Abstract
To identify rheumatoid arthritis risk loci in European populations, we conducted a meta-analysis of two published genome-wide association (GWA) studies totaling 3,393 cases and 12,462 controls1,2. We genotyped 31 top-ranked SNPs not previously associated with rheumatoid arthritis in an independent replication of 3,929 autoantibody-positive rheumatoid arthritis cases and 5,807 matched controls from eight separate collections. We identified a common variant at the CD40 gene locus (rs4810485, P = 0.0032 replication, P = 8.2 × 10−9 overall, OR = 0.87). Along with other associations near TRAF1 (refs. 2,3) and TNFAIP3 (refs. 4,5), this implies a central role for the CD40 signaling pathway in rheumatoid arthritis pathogenesis. We also identified association at the CCL21 gene locus (rs2812378, P = 0.00097 replication, P = 2.8 × 10−7 overall), a gene involved in lymphocyte trafficking. Finally, we identified evidence of association at four additional gene loci: MMEL1-TNFRSF14 (rs3890745, P = 0.0035 replication, P = 1.1 × 10−7 overall), CDK6 (rs42041, P = 0.010 replication, P = 4.0 × 10−6 overall), PRKCQ (rs4750316, P = 0.0078 replication, P = 4.4 × 10−6 overall), and KIF5A-PIP4K2C (rs1678542, P = 0.0026 replication, P = 8.8 × 10−8 overall).
Rheumatoid arthritis is a systemic autoimmune disease with intra-articular inflammation as a dominant feature that affects up to 1% of the population. It can be subdivided clinically by the presence or absence of autoantibodies (antibodies to cyclic citrullinated peptide (CCP) or rheumatoid factor (RF), both of which are highly correlated to each other). Previous genetic studies have identified and validated five risk loci for autoantibody-positive RA: multiple alleles within the MHC region6; a missense allele in the PTPN22 gene7; two alleles at the 6q23 locus near the TNFAIP3 gene4,5; and single alleles in the STAT4 locus8 and TRAF1-C5 loci2. Additional alleles at 4q27 (ref. 9), CTLA4 (ref. 10) and PADI4 (ref. 11) have suggestive associations, but have not yet been widely replicated in individuals of European ancestry.
To identify a collection of unbiased candidate rheumatoid arthritis risk loci for further investigation, we carried out a meta-analysis of SNP data from three case-control collections of European individuals from two published GWA studies1,2 (Table 1, see Methods for details). We investigated a common set of ~340,000 SNPs genotyped by the Wellcome Trust Case Control Consortium (WTCCC) with an Affymetrix 500K platform that (i) passed strict quality control criteria and (ii) were also present in the Phase II HapMap. We used the software package IMPUTE12 to determine genotypes of these SNPs in individuals from Sweden (Epidemiological Investigation of Rheumatoid Arthritis, EIRA) and North America (North American Rheumatoid Arthritis Consortium, NARAC) on the basis of available Illumina platform SNP data (Supplementary Fig. 1 online). To conduct a meta-analysis of SNP association with rheumatoid arthritis risk, we used the Cochran-Mantel-Haenszel (CMH) statistical test using genotype counts from the WTCCC and imputed probabilistic allele dosages in EIRA and NARAC. The CMH test allowed us to conduct a stratified analysis that maintained the three case-control collections as separate strata. CMH also allowed for further sub-stratification of EIRA and NARAC individuals into more homogenous subgroups using identity-by-state similarity for SNPs across the genome2 to correct for residual population stratification. This resulted in improved genomic control inflation for both EIRA (λGC = 1.03) and NARAC (λGC = 1.20). As there was little evidence of population stratification in the WTCCC (λGC = 1.06), we did not further sub-stratify those individuals.
Table 1.
Sample collections
| Case collection | Control collection | Origin | Antibody status | Cases | Controls | Genotyping platform | Case-control stratification | |
|---|---|---|---|---|---|---|---|---|
| Meta-analysis | North American Rheumatoid Arthritis Consortium (NARAC) | New York Cancer Project, New York City | North America | 100% CCP+ | 873 | 1,196 | Illumina 550K | Identity-by-state clustering |
| 3,393 cases; 12,460 controls | Epidemiological Investigation of Rheumatoid Arthritis (EIRA) | EIRA | Sweden | 100% CCP+ | 660 | 658 | Illumina 317K | Epidemiologically matched, identity- by-state clustering |
| Wellcome Trust Case Control Consortium (WTCCC) | Shared controls, multiple non-autoimmune diseases | United Kingdom | 80% CCP+, 84% RF+ | 1,860 | 10,606 | Affymetrix 500K | Geographically matched | |
| Stage 1 replication | Nurses Health Study (NHS) | NHS | North America | 100% RF+ or CCP+ | 257 | 411 | Sequenom iPlex | Epidemiologically matched |
| Brigham Rheumatoid Arthritis Sequential Study (BRASS) | National Institutes of Mental Health (NIMH) | Boston, USA | 100% CCP+ | 407 | 814 | Sequenom iPlex, Affymetrix 500K | Case-control matching with GWAS data | |
| 1,089 cases; 1,862 controls | NARAC II | New York Cancer Project, New York City | North America | 100% CCP+ | 425 | 637 | Sequenom iPlex | Case-control matching with ancestry informative markers |
| NARAC III | Publicly available Shared controls | North America | 100% CCP+ | 869 | 1,303 | Illumina 317K | Case-control matching with GWAS data | |
| Stage 2 replication | Genomics Collaborative Initiative (GCI) | GCI | North America | 100% RF+ | 457 | 460 | Kinetic PCR | Epidemiologically matched |
| Leiden University Medical Center (LUMC) | LUMC | Leiden, The Netherlands | 100% RF+ or CCP+ | 528 | 540 | Kinetic PCR | Geographically matched | |
| 2,840 cases; 3,945 controls | EIRA-II | EIRA-II | Sweden | 100% CCP+ | 435 | 412 | Sequenom iPlex | Epidemiologically matched |
| Genetics Network Rheumatology Amsterdam (GENRA) | GENRA | Amsterdam, The Netherlands | 100% CCP+ | 551 | 1,230 | Sequenom iPlex | Geographically matched | |
GWA data from three meta-analysis collections were used to identify candidate SNPs for replication. The replication set was divided into two stages: stage 1 replication (three collections) and stage 2 replication (five collections). For each collection we list the geographic origin, the source of the controls, the autoantibody status of cases, and the number of cases and controls. We also list the genotyping technology used to type SNPs of interest. Finally, we specify the strategy used to correct for case-control population stratification.
After calculating case-control CMH association statistics in the GWA meta-analysis, we observed minimal inflation for SNPs outside the MHC region (λGC = 1.09, λGC = 1.02 after normalizing to a 1,000 case and control collection, Supplementary Fig. 2 online). Thus, there was little evidence of bias due to technical artifact or population stratification. In Table 2 we present association statistics for validated and suggestive rheumatoid arthritis risk loci in European populations. Of the confirmed non-MHC risk loci, we observed association at PTPN22, 6q23/(containing TNFAIP3), STAT4 and TRAF1-C5. We also observed evidence of association at 4q27 (containing the IL2 and IL21 genes) and CTLA4, but not PADI4. The 4q27 result is an independent replication, suggesting that this is a true-positive association. CTLA4 replicates in the WTCCC (P = 0.026), providing further support for the role of this locus in rheumatoid arthritis, as suggested by previous studies in EIRA and NARAC10.
Table 2.
Meta-analysis results from regions previously associated with rheumatoid arthritis
| SNP |
EIRA |
NARAC |
WTCCC |
Meta-analysis |
Minor allele frequency |
Meta-analysis odds ratio (95% CI) |
Published odds ratio |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Prior study | Meta-analysis | r2 | Locus | Region | χ2 | P | χ2 | P | χ2 | P | χ2 | P | RA | Non-RA | ||
| rs2240340 | rs11203367 | 0.97 | PADI4 | 1p36 | 3.6 | 0.058 | 0.4 | 0.55 | 0.2 | 0.64 | 0.3 | 0.59 | 0.42 | 0.42 | 1.02 (0.96–1.08) | 1.10 |
| rs2476601 | rs6679677 | 1 | PTPN22 | 1p13 | 13.1 | 0.00029 | 30.2 | 3.8 × 10−08 | 151.8 | 7.1 × 10−35 | 184.3 | 5.7 × 10−42 | 0.17 | 0.10 | 1.79 (1.65–1.94) | 1.75 |
| rs7574865 | rs11893432 | 0.74 | STAT4 | 2q32 | 2.7 | 0.10 | 3.9 | 0.050 | 6.0 | 0.014 | 11.9 | 0.00055 | 0.21 | 0.18 | 1.14 (1.06–1.23) | 1.32 |
| rs3087243 | rs3087243 | n.a. | CTLA4 | 2q33 | 4.2 | 0.041 | 4.3 | 0.037 | 4.9 | 0.026 | 11.8 | 0.00060 | 0.40 | 0.44 | 0.90 (0.85–0.95) | 0.88 |
| rs6822844 | rs4572894 | 0.47 | IL2, IL21 | 4q27 | 1.8 | 0.18 | 4.8 | 0.029 | 1.8 | 0.18 | 6.4 | 0.011 | 0.27 | 0.29 | 0.92 (0.86–0.97) | 0.72 |
| HLA-DRB1*04 | rs6457620 | n.a. | HLA-DRB1 | 6p21 | 145.1 | 2.0 × 10−33 | 321.6 | 6.4 × 10−72 | 439.5 | 1.4 × 10−97 | 846.8 | 3.6 × 10−186 | 0.72 | 0.50 | 2.55 (2.40–2.71) | ~3 |
| rs6920220 | rs6920220 | n.a. | OLIG3, TNFAIP3 | 6q23 | 7.6 | 0.0059 | 7.8 | 0.0053 | 22.6 | 2.0 × 10−06 | 36.5 | 1.5 × 10−09 | 0.26 | 0.22 | 1.24 (1.16–1.32) | 1.22 |
| rs10499194 | rs1167223 | 0.5 | OLIG3, TNFAIP3 | 6q23 | 1.9 | 0.17 | 4.9 | 0.026 | 10.7 | 0.0011 | 17.0 | 0.000038 | 0.36 | 0.39 | 0.88 (0.83–0.93) | 0.75 |
| rs3761847 | rs10118357 | 0.97 | TRAF1, C5 | 9q33 | 7.9 | 0.0050 | 22.1 | 2.6 × 10−06 | 0.0 | 0.96 | 9.3 | 0.0023 | 0.46 | 0.44 | 1.10 (1.04–1.16) | 1.32 |
For each SNP, we identified the best proxy in our study and the calculated LD to that proxy on the basis of CEU HapMap (reported as r2). We present the χ2 score and two-tailed P value in EIRA, NARAC and WTCCC, and in the meta-analysis.
After excluding published risk loci (including those in the MHC region) and correcting for residual inflation by λGC, we found that 78 SNPs remained with possible associations at a P < 10−4 threshold (Supplementary Table 1 online). These SNPs were grouped into 38 independent regions on the basis of pairwise linkage disequilibrium (LD) estimates derived from CEU HapMap (where SNPs were grouped together if r2 > 0.1). We tested the single most significant SNP from each region in a two-staged replication.
Our replication collection consisted of eight independent case-control collections totaling 3,929 autoantibody (either CCP or RF) positive rheumatoid arthritis cases and 5,807 matched controls, all self-described as white and of European ancestry (Table 1). The presence of CCP or RF autoantibodies assures specificity for the diagnosis of rheumatoid arthritis and helps to minimize clinical heterogeneity across the eight collections. For each of the collections we further addressed potential case-control population stratification by either (i) using epidemiologically matched samples or (ii) matching cases and controls with ancestry informative genetic data; detailed strategies for each collection are described in the Supplementary Note online.
A total of 31 of these SNPs were successfully genotyped in all three stage 1 replication collections. The 17 most significant SNPs were genotyped in the stage 2 replication collections. For each SNP we calculated the replication P value using a one-tailed CMH statistic across the replication collections, and for those that replicated with P < 0.05, we calculated an overall P value (replication and the three meta-analysis collections) using a two-tailed CMH statistic.
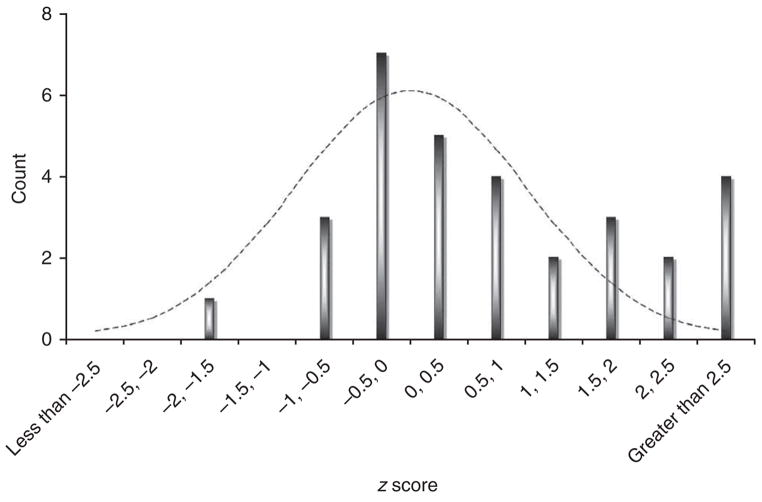
Testing in our complete replication set identified rheumatoid arthritis–associated alleles (Table 3 and Supplementary Tables 2 and 3 online). In replication genotyping, we observed that 6 out of 31 SNPs obtained P ≤ 0.01; this is significantly more than expected by chance alone (P = 9 × 10−7 by Poisson). Figure 1 illustrates the observed one-tailed CMH replication z scores, which clearly show that our results are enriched for z > 2 values (which corresponds to P < 0.023). Also, 4 of the 340,000 SNPs tested initially in the meta-analysis are associated with P < 5 × 10−7 in joint analysis; this is also significantly more than expected by chance alone (P = 3 ×10−5 by Poisson).
Table 3.
Newly identified SNPs associated with rheumatoid arthritis susceptibility
| SNP |
Meta-analysis |
Replication |
Joint | Breslow-Day test |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ID | Chr. | Position | Locus | Allele |
Frequency |
Frequency |
||||||||||
| Major | Minor | Control | Case | p | OR | Control | Case | p | OR | p | OR | p | ||||
| rs3890745 | 1p36 | 2585786 | MMEL1-TNFRSF14 | T | C | 0.329 | 0.295 | 4.3 × 10−6 | 0.86 | 0.321 | 0.301 | 0.0035 | 0.92 | 1.1 × 10−7 | 0.89 | 0.31 |
| rs42041 | 7q21 | 91891395 | CDK6 | C | G | 0.243 | 0.274 | 5.5 × 10−5 | 1.15 | 0.261 | 0.277 | 0.010 | 1.08 | 4.0 × 10−6 | 1.11 | 0.43 |
| rs2812378 | 9p13 | 34700260 | CCL21 | A | G | 0.339 | 0.364 | 6.9 × 10−5 | 1.13 | 0.334 | 0.355 | 0.00097 | 1.10 | 2.8 × 10−7 | 1.12 | 0.67 |
| rs4750316 | 10p15 | 6433266 | PRKCQ | G | C | 0.194 | 0.171 | 9.9 × 10−5 | 0.86 | 0.197 | 0.183 | 0.0078 | 0.91 | 4.4 × 10−6 | 0.88 | 0.77 |
| rs1678542 | 12q13 | 56254982 | KIF5A-PIP4K2C | C | G | 0.373 | 0.352 | 5.4 × 10−6 | 0.87 | 0.366 | 0.351 | 0.0026 | 0.92 | 8.8 × 10−8 | 0.89 | 0.62 |
| rs4810485 | 20q13 | 44181354 | CD40 | G | T | 0.254 | 0.221 | 2.4 × 10−7 | 0.83 | 0.246 | 0.231 | 0.0032 | 0.91 | 8.2 × 10−9 | 0.87 | 0.37 |
We list results of 6 SNPs (out of 31 tested) that replicated with P ≤ 0.01. The first six columns list SNP characteristics. The next four columns list GWA meta-analysis results including allele frequencies, a two-tailed P value for SNP association, and an odds ratio. The next four columns list similar results for replication genotyping; significance is reported on the basis of a stratified one-tailed CMH statistic. The next two columns summarize joint (overall) analysis results. Significance is reported on the basis of a stratified two-tailed CMH statistic across all 11 sample collections. The final column lists the Breslow-Day test for heterogeneity of odds ratios across all 11 collections.
Figure 1.
Enrichment of SNPs with z scores >2 in replication samples. For each of the 31 SNPs tested, we calculated a one-sided CMH z-score statistic from our two-staged replication data. Results were calculated using either stage 1 replication samples only (n = 14 SNPs) or using both stage 1 and 2 replication samples (n = 17 SNPs). A z score of 0 corresponds to a P value of 0.5, and a z score of 1.65 corresponds to a P value of 0.05. For a random collection of unassociated SNPs, this histogram should approximate a normal distribution (dotted line).
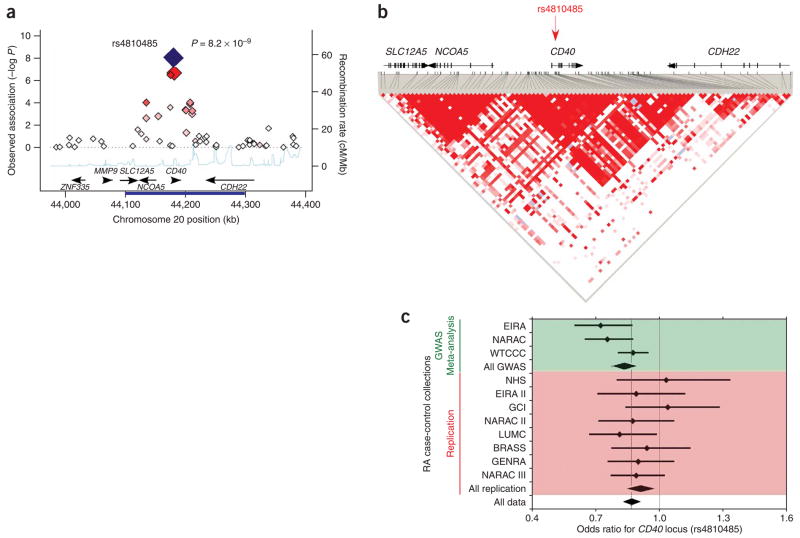
One SNP, rs4810485 in the 20q13 region, surpasses a conservative level of significance in joint analysis (P = 8.2 × 10−9) and thus represents a confirmed rheumatoid arthritis risk variant (Fig. 2). This SNP is located in the second intron of CD40 and is within an LD block that contains a large portion of the CD40 gene and its 5′ intergenic region. A SNP in near-perfect LD with rs4810485 (r2 = 0.95, rs1883832) has been associated with autoimmune thyroid disease13, although the association has not been confirmed unequivocally14,15. The same allele contributes to risk in both diseases. The rs1883832 variant has been shown to influence the efficiency of CD40 protein translation by disrupting a Kozak sequence13. The CD40 protein is expressed on the surface of multiple immune cells, including B cells, monocytes and dendritic cells, whereas its ligand, CD154, is expressed by activated CD4+ T cells. CD40-CD154 interactions play a pivotal role in provision of helper activity by CD4+ T cells in immune reactions including immunoglobulin class switching, memory B-cell development and germinal center formation16. Null mutations in CD40 are known to cause a rare B cell–dependent hyper-IgM immunodeficiency syndrome17.
Figure 2.
CD40 region and association with rheumatoid arthritis. (a) Observed association within a 400-kb region surrounding the CD40 locus in meta-analysis of three GWA datasets. We plot the meta-analysis P value on the y axis versus genomic position on chromosome 20 on the x axis. Best associated SNP was rs4810485 (large red diamond). Other data points are shaded with increasing red intensity to suggest LD to rs4810485: dark red signifies r2 ≥ 0.8, orange 0.8 > r2 ≥ 0.5, gray 0.5 > r2 ≥ 0.2, and white r2 < 0.2. In two-tailed joint analysis including all 11 collections, the SNP was strongly associated with risk of rheumatoid arthritis (P = 8.2 × 10−9, large blue diamond). Along the bottom of the figure we plot the recombination rate with the CEU HapMap (light blue line) and have indicated known human genes (black arrows). (b) The LD structure for a 200-kb region (indicated by dark blue bar along the x axis in a) surrounding the rs4810485 SNP (red arrow). The rs4810485 SNP is located in the second intron of CD40. LD between SNPs is indicated by the redness of individual spots in the triangular graphic, the most intense red spots have a D′ = 1. (c) Forest plot for rs4810485 association across 11 collections. For each sample collection we plot the odds ratio (small diamond) and the 95% CI. A light dashed line indicates the final odds ratio across all collections. The top three bars (green) represent GWA data and the diamond below them summarizes their meta-analyzed effect. The next eight bars (red) represent replication data and the diamond below them summarizes their meta-analyzed effect. The final diamond at the bottom of the page (clear) represents the final meta-analysis odds ratio and the 95% CI for all 11 collections.
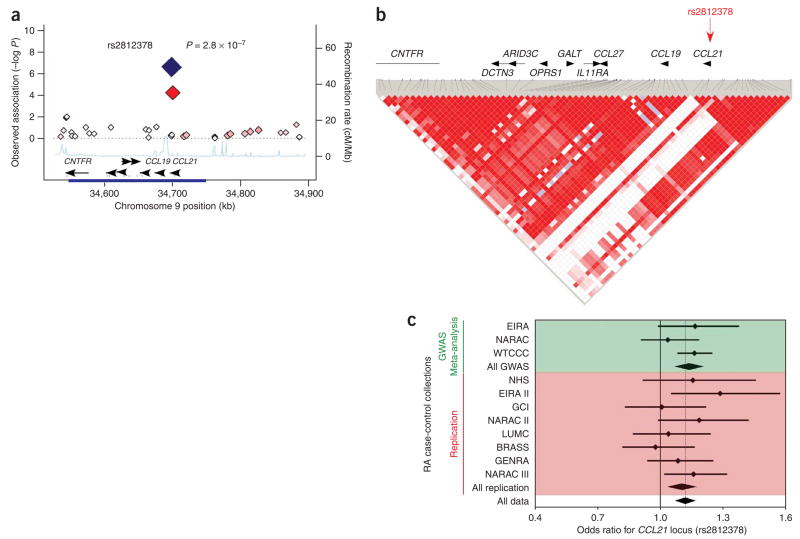
The rs2812378 SNP in the 9p13 region replicates convincingly (P = 0.00097) and has a highly suggestive level of significance with an overall P = 2.8 × 10−7 (Fig. 3). The SNP is located ~0.1 kb from the 5′ untranslated region of the CCL21 gene, and is near a cluster of other genes including CCL19 and CCL27. However, it is in an LD block that fully includes the CCL21 coding sequence and not the other genes. The CCL21 protein is a chemokine that is involved in homing lymphocytes to secondary lymphoid organs. Expression of this chemokine is associated with ectopic lymphoid structures and has been implicated in the organization of lymphoid tissue affected by rheumatoid arthritis18.
Figure 3.
CCL21 region and association with rheumatoid arthritis. (a) Observed association in the 400-kb region surrounding the CCL21 locus in meta-analysis of three GWA datasets; plot characteristics are similar to those in Figure 2. Best associated SNP was rs2812378 (large red diamond). In two-tailed joint analysis including all 11 collections this SNP was associated with rheumatoid arthritis at P = 2.8 × 10−7 (large blue diamond). (b) The LD structure for a 200-kb region surrounding the rs2812378 SNP (red arrow). The rs2812378 SNP is located close to the 5′end of the CCL21. (c) Forest plot for rs2812378 association across 11 collections.
Most of the four other SNPs with P ≤ 0.01 in our combined stage 1 and 2 genotyping probably represent true rheumatoid arthritis susceptibility alleles, although additional validation in large sample collections is required. Of the four regions, each contains genes that are known to be critical to the immune system. The rs42041 SNP on chromosome 7 maps to a CDK6 intron; CDK6 is a ubiquitous cyclin-dependent kinase that regulates cell cycle progression, and it has been identified as a key mediator in the rapid proliferation of B cells and CD8 memory cells19,20. The rs1678542 SNP on chromosome 12 is ~20 kb away from the PIP4K2C locus, which has been implicated in signaling through the B-cell antigen receptor21. The rs3890745 SNP on chromosome 1 is ~60 kb away from TNFRSF14, which is similar to CD40 in that it is a member of the TNF receptor super-family; it is known to bind TRAF family members including TRAF1 and is involved in activation of the transcription factors NF-κB and AP-1 (ref. 22). The rs4750316 SNP on chromosome 10 is ~100 kb away from the 3′ end of the PRKCQ gene, which encodes a kinase required for the activation of the transcription factors NF-κB and AP-1, and may link the T-cell receptor signaling complex to the activation of the transcription factors23.
A parallel UK study in this issue investigating the most significantly associated SNPs within the WTCCC study1 provides additional evidence for associations with rs4750316 (PRKCQ), rs1678542 (KIF5A-PIP4K2C) and rs10910099 (with r2 = 0.96 to rs3890745 in the MMEL1-TNFRSF14 locus)24.
Population stratification is unlikely to account for these observed effects, despite the modest effect sizes observed for rheumatoid arthritis risk (0.87 ≤ OR ≤ 1.12). We were careful to control for stratification individually in each of the meta-analysis GWA studies and also in each of the eight replication collections. Furthermore, the WTCCC collection contributed the greatest number of samples to the meta-analysis, and careful investigation across 12 subregions in the UK showed little evidence of case-control stratification1. Each of the associations presented here is notably significant in the WTCCC alone (P < 0.001). We found no evidence that different effects were present for these six loci across the five collections with genetically matched controls and the six collections with epidemiologically matched samples (Breslow-Day P > 0.31, Table 2). Technical artifact cannot explain the associations, as all SNPs passed strict quality control criteria (Supplementary Table 4 online).
These associations provide strong evidence for the importance of the CD40 signaling pathway in autoantibody-positive rheumatoid arthritis. Our study implicates a putative functional variant that affects protein translation of the CD40 receptor. Established associations near TRAF1 and TNFAIP3 (also known as A20) already suggest the possibility that the CD40 signaling pathway mediates rheumatoid pathogenesis through NF-κB activation25, although the rheumatoid arthritis risk variants have not yet been proven to modulate function of these genes. In particular, TRAF1 binds the CD40 receptor and cooperates with TRAF2 to activate NF-κB26. TRAF1 also binds TNFAIP3, which is a negative regulator of NF-κB signaling27. Furthermore, CD40 stimulation results in B-cell proliferation through regulation of CDK6 expression19. The CD40 signaling pathway has been investigated in drug development, and mouse models have demonstrated that its disruption could prevent development of immune-mediated arthritis and diabetes28,29.
In conclusion, our study has identified an rheumatoid arthritis–associated variant for European populations at the CD40 locus, provided strong evidence for association at the CCL21 locus, and also suggests association at four other loci. It also provides empirical data suggesting that additional common alleles with odds ratios ~1.15 remain to be discovered. Even under the assumption that all of these variants are true risk factors, their total percent variance explained is only 0.6% (Supplementary Note). In this study, we estimate that the total percent variance explained for all known non-MHC common genetic variants is just 3.6%. Considering that ~60% of rheumatoid arthritis risk is thought to be genetic, and one-third of this risk is from the MHC locus30, this indicates that less than half of genetic variation can be explained by the known rheumatoid arthritis risk alleles. One possibility is that there are other non-MHC common variants that have not yet been detected. All of the variants identified in our study have very modest effects and the rheumatoid arthritis case collections used in the meta-analysis were underpowered to screen for these effects at a modest level of significance (P < 10−4). For example, assuming the observed odds ratios and allele frequencies, simulations show that the rs4810485 SNP in the CD40 gene only had a 53% chance of meeting the P < 10−4 significance criteria that we used to initially select SNPs. The other SNPs that replicate would have had only a 19–36% chance of being selected for further replication. Together, this suggests that other common alleles of modest effect size should be identified with additional GWA studies and deeper replication in large autoantibody-positive rheumatoid arthritis sample collections.
METHODS
Subject groups
Subject groups are described in detail in Table 1 and in the Supplementary Note. Subjects were subdivided into three separate sets: (i) meta-analysis set, (ii) stage 1 replication set, and (iii) stage 2 replication set. Each collection consisted only of individuals that were self-described white and of European descent, and all cases either met 1987 ACR diagnostic criteria or were diagnosed by board-certified rheumatologists. Informed consent was obtained from each individual, and the institutional review board at each collecting site approved the study.
We used three subject groups to conduct the rheumatoid arthritis GWA meta-analysis. The groups used in the meta-analysis have been described elsewhere, and include those from the WTCCC, NARAC and EIRA. All of the cases in EIRA and NARAC and >80% of cases in WTCCC are CCP positive. For the WTCCC collection, we used an expanded collection of controls drawing from five non-autoimmune diseases that were genotyped as part of the larger study.
We used eight subject groups in our replication set. These collections consisted entirely of cases that were autoantibody positive (CCP or RF). For most of the collections, control samples were collected along with case samples as part of the same study. For some of the collections, control samples were unavailable; we matched these case collections to publicly available shared controls that had been genotyped on compatible platforms. The stage 1 replication set consisted of three subject groups: (i) CCP- or RF-positive cases identified by chart review from the Nurses Health Study (NHS) and matched controls based on age, gender, menopausal status, and hormone use; (ii) CCP-positive cases from the Brigham Rheumatoid Arthritis Sequential Study (BRASS) and controls from the National Institutes of Mental Health (NIMH); and (iii) CCP-positive cases drawn from North American clinics and controls from the New York Cancer Project (together this collection is called NARAC-II). The stage 2 replication set consisted of five collections: (i) CCP-positive cases drawn from North American clinics (NARAC-III) (P.K.G., unpublished data) and publicly available controls taken from a Parkinson’s study and study 66 and 67 of the Illumina Genotype Control Database; (ii) North American RF-positive cases and controls matched on gender, age and grandparental country of origin from the Genomics Collaborative Initiative; (iii) CCP- or RF-positive Dutch cases and controls from Leiden University Medical Center (LUMC); (iv) CCP-positive cases from Sweden and epidemiologically matched controls (EIRA-II); and (v) CCP-positive Dutch cases and controls collected from the greater Amsterdam region (GENRA).
Genotyping
Detailed description of genotyping is provided in the Supplementary Note. All GWA meta-analysis genotyping was previously described. We directly genotyped 38 SNPs in stage 1 replication samples with the Sequenom iPlex platform at the Broad Institute (for NHS case-control samples and BRASS case samples) and National Institutes of Health (for NARAC-II and NYCP samples). We obtained NIMH genotypes from previously generated GWA data on the Affymetrix 500K platform through a formal application process. We genotyped stage 2 replication samples with (i) the Illumina 317K array at the Feinstein Institute (for the NARAC-III samples; unpublished data); (ii) using the kinetic PCR platform at Celera Diagnostics (for the GCI and LUMC samples); and (iii) with the Sequenom iPlex platform at the Broad Institute (for the EIRA-II and AMC/UVA samples). We obtained publicly available genotype data for shared controls for NARAC-III cases after an official application to a Parkinson’s Disease consortium and Illumina Genotype Control Database. All stage 2 SNPs were directly genotyped in the GCI, LUMC, EIRA-II and GENRA samples, and individually imputed in NARAC-III case-control samples to determine genotype probability, as in our GWAS meta-analysis (see below).
In stages 1 and 2 we required that each SNP pass the following criteria for each collection separately: (i) genotype missing rate <10%, (ii) minor allele frequency >1%, and (iii) Hardy-Weinberg equilibrium with P > 10−3. We also excluded individuals with data missing for >10% of SNPs. Of the 38 SNPs advanced into stage 1, 6 SNPs failed genotyping in Sequenom iPlex at either the Broad or NIH, and 2 failed in the NIMH dataset. The remaining SNPs had <4% missing data for each collection. All 17 SNPs passed stage 2 replication in NARAC-III, 2 failed in GCI and LUMC, and 4 failed in EIRA-II and GENRA.
Imputation and GWA meta-analysis
We conducted a GWA meta-analysis on a set of SNPs genotyped in the WTCCC study (Supplementary Note). We selected WTCCC SNPs on the basis of strict quality control criteria: (i) genotype missing rate <1% in cases and controls separately, (ii) minor allele frequency >1% in cases and controls separately, (iii) Hardy-Weinberg equilibrium with P > 10−4 by a 2 degree-of-freedom test in cases and controls separately and (iv) availability of Phase II HapMap data. This resulted in a total of 336,721 SNPs. We imputed these SNPs in the EIRA and NARAC collections with IMPUTE. We used EIRA and NARAC data that had been filtered and imputed genotypes separately. We conducted separate runs for each chromosome using default parameters. As IMPUTE provides probabilistic confidence scores that track with prediction accuracy, we elected to use probabilistic dosages in our statistical analysis rather than hard genotype calls. This approach accounts for some uncertainty in imputation, and avoids bias.
To address case-control stratification we used identity-by-state to cluster EIRA cases and controls on the basis of on Illumina 317K SNP data into 165 substrata, and then to cluster NARAC cases and controls similarly on the basis of Illumina 550K SNP data into 396 substrata. This strategy was identical to that used to effectively control stratification previously in this dataset. As previous investigations revealed minimal case-control stratification in the WTCCC data, we placed all cases and controls from the WTCCC into a single stratum. We calculated association statistics using genotype counts available online for the WTCCC (see URLs section below) and probabilistic allele dosages for EIRA and NARAC. We calculated a CMH 1 degree-of-freedom test on the basis of allele frequency across 562 strata, and then after correcting χ2 scores by genomic control inflation (λGC), we assigned P values.
Population stratification in replication samples
For each replication collection we corrected for possible case-control stratification by either (i) using only epidemiologically matched samples when cases and controls were drawn from the same population, or (ii) matching at least one control for each case on the basis of ancestry informative markers (see Supplementary Note for details). As the cases in the NHS, GCI, LUMC, EIRA-II, and GENRA collections were well matched to controls, we did not pursue further strategies to correct for case-control stratification. For the BRASS, NARAC-II and NARAC-III collections, we matched cases and controls with ancestry informative markers and placed them into a single stratum. For the BRASS cases and NIMH controls, GWA data on Affymetrix 6.0 (unpublished data) and Affymetrix 500K platforms were available, respectively. A total of 57,417 SNPs overlapped both datasets that had 0% missing data across all individuals; we used these as SNPs to derive ancestry information. For NARAC-II cases and NYCP controls, cases and controls were matched using genotype data on 760 ancestry informative markers. Finally for the NARAC-III cases (unpublished data) and shared controls, we used available Illumina 317K GWA data for 269,771 SNPs passing stringent quality control criteria. For each case-control collection, we used these SNPs to define the top ten principal components and to remove genetically distinct outliers (σ threshold = 6 with five iterations) with the software program EIGENSTRAT. We eliminated vectors that correlated with known structural variants on chromosomes 8 and 17, demonstrated minimal variation, or did not stratify cases and controls. After mapping cases and controls in the space of eigenvectors, we matched cases to controls that were nearest in Euclidean distance. A total of 814 of the available 1,498 NIMH controls were included (matching along the top two principal components), a total of 637 of the available 1,153 NARAC-II controls were included (matching along the top principal component), and a total of 1,303 of the available 2,189 NARAC-III shared controls were included (matching along the top two principal components).
Stage 1 and 2 replication analysis
We selected SNPs for replication by (i) identifying all SNPs that achieve P < 10−4 significance in meta-analysis, (ii) grouping SNPs with r2 < 0.1 into regions, and (iii) forwarding the SNP showing strongest association from each region for replication (Supplementary Note). We excluded SNPs from regions that had already demonstrated association in other studies. We forwarded 38 SNPs for stage 1 replication. The most significant SNPs from a preliminary statistical analysis conducted without correcting for possible case-control stratification were forwarded for stage 2 replication. For only those SNPs that replicated with P < 0.05, we genotyped EIRA samples and replaced imputed genotypes with genotype data for our final analysis (Supplementary Table 5 online).
For each SNP we conducted three statistical tests. First, we conducted a one-sided CMH statistical test across eight strata to assess whether rheumatoid arthritis association was reproducible in the replication collections in the same direction as the GWAS meta-analysis used to select the SNPs of interest. Second, we calculated a 570 strata joint analysis across all meta-analysis strata and substrata and replication strata; the eight replication collections were each placed into their own strata and the GWAS samples were partitioned into 562 strata, as described above. We considered P < 5 × 10−8 as a reproducible level of significance. Third, we calculated a Breslow-Day test of heterogeneity of odds ratios. We performed all analyses in MATLAB.
URLs
WTCCC genotype data, http://www.wtccc.org.uk/info/access_to_data_samples.shtml.
Supplementary Material
Acknowledgments
We thank the WTCCC for making access to genotype data available online, and for providing autoantibody status of the WTCCC rheumatoid arthritis cases. We thank S. Myers and J. Marchini for help with IMPUTE. S.R. is supported by a T32 NIH training grant (AR007530-23), a K08 grant from the NIH (KAR055688A), and through the BWH Rheumatology Fellowship program, directed by S. Helfgott. R.M.P. is supported by a K08 grant from the NIH (AI55314-3), a private donation from the Fox Trot Fund, the William Randolph Hearst Fund of Harvard University, and holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund. M.J.D. is supported by a UO1 NIH grant (UO1 HG004171). The BRASS Registry is supported by a grant from Millennium Pharmaceuticals and Biogen-Idec. The Broad Institute Center for Genotyping and Analysis is supported by grant U54 RR020278 from the National Center for Research Resources. The NARAC is supported by NIH grants RO1-AR44422 and NO1-AR-2-2263 (P.K.G.). This work was also supported in part by the Intramural Research Program of the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health. The EIRA study is supported by grants from the Swedish Medical Research Council, the Swedish Council for Working Life and Social Research, King Gustaf V’s 80-year Foundation, the Swedish Rheumatic Foundation, the Stockholm County Council, the insurance company Arbetsmarknadens Försäkringsaktiebolag, and the County of Sörmland Research and Development Center. Genotyping of the EIRA cohort was supported by the Agency for Science Technology and Research (Singapore). Genotyping of the GCI and LUMC samples was funded by Celera. D.A. is a Burroughs Wellcome Fund Clinical Scholar in Translational Research, and a Distinguished Clinical Scholar of the Doris Duke Charitable Foundation. B.D.K. was supported by the NIH Clinical Research Training Program, a public-private partnership between the Foundation for the National Institutes of Health and Pfizer Inc. E.W.K. is supported by NIH grants R01 AR49880, CA87969, P60 AR047782, K24 AR0524-01 and BIRCWH K12 HD051959 (supported by US National Institute of Mental Health, National Institute of Allergy and Infectious Diseases, National Institute of Child Health and Human Development and the Office of the Director). K.H.C. is the recipient of an Arthritis Foundation/American College of Rheumatology Arthritis Investigator Award and a Katherine Swan Ginsburg Memorial Award. L.A.C. is a Kirkland Scholar Awardee and her work on this project was supported by K24 AR02175, R01 AI065841 and 5 M01 RR-00079. P.P.T. and N.d.V. were supported by European Community’s FP6 funding (Autocure). We acknowledge the help of C. Ellen van der Schoot for healthy control samples for GENRA and the help of B.A.C. Dijkmans, D. van Schaardenburg, A.S. Peña, P.L. Klarenbeek, Z. Zhang, M.T. Nurmohammed, W.F. Lems, R.R.J. van de Stadt, W.H. Bos, J. Ursum, M.G.M. Bartelds, D.M. Gerlag, M.G.H. van der Sande, C.A. Wijbrandts and M.M.J. Herenius in gathering GENRA subject samples and data. We thank Y. Li, S. Schrodi, and J. Sninsky (Celera); and B. Voight and C. Cotsapas (Broad Institute) for comments on the manuscript.
Footnotes
Note: Supplementary information is available on the Nature Genetics website.
References
- 1.The Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Plenge RM, et al. TRAF1–C5 as a risk locus for rheumatoid arthritis–a genomewide study. N Engl J Med. 2007;357:1199–1209. doi: 10.1056/NEJMoa073491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kurreeman FA, et al. A candidate gene approach identifies the TRAF1/C5 region as a risk factor for rheumatoid arthritis. PLoS Med. 2007;4:e278. doi: 10.1371/journal.pmed.0040278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Plenge RM, et al. Two independent alleles at 6q23 associated with risk of rheumatoid arthritis. Nat Genet. 2007;39:1477–1482. doi: 10.1038/ng.2007.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thomson W, et al. Rheumatoid arthritis association at 6q23. Nat Genet. 2007;39:1431–1433. doi: 10.1038/ng.2007.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stastny P. Association of the B-cell alloantigen DRw4 with rheumatoid arthritis. N Engl J Med. 1978;298:869–871. doi: 10.1056/NEJM197804202981602. [DOI] [PubMed] [Google Scholar]
- 7.Begovich AB, et al. A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am J Hum Genet. 2004;75:330–337. doi: 10.1086/422827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Remmers EF, et al. STAT4 and the risk of rheumatoid arthritis and systemic lupus erythematosus. N Engl J Med. 2007;357:977–986. doi: 10.1056/NEJMoa073003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhernakova A, et al. Novel association in chromosome 4q27 region with rheumatoid arthritis and confirmation of type 1 diabetes point to a general risk locus for autoimmune diseases. Am J Hum Genet. 2007;81:1284–1288. doi: 10.1086/522037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plenge RM, et al. Replication of putative candidate-gene associations with rheumatoid arthritis in >4,000 samples from North America and Sweden: association of susceptibility with PTPN22, CTLA4, and PADI4. Am J Hum Genet. 2005;77:1044–1060. doi: 10.1086/498651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki A, et al. Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nat Genet. 2003;34:395–402. doi: 10.1038/ng1206. [DOI] [PubMed] [Google Scholar]
- 12.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 13.Jacobson EM, et al. A CD40 Kozak sequence polymorphism and susceptibility to antibody-mediated autoimmune conditions: the role of CD40 tissue-specific expression. Genes Immun. 2007;8:205–214. doi: 10.1038/sj.gene.6364375. [DOI] [PubMed] [Google Scholar]
- 14.Heward JM, et al. A single nucleotide polymorphism in the CD40 gene on chromosome 20q (GD-2) provides no evidence for susceptibility to Graves’ disease in UK Caucasians. Clin Endocrinol. 2004;61:269–272. doi: 10.1111/j.1365-2265.2004.02099.x. [DOI] [PubMed] [Google Scholar]
- 15.Houston FA, et al. Role of the CD40 locus in Graves’ disease. Thyroid. 2004;14:506–509. doi: 10.1089/1050725041517039. [DOI] [PubMed] [Google Scholar]
- 16.Kawabe T, et al. The immune responses in CD40-deficient mice: impaired immunoglobulin class switching and germinal center formation. Immunity. 1994;1:167–178. doi: 10.1016/1074-7613(94)90095-7. [DOI] [PubMed] [Google Scholar]
- 17.Lougaris V, Badolato R, Ferrari S, Plebani A. Hyper immunoglobulin M syndrome due to CD40 deficiency: clinical, molecular, and immunological features. Immunol Rev. 2005;203:48–66. doi: 10.1111/j.0105-2896.2005.00229.x. [DOI] [PubMed] [Google Scholar]
- 18.Manzo A, et al. Systematic microanatomical analysis of CXCL13 and CCL21 in situ production and progressive lymphoid organization in rheumatoid synovitis. Eur J Immunol. 2005;35:1347–1359. doi: 10.1002/eji.200425830. [DOI] [PubMed] [Google Scholar]
- 19.Ishida T, et al. CD40 signaling-mediated induction of Bcl-XL, Cdk4, and Cdk6. Implication of their cooperation in selective B cell growth. J Immunol. 1995;155:5527–5535. [PubMed] [Google Scholar]
- 20.Veiga-Fernandes H, Rocha B. High expression of active CDK6 in the cytoplasm of CD8 memory cells favors rapid division. Nat Immunol. 2004;5:31–37. doi: 10.1038/ni1015. [DOI] [PubMed] [Google Scholar]
- 21.Carpenter CL. Btk-dependent of phosphoinositide synthesis. Biochem Soc Trans. 2004;32:326–329. doi: 10.1042/bst0320326. [DOI] [PubMed] [Google Scholar]
- 22.Marsters SA, et al. Herpesvirus entry mediator, a member of the tumor necrosis factor receptor (TNFR) family, interacts with members of the TNFR-associated factor family and activates the transcription factors NF-kappaB and AP-1. J Biol Chem. 1997;272:14029–14032. doi: 10.1074/jbc.272.22.14029. [DOI] [PubMed] [Google Scholar]
- 23.Gruber T, et al. PKCtheta cooperates with atypical PKCzeta and PKCiota in NF-kappaB transactivation of T lymphocytes. Mol Immunol. 2008;45:117–126. doi: 10.1016/j.molimm.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 24.Barton A, et al. Rheumatoid arthritis susceptibility loci at chromosomes 10p15, 12q13 and 22q13. Nat Genet. 2008 September 14; doi: 10.1038/ng.218. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harnett MM. CD40: a growing cytoplasmic tale. Sci STKE. 2004:pe25. doi: 10.1126/stke.2372004pe25. [DOI] [PubMed] [Google Scholar]
- 26.Xie P, Hostager BS, Munroe ME, Moore CR, Bishop GA. Cooperation between TNF receptor-associated factors 1 and 2 in CD40 signaling. J Immunol. 2006;176:5388–5400. doi: 10.4049/jimmunol.176.9.5388. [DOI] [PubMed] [Google Scholar]
- 27.Song HY, Rothe M, Goeddel DV. The tumor necrosis factor-inducible zinc finger protein A20 interacts with TRAF1/TRAF2 and inhibits NF-kappaB activation. Proc Natl Acad Sci USA. 1996;93:6721–6725. doi: 10.1073/pnas.93.13.6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balasa B, et al. CD40 ligand-CD40 interactions are necessary for the initiation of insulitis and diabetes in nonobese diabetic mice. J Immunol. 1997;159:4620–4627. [PubMed] [Google Scholar]
- 29.Durie FH, et al. Prevention of collagen-induced arthritis with an antibody to gp39, the ligand for CD40. Science. 1993;261:1328–1330. doi: 10.1126/science.7689748. [DOI] [PubMed] [Google Scholar]
- 30.MacGregor AJ, et al. Characterizing the quantitative genetic contribution to rheumatoid arthritis using data from twins. Arthritis Rheum. 2000;43:30–37. doi: 10.1002/1529-0131(200001)43:1<30::AID-ANR5>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.