figure 4F). Taken together, these results suggest that in vivo, the centromeric Cse4 nucleosome contains histones H2A, H2B, and H4, and excludes H3 and Scm3.
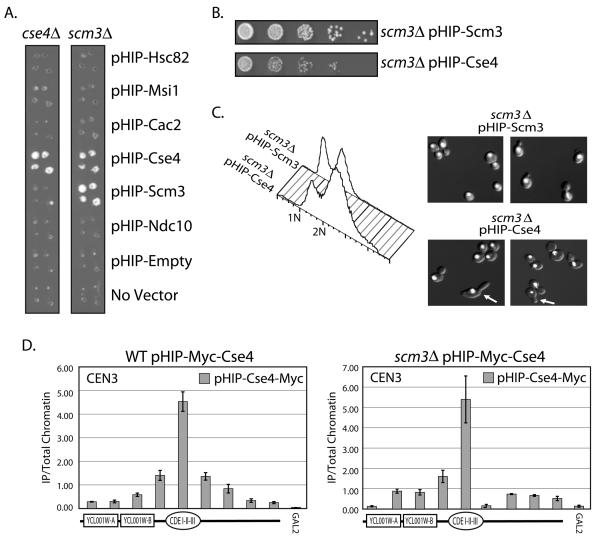
Overexpression of Cse4 can rescue a scm3Δ strain
Deletion of SCM3 in budding yeast is lethal (Giaever et al., 2002). We tested whether overexpression of other proteins could rescue the viability of a scm3Δ strain. Both CSE4/cse4Δ and SCM3/scm3Δ heterozygous diploid knockout strains were transformed with GAL1-10 overexpression plasmids from the yeast Harvard Institute of Proteomics ORF collection (pHIP), sporulated, and then pinned in quadruplicate to medium that allows the recovery of haploid yeast that contain both plasmid and the scm3 or cse4 deletion. As expected, overexpression of either Cse4 (pHIP-CSE4) or Scm3 (pHIP-SCM3) allows growth in each of their respective knock-out strains (Figure 5A). Surprisingly, we found that overexpression of Cse4 could rescue a scm3Δ strain. The scm3Δ strain covered by the Cse4 overexpression plasmid grows slightly slower than scm3Δ covered by a plasmid overexpressing Scm3 (Figure 5B). To confirm this finding we also transformed the galactose/glucose regulatable Scm3on/off strain (Camahort et al., 2007) with a plasmid containing Cse4 expressed under the control of the CUP1 (copper-inducible, pCUP1-CSE4) promoter. We found that with the addition of copper (Cu++) to the growth medium, pCUP1-CSE4 could rescue growth on medium containing glucose (Scm3off) (Supplementary Figure 4). This result is consistent with a previous report in S. pombe in which overexpression of Cnp1/CENP-A has been shown to rescue a mutant allele of SCM3sp (Pidoux et al., 2003).
Figure 5.
Suppression of the scm3Δ strain. The SCM3/scm3Δ (RC204) or CSE4/cse4Δ (RC203) heterozygous diploid knockout “Magic Marker” strain was transformed with plasmids from the HIP overexpression library. A. Sporulated cultures were pinned in quadruplicate onto medium which allowed the recovery of haploid yeast containing either scm3Δ or cse4Δ and the plasmid indicated. B. Dilution assay comparing the growth of scm3Δ covered by either pHIP-Scm3 or pHIP-Cse4 on Gal-Ura medium. C. FACscan analysis and DAPI staining of the scm3Δ strain covered by pHIP-Scm3 and scm3Δ covered by pHIP-Cse4. DAPI stained DNA is shown in white. Arrows indicate cells with abnormal morphology (elongated buds). D. qPCR analysis of pHIP-Myc-Cse4 localization was performed in both a wild type (WT) Scm3 strain containing the pHIP-Myc-Cse4 plasmid (RC205) and a scm3Δ strain containing the pHIP-Myc-Cse4 plasmid (RC192). The primers used amplify a +/− 2kb region around CEN3. A depiction of the features within the region is shown below each histogram. GAL2 serves as a negative control for Cse4 localization. Error bars represent +/− the average deviation of biological replicates.
To test for mitotic defects, we performed FACscan analyses (FACs) to look at DNA content in scm3Δ cells that are rescued by pHIP-CSE4 (Figure 5C). We find that when we analyze DNA content by cytometry, scm3Δ cells rescued by pHIP-CSE4 have a mitotic delay, with a larger proportion of the cells exhibiting 2N DNA content, as compared to scm3Δ cells rescued by pHIP-SCM3. Additionally, we have also performed DAPI staining to visualize the DNA (Figure 5C). The DAPI staining shows that DNA segregation is occurring, although the preponderance of the cells are large budded, with some exhibiting morphology defects (elongated buds). We therefore surmise that the kinetochore is minimally functional for chromosome segregation in the absence of Scm3, but kinetochore defects are likely. To confirm that Cse4 still localized properly to the centromere in the absence of Scm3, we performed XChIP using a Myc epitope tagged pHIP-CSE4 (pHIP-Myc-CSE4) in the scm3Δ strain. Using XChIP/qPCR and sheared chromatin we find that in the absence of Scm3, Cse4 localizes to the centromere in a pattern consistent with single nucleosome (Figure 5D). Although the scm3Δ pHIP-CSE4 strain does have some growth defects, we conclude that functional Cse4-containing centromeric chromatin can be formed in the absence of Scm3 when we overexpress Cse4, and a specialized Scm3-containing nucleosome is not essential for kinetochore function or chromosome segregation in budding yeast.
Cse4-containing octameric nucleosomes can be assembled in vitro
Our in vivo data suggested that we should be able to reconstitute octameric Cse4-containing nucleosomes in vitro. To this end, we purified recombinant Cse4 and canonical histones (H2A, H2B, H3, and H4), and reconstituted Cse4-containing octamers and canonical octamers (Supplementary Figure 5A). Recombinant yeast canonical histones were reconstituted into octamers (10% octamer, 20% tetramer; 40% dimer with the remainder being aggregates, Supplementary Figure 5B). Based on apparent molecular weight, the reconstituted Cse4 octamers contained two copies of Cse4, H2A, H2B, H4 and no H3. By comparison to canonical octamers, reconstitution of Cse4-containing octamers is very efficient. There is little protein aggregation and >95% of histones are in octamer form (Supplementary Figure 5B). Additionally, reconstitution of Cse4-H4 tetramers is dramatically inefficient, with ~.05% of the total input protein actually forming a tetramer (data not shown).
It was previously reported that when recombinant Scm3 was added to Cse4-containing octamers, H2A-H2B dimers were evicted, and a hexameric Scm3-Cse4-H4 complex was formed (Mizuguchi et al., 2007). Using recombinant histidine-tagged Scm3 (6HIS-Scm3) purified from E. coli, and pre-assembled Cse4 octamers, we have confirmed this finding (Supplementary Figure 6A-B). Surprisingly, we also found that when recombinant 6HIS-Scm3 was incubated with canonical octamers, and this reaction was subjected to gel filtration chromatography, the canonical octamers are split into two distinct populations, one that contains Scm3/H3/H4 and one that contains Scm3/H2A/H2B (Supplementary Figure 6C). This finding indicates a lack of specificity of Scm3 for interaction with Cse4 octamers in vitro.
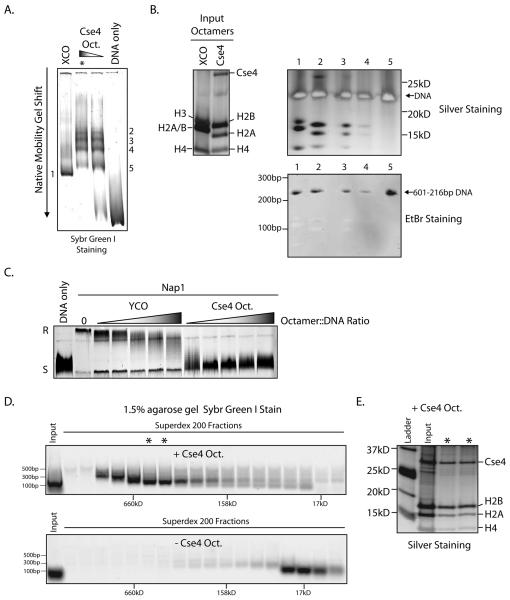
Using purified Cse4-containing octamers and Xenopus laevis canonical octamers we attempted to reconstitute nucleosomes with the “601” 216 base pair nucleosome positioning sequence (Anderson and Widom, 2000) using salt dilution. On a native polyacrylamide gel we find that when reconstitute with 601-216bp DNA, Cse4 containing octamers create a shift in the DNA migration, indicative of formation of a nucleosome (Figure 6A). Unlike the Xenopus octamer reconstitution, we observed that the Cse4 reconstitution gave rise to several shifted bands. To ascertain the protein composition of these shifted species, each band was excised from the native gel and its contents were electrophoresed directly onto a denaturing SDS-PAGE gel and silver stained to visualize the proteins. We found that the top two gel-shifted bands contained a full complement of histones and also DNA (Figure 6B), indicative of a reconstituted nucleosome. Additionally, following silver staining, the gel was stained with ethidium bromide to verify the identity of the DNA band (Figure 6B). All attempts to reconstitute Cse4 nucleosomes using a centromeric DNA sequence and salt dialysis failed (data not shown). This will be an important goal for future studies. Next, using purified Scm3-Cse4-H4 hexamers (Supplementary Figure 6E) we attempted to reconstitute a hexameric nucleosome. Using salt dialysis and both 601 and CEN3 DNA we were unable to reconstitute hexameric nucleosomes in vitro (data not shown).
Figure 6.
Reconstitution of Cse4 nucleosomes. A. Cse4 and Xenopus canonical octamers (XCO) were combined with DNA containing the 601-216bp nucleosome positioning sequence and nucleosomes were assembled by salt dilution. The resulting products were resolved on a 5% native poly-acrylamide gel. B. The shifted native gel band from the XCO reconstitution (6A,1) and the bands from the Cse4 reconstitution (6A, 2-5, asterisk) were excised and placed directly into the wells of a denaturing SDS-PAGE gel. This gel was subsequently silver stained and ethidium bromide stained. The top two bands (2-3) from the Cse4 reconstitution contain a full complement of histones and also DNA. C. Both Cse4 and yeast canonical octamers (YCO) were used in a DNA supercoiling assay with recombinant yNap1 as the histone chaperone. Addition of Nap1 and Cse4 octamers to fully relaxed plasmid DNA (R) leads to reversion to a fully supercoiled state (S), an indication of nucleosome deposition onto the plasmid template. D. A supercoiling assay was performed as in 6C, both with and without Cse4 octamers. The reaction was then treated with MNase and loaded onto a Superdex 200 gel filtration column. Fractions were collected and analyzed for DNA content by agarose gel - electrophoresis and Sybr Green I staining. E. Superdex 200 fractions containing ~150bp DNA (6D, asterisks) were treated with Benzonase to digest DNA. These fractions were then analyzed by SDS-PAGE followed by silver staining to examine protein content.
To further explore the composition of Cse4 nucleosomes assembled in vitro, we used a plasmid supercoiling assay to test for the formation of Cse4-containing nucleosomes onto a relaxed, circular, plasmid DNA template (Ito et al., 1997). In this assay, addition of a nucleosome onto the relaxed plasmid template DNA induces a positive supercoil. Using recombinant yeast Nap1 (yNap1) as the chaperone we found that Cse4 octamers efficiently supercoil the plasmid template (Figure 6C). To elucidate the composition of these nucleosomes, we treated both the Cse4 supercoiling reaction and the negative control supercoiling reaction (−Cse4 octamers) with MNase. After a short incubation, the reaction was then subjected to gel filtration chromatography to isolate distinct protein and DNA populations. We found that when we examined the DNA content of each fraction, the Cse4-containing sample yielded a distinct peak which contained DNA that was ~150bp, and whose apparent molecular weight was consistent with that of octameric mononucleosomes (Figure 6D). This is compared to the control reaction lacking Cse4 octamers, which yielded only small fragments of DNA (<100bp) of low molecular weight (Figure 6D). When the putative mononucleosome-containing fractions were subjected to SDS-PAGE followed by silver staining, we find that they contained a complete complement of histones, a strong indication that this fraction contained bona fide nucleosomes (Figure 6E). Coupled with the in vivo co-precipitation data, our in vitro data supports the model that Cse4 can form an octameric nucleosome, whose overall structure resembles that of a canonical nucleosome.
Dimerization of Cse4
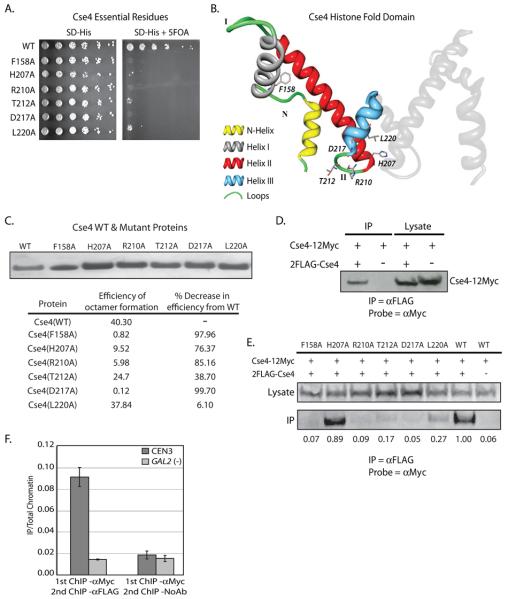
To determine which individual amino acids are essential for Cse4 function we have performed single residue alanine scanning mutagenesis across the entire CSE4 open reading frame. Each mutant was tested for its ability rescue growth in a cse4Δ strain. Using this unbiased approach we have identified 6 single amino acid residues that are essential for Cse4 function in vivo (Figure 7A). All 6 lethal mutants are located at the C-terminal end of Cse4, in the evolutionarily conserved HFD. This finding is consistent with previous reports that the N-terminus of Cse4 is dispensable for Cse4 function (Morey et al., 2004). Using the predicted crystal structure of the Cse4-containing nucleosome as a guide (Bloom et al., 2006), we have mapped the location of each of the 6 essential residues (Figure 7B). We find that 5 of the 6 essential residues are located in close proximity to the Loop II-Helix III transition. Mutations in this region would be predicted to interfere with the structure and folding of Helix III, disrupting the dyad axis and subsequently the Cse4-Cse4 dimer interface at the four-helix bundle. It is important to note that 4/5 of the identified Cse4 lethal point mutants at the Loop II-Helix III transition (H207A, R210A, T212A, D217A) have analogous lethal mutations in the H3-H3 dimer interface (Nakanishi et al., 2008). This underscores the importance of this structure in the function of both Cse4 and histone H3. We next purified recombinant Cse4 point mutants that we identified in 7A. Using these recombinant Cse4 mutant proteins and histones H2A, H2B, and H4, we assembled octamers in vitro. With the exception of L220A, we find that all of the point mutants exhibit a significant decrease in octamer reconstitution efficiency when compared to octamers reconstituted with WT Cse4 (Figure 7C).
Figure 7.
Dimerization of Cse4. Cse4 was cloned into pRS413 and subjected to site directed mutagenesis which mutated each individual amino acid to an alanine. Each mutated plasmid in the collection was transformed into a haploid cse4Δ strain containing another plasmid with a wild type copy of Cse4, and a plasmid-shuffle assay was performed. A. Growth on 5-FOA identified six single alanine substitutions that do not support growth in the cse4Δ background. B. Using the modeled Cse4 crystal structure as a guide (Bloom et al., 2006), the location of each of the 6 lethal point mutants was mapped. One molecule of Cse4 histone fold domain from the predicted Cse4 octamer crystal structure is shown in color and the essential residues are indicated. The second molecule of Cse4 is shown in grey. Five of six of the lethal point mutations lie in close proximity in either Loop II or Helix III. C. Each of the Cse4 mutants identified in our alanine scanning mutagenesis (7A) was purified from E. coli inclusion bodies. We then used these recombinant Cse4 point mutants to reconstitute octamers in vitro. Octamers were subjected to gel filtration chromatography. The efficiency of octamer formation was calculated by dividing the amount (mg) of octamer recovered by the amount of input histones (Cse4, H2A, H2B, H4). D. Co-immunoprecipitation was performed using whole cell extracts (WCE) isolated from a strain which expresses both Cse4-12Myc and 2FLAG-Cse4 (MM118). αFLAG-conjugated beads were used to pulldown 2FLAG-Cse4 from WCE, and the pulldown was probed by Western blotting with the αMyc antibody. The negative control pulldown was performed using WCE from a strain lacking the 2FLAG tag on Cse4 (MM117). E. Co-immunoprecipitation was performed from WCE isolated from a strain which expresses both Cse4-12Myc and a 2FLAG-Cse4 point mutant identified in the alanine scanning mutagenesis (MM111-116), WT 2FLAG-Cse4 (MM118), or Cse4 without a FLAG tag (MM117). αFLAG-conjugated beads were used to pulldown 2FLAG-Cse4 from WCE, and the pulldown was probed by Western blotting with the αMyc antibody. The values below each lane of the blot represent quantification of the IP band over the lysate band, with the WT sample set to 1. E. SeqXChIP was performed using sheared chromatin isolated from MM118. αMyc antibody was used for the 1st round of XChIP, followed by XChIP using either the αFLAG antibody or no antibody. The signal from each XChIP has been divided by the signal obtained with total chromatin. The centromeric primer pair spans ~ 350bp across CEN3. The GAL2 gene serves as a negative control for Cse4 localization. Error bars represent +/− the average deviation of biological replicates.
Although the results our in vitro reconstitutions and the alanine scanning mutagenesis strongly suggest that Cse4-Cse4 dimers are required for Cse4 function, these experiments do not directly address whether multimers of Cse4 are actually found in vivo. In order to do this, we created a yeast strain that co-expressed two differentially epitope-tagged Cse4 proteins, Cse4-12Myc and 2FLAG-Cse4. Using this strain we performed co-immunoprecipitation to detect an interaction between the two epitope-tagged Cse4 proteins. We found that in whole cell extracts we were able to detect a physical interaction between 2FLAG-Cse4 and Cse4-12Myc (Figure 7D). To test whether this interaction occurred in the context of the Cse4 point mutants identified in our alanine scanning mutagenesis, we constructed a strain which carried both Cse4-12Myc and a 2FLAG epitope tagged lethal Cse4 point mutant. We found that for five of the six point mutants identified in the alanine scanning mutagenesis, we lost the ability to coimmunoprecipitate the WT Cse4-12Myc protein with the mutant copy to a significant degree (Figure 7E).
To test for Cse4-Cse4 interaction in the context of the centromeric nucleosome we performed quantitative sequential chromatin immunoprecipitation (SeqXChIP) (Geisberg and Struhl, 2004) using the differentially-tagged Cse4 strain. We find that when we XChIP with an antibody against the Myc epitope, followed by XChIP using an antibody against the FLAG epitope, we observe significant enrichment of Cse4 specifically at the centromere (CEN3) (Figure 7F). Coupled with the alanine scanning mutagenesis data and the Cse4-Cse4 co-immunoprecipitation, these compelling results strongly suggest that Cse4-Cse4 dimers are required for Cse4 function, and indicate that the centromeric nucleosome contains two copies of Cse4.
Discussion
To address the composition of centromeric chromatin in budding yeast, we have performed a variety of experiments. When we treat bulk chromatin with MNase in vivo we find that unlike Cse4 and the core histones, Scm3 is not solubilized. Co-immunoprecipitation from the MNase-solubilized chromatin reveals that Cse4 physically associates with histones H2A, H2B, and H4. This result indicates that Cse4, H2A, H2B, and H4 can be found in the same nucleosome, although we cannot rule out the possibility that this interaction is an artifact of the MNase solubilization process. In MNase-solubilized chromatin we cannot detect an interaction between Cse4 and H3. Also, relative to Cse4, the position of Scm3 is shifted towards the CDE III end of the centromere when we examined localization by high resolution XChIP/qPCR. These results strongly suggest that the Cse4-containing nucleosome contains Cse4, H2A, H2B, and H4, but not H3 or Scm3. Overexpression of Cse4 can bypass the requirement for Scm3. Furthermore, Cse4, when overexpressed, can localize properly to the centromere even in the absence of Scm3. These results are a strong indication that a Scm3-containing nucleosome is not essential for chromosome segregation in budding yeast.
To further study the properties of the Cse4-containing nucleosome we have reconstituted Cse4 mononucleosomes in vitro. We can efficiently reconstitute Cse4 octamers and use these octamers to reconstitute nucleosomes. Using comprehensive alanine scanning mutagenesis we have identified a number of Cse4 residues that are essential for protein function in vivo. The large majority of these mutations are located at the predicted Cse4-Cse4 dimer interface, underscoring the importance of this domain for Cse4 function in vivo. Additionally, using co-expression of differentially epitope-tagged Cse4 proteins and sequential XChIP, we have verified the presence of a Cse4 nucleosome at the centromere which contains multiple copies of Cse4. Taken together our results suggest that Cse4 is found in an octameric nucleosome in vivo, which also contains histones H2A, H2B, and H4.
The Implications of Non-Centromeric Cse4 Localization
We observe that small amounts of Cse4 localize to non-centromeric repetitive sites in the yeast genome. It has also been reported recently that Cse4 localizes to certain highly expressed genes in rich media by ChIP-Seq (Lefrancois et al., 2009). We do not observe localization of other kinetochore proteins (Mif2, Mtw1) to repetitive DNA elements in yeast (data not shown). Therefore, the localization of Cse4 to non-centromeric loci suggests that Cse4 alone is not sufficient to nucleate a kinetochore. This is unlike mislocalization of Drosophila CID/CENP-A, which can promote formation of ectopic kinetochores and multicentric chromosomes (Heun et al., 2006). It is also important to note that the enrichment of Cse4 observed by qPCR at non-centromeric loci by either XChIP or NChIP is many fold less than what is observed at the centromere (6-fold at the rDNA vs. >100-fold at the centromere for XChIP, and 2-fold at the rDNA vs. 370-fold at the centromere for NChIP), an indication of the paucity of non-centromeric Cse4 nucleosomes. While yeast mutants that have high levels of Cse4 at non-centromeric locations have chromosome segregation defects (Collins et al. 2004) it appears that a low level of Cse4 localization to non-centromeric positions is not only tolerated, it is normal. At this time it is unclear what functional role, if any, non-centromeric Cse4 nucleosomes may play in budding yeast chromatin structure/dynamics.
Putting the hexamer to the test
It has been proposed that in budding yeast, Scm3 is a component of a specialized hexameric centromeric nucleosome (Mizuguchi et al., 2007) which lacks histones H2A and H2B. This model is based partly on experiments in vitro in which Scm3 was shown to evict H2A and H2B from preassembled Cse4-containing octamers. Based on our experimental evidence we hypothesize that Scm3-Cse4-H4 hexamer may be an intermediate in the formation of the centromeric nucleosome. We also observed that Cse4/H4 tetramers are poorly reconstituted. Perhaps Scm3 stabilizes the Cse4/H4 tetramer in vivo but is not maintained as an actual component of the centromeric nucleosome. The hexameric nucleosome model is also difficult to reconcile with previous work that shows H2A is required for proper centromere function (Pinto and Winston, 2000), and that the inner kinetochore protein Mif2 physically interacts with Cse4, H2A, H2B, and H4, but not H3 (Pinto and Winston, 2000; Westermann et al., 2003). Additionally, recent studies of the Scm3 homologue in S. pombe have reported a key finding that Scm3sp disassociates from centromeres in early mitosis, while Cnp1/CENP-A appears to remain stably associated with the centromere throughout mitosis (Pidoux et al., 2009; Williams et al., 2009). The authors of these two studies therefore favor a model in which Scm3sp is required for stable incorporation of S. pombe Cnp1 into centromeric nucleosomes, as opposed to Scm3sp being a constitutive component of centromeric chromatin.
Another compelling result is that overexpression of Cse4 can rescue a complete deletion of SCM3. Although unexpected, this result is consistent with previous work done in S. pombe that demonstrated that overexpression of Cnp1/CENP-A could rescue a lethal Scm3sp mutant (Pidoux et al., 2003). Based on these results it seems likely that Scm3 is required when Cse4 protein levels are limited, but Scm3 is not an essential component of Cse4-containing centromeric chromatin. We have tested 8 of the Cse4 point mutants from the alanine scanning mutagenesis (5 non-lethal and 3 lethal) for the ability to suppress scm3Δ when overexpressed. None are able to rescue scm3Δ (data not shown). Additional analysis of both lethal and non-lethal Cse4 point mutants will be helpful in further elucidating the structure/function relationship of Cse4 and Scm3.
In the experiments we have conducted, we were unable to find evidence for a Scm3-containing centromeric nucleosome. While we cannot completely rule out the possibility that a Scm3-containing centromeric nucleosome exists, we favor a model in which Scm3 is not an actual component of the centromeric nucleosome, but rather intimately associates with it at the inner kinetochore.
Cse4 nucleosomes are likely octamers in vivo
Another proposed model for centromeric chromatin is that a single molecule of each CENP-A (Cse4), H2A, H2B, and H4, form a structure at centromeres called a hemisome (Dalal et al., 2007a). Hemisomes were observed in interphase Drosophila cells (Dalal et al., 2007b), but have not been observed in any other organism. Unlike both CENP-A and CID (Furuyama et al., 2006; Yoda et al., 2000), Cse4-containing nucleosomes had not been reconstituted in vitro. When we reconstituted Cse4 complexes in vitro, we found that Cse4 readily forms an octameric complex with the core histones. We did not detect any species by gel-filtration chromatography whose size would be consistent with a half octamer, the pre-requisite of a hemisome. We also demonstrate that the reconstituted Cse4 octamer can wrap DNA to form a nucleosome which contains Cse4, H2A, H2B, and H4, and is most likely octameric in nature.
In addition, using comprehensive alanine scanning mutagenesis we find 6 single amino acids required for Cse4 function in vivo. Unlike previous Cse4 mutagenesis studies that identified functional regions of the HFD in the context of multiple mutations across the region (Chen et al., 2000; Keith et al., 1999), we have identified single residues essential for Cse4 function. The fact that 5 of 6 lethal mutants fall either in Loop II or Helix III is indicative of the importance of this region in Cse4 function. By analogy to H3, the Loop II-Helix III transition is the location of the dyad axis, which forms the Cse4-Cse4 four-helix bundle. Mutation in this region would be predicted to disrupt two heterodimers of Cse4-H4 coming together to form a tetramer, which would occur at the Cse4-Cse4 dimerization interface. If Cse4 was found in a hemisome, one might predict that this dimerization domain would be unnecessary. Octamer reconstitution using the Cse4 point mutants also demonstrates that residues essential for Cse4 function in vivo are required for optimal octamer reconstitution in vitro. Additionally, the results of co-IP and sequential XChIP using a strain expressing two differentially-tagged Cse4 proteins provides direct in vivo evidence that the budding yeast centromeric nucleosome contains more than one copy of Cse4 and the interaction between these molecules of Cse4 requires a fully functional four-helix bundle. Therefore, it is hard to reconcile the above data with a Cse4-containing hemisome. The sum of all of our experiments strongly suggests that the CenH3 variant Cse4 is found at a single octameric nucleosome at the budding yeast centromere, along with histones H2A, H2B, and H4.
Materials and Methods
Yeast strains and culturing
All yeast strains used were from a W303 isogenic background unless otherwise noted. All strains used are listed in Supplementary Table 1. The yeast HIP overexpression library was obtained from Harvard Proteomics and transformed into the “Magic Marker” yeast strain (Open Biosystems) as per standard high-throughput 96-well yeast protocols. Transformed diploids were selected on synthetic dropout medium lacking uracil (SD-Ura), sporulated, and then pinned in quadruplicate by hand to medium that allowed the recovery of haploid yeast that contain both the GAL1-10 overexpression plasmid and the deletion of interest (SGal glutamate, -Leu, -His-,-Ura, -Arg, +G418, 6 μg/ml +L-Canavanine) (Pan et al., 2004).
Chromatin Fractionation and Co-IPs
Chromatin fractionation was performed as previously described (Zhang et al., 2005). MNase (Worthington) was used at 100-500 units and incubated at 37° C for 10 min. Gel filtration chromatography of MNase solubilized chromatin was performed on an AKTA FPLC equipped with a Superose 6 column (GE Healthcare). Denaturing polyacrylamide gel electrophoreses (PAGE) was performed using the Novex 16% tricine pre-cast gels (Invitrogen) as per the manufacturer's protocol. Samples were electrophoresed, transferred to a nitrocellulose membrane and Western blots were performed using standard molecular biology protocols (Sambrook and Russell, 2001). Quantification of blots was performed using ImageQuant TL (Amersham). Primary antibodies for Western blots were as follows: αMyc antibody (Santa Cruz, 9E10, 1:5000), αFLAG M2 (Sigma, 1:5000), αH2A (Lake Placid Biologicals, 1:1000), αH2B (Lake Placid Biologicals, 1:5000), αH3 (Abcam 1791, 1:1000), αH4 (Abcam 31827, 1:1000, Millipore 05-858, 1:1000). Polyclonal rabbit antibodies against a C-terminal Scm3 peptide (aa 210-223) were generated and affinity purified by YenZym Antibodies, and used at 1:5000.
Chromatin immunoprecipitation with formaldehyde crosslinking
All cultures were grown to mid-log phase. Nocodazole (6.6 μg/ml, Sigma) was used to achieve metaphase arrest (verified by FACScan analysis, data not shown). Cultures were fixed with formaldehyde (1% final) for 10 minutes and chromatin was harvested by beadbeating in the presence of lysis buffer (100 mM Tris pH 7.5, 150 mM NaCl, 0.1 mM EDTA, 1 mM DTT, 0.1% NP-40, 10% glycerol, protease inhibitors). For sheared DNA, chromatin was sonicated to obtain fragments ~300 bps - 1 kb in size. The remainder of the ChIP protocol and sample preparation for DNA microarrays has been previously described (Glynn et al., 2004). The antibodies used for ChIP are as follows and were all used at 1:500 unless otherwise noted: αMyc (Santa Cruz, 9E10), αFLAG M2 (Sigma), αHA (Roche, 12CA5), αH2B (Gift of Carl Wu- NIH, Lake Placid Biologicals), αH3 (Abcam 1791), αH4 (Abcam 31827, Millipore 05-858). Controls omitting antibody were done for all ChIPs. For mononucleosome ChIPs, CaCl2 (3 mM final) was added to the chromatin after beatbeating. Micrococcal nuclease was added (500 units, Worthington), and chromatin was incubated for 30 minutes at 37°C. The MNase reaction was stopped by addition of EDTA and EGTA to a final concentration of 25 mM. For sequential ChIP, eluates from the αMyc ChIP were diluted to 0.05% SDS with lysis buffer and an αFLAG ChIP was performed.
Quantitative PCR
qPCR was performed on an iCycler real-time PCR machine using IQ Sybr Green Supermix (Bio-Rad). Primers sets for qPCR can be found in Supplementary Table 2. PCR of ChIP DNA was quantified for biological replicates by comparing immunoprecipitated samples against a standard curve established with PCRs of serial 10-fold dilutions of a DNA standard. Occupancy level was determined by dividing the average of the ChIP DNA by the relative abundance of a control total chromatin sample. This ratio represents the enrichment of immunoprecipitated DNA over the input DNA for a specific target.
Microarrays and statistical data analysis
Microarrays were printed in house and consist of over 13,000 PCR fragments representative of the entire yeast genome. Microarrays were competitively hybridized with fluorescently labeled immunoprecipitated and input DNA as previously described (Glynn et al., 2004). Microarray data analysis was carried out in Acuity, Microsoft Excel, and PeakFinder (Glynn et al., 2004). Supplementary Table 3 contains the entire dataset (log2 IP/input) for Cse4-12Myc replicates and Supplementary Table 4 contains the entire dataset (log2 IP/input) for pGAL1-10-Cse4-12Myc replicates. For statistical analysis we first used Levene's test to determine if the variance of centromere or repetitive element probes were equal to the background. From this, no population variances (centromere or repetitive elements) tested were equal to the background. Correspondingly, we applied four different statistical tests (student t-test, welch t-test, Wilcox test and Kolmogorov-Smirnov test) for each comparison, with or without assuming equal variance. The results of the statistical tests are consistent with each other.
In vitro nucleosome reconstitutions and supercoiling assay
Yeast recombinant histones (H3, H4, H2A, H2B, Cse4, Cse4 point mutants) were individually expressed in E. coli and purified from inclusion bodies as previously described (Luger et al., 1997). 6HIS-Scm3 was purified using Talon metal-affinity agarose and standard HIS-tag protein purification protocols (Sambrook and Russell, 2001). Assembly of histone octamers was carried out as in (Li et al., 2005) and octamers were purified on a HiLoad 16/600 Superdex 200 column (Amersham Biosciences) and an AKTA FPLC (GE Healthcare). Assembly of Scm3-Cse4-H4 hexamers was performed as previously described (Mizuguchi et al., 2007) and gel filtration was performed using a Superdex 200 PC 3.2/30 gel filtration column (Amersham Biosciences) on a SMART system (Pharmacia Biotech). PCR amplified DNA fragments (PCR primers are listed in Supplementary Table 2) for nucleosome reconstitution were mixed with histone octamers in high salt (2M) and subsequently subjected to serial dilution to physiological salt concentrations (100 mM) to form mononucleosomes. Mononucleosome bands were excised from the 5% non-denaturing poly-acrylamide gel and placed directly into the wells of to 16% tricine denaturing gel (Invitrogen). The plasmid supercoiling assay was performed as previously described (Ito et al., 1997). Recombinant Topoisomerase I was a kind gift from S. Venkatesh, Stowers Institute. Plasmid G5E4 used in the supercoiling assay contains 5 repeats of 5S flanking an E4 core promoter downstream of 5 Gal4 binding sites. For isolation of nucleosomes reconstituted in the supercoiling assay, the reaction was treated with 1 unit of MNase and incubated for 5 min. at 37°C. The entire reaction was concentrated and loaded onto a SMART FPLC system equipped with a Superdex 200 PC 3.2/30 gel filtration column.
Alanine scanning mutagenesis and structure analysis
Cse4 was cloned into pRS413 and site-directed mutagenesis was performed in 96-well plates using the QuickChange II Site-Directed Mutagenesis Kit (Stratagene) and plasmid manipulation was performed as previously described (Nakanishi et al., 2008). All mutants were confirmed by sequencing with T7 and T3 sequencing primers (Supplementary Table 2). Yeast were transformed using standard yeast protocols in 96-well format. Transformants were selected on SD-His-Ura. The plasmid shuffle to identify essential residues was performed as previously described (Nakanishi et al., 2008). 5-Fluoroortic Acid (5-FOA) was used at 500 μg/ml. Analysis of the predicted Cse4 crystal structure was performed with Sirius 1.2 structure visualization software (San Diego Supercomputer Center) and PDB 2FSB (Protein Data Bank) (Bloom et al., 2006).
Supplementary Material
Acknowledgements
We would like to thank Sue Biggins for strains and plasmids; Chris Seidel for assistance with design and printing of DNA microarrays; Carl Wu for strains and antibodies; and the Stowers Institute Molecular Biology Facility for help with mutagenesis. We would like to thank Laurence Florens for her help with proteomic analysis. We would like to thank Swami Venkatesh and Jeong-Hoon Kim for reagents, and helpful discussion. We would also like to thank Joan Conaway for thoughtful discussion and feedback, as well as all of the members of the Gerton lab for their help and support. Some of the experiments described herein were done to fulfill, in part, requirements for Manjunatha Shivaraju's PhD thesis research as a student registered with the Open University. This research was supported by NIH R01GM080477 and the Stowers Institute for Medical Research. B.L. is supported by a grant from the Welch Foundation (I-1713) and is a W.A. “Tex” Moncrief, Jr. Scholar in Medical Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmad K, Henikoff S. Histone H3 variants specify modes of chromatin assembly. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(Suppl 4):16477–16484. doi: 10.1073/pnas.172403699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almer A, Rudolph H, Hinnen A, Horz W. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. The EMBO journal. 1986;5:2689–2696. doi: 10.1002/j.1460-2075.1986.tb04552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JD, Widom J. Sequence and position-dependence of the equilibrium accessibility of nucleosomal DNA target sites. Journal of Molecular Biology. 2000;296:979–987. doi: 10.1006/jmbi.2000.3531. [DOI] [PubMed] [Google Scholar]
- Aravind L, Iyer LM, Wu C. Cell cycle. Vol. 6. Tex; Georgetown: 2007. Domain architectures of the Scm3p protein provide insights into centromere function and evolution; pp. 2511–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom K, Sharma S, Dokholyan NV. The path of DNA in the kinetochore. Current Biology. 2006;16:R276–R278. doi: 10.1016/j.cub.2006.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower MD, Sullivan BA, Karpen GH. Conserved Organization of Centromeric Chromatin in Flies and Humans. Developmental Cell. 2002;2:319–330. doi: 10.1016/s1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camahort R, Li B, Florens L, Swanson SK, Washburn MP, Gerton JL. Scm3 Is Essential to Recruit the Histone H3 Variant Cse4 to Centromeres and to Maintain a Functional Kinetochore. Molecular cell. 2007;26:853–865. doi: 10.1016/j.molcel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- Chen Y, Baker RE, Keith KC, Harris K, Stoler S, Fitzgerald-Hayes M. The N terminus of the centromere H3-like protein Cse4p performs an essential function distinct from that of the histone fold domain. Mol Cell Biol. 2000;20:7037–7048. doi: 10.1128/mcb.20.18.7037-7048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland DW, Mao Y, Sullivan KF. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- Cohen RL, Espelin CW, De Wulf P, Sorger PK, Harrison SC, Simons KT. Structural and functional dissection of Mif2p, a conserved DNA-binding kinetochore protein. Molecular biology of the cell. 2008;19:4480–4491. doi: 10.1091/mbc.E08-03-0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins KA, Furuyama S, Biggins S. Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Curr Biol. 2004;14:1968–1972. doi: 10.1016/j.cub.2004.10.024. [DOI] [PubMed] [Google Scholar]
- Dalal Y, Furuyama T, Vermaak D, Henikoff S. Structure, dynamics, and evolution of centromeric nucleosomes. Proceedings of the National Academy of Sciences. 2007a;104:15974–15981. doi: 10.1073/pnas.0707648104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalal Y, Wang H, Lindsay S, Henikoff S. Tetrameric Structure of Centromeric Nucleosomes in Interphase Drosophila Cells. PLoS Biology. 2007b;5:e218. doi: 10.1371/journal.pbio.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espelin CW, Kaplan KB, Sorger PK. Probing the Architecture of a Simple Kinetochore Using DNA-Protein Crosslinking. J. Cell Biol. 1997;139:1383–1396. doi: 10.1083/jcb.139.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald-Hayes M, Clarke L, Carbon J. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell. 1982;29:235–244. doi: 10.1016/0092-8674(82)90108-8. [DOI] [PubMed] [Google Scholar]
- Furuyama S, Biggins S. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:14706–14711. doi: 10.1073/pnas.0706985104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama T, Dalal Y, Henikoff S. Chaperone-mediated assembly of centromeric chromatin in vitro. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:6172–6177. doi: 10.1073/pnas.0601686103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisberg JV, Struhl K. Quantitative sequential chromatin immunoprecipitation, a method for analyzing co-occupancy of proteins at genomic regions in vivo. Nucleic acids research. 2004;32:e151. doi: 10.1093/nar/gnh148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, et al. Functional profiling of the Saccharomyces cerevisiae genome. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Glynn EF, Megee PC, Yu HG, Mistrot C, Unal E, Koshland DE, DeRisi JL, Gerton JL. Genome-Wide Mapping of the Cohesin Complex in the Yeast Saccharomyces cerevisiae. PLoS Biol. 2004;2:E259. doi: 10.1371/journal.pbio.0020259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegemann JH, Fleig UN. The centromere of budding yeast. Bioessays. 1993;15:451–460. doi: 10.1002/bies.950150704. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Dalal Y. Centromeric chromatin: what makes it unique? Current Opinion in Genetics & Development. 2005;15:177–184. doi: 10.1016/j.gde.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Heun P, Erhardt S, Blower MD, Weiss S, Skora AD, Karpen GH. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev Cell. 2006;10:303–315. doi: 10.1016/j.devcel.2006.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Bulger M, Pazin MJ, Kobayashi R, Kadonaga JT. ACF, an ISWI-Containing and ATP-Utilizing Chromatin Assembly and Remodeling Factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- Keith KC, Baker RE, Chen Y, Harris K, Stoler S, Fitzgerald-Hayes M. Analysis of Primary Structural Determinants That Distinguish the Centromere-Specific Function of Histone Variant Cse4p from Histone H3. Mol. Cell. Biol. 1999;19:6130–6139. doi: 10.1128/mcb.19.9.6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefrancois P, Euskirchen GM, Auerbach RK, Rozowsky J, Gibson T, Yellman CM, Gerstein M, Snyder M. Efficient yeast ChIP-Seq using multiplex short-read DNA sequencing. BMC genomics. 2009;10:37. doi: 10.1186/1471-2164-10-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Pattenden SG, Lee D, Gutierrez J, Chen J, Seidel C, Gerton J, Workman JL. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc Natl Acad Sci U S A. 2005;102:18385–18390. doi: 10.1073/pnas.0507975102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Meluh PB, Koshland D. Budding yeast centromere composition and assembly as revealed by in vivo cross-linking. Genes Dev. 1997;11:3401–3412. doi: 10.1101/gad.11.24.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- Morey L, Barnes K, Chen Y, Fitzgerald-Hayes M, Baker RE. The Histone Fold Domain of Cse4 Is Sufficient for CEN Targeting and Propagation of Active Centromeres in Budding Yeast. Eukaryotic Cell. 2004;3:1533–1543. doi: 10.1128/EC.3.6.1533-1543.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi S, Sanderson BW, Delventhal KM, Bradford WD, Staehling-Hampton K, Shilatifard A. A comprehensive library of histone mutants identifies nucleosomal residues required for H3K4 methylation. Nat Struct Mol Biol. 2008;15:881–888. doi: 10.1038/nsmb.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Yuan DS, Xiang D, Wang X, Sookhai-Mahadeo S, Bader JS, Hieter P, Spencer F, Boeke JD. A robust toolkit for functional profiling of the yeast genome. Molecular cell. 2004;16:487–496. doi: 10.1016/j.molcel.2004.09.035. [DOI] [PubMed] [Google Scholar]
- Pearson CG, Yeh E, Gardner M, Odde D, Salmon ED, Bloom K. Stable kinetochore-microtubule attachment constrains centromere positioning in metaphase. Curr Biol. 2004;14:1962–1967. doi: 10.1016/j.cub.2004.09.086. [DOI] [PubMed] [Google Scholar]
- Pidoux AL, Choi ES, Abbott JK, Liu X, Kagansky A, Castillo AG, Hamilton GL, Richardson W, Rappsilber J, He X, Allshire RC. Fission yeast Scm3: A CENP-A receptor required for integrity of subkinetochore chromatin. Molecular cell. 2009;33:299–311. doi: 10.1016/j.molcel.2009.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux AL, Richardson W, Allshire RC. Sim4: a novel fission yeast kinetochore protein required for centromeric silencing and chromosome segregation. The Journal of cell biology. 2003;161:295–307. doi: 10.1083/jcb.200212110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto I, Winston F. Histone H2A is required for normal centromere function in Saccharomyces cerevisiae. The EMBO journal. 2000;19:1598–1612. doi: 10.1093/emboj/19.7.1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning : a laboratory manual. 3rd edn Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y.: 2001. [Google Scholar]
- Stoler S, Rogers K, Weitze S, Morey L, Fitzgerald-Hayes M, Baker RE. Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:10571–10576. doi: 10.1073/pnas.0703178104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hooser AA, Ouspenski II, Gregson HC, Starr DA, Yen TJ, Goldberg ML, Yokomori K, Earnshaw WC, Sullivan KF, Brinkley BR. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J Cell Sci. 2001;114:3529–3542. doi: 10.1242/jcs.114.19.3529. [DOI] [PubMed] [Google Scholar]
- Waye JS, Willard HF. Structure, organization, and sequence of alpha satellite DNA from human chromosome 17: evidence for evolution by unequal crossing-over and an ancestral pentamer repeat shared with the human X chromosome. Mol Cell Biol. 1986;6:3156–3165. doi: 10.1128/mcb.6.9.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S, Cheeseman IM, Anderson S, Yates JR, III, Drubin DG, Barnes G. Architecture of the budding yeast kinetochore reveals a conserved molecular core. J. Cell Biol. 2003;163:215–222. doi: 10.1083/jcb.200305100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland G, Orthaus S, Ohndorf S, Diekmann S, Hemmerich P. Functional Complementation of Human Centromere Protein A (CENP-A) by Cse4p from Saccharomyces cerevisiae. Mol. Cell. Biol. 2004;24:6620–6630. doi: 10.1128/MCB.24.15.6620-6630.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JS, Hayashi T, Yanagida M, Russell P. Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Molecular cell. 2009;33:287–298. doi: 10.1016/j.molcel.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda K, Ando S, Morishita S, Houmura K, Hashimoto K, Takeyasu K, Okazaki T. Human centromere protein A (CENP-A) can replace histone H3 in nucleosome reconstitution in vitro. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:7266–7271. doi: 10.1073/pnas.130189697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Roberts DN, Cairns BR. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell. 2005;123:219–231. doi: 10.1016/j.cell.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlatanova J, Bishop TC, Victor JM, Jackson V, van Holde K. The nucleosome family: dynamic and growing. Structure. 2009;17:160–171. doi: 10.1016/j.str.2008.12.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.