Abstract
Successes in biomedical research and state-of-the-art medicine have undoubtedly improved the quality of life. However, a number of diseases, such as cancer, immunodeficiencies, and neurological disorders, still evade conventional diagnostic and therapeutic approaches. A transformation towards personalized medicine may help to combat these diseases. For this, identification of disease molecular fingerprints and their association with prognosis and targeted therapy must become available. Quantum dots (QDs), semiconductor nanocrystals with unique photo-physical properties, represent a novel class of fluorescence probes to address many of the needs of personalized medicine. This review outlines the properties of QDs that make them a suitable platform for advancing personalized medicine, examines several proof-of-concept studies showing utility of QDs for clinically relevant applications, and discusses current challenges in introducing QDs into clinical practice.
Keywords: Quantum dots, Nanoparticles, Molecular profiling, Fluorescence, Multiplexing, Pathology, Diagnosis, Targeted therapy, Drug delivery, Personalized medicine
Introduction
State-of-the-art medicine is an indispensable part of the human society. Wealth of medical knowledge accumulated over centuries of observation and experimentation, advanced diagnostic techniques made possible by the technological revolution, and innovative biomedical research done on the cellular and molecular levels provide a formidable weapon against nearly any threat to human health. However, the most devastating diseases, such as cancer, immunodeficiencies and neurological disorders to name a few, are notorious for their ability to evade current diagnostic methods and resist therapy. It is not easy to pinpoint the main reasons for poor success in combating these diseases, as they might range from a lack of understanding of patho-physiology to the absence of appropriate diagnostic techniques capable of addressing the complexity of these diseases. One potential issue is that utilization of generalized diagnostic and treatment approaches based on identifying and targeting disease symptoms (often with limited information about the underlying cause) is inefficient in addressing the great genetic and phenotypic variability of cancer and immune system disorders. Significant heterogeneity on molecular level, complex interlinking of subcellular mechanisms along with integrated pathophysiological effects on organs and systems of the human body, and an often unclear origin and cause of the disease represent major challenges for current biomedical research and clinical practice.
Personalized medicine, a practice of addressing individual diseases in a pathology-specific and patient-specific manner spanning all levels from whole-body symptoms down to molecular signatures of the disease, is an emerging field of medicine promising to provide efficient tools against cancer and other challenging diseases. A personalized approach offers unique opportunities to accurate diagnosis (i.e. pinpoint exact changes that occurred within healthy cells and tissues), prognosis (i.e. predict progression of a disease based on these changes), and treatment (i.e. specifically reverse the changes or, if not possible, target and kill the diseased cells without affecting healthy ones). Such an approach relies on advances in basic research as well as integration of novel diagnostic and therapeutic techniques into clinical practice.
Currently, attempts of introducing personalized approach in medicine rely on screening for genetic alterations in diseased cells; yet diagnostic and predictive power of genetic screening alone is questionable due to insufficient knowledge of how certain alterations on the DNA level propagate along the DNA-RNA-protein chain [1, 2] and the requirement of performing analysis on a homogenized mixture of different cell types, including a variety of healthy cells [3]. Therefore, complementary analysis of phenotypic changes (i.e. changes in protein expression) as well as assessment of the effect of diseased cells on the healthy tissues (e.g. activation of angiogenesis in tumors) is necessary for comprehensive analysis of a pathological process. Compilation of a database of genetic and phenotypic signatures of individual diseases will provide an access to a more accurate prognosis and personalized treatment targeted directly against the biomarkers expressed. Realizing this, significant research effort is being focused at understanding the physiology of normal cellular processes as well as patho-physiology of diseases in order to determine specific disease-causing changes in individual cells, organs, and systems.
A key challenge is presented by the complexity of inter- and intracellular networks with multiple inputs, controllers, and feedback loops, which is hard to assess using conventional biomedical techniques (such as immunohistochemistry, Western blot, ELISA, etc) that suffer from a limitation in the number of biomarkers that can be analyzed simultaneously, lack real-time monitoring capacity for intracellular processes, provide limited single-cell information resulting from the need to analyze signals averaged over many cells, and utilize qualitative rather than quantitative analytical techniques [4–6]. Consequently, diagnosis and prognosis are limited by the lack of knowledge about the predictive biomarkers that would unambiguously discriminate between disease and normal function as well as distinguish different disease types and provide information about possible progression of the pathological process.
Advances in nanotechnology have enabled the design of nanoparticle-based tools for improved diagnosis and personalized treatment of many complex diseases. In particular, semiconductor QDs have emerged as a new platform for high-throughput quantitative characterization of multiple biomarkers in cells and clinical tissue specimens ex vivo, detection of diseased cells in vivo, and potentially targeted and traceable drug delivery [3, 7, 8].
Properties of quantum dots for addressing the needs of personalized medicine
QDs are semiconductor nanoparticles with size ranging between 2 and 10 nm in diameter (hydrodynamic size often larger). Restricting the mobility of charge carriers (electrons and holes) within the nanoscale dimensions generates the quantum confinement effect responsible for unique size-dependent photo-physical properties of QDs [9–11]. Additionally, nanometer-scale size of QDs comparable with the size of large proteins enables integration of nanoparticles and biomolecules yielding biologically functional nanomaterials suitable for probing physiological processes on a molecular level [12–14]. While a relatively large size (compared to small drug molecules or organic fluorescent dyes) might be associated with slower diffusion, limited permeability, complex bio-distribution, and possible interference with intracellular processes [15], QDs possess a wide range of features essential for addressing the most urgent needs of personalized medicine. Among such features are size-tunable and spectrally narrow light emission, simultaneous excitation of multiple colors, improved brightness, resistance to photobleaching, and an extremely large Stokes shift.
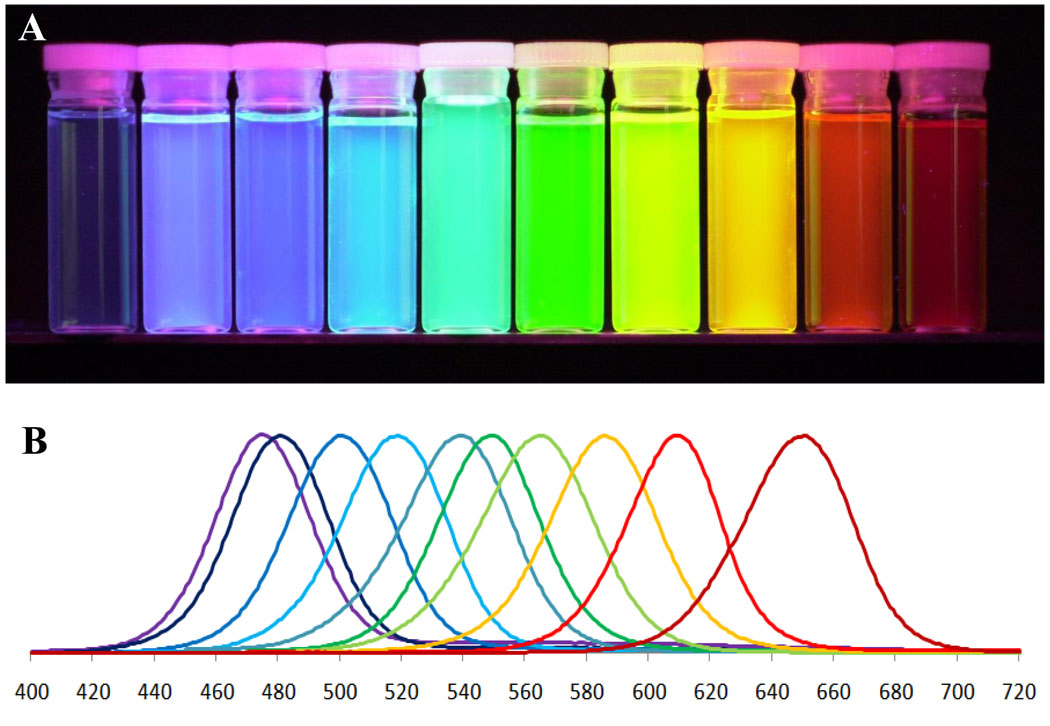
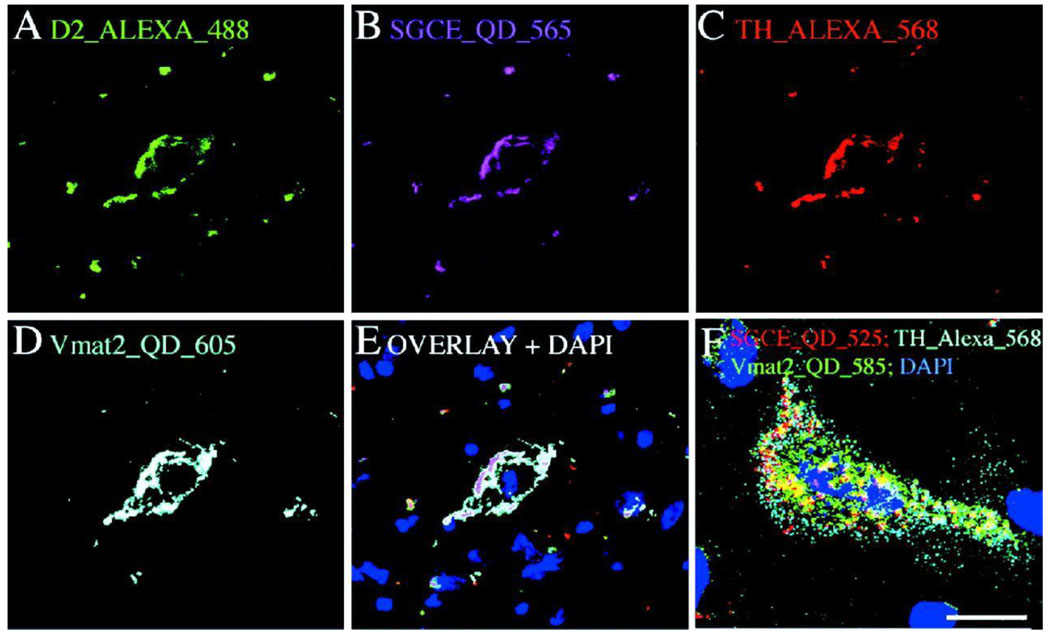
The cornerstone of personalized medicine is the ability to uniquely identify the disease by its “molecular fingerprint” (i.e. pattern of biomarker expression), associate the fingerprint with possible progression of the disease, and assign a treatment which targets diseased cells with the identified fingerprint. Achieving this goal is not a trivial task – many diseased cells look very much like the healthy ones (especially in case of cancer), and screening for a large panel of biomarkers is required. It is quite possible that certain diseases have one or few biomarkers specific enough for unique identification, yet finding these biomarkers de novo using low-throughput conventional approaches is like looking for a needle in a haystack. QDs open access to a multi-parameter biomarker screening on intact specimens via multiplexed detection [16]. This feature is based on two properties of QDs: spectrally narrow size-tunable light emission [17–19] and effective light absorption throughout a wide spectrum [12] (Fig. 1). Excitation of multiple QD probes with a single light source (e.g. laser) significantly reduces the complexity and cost of imaging instrumentation and simplifies data analysis. Utilization of hyperspectral imaging, a technique that allows deconvolution of an image into spectral components, further improves the multiplexing capabilities of QD technology (Fig. 2) [20]. It is worth mentioning that highly multiplexed molecular analysis would be limited if hyperspectral imaging or QDs are used separately. Combination of these two complementary technologies enhances each other’s capability.
Figure 1.
Quantum dots possess unique photo-physical properties suitable for addressing the needs of personalized medicine. The ability to utilize multicolor QD probes (A) and tune the emission color by the particle size allows multiplexed biomarker detection. Narrow emission spectra (B) along with efficient light absorption throughout a wide spectrum enable simultaneous imaging of several biomarkers critical for molecular profiling of diseases.
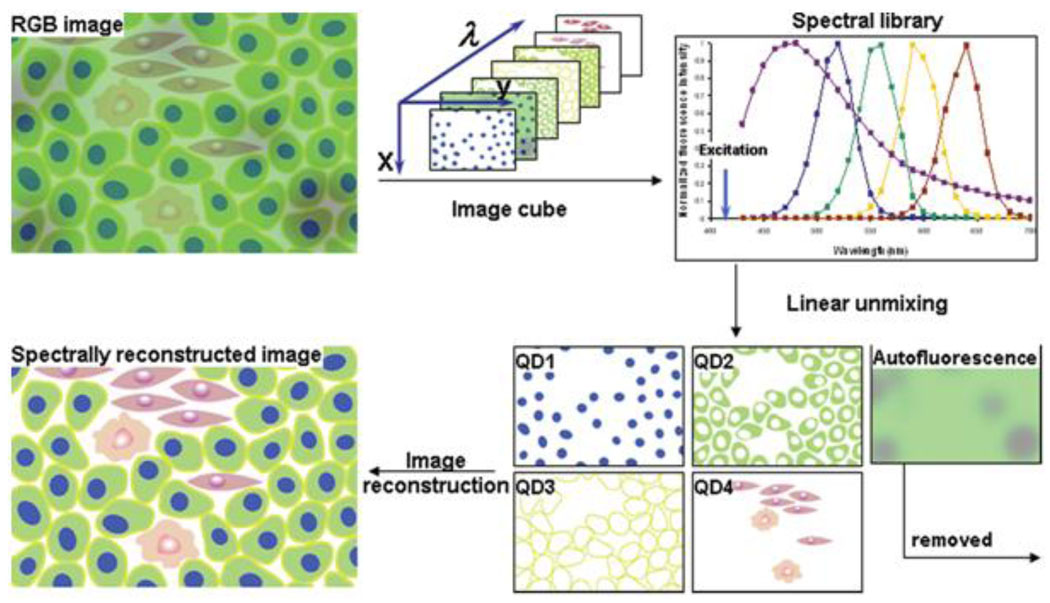
Figure 2.
Hyperspectral imaging represents a powerful technique for analysis of multiple QD-labeled biomarkers within a single specimen. While standard RGB camera cannot distinguish spectrally overlapping probes and is limited to analysis of few biomarkers, hyperspectral imaging applies narrow band-pass filters and takes a series of images for each wavelength, thus providing spectral information for each pixel of an image. Further application of spectral library allows accurate unmixing of individual spectrally-distinct components, enhancing the ability for molecular profiling [3].
An indispensable part of disease molecular profiling is the ability to quantify biomarker expression in an accurate and consistent manner. So far, this requirement has been only partially fulfilled. The problem lies in the fact that colorimetric assays usually rely on amplification mechanisms, which are difficult to control, thus providing inconsistent and mostly qualitative information about the biomarker expression. Quantitative analysis with fluorescence imaging using organic fluorophores is often compromised by the quick photobleaching of the dyes and unstable signal intensity. Destructive techniques, while allowing protein quantification (e.g. Western blot, RT-PCR, protein chips), do not preserve tissue morphology and cannot properly address the heterogeneity of specimens. QD probes, on the other hand, are well-suited for addressing these issues. First of all, QDs are highly resistant to photobleaching and photodegradation: in one example QDs retained constant signal intensity for over 30 minutes of illumination, while organic dyes faded by more than 90% in less than one minute under identical experimental conditions [21]. Second, QDs do not rely on chemical amplification (in contrast to assays such as horse radish peroxidase mediated color development and Au catalyzed Ag-enhancement) and have a promise of providing imaging probes with a 1:1 stoichiometry. It is necessary to note, though, that the intensity of different color QDs under identical illumination conditions differ significantly, showing enhancement of red QD signal over green/blue QDs. Such discordance has been observed by Ghazani and coworkers in a three-color staining of lung carcinoma xenografts for epidermal growth factor receptor, E-cadherin, and cytokeratin using QDs emitting at 655, 605, and 565 nm [22]. While quantitative analysis of individual QD signals was readily achievable, comparison between different QD signals was not possible through this study. The discordance in fluorescence intensity of individual probes directly relates to light absorption properties and the quantum yield of QDs (i.e. red particles having larger cross-section absorb light more efficiently) and can be accounted for in signal analysis algorithm. For example, Yezhelyev et al used bulk fluorescence measurement of equal concentrations of QDs and determined that QD655 were 8 times as bright and QD605 4 times as bright as QD565 [23]. However, other effects associated with high QD concentration, such as steric hindrance between the probes, self-quenching, and fluorescence resonance energy transfer (FRET) from smaller to larger particles [22], might be possible in cases of high biomarker density and deserves further investigation for achieving accurate quantitative analysis.
Studying patho-physiology with QD probes
A variety of nanomaterials have already shown utility in addressing tough questions posed by unmet clinical needs. In particular, QDs have proven to be well suited for sensitive quantitative molecular profiling of cells and tissues, holding tremendous promise for unraveling the complex gene expression profiles of diseases, accurate clinical diagnosis and personalized treatment of patients [3, 24]. Possessing advantageous photo-physical properties and being compatible with conventional biomedical assays, QDs have found use in most techniques where fluorescence or colorimetric imaging of target biomarker is utilized (e.g. cell and tissue staining, Western blot, ELISA, etc.) and have launched many novel applications (e.g. targeted in vivo imaging, single-molecule tracking, traceable drug delivery, etc.). The number of biomedical applications of QDs continues growing, ranging from ultrasensitive detection in vitro to targeted drug delivery and imaging in vivo.
Identification of molecular fingerprints of diseases
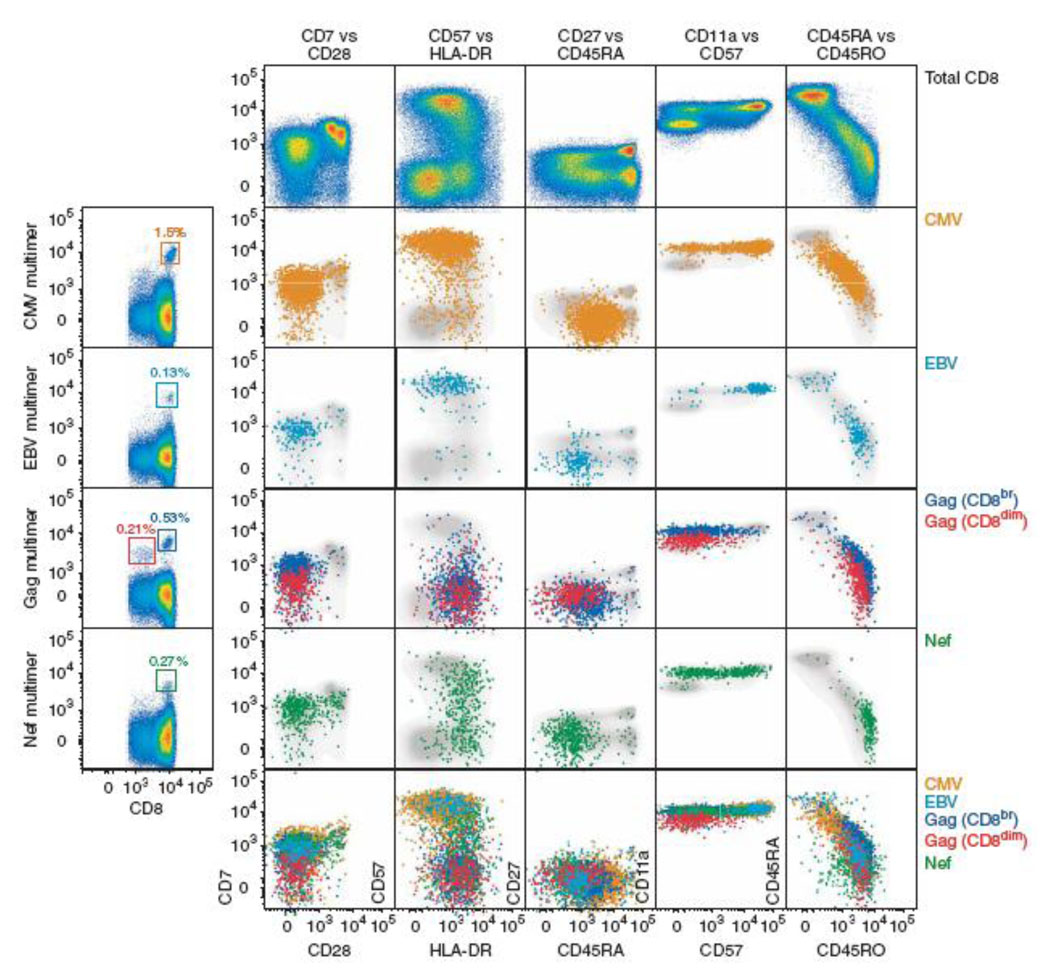
Molecular fingerprinting of diseases implies characterization of biomarker expression schemes in diseased cells in comparison to healthy ones. QD-based probes are uniquely suited for this task when employed by both multi-parameter flow-cytometry analysis of cell populations and quantitative multiplexed analysis of biomarker expression in intact tissue specimens. For example, Chattopadhyay et al, by utilizing a 17-parameter flow-cytometry (based on 8 QD probes and 9 organic fluorophores), revealed significant phenotypic differences between T-cells specific to distinct epitopes of the same pathogen (Fig. 3) [25]. Access to molecular profiles of individual cell populations not only improves our understanding of immune response, but also enables analysis of changes occurring during immune system disorders, sensitive detection of metastasizing cancer cells in a bloodstream, and accurate phenotyping of heterogeneous cell populations.
Figure 3.
Seventeen-parameter flow-cytometry analysis of antigen-specific T-cell populations was achieved using 8 QD probes and 9 organic fluorophores. Significant heterogeneity in biomarker expression within a CD8+ T-cell population (shown in gray) emphasizes the need for multi-parameter analysis in studying immune response and other complex systems [25].
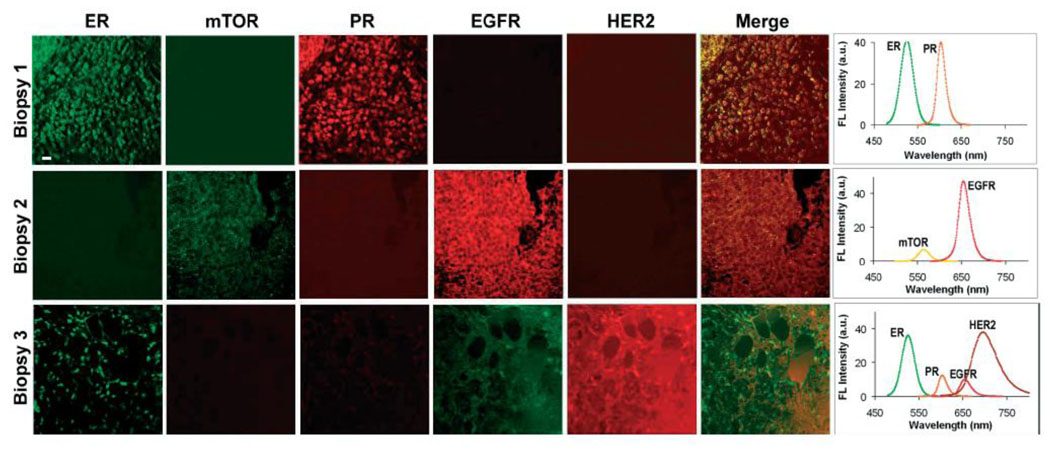
Moving towards introducing QD technology into clinical diagnostics, five-parameter characterization of breast cancer tissue specimens obtained from biopsies has been demonstrated [23]. Comparison of the three specimens revealed distinct molecular profiles, where one tumor over-expressed such biomarkers as ER and PR, another tumor primarily expressed EGFR, and third tumor showed abundance of ER and HER2 (Fig. 4). Besides diagnostic and prognostic value of such analysis, potential targets for anti-cancer treatment can also be identified, thus enabling a “personalized” approach in therapy.
Figure 4.
Five-parameter quantitative analysis of the three tissue specimens obtained from tumor biopsies clearly identified the differences in biomarker expression profiles between different types of breast cancers. Molecular fingerprinting might not only provide more accurate diagnosis and prognosis, but also identify suitable molecular targets for anti-cancer therapy [23].
Accuracy of molecular fingerprinting based on protein expression can be further improved by analysis of gene expression via quantification of mRNA using fluorescent in situ hybridization (FISH). Relying on binding of oligonucleotide probes to complimentary mRNA molecules in 1:1 probe-to-target ratio, this technique offers high level of specificity, yields direct quantitative correlation between gene amplification (i.e. number of mRNA molecules present) and signal intensity, and provides accurate information about mRNA localization within the cell. Similar to protein-based staining, quantitative potential and sensitivity of FISH might be significantly improved by utilization of QD probes [14]. In early proof-of-concept studies Xiao and Barker have used highly stable QD-Streptavidin bioconjugates for monochromatic visualization of biotinylated oligonucleotide probes in FISH analysis of amplification of clinically important erbB2 gene [26]. Using a slightly modified procedure, Tholouli et al have achieved multiplexed staining of 3 mRNA targets within one specimen [27]. In order to reduce the size of imaging probe and improve binding stoichiometry, Chan et al have developed a monovalent FISH probe by blocking extra streptavidin sites with biocytin (water-soluble biotin derivative) [28]. High-resolution multiplexed FISH has been demonstrated in simultaneous detection of four mRNA targets using two different QD probes and two different organic fluorophore probes within a single mouse midbrain neuron (Fig. 5). Notably, reduced size of FISH probes enabled staining in milder, protein-compatible specimen permeabilization conditions, which is essential for combined QD-based FISH and QD-based immunohistochemistry (IHC), thus offering the possibility of correlating gene expression at the mRNA level with the number of corresponding protein copies in diseased cells or tissue specimens [14].
Figure 5.
Multi-parameter FISH using QD probes and organic fluorophores enables high-resolution imaging of different mRNA molecular within single cells, thus providing information about relative gene expression levels, localization of mRNA within cellular compartments, and co-localization of different mRNA molecules and other biomarkers [28].
Probing intracellular pathways
While molecular fingerprinting of diseases holds tremendous diagnostic and therapeutic value, uncovering intracellular pathways leading to disorder is essential for understanding the patho-physiology of a disease, identification of an underlying cause of the pathologic changes, and design of therapies targeting dysfunctional pathways on a molecular level. Study of patho-physiology on sub-cellular level involves the characterization of intracellular distribution and relative expression of biomarkers (proteins, mRNA, etc.), analysis of phenotypic changes in cells upon certain stimulation, and real-time monitoring of changes in intracellular processes (e.g. phagocytosis, intracellular trafficking, and cell motility) in live cells.
One interesting study of intracellular morphology was demonstrated by Matsuno et al who combined QD-based FISH and IHC along with confocal laser scanning microscopy for three-dimensional imaging of the intracellular localization of growth hormone (GH), prolactin (PRL), and of their mRNAs within tissue specimens [29]. With further improvements in design of QD probes suitable for multiplexed FISH and IHC, this technology will allow three-dimensional mapping of the relative position of biomarkers and corresponding mRNAs inside cells and tissues with high resolution and sensitivity, thus providing access to studies of intricate signaling pathways and mechanisms of pathogenesis.
Further improvement in imaging resolution can be achieved by utilization of transmission electron microscopy (TEM). For example, relatively high electron density of QDs was successfully employed by Giepmans et al for high-resolution study of intracellular biomarker distribution [30]. In this study initial optimization of staining conditions was achieved using fluorescence imaging, while further examination with TEM revealed intracellular localization of QD probes (and corresponding biomarkers) with respect to sub-cellular structures. Due to direct correlation between fluorescence emission color and QD size, detection of three QD-labeled biomarkers distinguishable at both fluorescence (by color) and TEM (by size) levels was achieved [30]. Enhancement in multiplexing functionality of this technique can be obtained from discrimination of QDs based on their elemental composition. Nisman et al have proposed the use of electron spectroscopic imaging (ESI, a technique for generating elemental maps of materials with high resolution and detection sensitivity) for mapping the distribution of QDs in cells and tissues based on QD internal chemistry in addition to discriminating probes by size [31].
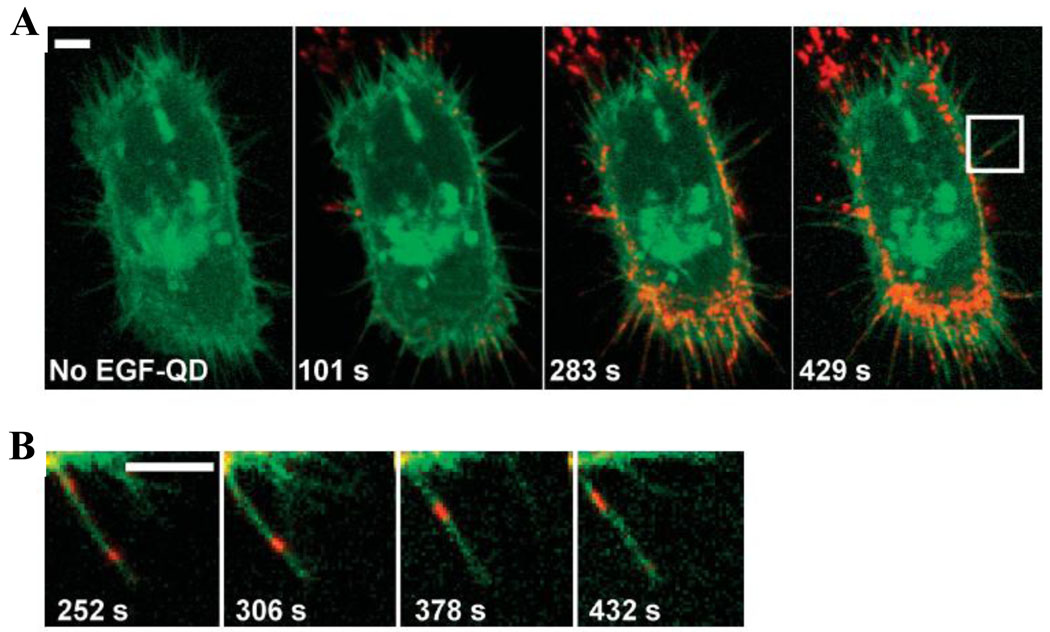
Monitoring of intracellular processes in live cells, although more difficult and less flexible in terms of multiplexing, provides information about dynamics of cellular functioning and real-time cellular response to applied stimuli. Design of biocompatible coatings and unprecedented photostability render QDs well-suited for this task, as long exposure to excitation source and constant signal intensity are often not achievable with conventional techniques. The relatively large size of QD probes creates a barrier for intracellular targeting, yet biomarkers expressed on the cell membrane are readily accessible. As a result the majority of reports on real-time tracking describe dynamics of membrane proteins rather than intracellular targets. For example, Lidke et al used QDs conjugated to epidermal growth factor (EGF) to study erbB/HER receptor-mediated cellular response to EGF in living human epidermoid carcinoma A431 cells, assigning the mechanism of EGF-induced signaling to heterodimerization of erbB1 and erbB2 monomers and uncovering retrograde transport of endocytosed QD probes (Fig. 6) [32]. Murcia et al utilized QD-lipid bioconjugates for high-speed tracking of single-probe movement on cell surface and accurate measurement of diffusion coefficient [33], while Roullier et al labeled two subunits of type I interferon receptor with QD probes and monitored diffusion and interaction of these subunits in real-time [34].
Figure 6.
Outstanding photostability and high brightness of QD probes enable long-term real-time monitoring of erbB receptor activation by QD-EGF and study the retrograde transport of these probes along the filopodia towards the cell body. Scale bars 5 um [32].
One highly informative method of intracellular tracking involves endocytosis of QD probes with consequent monitoring of endosome dynamics. Cui et al utilized pseudo-TIRF (total internal reflection fluorescence) microscopy for long-term real-time tracking of intracellular transport of QD-labeled nerve growth factor (NGF) along axons of rat dorsal root ganglion neurons and described the dynamics of axonal internalization and neuronal retrograde transport of QD-NGF [35]. In another example, Zhang et al induced single QD uptake into synaptic vesicles and monitored fluorescence of each QD probe to discriminate between complete vesicle fusion (full-collapse fusion) and transient fusion (so-called kiss-and-run behavior), thus characterizing dynamics of neuronal transmission with respect to time and frequency of impulse firing [36].
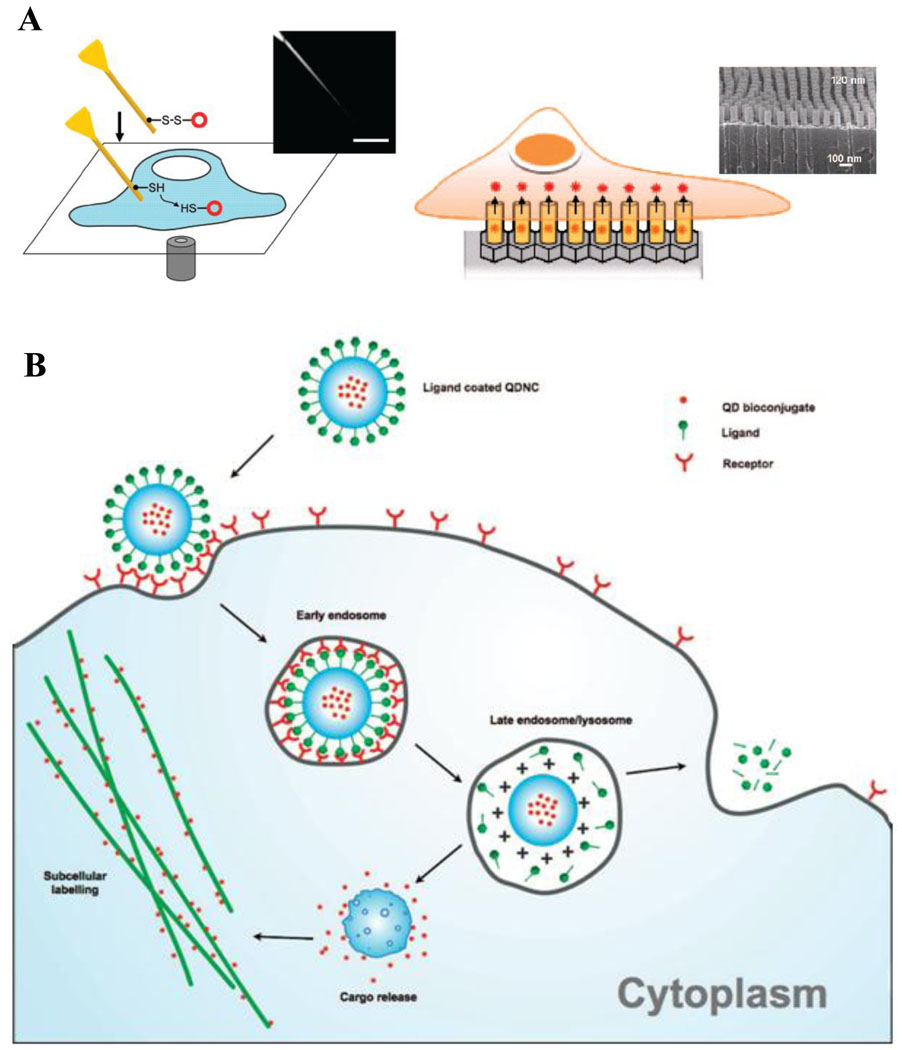
The challenge that is yet to be overcome is labeling of intracellular components in live cells. Integrity of cellular membrane and crowded intracellular environment have proven to be an obstacle for QD entry into live cells. While endosomal uptake of bare QDs is readily achievable, escape from endosomal compartments and labeling of specific components is challenging. Further, elimination of unbound probes from intracellular compartments to avoid false positive detection is often hampered, because, unlike fixed cells, unbound nanoparticles cannot be washed away. Recently a few reports on delivery of nanoparticles within live cells have been published. In mechano-chemical approach, Yum et al utilized gold-coated boron nitride nanotubes (with a diameter of 50 nm) to deliver QDs within the cytoplasm or nucleus of live HeLa cells with consequent 30-minute monitoring of QD diffusion within those compartments (Fig. 7A) [37]. Park et al engineered arrays of vertically aligned carbon nanosyringes for intracellular delivery of QDs and therapeutic agents (Fig. 7B) [38]. While efficiently delivering nanoparticles within cells, both techniques are quite labor-intensive and low-throughput. Design of nanoparticles capable of escaping endosomes or entering cells without inducing endocytosis remains the most promising approach for intracellular delivery [39–42]. For example, Kim et al encapsulated multiple QDs within the biodegradable polymer poly(D,L-lactide-co-glycolide) (PLGA) that induced cellular uptake, endosomal escape, and release of QD load within the cytoplasm (Fig. 7C), providing efficient high-throughput method for intracellular delivery of multicolor QDs and enabling multiplexed staining within live cells [40]. In a single-particle approach, Qi and Gao coated individual QDs with a pH-responsive amphyphilic polymer [42]. Besides achieving efficient cellular uptake and endosomal escape facilitated by a proton sponge effect, polymer-coated QDs allowed delivery of intact siRNA inside the cells and monitoring of siRNA release within the cytoplasm.
Figure 7.
Delivery of QD probes inside cells represents a challenge for labeling intracellular targets. Different modes of delivery are being developed to overcome this issue. A) Mechano-chemical modes of QD delivery involve utilization of mechanically strong materials capable of puncturing cell membrane and reaching into intracellular compartments. Delivery using nanoneedle (left) involves attachment of QDs on the outer surface of a stiff nanotube and manual manipulation of the needle on the cell-by-cell basis [37], while delivery platform based on nanosyringes (right) utilizes arrays of hollow vertically aligned nanotubes that enable intracellular release of QD probes upon cell growth on top of these arrays [38]. B) Encapsulation of QD probes within materials capable of endosomal escape represents a promising high-throughput technique for intracellular QD delivery. A general approach involves coating of QDs with materials possessing proton sponge functionality or other means of destabilizing endosome membrane and functionalizing the surface with targeting ligand. Once ligand binds to a receptor on the cell surface, nanoparticles are uptaken by endocytosis. Decrease in pH inside the endosome causes physical changes in QD coating (usually in surface charge), which triggers mechanisms for cytosolic release of QDs and enables targeting of intracellular components [40].
In vivo molecular imaging and profiling using quantum dots
In vivo imaging of diseased cells and tissues provides many benefits for personalized medicine, including high-throughput screening and potential for diagnosis at early stages of disease, obtaining patient-specific information about the localization and size of the disease core, assessment of adverse effects on healthy tissues, and monitoring of disease progression and response to therapy. Therefore, non-invasive in vivo imaging represents one of the major goals of current biomedical research. Conventional medical imaging techniques, such as ultrasound imaging, magnetic resonance imaging (MRI), and positron emission tomography (PET), in most cases do not offer sensitivity and resolution simultaneously for early-stage diagnosis (e.g. MRI provides high resolution, yet poor sensitivity; while PET offers high sensitivity with low resolution) as well as specificity for conveying disease molecular information.
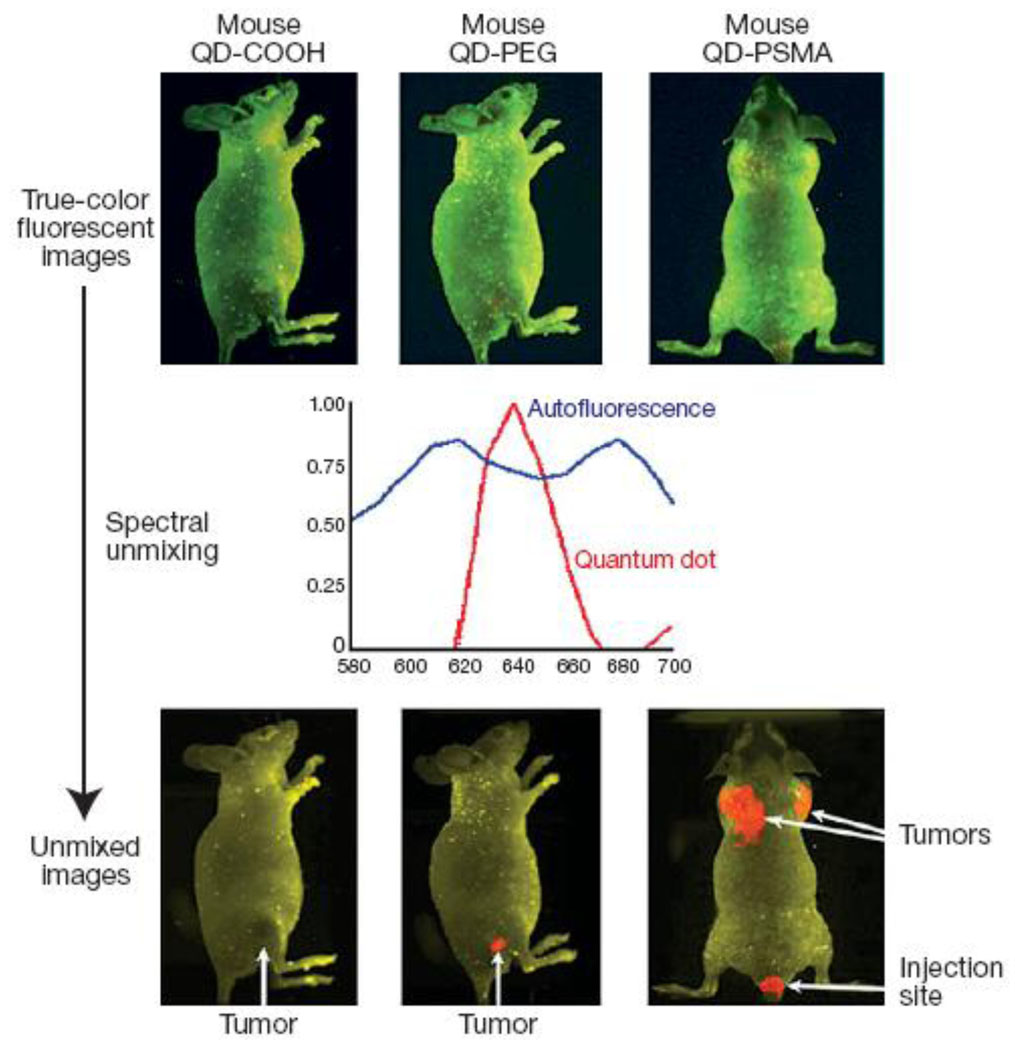
Fluorescence microscopy remains the most potent technique for molecular profiling of diseased cells. However, presence of tissue barriers between disease sites and imaging equipment complicates the utilization of fluorescence microscopy for in vivo imaging, as biological tissues efficiently absorb and scatter visible light along with producing intense autofluorescence over a broad spectrum. Unlike organic fluorophores, QDs possess high brightness and multiplexing capabilities along with large Stokes shifts, thus representing a promising tool for in vivo molecular imaging and profiling [16, 22, 29–31, 43–45]. In particular, the spectral gap between excitation and emission for QDs is significantly larger than that of organic fluorophores and can be as large as 300–400 nm, depending on the wavelength of the excitation light [46, 47], thus moving QD signal into a region with reduced tissue autofluorescence. For example, in early studies Akerman et al demonstrated targeted imaging of tumor vasculature using QD-peptide bioconjugates [48]. However, utilization of green and red QDs limited deep-tissue imaging in live animals, and post-mortem histological examination of tissue specimens was used to evaluate QD biodistribution. Taking advantage of large stokes shift, Gao et al have demonstrated the utility of PEG-coated red QDs (emission peak around 640 nm) conjugated to antibodies against prostate-specific membrane antigen (PSMA) for in vivo tumor imaging in mice [49]. Further signal processing with spectral unmixing algorithm allowed clear separation of QD signal from the background fluorescence (Fig. 8).
Figure 8.
Utilization of large Stokes shift produced by red and NIR QD probes enables targeted in vivo imaging of subcutaneous tumors. Further image processing with spectral demixing allows efficient removal of tissue autofluorescence [49].
Utilization of near-infrared imaging probes might further reduce interference from tissue autofluorescence and enable in vivo imaging with deeper penetration and better resolution. Modeling studies performed by Lim et al have identified two spectral windows in far-red (700–900 nm) and infrared (1200–1600 nm) regions suitable for nearly background-free deep-tissue imaging [50]. Kim et al utilized model predictions on practice in mapping sentinel lymph nodes (SLN) with NIR QDs, providing accurate identification and image-guided resection of SLN – an indispensable tool in surgical treatment of metastatic cancers [51]. Targeted in vivo imaging of human glioblastoma vasculature in mouse model was demonstrated by Cai et al, who used NIR CdTe/ZnS QDs conjugated to targeting peptide against integrin αvβ3, which is significantly up-regulated in tumors [52]. Recently, Diagaradjane et al reported on in vivo imaging and quantitative analysis of EGFR with NIR QDs (emission peak at 800 nm), showing QD capability to distinguish EGFR over-expression in tumor site compared to normal expression levels in surrounding tissues [53]. Meanwhile Shi et al used NIR QDs to achieve a deep-tissue high-sensitivity detection of prostate cancer xenografts grown in mouse tibia [54].
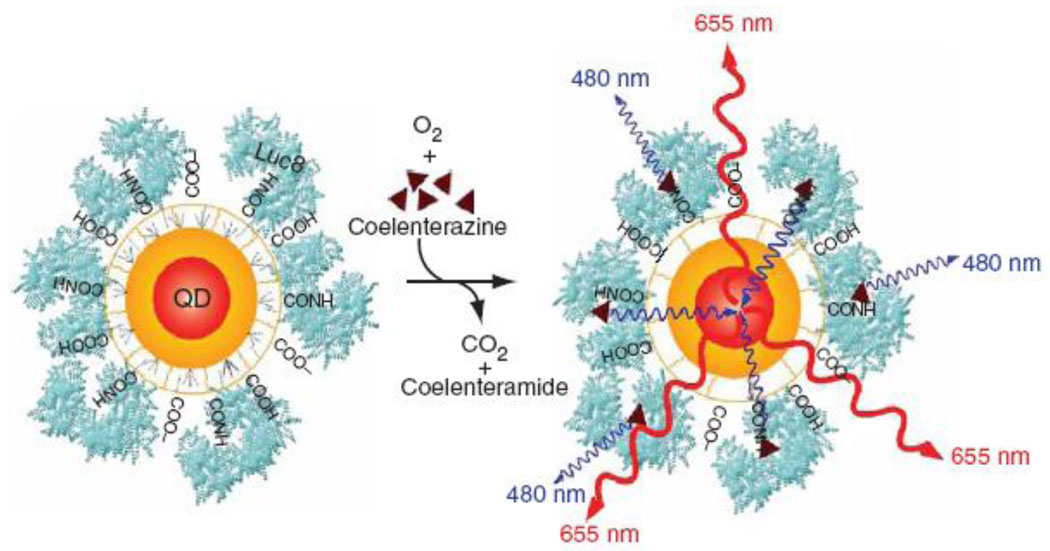
An alternative NIR QD probe was developed by So et al, who conjugated luciferase to QD surface to yield self-illuminating fluorescent probes via bioluminescence resonance energy transfer process (Fig. 9) [55]. By making external excitation unnecessary, bioluminescent QDs completely eliminated the tissue autofluorescence and provided higher sensitivity of detection. Increased size of luciferase-QD bioconjugates and requirement for supplying the substrate coelenterazine put certain limitations on in vivo imaging applications. Therefore, development of compact self-illuminating QDs utilizing naturally occurring biomolecules as a substrate might further advance this technology and provide high-sensitivity in vivo imaging probes.
Figure 9.
Shallow depth of in vivo imaging with QD probes imposes significant limits on utilization of this technology for deep-tissue imaging. One way to improve the depth and sensitivity of imaging is to use self-illuminating QDs. QD probes conjugated with luciferase convert bioluminescense produced by the enzyme into QD fluorescence emitted in NIR region, thus eliminating autofluorescence and signal intensity attenuation by tissues [55].
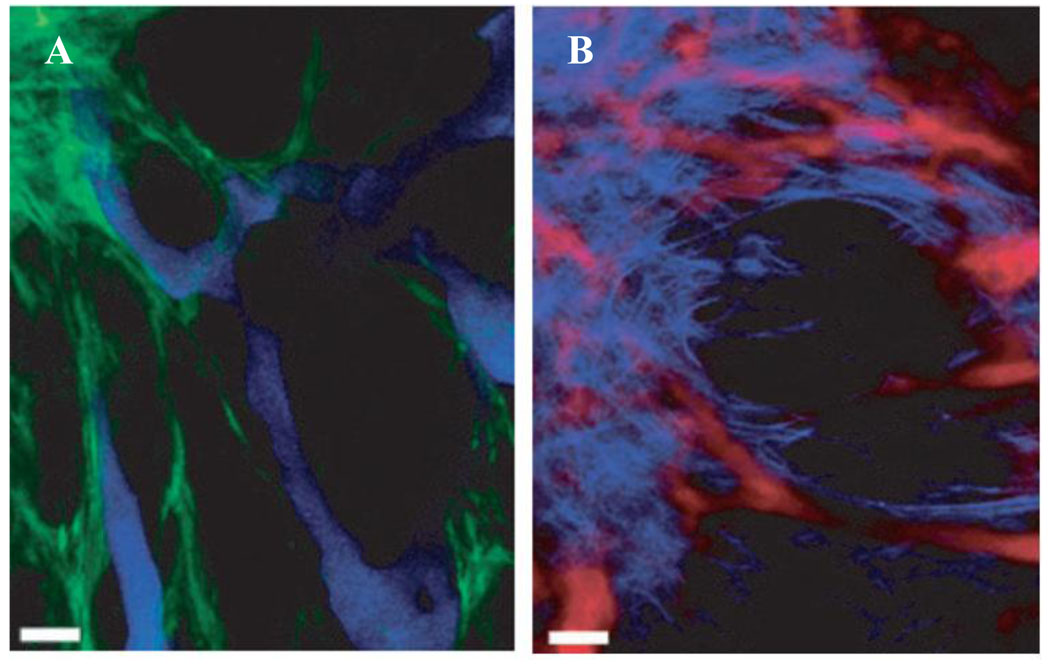
Two-photon microscopy represents another promising alternative to standard in vivo fluorescence imaging. Despite some technical limitations, two-photon microscopy represents a powerful tool for multiplexed and highly sensitive in vivo imaging. This technique uses low-energy photons (in red and infrared regions) for excitation of QDs emitting in visible range, achieving dramatically reduced attenuation of excitation light by tissues along with reducing the autofluorescense, while allowing utilization of QDs emitting over a full visible spectrum. Moreover, the high two-photon cross-section of QDs enables deeper-tissue imaging with higher sensitivity. The first study of QD-based multiphoton fluorescence in vivo imaging was reported by Larson et al, when green CdSe/ZnS QDs were used for imaging of capillaries under the dermis layer of skin [56]. Levene et al have combined needle-like gradient index lens imaging setup together with multiphoton microscopy to obtain high-resolution microangiographs of deep brain blood vessels labeled with QDs [57]. In a recent in vivo study of tumor morphology Stroh et al utilized two-photon microscopy for simultaneous imaging of tumor vessels (stained with blue QDs) and perivascular cells (expressing GFP) [58]. Further incorporation of second harmonic generation signal emanating from collagen provided information about the distribution and morphology of extracellular matrix (Fig. 10).
Figure 10.
Multi-photon microscopy represents a powerful tool for multiplexed in vivo imaging. By utilizing low-energy photons (minimally absorbed by tissues) for excitation of multicolor QD probes, this technique provides deeper tissue penetration and higher sensitivity of imaging. Application of this technique enabled the study of tumor morphology using QDs for labeling of tumor vasculature (blue QDs in (A) and red QDs in (B)), further enhanced by GFP labeling of perivascular cells (green in (A)) and detection of second harmonic generation signal from collagen to visualize extracellular matrix (light-blue in (B)) [58].
Overall, NIR QDs have already proven to be a great tool for characterization of disease models in small animals and post-mortem evaluation of tissue specimens. Moving towards in vivo imaging in human subjects, mapping of lymph nodes and image-guided resection of tumors represent most promising clinical applications of QD probes. Additionally, recent reports on decorating QD probes with TAT peptide [59] and wheat germ agglutinin [60] for improving QD transport over a blood-brain barrier and targeting cells of the central nervous system might enable highly accurate and conservative image-guided surgeries on brain tissue.
Targeted and traceable drug delivery
Accurate identification of key molecular targets distinguishing diseased cells from healthy ones enables targeted drug delivery with minimal side-effects. Nanoparticle-based drug carriers show great potential for efficient targeted delivery applications, as they can provide sufficiently long blood circulation, protect the cargo from degradation, possess large drug loading capacity and controlled drug release profile, and integrate multiple targeting ligands on their surface. Additionally, QD probes provide unique functionality of traceable drug delivery, as biodistribution of carriers and intracellular uptake can be monitored via fluorescence.
Several drug delivery applications employing tracing functionality of QDs have been developed recently. For example, Chen et al co-transfected QDs and siRNA using Lipofectamine 2000 and monitored transfection efficiency via QD fluorescence [61]. Mixing QDs with transfection reagent in 1:1 mass ratio provided correlation between the QD signal intensity and the degree of gene silencing. Such an approach enabled the collection of uniformly silenced cell population by fluorescence-activated cell sorting, while introducing minimal modifications into standard siRNA transfection protocol and requiring no chemical modifications of siRNA. Interestingly, additional co-transfection of different siRNA molecules with different QD colors might allow multiplexed monitoring of gene silencing. Yet, indirect link between siRNA and QD transfection imposes certain limitations on this technology, as there is a possibility of interference between QD probes and siRNA transfection resulting in inaccurate correlation of fluorescence signal with the degree of gene silencing. More reliable quantitative information about the number of siRNA molecules delivered into cells can be achieved by using QD-doped chitosan nanobeads developed by Tan et al [62]. In such an approach siRNA molecules are deposited on the surface of nanobeads, and intracellular delivery can be directly monitored by the nanobead fluorescence. Further improvement can be gained from a direct labeling approach demonstrated by Jia et al, who used carbon nanotubes for intracellular delivery of antisense oligonucleotides tagged with QDs [63]. This technology might not only enable a reliable method of quantification of intracellular oligonucleotide concentration, but also provide spatial information about the localization of oligonucleotides within the cell. For example, direct labeling of plasmid DNA with QDs followed by Lipofectamine-mediated transfection enabled long-term study of intracellular and intranuclear localization and transport of plasmid DNA, while preserving the ability of expressing reporter protein encoded by the plasmid [64].
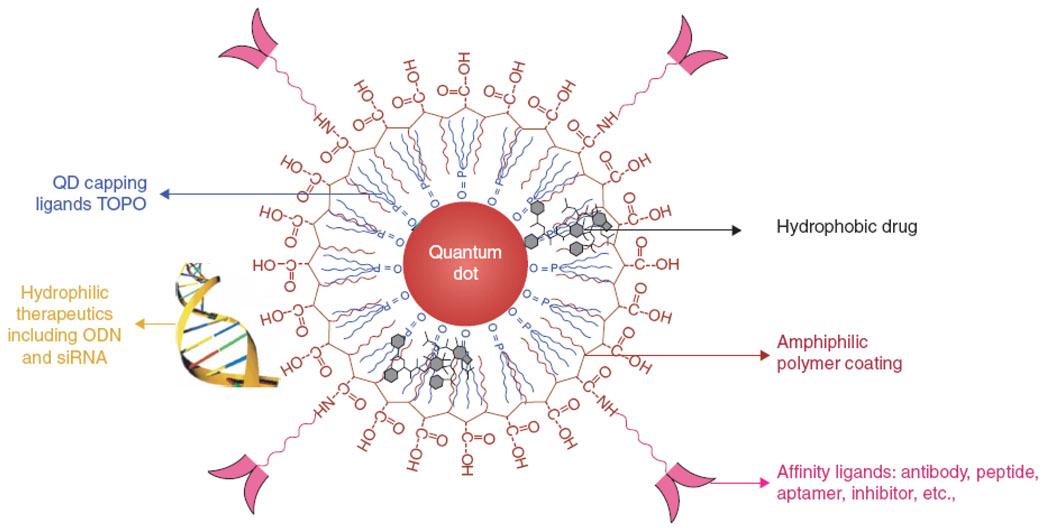
Initial success of highly efficient and traceable intracellular drug delivery utilizing supplementary transfection reagents inspired the design of compact single-QD based carriers with integrated functionalities. Utilization of single-QD drug delivery vehicles for in vivo applications is desirable, as intermediate size of such carriers (~10–20 nm in diameter) reduces the renal clearance as well as uptake by reticulo-endothelial system (RES), thus increasing the blood circulation time and improving the delivery efficiency. Further, QD core can serve as a structural scaffold for loading of various types of drug molecules. For example, small-molecule hydrophobic drugs can be embedded between the inorganic core and the amphiphilic polymer coating layer [65], while hydrophilic therapeutic agents (such as siRNA and antisense oligonucleotides) can be deposited onto the hydrophilic exterior of the polymeric shell (Fig. 11) [41]. Flexibility of the shell design enables engineering of drug carriers with different physical properties (e.g. size, charge, biodegradability, etc), thus providing a large platform for a variety of specific applications.
Figure 11.
QD-based drug carriers integrate drug delivery tracing, loading of various types of drugs (e.g. hydrophobic small-molecule drugs between the QD core and polymer coating or hydrophilic drugs on the exterior surface of the polymeric shell), and targeting functionality. Intermediate size of such carriers ensures slower renal filtration as well as RES uptake, thus increasing blood circulation time and improving delivery efficiency [41].
Integration of functionality for enhanced cellular uptake and endosomal escape within single-QD probes was demonstrated by Qi and Gao [42]. Encapsulation of QDs with zwitterionic amphipols enabled non-covalent deposition of up to 10 siRNA molecules on the surface of each particle via electrostatic interaction. Efficient endosomal uptake of such particles followed by decrease in pH and shift in particle surface charge resulted in endosomal escape and release of intact siRNA within the cells. While outperforming transfection efficiency of common reagents (such as PEI and Lipofectamine), QD carriers showed substantially lower toxicity in cell cultures. Further, real-time monitoring of particle uptake (via QD fluorescence) and release of siRNA (via labeling of individual siRNA molecules with FITC) was achieved. Targeted siRNA transfection to tumor cells was demonstrated by Defrus et al, who used PEG-coated QDs as a platform for deposition of siRNA and tumor-homing peptide [66]. Attachment of siRNA molecules via cleavable chemical bonds ensured efficient intracellular release of active siRNAs. However, deposition of targeting ligands and cargo molecules in a “parallel” manner introduced competition between the loading capacity and targeting capabilities of the delivery vehicles. In light of successful RNAi experiments with aptamer-siRNA chimeras performed by McNamara et al [67] it is reasonable to expect higher efficiency from vehicles with “serial” attachment of therapeutic molecule and targeting ligand. For small-molecule drug delivery, Bagalkot et al functionalized the QDs with targeting RNA aptamers and loaded anti-cancer drug Doxorubicin via intercalation within the aptamer [68]. Notably, bi-FRET from QD core to Doxorubicin and then to aptamer enabled monitoring of the vehicle disintegration and drug release within the cells via restoration of QD fluorescence.
In vivo drug delivery with QD carriers was demonstrated by Manabe et al [69]. Conjugation of an antihypertensive drug captopril to the QD surface provided the therapeutic effect similar to that of the free drug, while also enabling the monitoring of QD-drug biodistribution over a 96-hour period. With advancements in design of biocompatible QD surface coatings and identification of suitable molecular targets for therapy, QD-based drug delivery vehicles promise to provide an indispensable tool for modeling of pharmacokinetics and pharmacodynamics of nanoparticle-drug carriers.
Challenges of integrating QD technology into clinical practice
Nanotechnology represents a highly dynamic field of research developing novel platforms for a variety of applications. Unique properties of nanomaterials inspire enthusiasm for overcoming limitations of current technology and hold promise of advancing the field of personalized medicine. An increasing number of proof-of-concept studies along with more applied and clinically relevant QD-based tools appearing in a variety of fields ranging from ex vivo molecular fingerprinting of individual cells to in vivo diagnostics and image-guided therapy will undoubtedly make their way into clinical practice. However, there are still a number of challenges on the way of integrating QD technology into biomedical applications.
Unique behavior of nanomaterials compared with small molecules and lack of clinical experience of utilizing nanoparticle-based assays often raise concerns of result reproducibility, reliability, and comparability between each other and conventional techniques. However, increasing numbers of proof-of-concept studies are actively exploring a wide range of possible areas of QD applications. A forthcoming leap towards technologies working in clinical settings along with wide-scale “test-drives” of QD tools and training of technical personnel should encourage interest in QD-based tools, increase familiarity and hands-on experience working with QD probes, and establish confidence in this technology within scientific and medical communities. Among first steps towards this goal, standardization of QD-based assays will be beneficial for making data from different research centers comparable and enabling large-scale clinical studies.
Increasing efforts are focused on the study of the effect of QDs on human health and environment. Partially due to the novelty of nanotechnology, there is not much information about these effects available yet. Short-term and long-term toxicity and immunogenicity of nanoparticles as well as disposal of nanoparticle-containing waste remain a highly debatable area of research and deserve thorough investigation to ensure safety of QD technology in clinical practice [70–72]. While early studies of QD toxicity by Derfus et al indicated significant cytotoxicity of unprotected CdSe QDs due to nanoparticle photo-oxidation upon exposure to UV light and release of toxic Cd2+ ions [73, 74], capping of CdSe core with ZnS layer and deposition of a stable coating seemed to dramatically reduce QD toxicity in cell cultures. In a more adequate model based on 3D cell culture (liver tissue spheroids) Lee et al observed substantially decreased nanoparticle-induced toxicity compared to 2D cell cultures, emphasizing the impact of tissue morphology on toxicity [75]. Sometimes conflicting toxicity data might also result from significant over- or under-estimation of cell toxicity determined with standard cell viability assays [76]. In addition to in vitro assays, greater complexity of live organisms with plethora of mechanisms for QD accumulation, degradation, and excretion might require more thorough in vivo toxicity studies. For example, Mancini et al suggested that hypochlorous acid together with hydrogen peroxide produced by phagocytes can diffuse through an otherwise stable secondary coating, causing solubilization of the QD core and release of toxic ions [77]. Additionally, the QD surface coating and particle size play important role in the particle biodistribution and toxicity [71, 78, 79]. Pharmacokinetics studies performed on rat models by Fischer et al have shown that QDs coated with bovine serum albumin (BSA) are efficiently eliminated from the bloodstream by liver uptake, while QDs lacking BSA on their surface are cleared at slower rate [80]. As each QD probe appears to be unique, development of comprehensive assays for QD toxicity assessment should improve our understanding of potential risks of this technology, provide guidance for design of QD probes with minimized adverse effects, and increase the public confidence in QD-based diagnostics and therapeutics.
As promising benefits of QD technology might be hampered by potential adverse effects, design of biocompatible and non-toxic QD probes has become an active area of research. One way of resolving an issue of heavy metal toxicity involves utilization of QD probes made of benign materials. For example, Yong et al recently prepared Cd-free InP/ZnS QDs and utilized these probes for targeting of pancreatic cancer cell lines [81]; however, low quantum yield (~30%) and large size (~30 nm in diameter) might limit utility of such probes for in vivo imaging. Higher-quality probes with quantum yield of up to ~60% and hydrodynamic diameter of 17 nm were developed by Li et al on the basis of CuInS2/ZnS QDs [82]. Further, engineering of low-cost, non-toxic, and potentially biodegradable in vivo imaging probes might become available through utilization of recently developed technology for preparation of water-soluble QDs made of silicon [83, 84] – intert, biocompatible, and abundant material.
While being an attractive approach, Cd-free QDs still suffer from poor stability and inferior photo-physical properties compared to high-quality QDs made of toxic materials (such as CdSe). Therefore, improving biocompatibility of potentially toxic QD probes remains a sound and highly promising alternative, and elimination or reduction of cadmium interaction with live cells seem to be the cornerstone of such approach. There are several feasible strategies to achieving this goal. The toxicity associated with cadmium poisoning comes from a quick release of large amounts of this metal into a bloodstream, its preferential accumulation in kidneys, and consequent nephrotoxicity. However, up to 30 ug/day of dietary Cd (coming from fish, vegetables, and other sources) can be consumed by a healthy adult without adverse effects on kidney function [85]. Therefore, slow degradation of QD probes within a human body followed by urinary excretion might offer a way of safe and efficient elimination of QDs. Adapting technology developed for controlled drug release and coating nanoparticles with biodegradable polymers might provide one strategy for gaining control over QD degradation and Cd release in vivo.
Complete and quick elimination of intact QD probes from the body via renal excretion represents another approach to overcoming possible toxicity. This approach seems especially favorable in light of sparse information on in vivo QD degradation mechanisms and long-term effect of QD accumulation in organs. Systematic investigation of the renal clearance of QDs on rat and mice models done by Choi et al has defined the renal clearance threshold of 5.5 nm and emphasized the role of zwitterionic surface coatings in preventing protein absorption and retaining the original nanoparticles size [79]. Working along this direction, Law et al prepared ultrasmall (3–5 nm in diameter) cysteine-coated CdTe/ZnTe QDs and tested biodistribution of these probes in mice, finding no QDs in liver and spleen 2 weeks post-injection [86]. However, bio-functionalization of QDs, required for targeted imaging and therapy, increases the QD size, thus making renal clearance of functional QD probes difficult. Further, quick renal clearance is often undesirable, as prolonged QD circulation is required for specific targeting, high-sensitivity imaging, and therapeutic efficiency. Therefore, high molecular weight coatings are routinely applied to QD probes to increase their circulation time and improve bioavailability. Ballou et al emphasized the importance of coating with high molecular weight PEG to reduce accumulation of QDs in liver and bone marrow [87], and Prencipe et al achieved remarkably long blood circulation of nanomaterials encapsulated with branched PEG [88]. Utilization of biodegradable ligands that would detach from QD probes after prolonged circulation in blood or due to degradation in target cells, thus releasing single nanoparticles with original size below 5.5 nm, might render renal excretion of functional QD probes feasible.
In some cases complete elimination of QD probes from the body via renal excretion or other means might prove challenging or undesirable. Engineering of ultra-stable QDs encapsulated with inert biocompatible materials might provide an alternative strategy for addressing Cd toxicity issue. If QD integrity within a human body can be retained for many years, biological systems might never be exposed to heavy metal components of the QD core. For example, Ballou et al indicating that intact PEG-coated QDs remained in bone marrow and lymph nodes of mice for several months after injection [87]. While organic coatings, such as polymers and lipids, might still degrade due to exposure to biological environment, utilization of more stable inorganic materials should protect the cores of QD probes for extended periods of time.
Summary
Advancement of personalized medicine is essential for making progress towards combating such complex diseases as cancer and immune system disorders, and incorporation of novel QD-based tools will undoubtedly play a major role in this process. Design of compact, stable, and biocompatible coatings functionalized with targeting agents have already converted QDs into multifunctional nanodevices suitable for in vitro as well as in vivo applications. While certain challenges and concerns regarding QD incorporation into clinical practice remain, and cautiously enthusiastic attitude towards QD-based tools prevails in scientific community, the benefits of this technology will ensure the increasing interest in QDs as more practical and clinically relevant applications are demonstrated and comprehensive toxicity data is made available. With further advances in design and engineering of biocompatible QD probes such applications as image-guided surgery, molecular fingerprinting of diseases, and personalized diagnosis and therapy will become widely available.
Acknowledgments
This work is support in part by the National Science Foundation for a Faculty Early Career Development award (CAREER 0645080), and the National Institute of Health (R01 CA131797). P.Z. thanks the UW Center for Nanotechnology for a UIF fellowship. We are also grateful to Prof. Larry True for fruitful discussion and technical assistance in pathology studies of prostate cancer, and Prof. Terry Kavanagh for discussion on nanotoxicity.
Biographies

Pavel Zrazhevskiy
Department of Bioengineering, University of Washington, Seattle, WA, USA
Pavel Zrazhevskiy received his Bachelor of Science degree in Bioengineering from the University of Washington in 2006. In 2007 he became a Graduate student in Dr. Xiaohu Gao’s laboratory at the Department of Bioengineering, University of Washington. His research interests span fields of nanotechnology, molecular engineering, and medicine with focus on engineering of quantum dot based tools for biomedical applications. Pavel is a recipient of the UIF Fellowship from the UW Center for Nanotechnology, NSF Graduate Research Fellowship, and is a member of Biomedical Engineering Society (BMES) and American Association for the Advancement of Science (AAAS).

Xiaohu Gao, PhD
Department of Bioengineering, University of Washington, Seattle, WA, USA
Prof. Xiaohu Gao received his B.S. degree in chemistry from Nankai University, China in 1998, his Ph.D. degree in chemistry from Indiana University, Bloomington in 2004, and his postdoctoral training from the Department of Biomedical Engineering at Georgia Tech and Emory University. In 2005, he became a faculty member in the Department of Bioengineering and the Center for Nanotechnology at the University of Washington, Seattle. His research is focused on biomedical nanotechnology, molecular engineering, molecular imaging, and therapeutics. He has authored or coauthored over 50 papers and book chapters; and he is also a recipient of the NSF CAREER Award and the CDMRP New Investigator Award. He has been a member of the American Chemical Society (ACS) and Biomedical Engineering Society (BMES) since 2003.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chamary JV, Parmley JL, Hurst LD. Nat. Rev. Genet. 2006;7:98. doi: 10.1038/nrg1770. [DOI] [PubMed] [Google Scholar]
- 2.Sauna ZE, Kimchi-Sarfaty C, Ambudkar SV, Gottesman MM. Cancer Res. 2007;67:9609. doi: 10.1158/0008-5472.CAN-07-2377. [DOI] [PubMed] [Google Scholar]
- 3.True LD, Gao X. J. Mol. Diagn. 2007;9:7. doi: 10.2353/jmoldx.2007.060186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pedersen L, Holck S, Schiodt T, Zedeler K, Mouridsen HT. Breast Cancer Res. Treat. 1989;14:91. doi: 10.1007/BF01805979. [DOI] [PubMed] [Google Scholar]
- 5.Thomson TA, Hayes MM, Spinelli JJ, Hilland E, Sawrenko C, Phillips D, et al. Mod. Pathol. 2001;14:1079. doi: 10.1038/modpathol.3880440. [DOI] [PubMed] [Google Scholar]
- 6.True LD. Am. J. Clin. Pathol. 1988;90:324. doi: 10.1093/ajcp/90.3.324. [DOI] [PubMed] [Google Scholar]
- 7.Alivisatos AP, Gu W, Larabell C. Annu. Rev. Biomed. Eng. 2005;7:55. doi: 10.1146/annurev.bioeng.7.060804.100432. [DOI] [PubMed] [Google Scholar]
- 8.Geho DH, Luchini A, Garaci E, Belluco C, Petricoin E, Liotta LA. Nanomed. 2007;2:1. doi: 10.2217/17435889.2.1.1. [DOI] [PubMed] [Google Scholar]
- 9.Alivisatos AP. J. Phys. Chem. 1996;100:13226. [Google Scholar]
- 10.Alivisatos AP. Science. 1996;271:933. [Google Scholar]
- 11.Smith SM, Gao X, Nie S. Photochem. Photobiol. 2004;80:377. doi: 10.1562/0031-8655(2004)080<0377:QDNFIV>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 12.Chan WC, Maxwell DJ, Gao X, Bailey RE, Han M, Nie S. Curr. Opin. Biotechnol. 2002;13:40. doi: 10.1016/s0958-1669(02)00282-3. [DOI] [PubMed] [Google Scholar]
- 13.Gao X, Yang L, Petros JA, Marshall FF, Simons JW, Nie S. Curr. Opin. Biotechnol. 2005;16:63. doi: 10.1016/j.copbio.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 14.Yezhelyev MV, Gao X, Xing Y, Al-Hajj A, Nie S, O'Regan M. Lancet Oncol. 2006;7:657. doi: 10.1016/S1470-2045(06)70793-8. [DOI] [PubMed] [Google Scholar]
- 15.Resch-Genger U, Grabolle M, Cavaliere-Jaricot S, Nitschke R, Nann T. Nat. Methods. 2008;5:763. doi: 10.1038/nmeth.1248. [DOI] [PubMed] [Google Scholar]
- 16.Wu X, Liu H, Liu J, Haley KN, Treadway JA, Larson JP, et al. Nat. Biotechnol. 2003;21:41. doi: 10.1038/nbt764. [DOI] [PubMed] [Google Scholar]
- 17.Bruchez M, Jr, Moronne M, Gin P, Weiss S, Alivisatos AP. Science. 1998;281:2013. doi: 10.1126/science.281.5385.2013. [DOI] [PubMed] [Google Scholar]
- 18.Lin SL, Pradhan N, Wang YJ, Peng XG. Nano Lett. 2004;4:2261. [Google Scholar]
- 19.Zhong XH, Feng YY, Knoll W, Han MY. J. Am. Chem. Soc. 2003;125:13559. doi: 10.1021/ja036683a. [DOI] [PubMed] [Google Scholar]
- 20.Hiraoka Y, Shimi T, Haraguchi T. Cell Struct. Funct. 2002;27:367. doi: 10.1247/csf.27.367. [DOI] [PubMed] [Google Scholar]
- 21.Gao X, Nie S. Trends Biotechnol. 2003;21:371. doi: 10.1016/S0167-7799(03)00209-9. [DOI] [PubMed] [Google Scholar]
- 22.Ghazani AA, Lee JA, Klostranec J, Xiang Q, Dacosta RS, Wilson BC, et al. Nano Lett. 2006;6:2881. doi: 10.1021/nl062111n. [DOI] [PubMed] [Google Scholar]
- 23.Yezhelyev MV, Al-Hajj A, Morris C, Marcus AI, Liu T, Lewis M, et al. Advanced Materials. 2007;19:3146. [Google Scholar]
- 24.Jain KK. Expert Opin. Pharmacother. 2005;6:1463. doi: 10.1517/14656566.6.9.1463. [DOI] [PubMed] [Google Scholar]
- 25.Chattopadhyay PK, Price DA, Harper TF, Betts MR, Yu J, Gostick E, et al. Nat. Med. 2006;12:972. doi: 10.1038/nm1371. [DOI] [PubMed] [Google Scholar]
- 26.Xiao Y, Barker PE. Nucleic Acids Res. 2004;32:e28. doi: 10.1093/nar/gnh024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tholouli E, Hoyland JA, Di Vizio D, O'Connell F, Macdermott SA, Twomey D, et al. Biochem. Biophys. Res. Commun. 2006;348:628. doi: 10.1016/j.bbrc.2006.07.122. [DOI] [PubMed] [Google Scholar]
- 28.Chan P, Yuen T, Ruf F, Gonzalez-Maeso J, Sealfon SC. Nucleic Acids Res. 2005;33:e161. doi: 10.1093/nar/gni162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsuno A, Itoh J, Takekoshi S, Nagashima T, Osamura RY. Histochem. Cytochem. 2005;53:833. doi: 10.1369/jhc.4A6577.2005. [DOI] [PubMed] [Google Scholar]
- 30.Giepmans BN, Deerinck TJ, Smarr BL, Jones YZ, Ellisman MH. Nat. Methods. 2005;2:743. doi: 10.1038/nmeth791. [DOI] [PubMed] [Google Scholar]
- 31.Nisman R, Dellaire G, Ren Y, Li R, Bazett-Jones DP. J. Histochem. Cytochem. 2004;52:13. doi: 10.1177/002215540405200102. [DOI] [PubMed] [Google Scholar]
- 32.Lidke DS, Nagy P, Heintzmann R, Arndt-Jovin DJ, Post JN, Grecco HE, et al. Nat. Biotechnol. 2004;22:198. doi: 10.1038/nbt929. [DOI] [PubMed] [Google Scholar]
- 33.Murcia MJ, Minner DE, Mustata GM, Ritchie K, Naumann CA. J. Am. Chem. Soc. 2008;130:15054. doi: 10.1021/ja803325b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roullier V, Clarke S, You C, Pinaud F, Gouzer GG, Schaible D, et al. Nano Lett. 2009;9:1228. doi: 10.1021/nl9001298. [DOI] [PubMed] [Google Scholar]
- 35.Cui B, Wu C, Chen L, Ramirez A, Bearer EL, Li WP, et al. Proc. Natl. Acad. Sci. U. S. A. 2007;104:13666. doi: 10.1073/pnas.0706192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Q, Li Y, Tsien RW. Science. 2009;323:1448. doi: 10.1126/science.1167373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yum K, Na S, Xiang Y, Wang N, Yu MF. Nano Lett. 2009;9:2193. doi: 10.1021/nl901047u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park S, Kim YS, Kim WB, Jon S. Nano Lett. 2009;9:1325. doi: 10.1021/nl802962t. [DOI] [PubMed] [Google Scholar]
- 39.Yezhelyev MV, Qi L, O'Regan RM, Nie S, Gao X. J. Am. Chem. Soc. 2008;130:9006. doi: 10.1021/ja800086u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim BYS, Jiang W, Oreopoulos J, Yip CM, Rutka JT, Chan WCW. Nano Lett. 2008;8:3887. doi: 10.1021/nl802311t. [DOI] [PubMed] [Google Scholar]
- 41.Qi L, Gao X. Expert Opin. Drug Deliv. 2008;5:263. doi: 10.1517/17425247.5.3.263. [DOI] [PubMed] [Google Scholar]
- 42.Qi L, Gao X. ACS Nano. 2008;2:1403. doi: 10.1021/nn800280r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ness JM, Akhtar RS, Latham CB, Roth KA. J. Histochem. Cytochem. 2003;51:981. doi: 10.1177/002215540305100801. [DOI] [PubMed] [Google Scholar]
- 44.Tokumasu F, Dvorak J. J. Microsc. 2003;211:256. doi: 10.1046/j.1365-2818.2003.01219.x. [DOI] [PubMed] [Google Scholar]
- 45.Xing Y, Chaudry Q, Shen C, Kong KY, Zhau HE, Chung LW, et al. Nat. Protoc. 2007;2:1152. doi: 10.1038/nprot.2007.107. [DOI] [PubMed] [Google Scholar]
- 46.Klostranec JM, Chan WCM. Advanced Materials. 2006;18:1953. [Google Scholar]
- 47.Mews A, Kadavanich AV, Banin U, Alivisatos AP. Physical Review B. 1996;53:13242. doi: 10.1103/physrevb.53.r13242. [DOI] [PubMed] [Google Scholar]
- 48.Akerman ME, Chan WC, Laakkonen P, Bhatia SN, Ruoslahti E. Proc. Natl. Acad. Sci. U. S. A. 2002;99:12617. doi: 10.1073/pnas.152463399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao X, Cui Y, Levenson LM, Chung LW, Nie S. Nat. Biotechnol. 2004;22:969. doi: 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- 50.Lim YT, Kim S, Nakayama A, Stott NE, Bawendi MG, Frangioni JV. Mol. Imaging. 2003;2:50. doi: 10.1162/15353500200302163. [DOI] [PubMed] [Google Scholar]
- 51.Kim S, Lim YT, Soltesz EG, De Grand AM, Lee J, Nakayama A, et al. Nat. Biotechnol. 2004;22:93. doi: 10.1038/nbt920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cai W, Shin DW, Chen K, Gheysens O, Cao Q, Wang SX, et al. Nano Lett. 2006;6:669. doi: 10.1021/nl052405t. [DOI] [PubMed] [Google Scholar]
- 53.Diagaradjane P, Orenstein-Cardona JM, Colon-Casasnovas NE, Deorukhkar A, Shentu S, Kuno N, et al. Clin. Cancer Res. 2008;14:731. doi: 10.1158/1078-0432.CCR-07-1958. [DOI] [PubMed] [Google Scholar]
- 54.Shi C, Zhu Y, Xie Z, Qian W, Hsieh CL, Nie S, et al. Urology. 2009 doi: 10.1016/j.urology.2009.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.So MK, Xu C, Loening AM, Gambhir SS, Rao J. Nat. Biotechnol. 2006;24:339. doi: 10.1038/nbt1188. [DOI] [PubMed] [Google Scholar]
- 56.Larson DR, Zipfel WR, Williams RM, Clark SW, Bruchez MP, Wise FW, et al. Science. 2003;300:1434. doi: 10.1126/science.1083780. [DOI] [PubMed] [Google Scholar]
- 57.Levene MJ, Dombeck DA, Kasischke KA, Molloy RP, Webb WW. J. Neurophysiol. 2004;91:1908. doi: 10.1152/jn.01007.2003. [DOI] [PubMed] [Google Scholar]
- 58.Stroh M, Zimmer JP, Duda DG, Levchenko TS, Cohen KS, Brown EB, et al. Nat. Med. 2005;11:678. doi: 10.1038/nm1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Santra S, Yang H, Stanley JT, Holloway PH, Moudgil BM, Walter G, et al. Chem Commun (Camb) 2005:3144. doi: 10.1039/b503234b. [DOI] [PubMed] [Google Scholar]
- 60.Gao X, Chen J, Wu B, Chen H, Jiang X. Bioconjug. Chem. 2008;19:2189. doi: 10.1021/bc8002698. [DOI] [PubMed] [Google Scholar]
- 61.Chen AA, Derfus AM, Khetani SR, Bhatia SN. Nucleic Acids Res. 2005;33:e190. doi: 10.1093/nar/gni188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tan WB, Jiang S, Zhang Y. Biomaterials. 2007;28:1565. doi: 10.1016/j.biomaterials.2006.11.018. [DOI] [PubMed] [Google Scholar]
- 63.Jia N, Lian Q, Shen H, Wang C, Li X, Yang Z. Nano Lett. 2007;7:2976. doi: 10.1021/nl071114c. [DOI] [PubMed] [Google Scholar]
- 64.Srinivasan C, Lee J, Papadimitrakopoulos F, Silbart LK, Zhao M, Burgess DJ. Mol. Ther. 2006;14:192. doi: 10.1016/j.ymthe.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 65.Jain TK, Morales MA, Sahoo SK, Leslie-Pelecky DL, Labhasetwar V. Mol. Pharm. 2005;2:194. doi: 10.1021/mp0500014. [DOI] [PubMed] [Google Scholar]
- 66.Derfus AM, Chen AA, Min DH, Ruoslahti E, Bhatia SN. Bioconjug. Chem. 2007;18:1391. doi: 10.1021/bc060367e. [DOI] [PubMed] [Google Scholar]
- 67.McNamara JO, 2nd, Andrechek ER, Wang Y, Viles KD, Rempel RE, Gilboa E, et al. Nat. Biotechnol. 2006;24:1005. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- 68.Bagalkot V, Zhang L, Levy-Nissenbaum E, Jon S, Kantoff PW, Langer R, et al. Nano Lett. 2007;7:3065. doi: 10.1021/nl071546n. [DOI] [PubMed] [Google Scholar]
- 69.Manabe N, Hoshino A, Liang YQ, Goto T, Kato N, Yamamoto K. IEEE Trans. Nanobioscience. 2006;5:263. doi: 10.1109/tnb.2006.886569. [DOI] [PubMed] [Google Scholar]
- 70.Dobrovolskaia MA, McNeil SE. Nat. Nanotechnol. 2007;2:469. doi: 10.1038/nnano.2007.223. [DOI] [PubMed] [Google Scholar]
- 71.Hardman R. Environ. Health Perspect. 2006;114:165. doi: 10.1289/ehp.8284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mahendra S, Zhu H, Colvin VL, Alvarez PJ. Environ. Sci. Technol. 2008;42:9424. doi: 10.1021/es8023385. [DOI] [PubMed] [Google Scholar]
- 73.Derfus AM, Chan WCM, Bhatia SN. Nano Lett. 2004;4:11. doi: 10.1021/nl0347334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang T, Stilwell JL, Gerion D, Ding L, Elboudwarej O, Cooke PA, et al. Nano Lett. 2006;6:800. doi: 10.1021/nl0603350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee J, Lilly GD, Doty RC, Podsiadlo P, Kotov NA. Small. 2009;5:1213. doi: 10.1002/smll.200801788. [DOI] [PubMed] [Google Scholar]
- 76.Monteiro-Riviere NA, Inman AO, Zhang LW. Toxicol. Appl. Pharmacol. 2009;234:222. doi: 10.1016/j.taap.2008.09.030. [DOI] [PubMed] [Google Scholar]
- 77.Mancini MC, Kairdolf BA, Smith AM, Nie S. J. Am. Chem. Soc. 2008;130:10836. doi: 10.1021/ja8040477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Aillon KL, Xie Y, El-Gendy N, Berkland CJ, Forrest ML. Adv Drug Deliv Rev. 2009;61:457. doi: 10.1016/j.addr.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Choi HS, Liu W, Misra P, Tanaka E, Zimmer JP, Itty Ipe B, et al. Nat. Biotechnol. 2007;25:1165. doi: 10.1038/nbt1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fischer HC, Liu L, Pang KS, Chan WCW. Adv. Funct. Mater. 2006;16:1299. [Google Scholar]
- 81.Yong KT, Ding H, Roy I, Law WC, Bergey EJ, Maitra A, et al. ACS Nano. 2009;3:502. doi: 10.1021/nn8008933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li L, Daou TJ, Texier I, Kim Chi TT, Liem NQ, Reiss P. Chem. Mater. 2009;21:2422. [Google Scholar]
- 83.Park JH, Gu L, von Maltzahn G, Ruoslahti E, Bhatia SN, Sailor MJ. Nat. Mater. 2009;8:331. doi: 10.1038/nmat2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhenhui Kang YL, Tsang Chi Him A, Duo Duo Ma Dorothy, Fan Xia, Wong Ning-Bew, Lee Shuit-Tong. Advanced Materials. 2009;21:661. [Google Scholar]
- 85.Satarug S, Haswell-Elkins MR, Moore MR. Br. J. Nutr. 2000;84:791. [PubMed] [Google Scholar]
- 86.Law WC, Yong KT, Roy I, Ding H, Hu R, Zhao W, et al. Small. 2009;5:1302. doi: 10.1002/smll.200801555. [DOI] [PubMed] [Google Scholar]
- 87.Ballou B, Lagerholm BC, Ernst LA, Bruchez MP, Waggoner AS. Bioconjug. Chem. 2004;15:79. doi: 10.1021/bc034153y. [DOI] [PubMed] [Google Scholar]
- 88.Prencipe G, Tabakman SM, Welsher K, Liu Z, Goodwin AP, Zhang L, et al. J. Am. Chem. Soc. 2009;131:4783. doi: 10.1021/ja809086q. [DOI] [PMC free article] [PubMed] [Google Scholar]