Abstract
We present an extension of the CHARMM hexopyranose monosaccharide additive all-atom force field to enable modeling of glycosidic-linked hexopyranose polysaccharides. The new force field parameters encompass 1→1, 1→2, 1→3, 1→4, and 1→6 hexopyranose glycosidic linkages, as well as O-methylation at the C1 anomeric carbon, and are developed to be consistent with the CHARMM all-atom biomolecular force fields for proteins, nucleic acids, and lipids. The parameters are developed in a hierarchical fashion using model compounds containing the key atoms in the full carbohydrates, in particular O-methyl-tetrahydropyran and glycosidic-linked dimers consisting of two molecules of tetrahyropyran or one of tetrahydropyran and one of cyclohexane. Target data for parameter optimization include full two-dimensional energy surfaces defined by the Φ/Ψ glycosidic dihedral angles in the disaccharide analogs as determined by quantum mechanical MP2/cc-pVTZ single point energies on MP2/6-31G(d) optimized structures (MP2/cc-pVTZ//MP2/6-31G(d)). In order to achieve balanced, transferable dihedral parameters for the Φ/Ψ glycosidic dihedral angles, surfaces for all possible chiralities at the ring carbon atoms involved in the glycosidic linkages are considered, resulting in over 5000 MP2/cc-pVTZ//MP2/6-31G(d) conformational energies. Also included as target data are vibrational frequencies, pair interaction energies and distances with water molecules, and intramolecular geometries including distortion of the glycosidic valence angle as a function of the glycosidic dihedral angles. The model-compound optimized force field parameters are validated on full disaccharides through comparison of molecular dynamics results to available experimental data. Good agreement is achieved with experiment for a variety of properties including crystal cell parameters and intramolecular geometries, aqueous densities, and aqueous NMR coupling constants associated with the glycosidic linkage. The newly-developed parameters allow for the modeling of linear, branched, and cyclic hexopyranose glycosides both alone and in heterogenous systems including proteins, nucleic acids and/or lipids when combined with existing CHARMM biomolecular force fields.
Introduction
Polysaccharide carbohydrates, composed of individual monosaccharide units that are connected together by glycosidic linkages, have numerous and varied roles in biology, where they serve as energy storage and transport molecules, structural scaffolds, and motifs for molecular recognition. An important subset of these glycosides has as its component monosaccharide hexopyranoses, such as glucose (Figure 1, compound 1), galactose, and mannose. Members of this subset include cellulose and starch, which are both composed exclusively of glucose, yet have dramatically different properties and functions arising from cellulose being composed exclusively of one epimer of glucose and starch exclusively of the other epimer.1 In addition to existing as isolated biological polymers, hexopyranose polysaccharides are commonly found covalently linked to other biomolecules. Attachment of hexopyranose polysaccharides to proteins yields glycoproteins and to lipids yields glycolipids, and these heterogeneous biomolecules play crucial roles in protein folding and molecular recognition.
Figure 1.
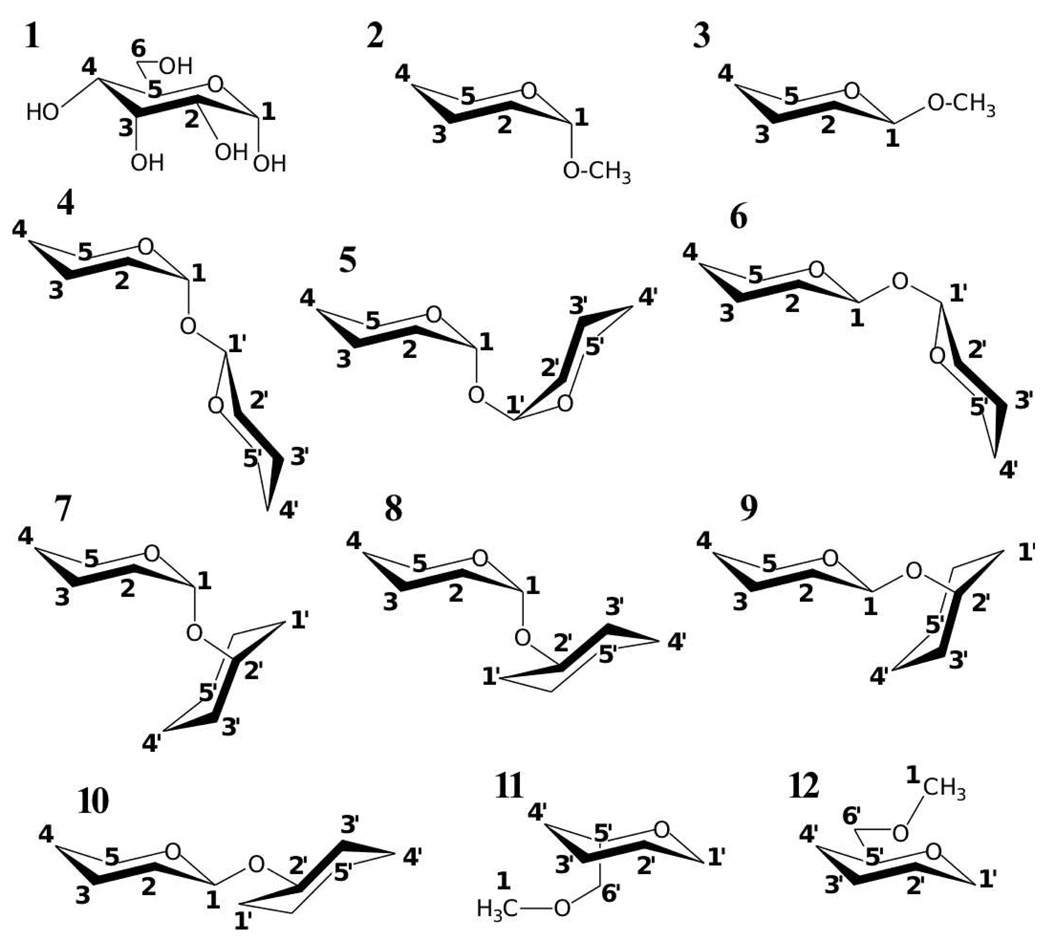
α-d-glucopyranose (1) and model compounds (2–12). Model compound atoms are numbered to correspond with the standard hexopyranose atom numbering as shown on α-d-glucopyranose.
Significant efforts spanning several decades have been made toward the development of molecular mechanics force field models for investigating the structure and energetics of polysaccharides.2–16 The large variety of component monosaccharides, the inherent conformational complexity of monosaccharides, and the additional degrees of freedom that need to be considered in parametrizing glycosidic linkages all present challenges to the development of comprehensive carbohydrate force fields. As such, the range of force field applicability (i.e. types of carbohydrates) is often limited, and the study of heterogeneous systems (e.g. glycolipids, glycoproteins, protein:carbohydrate complexes) is complicated by inconsistent treatment of 1,4 nonbonded interactions and differing force-field development protocols. Presently, the availability of high-quality experimental data and the ability to probe a larger variety of conformational energies on larger model systems using more accurate quantum mechanical methods allow for the inclusion of more and better target and validation data in the carbohydrate force field development process, as reflected in recent efforts.17–20 It is anticipated that such advances will lead to both more complete and more accurate carbohydrate force field models.
To enable the atomic-level modeling of a wide variety of carbohydrates in an aqueous environment and interacting with other biopolymers such as proteins, nucleic acids, and lipids, our laboratory has undertaken the development of a comprehensive force field for carbohydrates19,20 in the context of the CHARMM additive biomolecular force fields.21–33 The long-term goal of these efforts is to enable atomically-detailed investigation of carbohydrates in molecular recognition, including detailed understanding of the molecular basis of and physical insights into the interaction of carbohydrates with other biomolecules such as proteins and lipids. The present work builds on previously published work on model compounds, including linear and cyclic ethers,34 ethylene glycol,35 and 2-ethoxy tetrahydropyran,36 as well as all-atom pairwise additive force field development efforts for hexopyranose monosaccharides19 and acyclic polyols, acyclic carbohydrates, and inositol.20 Among the findings of these prior efforts is that, using the relatively simple functional form of the CHARMM all-atom additive force field equation, it is possible to develop carbohydrate force field parameters that give good agreement with gas phase, crystalline, and aqueous conformational properties as well as aqueous phase densities of very dilute to very concentrated aqueous solutions. In the present study, the parameters developed for hexopyranoses have been extended to a comprehensive collection of glycosidic linkages, i.e. 1→1, 1→2, 1→3, 1→4, and 1→6 glycosidic links with all possible combinations of chiralities for the two ring carbon atoms in each glycosidic link, as well as to both α- and β-anomers of 1-O-methyl hexopyranoses. Therefore, these parameters allow for the modeling of all possible unsubstituted linear, cyclic, and branched hexopyranose polysaccharides.
The present work represents a departure from previous parametrizations of hexopyranose glycosidic linkages in that extensive high-level quantum mechanical data on relatively large model compound systems are used in the parametrization procedure. Specifically, two molecules of tetrahydropyran or one molecule each of tetrahydropyran and cyclohexane connected by a glyclosidic linkage are employed as model compounds for hexopyranose disaccharides. Optimized two-dimensional scans at the MP2/6-31G(d) level with a resolution of 15° for the full range of the Φ/Ψ dihedral angles of a glycosidic linkage are performed, and MP2/cc-pVTZ single point energies are computed for all of these optimized structures. In sum, over 5250 MP2/cc-pVTZ//MP2/6-31G(d) conformational energies are used in the parametrization process, and include consideration of all possible chiralities at the ring carbon atoms involved in the glycosidic linkages. Particular care is taken in the parametrization of the C1-Olink-Cn’ valence angle distortion, where C1 corresponds to the anomeric carbon in the first ring, Olink the oxygen in the glycosidic linkage, and Cn’ the glycosidic ring carbon atom in the second ring, such that the force field accurately reproduces the quantum mechanical distortion of this angle, ranging nearly 30°, over the full range of Φ/Ψ dihedral values. Extensive validation of the parameter set is done by comparing results from molecular dynamics simulations of disaccharide crystals and aqueous solutions to experimental disaccharide crystal geometries, NMR J-coupling values, and aqueous solution densities.
Methods
All molecular mechanics calculations were performed with the CHARMM program,21,37,38 and using the same potential energy function as for the CHARMM protein,23–25 nucleic acid,26–28 and lipid all-atom additive force fields:29–33
| (1) |
In Eqn. 1, Kb, Kθ, KUB, Kχ and Kimp are bond, valence angle, Urey-Bradley, dihedral angle, and improper dihedral angle force constants, respectively. b, θ, S, χ and φ are the bond distance, valence angle, Urey-Bradley 1,3-distance, dihedral angle, and improper dihedral angle values, and the subscript 0 represents an equilibrium value. Additionally, for the dihedral term, n is the multiplicity and δ is the phase angle as in a Fourier series. The nonbonded interaction energy between pairs of atoms i and j consists of the Lennard-Jones (LJ) 6–12 term and the Coulomb term. εij is the LJ well depth, Rmin, ij is the interatomic distance at the LJ energy minimum, qi and qj are the partial atomic charges, and rij is the distance between atoms i and j. The Lorentz-Berthelot combining rules are applied to determine LJ parameters between different atom types.39
A modified version of the rigid three-site TIP3P model was used to represent water,40,41 and the SHAKE algorithm42 was applied to keep water molecules rigid and to constrain covalent bonds between hydrogen and heavy atoms to their equilibrium values. Gas-phase energies were calculated using infinite nonbonded cutoffs and gas-phase energy minimizations were done to a tolerance of 10−6 kcal*mol−1*Å−1. Aqueous and crystal simulations were performed using periodic boundary conditions,39 with a force-switched smoothing function43 applied to LJ interactions in the range of 10 to 12 Å and particle mesh Ewald44 with a real-space cutoff of 12 Å to compute the Coulomb interactions. A timestep of 1 fs was used for integration of the equations of motion with the “leapfrog” integrator,45 and all molecular dynamics (MD) employed Nosé-Hoover thermostating,46,47 Langevin piston barostating,48 and a long-range correction to the pressure to account for LJ interactions beyond 12 Å.39 Condensed-phase simulations were done at experimental temperature and pressure, with aqueous simulations done using a cube as the periodic unit cell and crystal simulations employing the appropriate experimental unit cell geometries. For aqueous simulations with a cubic unit cell, the cell edge lengths were varied isotropically to maintain the target pressure during simulation, whereas unit cell edge lengths in crystal simulations were allowed to vary independently. Angular crystal cell parameters of 90° were constrained to this value while those not 90° were allowed to vary independently. Data for crystal simulations were collected for 4 ns following 1 ns of equilibration.
QM calculations were performed using the Gaussian 03 program.49 Geometry optimizations and vibrational calculations were performed using the MP2/6-31G(d) model chemistry,50,51 with tight tolerances applied when optimizing structures for vibrational calculations. A scale factor of 0.9434 was applied to the QM frequencies, as required to account for limitations in the level of theory and reproduce experimental frequencies.52 Potential energy decomposition analysis was performed using the MOLVIB utility in CHARMM using internal coordinates as per Pulay et al.53 Relaxed potential energy scans were obtained by optimizing the geometry at the MP2/6-31G(d) level followed by MP2/cc-pVTZ single point calculations (MP2/cc-pVTZ//MP2/6-31G(d)).50,51,54 All potential energy scans were performed in 15° increments with only the scanned dihedral angles (e.g. Φ/Ψ or Φ/Ψ for the 2D surfaces) constrained.
QM calculations for interaction pairs consisting of a water molecule with a particular model compound followed the standard procedure for the CHARMM force field, thereby ensuring consistency of the nonbonded parameters with the remainder of the CHARMM additive biomolecular force fields.22 Per this procedure, the solute:water interaction distance was optimized at the HF/6-31G(d) level, with constraints on all other degrees of freedom. Following optimization, HF/6-31G(d) interaction energy target data were calculated as 1.16*(Epair − Esolute − Ewater), with no basis-set superposition-error correction and the empirical scaling factor of 1.16 introduced to yield parameters appropriate for a condensed phase force field.24,55 Target data for interaction distances were the QM-optimized distances minus 0.2 Å, again to yield parameters appropriate for a condensed phase force field. The water intramolecular geometry in both QM and MM calculations of pair interaction data was that of the TIP3P water model,40 and the model compound geometry was one that was previously gas-phase optimized in the MP2/6-31G(d) or the CHARMM representation, respectively.
Relaxed QM potential energy surfaces at the MP2/cc-pVTZ//MP2/6-31G(d) level of theory were used as target data for fitting the dihedral force constants in an automated manner using the freely-available Monte Carlo simulated annealing (MCSA) dihedral parameter fitting program fit_dihedral.py56 (available for downloading at http://mackerell.umaryland.edu). For each dihedral being fit, three multiplicities n of 1, 2, and 3 were included and the corresponding Kχ values (Eqn. 1) were optimized to minimize the root mean square error between the empirical and QM energies. In the MCSA approach, the adiabatic empirical energy surfaces are initially obtained with the force constants Kχ on the dihedral parameters being parametrized set to zero; restraining potentials on dihedral angles are used to maintain the values of the dihedral angles that are being scanned. The energy difference between the resulting empirical surface and the target QM surface is then determined and the dihedral parameters fit to reproduce that energy difference. Kχ values were constrained to be no more than 3 kcal/mol, and phase angles δ were limited to 0 and 180° to maintain symmetry of the dihedral potentials about χ = 0°, allowing for applicability of the parameters to both enantiomers of a chiral species.
An extension to the MCSA fitting program fit_dihedral.py was introduced to allow for independent root mean squared alignment of multiple MM surfaces to their corresponding QM surfaces during the simultaneous fitting of these surfaces. Previously, the target function to be optimized was the root-mean squared error RMSE between the QM conformational energies and the MM conformational energies :
| (2) |
where wi is a weight factor for conformation i and the constant c, defined by
| (3) |
optimally aligns the QM and MM data.56 When simultaneously fitting the same dihedral parameters in two or more different molecules (e.g. configurational isomers), it is sometimes desirable to allow for independent root-mean squared alignment of the QM and MM data on a per-molecule basis. Thus the target function for MCSA fitting was expanded to
| (4) |
where RMSEg is the RMSE for a grouping of data and is defined by Eqns. 2 and 3 for that grouping g. Each grouping of data is aligned independently, that is the c values are computed separately for each grouping, and wg is a weight factor applied to a particular grouping. Thus, for example, data for configurational isomers can be placed in separate groupings so as to optimally fit the conformational energies of the individual configurational isomers with a single set of dihedral parameters without requiring fitting of the QM ΔE values between configurational isomers.
The aqueous solution density ρ was calculated from the average volume 〈V〉, obtained from the simulations, using Eqns. 5 and 6:
| (5) |
| (6) |
Here, Nwater, Nsolute, and NAvogadro are the number of water molecules, solute molecules, and Avogadro’s number, respectively, and 〈MW〉 is the average molecular weight of the solution. For all aqueous density simulations, Nwater = 1100, while Nsolute was adjusted to give the appropriate density for each simulated system. These aqueous simulations were equilibrated for 0.5 ns followed by 5 ns of data collection, with the volume of the system calculated every 10 ps and averaged over the simulation time to give 〈V〉.
NMR heteronuclear three-bond proton-carbon coupling constants (in Hz) for the glycosidic angle rotation 3 JCOCH were computed from the simulations using the modified Karplus equation developed by Tvaroska et al.57
| (7) |
as well as that by Cloran et al.58
| (8) |
where ϕ is the C-O-C-H dihedral angle of interest. Similarly, for three-bond carbon-carbon coupling constants 3 JCOCC, the modified Karplus equation developed by Bose et al.,59
| (9) |
was used, where ϕ is the C-O-C-C dihedral angle of interest. Bose et al. also developed a second simpler equation in which the cos ϕ term was excluded,59
| (10) |
and this second equation was also used to analyze the MD data. To compute J values from MD simulations the value of the dihedral angle of interest was tabulated for each snapshot of each disaccharide in the simulation. Each dihedral value was then used as ϕ in one of the above equations to get a J value for that disaccharide in that snapshot. The resultant J values for the dihedral angle of interest were then averaged over all disaccharides in the system and over all snapshots from the simulation to get an ensemble-averaged value for J for comparison with previously published experimental data. Simulations consisted of four disaccharides in 1100 molecules of water, corresponding to a concentration of 200 mM, which is approximately 10-fold more concentrated than the comparison NMR experiments though still sufficiently dilute so as minimize direct solute-solute interactions. Of the four disaccharide molecules, the chirality at the reducing-end anomeric carbon was α for one molecule and β for the remaining three, so as to reflect the anomeric ratio for glucopyranose in water (α:β = 1:260). In accord with the comparison NMR experiments performed using pure D2O, simulations were done at 298 K and 1 atm, and with no ions added. Simulations were equilibrated for 0.5 ns, followed by 20 ns of data collection during which a snapshot was taken every 1 ps.
Results and Discussion
Parameter optimization
The focus of the present work was the development of a transferable set of parameters for the glycosidic linkages between hexopyranoses. Applying parameters developed for hexopyranose monosaccharides and associated model compounds19 left as parameters to-be-determined only those involving the glycosidic linkage oxygen atom. Starting values for bond, angle, dihedral, Lennard-Jones and electrostatic parameters involving this atom were transferred from published ether parameters34 and further optimized as described below using the model compounds in Figure 1, and, to a limited extent, data from disaccharide crystals. Parametrization was done in a self-consistent fashion such that whenever one parameter was changed, properties were recomputed and additional parameters re-optimized if necessary21–23; all data presented throughout reflect the final set of self-consistently optimized parameters. The optimized parameters were subsequently applied to full disaccharides for validation by comparison of MD results to available experimental data.
Conformational energies for all compounds were studied using the MP2/ccpVTZ// MP2/6-31G(d) model chemistry, which provides a reasonable compromise between accuracy and computational expediency. Previously, it was seen that energies for hexopyranose monosaccharides using this model chemistry were in excellent agreement compared to MP2/cc-pVTZ optimized structures.19 Additional studies on ethylene glycol35 and 2-ethoxy tetrahydropyran36 using a variety of model chemistries support the use of the MP2/cc-pVTZ model chemistry for energy calculations on carbohydrates.
1-O-methylation
O-methylation at the C1 position in hexopyranoses leads to formation of an acetal, thereby preventing spontaneous isomerization between the α- and β-anomers, and is a common chemical modification in synthetic hexopyranose polysaccharides. Force field parameters were developed for this moiety because of its common occurrence and its similarity to glycosidic linkages between hexopyranoses. Compounds 2 and 3, which are O-methyl derivatives of tetrahydropyran, were used as model compound analogs for the α- and β-anomers of 1-O-methyl hexopyranoses. The location of O-methylation and the geometry of the tetrahydropyran ring were chosen to mimic a 1-O-methyl-d-hexopyranose molecule having the energetically-favored 4C1 ring conformation.
Tetrahydropyran parameters were those previously developed in the context of hexopyranoses19 and bond, angle, dihedral, Lennard-Jones, and partial charge parameters involving the O-methyl group were transferred from existing ether parameters.34 Following this transfer of parameters, those that remained missing included the Oring-C1-Omethyl valence angle and the Oring-C1-Omethyl-Cmethyl, C2-C1-Omethyl-Cmethyl, and C5-Oring-C1-Omethyl dihedral parameters. Also in question was the ability of the transferred nonbonded parameters on the Oring and Omethyl atoms to capture the energetics of hydrogen bonding in the context of these molecules.
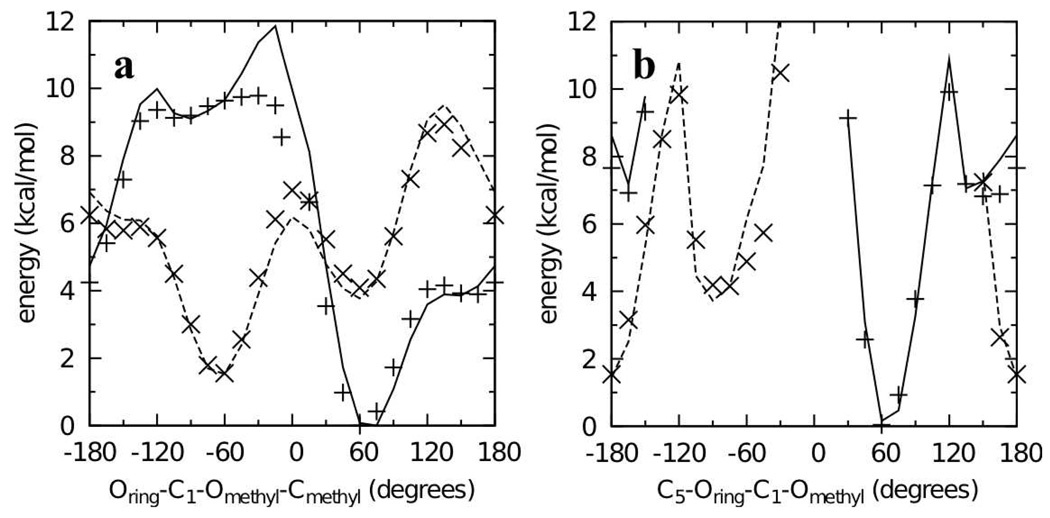
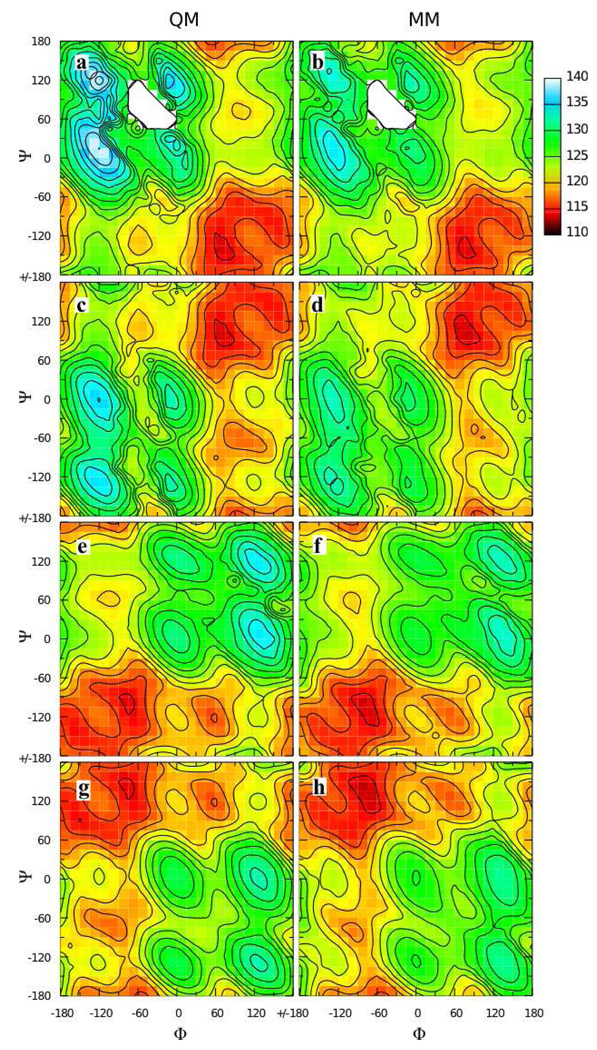
QM MP2/cc-pVTZ//MP2/6-31G(d) scans of the Oring-C1-Omethyl-Cmethyl dihedral in both the α-anomer 2 and β-anomer 3 were done to characterize the potential energy surfaces and determine the location of the minimum energy conformations of the two anomers (Figure 2). The global minimum for both surfaces was at Oring-C1-Omethyl-Cmethyl = 60° for the α-anomer. The minimum on the β-anomer surface was 1.55 kcal/mol higher in energy and located at Oring-C1-Omethyl-Cmethyl = −60°. Although the O-methyl moiety is located in the axial position in the α-anomer, it is energetically more favorable than the equatorial substituted β-anomer. This well-known result is counter to the general trend that equatorial substitutions on saturated six-membered rings are more favorable than axial ones, a phenomenon referred to as the “anomeric effect.” The energy difference between the two anomers in the molecular mechanics framework is determined by the C5-Oring-C1-Omethyl dihedral, and ring deformation potential energy scans of this dihedral were also done for inclusion as target data during dihedral parameter fitting.
Figure 2.
Dihedral potential energy scans in the QM (points) and MM (lines) representation for model compounds 2 and 3. Scans for the α- (crosses and solid lines) and β- (x’s and dashed lines) anomers (2 and 3 respectively) have all been offset to the same global minimum.
An initial set of parameters were developed by transferring existing ether C-C-O valence angle parameters to the Oring-C1-Omethyl valence angle and fitting the Oring-C1-Omethyl-Cmethyl, C2-C1-Omethyl-Cmethyl, and C5-Oring-C1-Omethyl dihedral parameters to reproduce the Oring-C1-Omethyl-Cmethyl and C5-Oring-C1-Omethyl QM potential energy scans. Using these initial parameters, both anomers were then fully geometry-optimized in the QM MP2/6-31G(d) and MM representations starting from Oring-C1-Omethyl-Cmethyl = 60° for the α-anomer and −60° for the β-anomer. Comparison of the MM geometries to the QM geometries showed errors in the C1-Omethyl bond and the Oring-C1-Omethyl and C2-C1-Omethyl angles. Additionally, pair interaction energies with water for these two conformations using the transferred partial charge of –0.34 e on Omethyl were consistently underestimated in the MM representation compared to the QM representation. Because of the errors in geometries and water pair interaction energies using transferred parameters from the linear ethers, further parameter optimization was undertaken.
A second iteration of parameter optimization was done to correct the QM water pair interaction energies while ensuring the molecular geometries and conformational energies were well reproduced. The C1-Omethyl geometry was improved by decreasing the transferred equilibrium bond length by 0.020 Å, and the Oring-C1-Omethyl and C2-C1-Omethyl geometries were improved by decreasing the transferred equilibrium value parameters by 1.5° to 110.0° and by 2.5° to 109.0°, respectively. Reproduction of the target vibrational frequencies was improved by increasing the Oring-C1-Omethyl force constant from 45 to 90 kcal*mol−1*radian−2. The partial charge on Omethyl was adjusted to −0.36 e, with +0.01 e added to the partial charges on C1 and Cmethyl to maintain charge neutrality, and the dihedral parameters were re-fit. With optimized partial charges, water pair interaction energies and distances in the MM representation compared very favorably to the QM target data, both for Omethyl and for Oring, which retained its initial partial charge of −0.40 e transferred from tetrahydropyran (Table 1, Figure 3). In order to use the same bonded parameters for both anomers, it was necessary to have the force field slightly underestimate Oring-C1-Omethyl and overestimate C2-C1-Omethyl for the α-anomer while doing the reverse for the β-anomer (Table 2). The valence angle parameters also yielded good agreement between the QM and MM vibrational frequencies having contributions from these degrees of freedom (Table 3). Conformational energies as well as the energy difference between the two anomers were very well reproduced with the fit dihedral parameters (Figure 2). In the conformational energy scans, the force field gives a ΔE of 1.53 kcal/mol between the minimum-energy conformations of the two anomers, in comparison to the reference MP2/cc-pVTZ//MP2/6-31G(d) value of 1.55 kcal/mol. In this case, the force field representation gives a significantly more accurate ΔE than the MP2/6-31G(d) value (2.25 kcal/mol), highlighting the importance of the choice of QM model chemistry for generation of target data.
Table 1.
Solute-water pair interaction energies and distances for model compounds 2 and 3.
| water orientationa |
energy (kcal/mol) |
distance (Å) |
||||||
|---|---|---|---|---|---|---|---|---|
| 1.16*HFb | MM | MM-QM | HF-0.20b | MM | MM-QM | |||
| 2 (α−) | Omethyl | ai | −4.87 | −4.50 | 0.37 | 1.90 | 1.90 | 0.00 |
| aii | −4.64 | −4.75 | −0.11 | 1.91 | 1.89 | −0.02 | ||
| bi | −4.20 | −3.32 | 0.88 | 1.90 | 1.87 | −0.03 | ||
| bii | −4.35 | −4.55 | −0.20 | 1.90 | 1.83 | −0.06 | ||
| Oring | ai | −5.88 | −5.70 | 0.18 | 1.81 | 1.72 | −0.09 | |
| aii | −5.53 | −5.79 | −0.26 | 1.84 | 1.72 | −0.12 | ||
| 3 (β−) | Omethyl | ai | −5.99 | −6.09 | −0.09 | 1.83 | 1.74 | −0.09 |
| aii | −5.40 | −5.56 | −0.16 | 1.86 | 1.75 | −0.11 | ||
| bi | −4.20 | −3.38 | 0.82 | 1.90 | 1.85 | −0.05 | ||
| bii | −4.62 | −4.83 | −0.21 | 1.89 | 1.81 | −0.08 | ||
| Oring | ai | −5.58 | −6.16 | −0.58 | 1.88 | 1.85 | −0.03 | |
| aii | −4.78 | −4.86 | −0.08 | 1.91 | 1.88 | −0.03 | ||
| average | 0.05 | −0.06 | ||||||
| standard deviation |
0.44 | 0.04 |
Molecular geometries are as illustrated in Figure 3.
HF target energies have been scaled by 1.16 and distances have been shortened by 0.20 Å.
Figure 3.
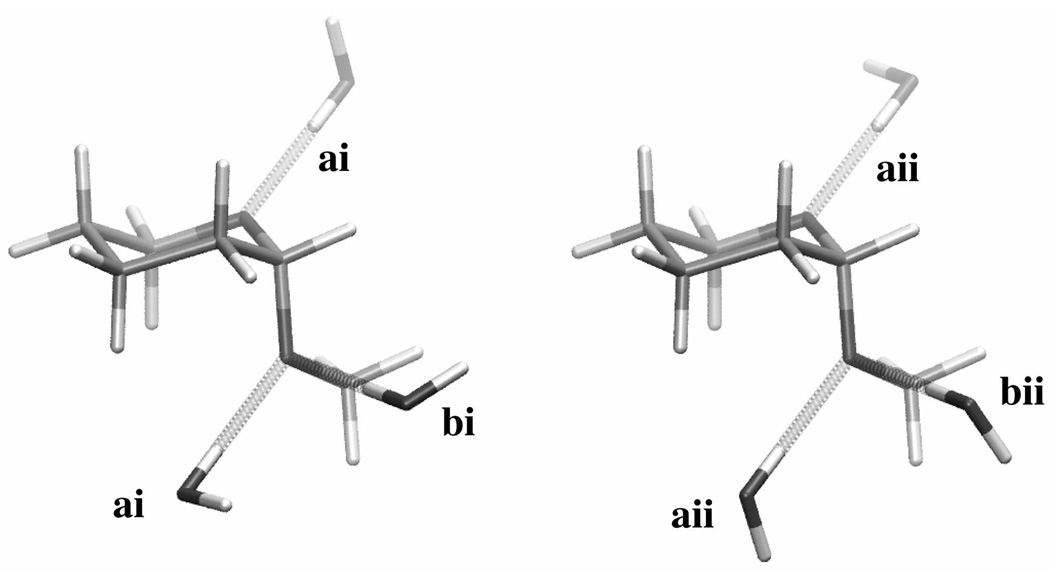
Water pair interaction geometries with model compound 2. For convenience, three molecules of water have been illustrated simultaneously interacting with the model compound, though in actuality all data (Table 1) are for a single water molecule interacting with a single model compound molecule. Interactions labeled “a” have the water H-O bond vector in the C-O-C angle plane and along the C-O-C bisector while interactions labeled “b” have the H-O bond vector in an ideal tetrahedral geometry (109.5°) with respect to the C-O-C angle. The water molecule in type i interactions lies perpendicular to the C-O-C plane. Type ii interactions differ from type i interactions by rotation of the non-interacting water hydrogen atom by 90° around the H-O bond vector. Molecular graphics prepared with VMD.66
Table 2.
Fully-optimized QM and MM geometries for model compounds 2 and 3.
| QM | MM | MM-QM | ||
|---|---|---|---|---|
| 2 (α−) | Oring-C1- Omethyla | 112.0 | 110.3 | −1.6 |
| C2-C1-Omethyl | 107.1 | 108.8 | 1.8 | |
| C1-Omethyl-Cmethyl | 112.3 | 112.0 | −0.3 | |
| Oring-C1-Omethyl- Cmethyl |
61.1 | 67.1 | 6.0 | |
| C1-Omethyl | 1.411 | 1.404 | −0.008 | |
| Omethyl-Cmethyl | 1.427 | 1.424 | −0.003 | |
| 3 (β−) | Oring-C1-Omethyl | 108.3 | 109.9 | 1.5 |
| C2-C1-Omethyl | 108.6 | 106.8 | −1.8 | |
| C1-Omethyl-Cmethyl | 112.7 | 111.9 | −0.7 | |
| Oring-C1-Omethyl- Cmethyl |
−62.6 | −65.3 | −2.8 | |
| C1-Omethyl | 1.392 | 1.401 | 0.009 | |
| Omethyl-Cmethyl | 1.427 | 1.423 | −0.005 |
Valence and dihedral angles are in degrees, bond lengths are in Å.
Table 3.
Vibrational frequencies for model compound 2 having contributions from the Oring-C1-Omethyl and C2-C1-Omethyl valence angles.
| frequency # |
frequency (cm−1) |
% XCZ contributiona,b |
% YCZ contributiona,b |
|||
|---|---|---|---|---|---|---|
| QM | MM | QM | MM | QM | MM | |
| 2 | 125.8 | 131.4 | 13 | 14 | ||
| 3 | 170.2 | 187.3 | 7 | 7 | 7 | 9 |
| 4 | 221.1 | 245.4 | 10 | |||
| 5 | 297.6 | 286.4 | 18 | 31 | ||
| 6 | 310.8 | 337.2 | 5 | 10 | 12 | 15 |
| 7 | 380.6 | 366.5 | 8 | |||
| 8 | 415.0 | 439.8 | 9 | |||
| 9 | 504.7 | 492.3 | 7 | 6 | ||
| 10 | 553.2 | 558.8 | 6 | 37 | 29 | |
| 11 | 661.2 | 651.0 | 5 | 12 | 9 | |
| 12 | 797.5 | 797.0 | ||||
| 13 | 805.0 | 827.0 | ||||
| 14 | 855.0 | 881.6 | ||||
| 15 | 869.6 | 893.8 | ||||
| 16 | 890.9 | 905.6 | ||||
| 17 | 946.3 | 949.5 | 7 | |||
| 18 | 1001.3 | 995.0 | ||||
| 19 | 1028.0 | 1041.1 | ||||
| 20 | 1032.9 | 1068.6 | ||||
| 21 | 1060.1 | 1075.4 | ||||
| 22 | 1107.9 | 1108.8 | 5 | |||
| 23 | 1123.7 | 1131.7 | 6 |
XCZ and YCZ are methine-associated internal coordinates per the definition of Pulay et al.53 such that X = C2, Y = Oring, C = C1, and Z = Omethyl.
Contributions from XCZ/YCZ of less than 5% to a particular frequency are not shown.
1→1 glycosidic linkages
Four different 1→1 glycosidic linkages can be formed between two hexopyranose monosaccharides depending on which anomers are linked together: α(1→1)α, α(1→1)β, β(1→1)α, and β(1→1)β. This number is reduced to three if the identities of the two component monosaccharides are the same, as the resultant α(1→1)β and β(1→1)α linked disaccharides constitute identical molecules. Accordingly, 4, 5, and 6 were used as model compound analogs of 1→1 linked hexopyranose disaccharides. These model compounds are dimers of tetrahydropyran connected by a glycosidic linkage and are analogous to α(1→1)β, α(1→1)α, and β(1→1)β hexopyranose disaccharides, respectively. Parameters to be determined for these linkages were those involving the glycosidic oxygen Olink. These included Olink-C1 bond parameters; C1-Olink-C1, Olink-C1-Oring, and Olink-C1-H valence angle parameters; C1-Olink-C1-X dihedral parameters (X = C2, Olink, or H), and nonbonded Olink parameters.
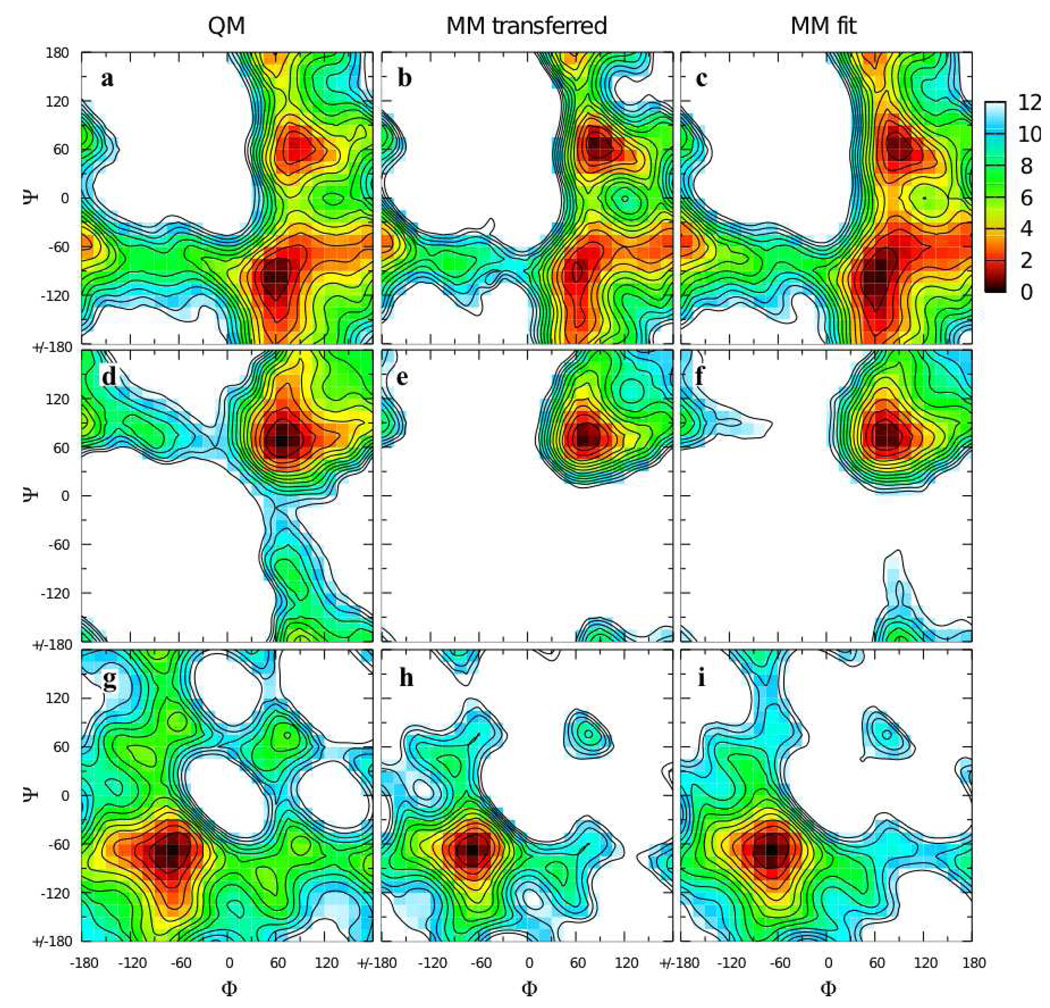
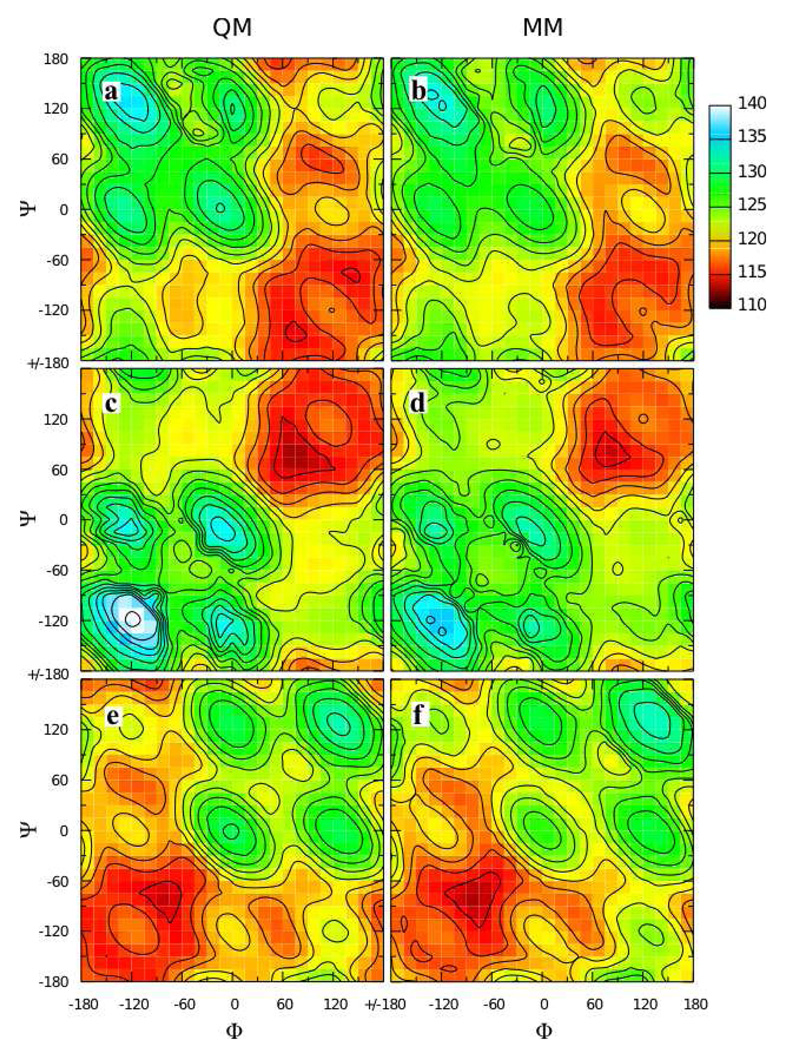
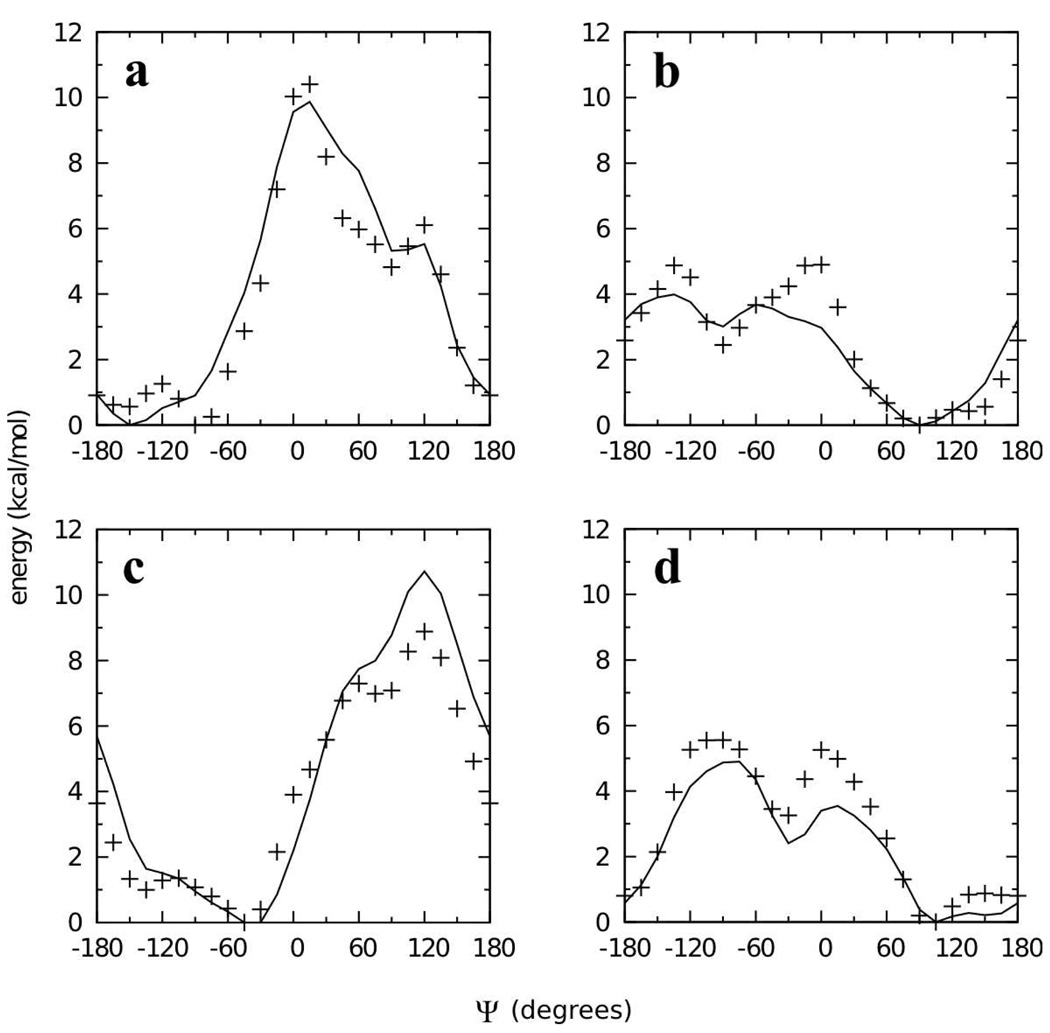
An attractive possibility for minimizing computer time and parametrization effort is to transfer analogous parameters from 2 and 3 to the full disaccharides, and this approach has been taken in other work. To test this approach, the QM Φ/Ψ two dimensional relaxed potential energy surface for the α(1→1)β analog 4 (Φ = Oring-C1-Olink-C1’, Ψ = C1-Olink-C1’-Oring’) was compared to the MM surface calculated using transferred parameters (Figure 4a,b). The obvious shortcoming of this approach is the incorrect ordering of the two minima on the QM surface at Φ/Ψ = 60°/−90° and 90°/60°; on the QM surface the global minimum is as 60°/−90° while on the MM surface it is at 90°/60°. Additional QM and MM Φ/Ψ scans done on 5 and 6 showed better agreement between the QM and MM surfaces, which is not surprising as they only have one low-lying minimum each (Figure 4d,e,g,h). However, in both instances the minima had notably steeper walls in the MM representation. Starting at the global minimum at 75°/75° for 5 (Figure 4d,e), the energy rises significantly faster moving along the valley toward 180°/90° in the MM as compared to the QM surface. Likewise for 6, starting at the global minimum at −75°/−75° and moving up the valley toward −180°/−60° the energy increases significantly more quickly in the MM as compared to the QM representation (Figure 4g,h). These problems reveal deficiencies in the transferred parameters when applied to disaccharide analogs.
Figure 4.
Φ/Ψ potential energy surfaces for model compounds 4 (top row), 5 (middle row), and 6 (bottom row) in the QM (first column), MM using transferred parameters (middle column), and MM using fit parameters (last column) representations. Energies are in kcal/mol with contours every 1 kcal/mol.
Given the shortcoming of transferring parameters developed for 2 and 3 to the disaccharide analogs, parameters were fit directly to target QM Φ/Ψ data. The Oring-C1-Olink-C1 and C2-C1-Olink-C1 dihedral parameters that determine Φ/Ψ energetics were fit to 4, with extra weighting given to points in the two minima and ignoring points greater than 12 kcal/mol above the global minimum (in Eqn. 2, wi=20 for points in the minima, 0 for points with energy greater than 12 kcal/mol, and 1 otherwise). The optimized parameters based on 4 were then directly transferred to 5 and 6. Additionally, the C1-Olink-C1 valence angle parameters were optimized to reproduce distortion of this angle as a function of Φ/Ψ for 4–6. This required reduction of the force constant transferred from C1-Omethyl-Cmethyl from 95 kcal*mol−1*radian−2 to a value of 50 kcal*mol−1*radian−2 and increasing the equilibrium value from 109.7° to 111.5°.
Very good energies and C1-Olink-C1 valence angle geometries as a function of Φ/Ψ were achieved by the self-consistent optimization of the dihedral and valence angle parameters. The Φ/Ψ energy surface for 4 using these optimized parameters gives the correct ordering of the two minima (Figure 4a,c). The MM model also satisfactorily reproduces the energy surfaces for 5 and 6, with the walls of the minima being less steep than with the transferred parameters, in better agreement with QM data (Figure 4d,f and 4g,i). The MM model does an excellent job of reproducing the change in C1-Olink-C1 valence angle geometry as a function of Φ/Ψ for all three model compounds (Figure 5). The C1-Olink-C1 geometry varies by more than 20° across the Φ/Ψ surface for each model compound. Quite remarkably, the simple form of the potential energy function, with a harmonic term for valence angle distortion, is able to correctly reproduce such large magnitude valence angle distortions.
Figure 5.
C1-Olink-C1’ valence angle as a function of Φ/Ψ in relaxed potential energy scans for model compounds 4 (top row), 5 (middle row), and 6 (bottom row) in the QM (first column) and MM using fit parameters (last column) representations. Angles are in degrees with contours every 2 degrees.
Water pair interactions energies and distances, intramolecular geometries of minimum energy conformations, and vibrational frequencies are also all well-reproduced using the optimized parameter set. The partial charge and Lennard-Jones parameters for Olink, transferred from the Omethyl atom in 2 and 3, capture the hydrogen bond energies and distances for 4–6 (Table 4). Of note is the fact that for the α(1→1)α analog 5, the hydrogen bond length is longer and the energy significantly less favorable than for the α(1→1)β and β(1→1)β analogs 4 and 6. This reflects the fact that water access to the Olink atom in the minimum energy geometry of 5 is sterically blocked by the two rings, whereas having at least one β anomer in the linkage opens up access to this atom. Minimum energy structures in the MM representation do a good job of reproducing bond, valence angle, and dihedral angle geometries relative to the QM MP2/6-31G(d) representation (Table 5); we note that the equilibrium length for the C1-Olink bonds in 4–6 was set to 1.415 Å, as is done for general linear ethers, compared to the value of 1.395 Å for compounds 2 and 3. The Φ and Ψ angles are particularly well represented by the MM model (Table 5), even though the same dihedral angle parameters are used for 4, 5, and 6. Finally, the good agreement between the MM and QM vibrational frequencies (Supplementary Material Figure S1) serves as additional confirmation of the appropriateness of the force field parameters.
Table 4.
Water pair interaction energies and distances for model compounds 4–6.
| model compound |
water orientationa |
energy (kcal/mol) |
distance (Å) |
||||
|---|---|---|---|---|---|---|---|
| 1.16*HFb | MM | MM- QM |
HF- 0.20b |
MM | MM- QM |
||
| 4 | −90 | −4.82 | −4.42 | 0.40 | 1.91 | 1.88 | −0.03 |
| 90 | −4.92 | −5.59 | −0.68 | 1.96 | 1.85 | −0.11 | |
| 5 | 90 | −1.93 | −2.72 | −0.79 | 2.51 | 2.49 | −0.02 |
| 6 | 90 | −5.75 | −5.64 | 0.10 | 1.85 | 1.77 | −0.09 |
| average | −0.24 | −0.06 | |||||
| standard deviation |
0.58 | 0.04 |
Dihedral angle (degrees) defined by C1-Olink…Owater-H2 where H2 is the non-interacting water hydrogen atom. 90 and −90 are equivalent in the cases of 5 and 6 due to the internal symmetry of these two model compounds. In all cases, the water H1-Owater bond vector is in the C1-Olink-C1’ angle plane and along the C1-Olink-C1’ bisector.
HF target energies have been scaled by 1.16 and distances have been shortened by 0.20 Å.
Table 5.
Optimized geometries for model compounds 4–6 in the MP2/6-31G(d) (QM) and MM representations.
| model compound |
valence angle / dihedral angle / bond lengtha |
QM | MM | MM-QM |
|---|---|---|---|---|
| 4 | Oring-C1-Olink | 112.7 | 110.9 | −1.8 |
| Oring’-C1’-Olink | 107.6 | 109.7 | 2.1 | |
| C2-C1-Olink | 106.2 | 108.7 | 2.6 | |
| C2’-C1’-Olink | 108.8 | 106.8 | −2.0 | |
| C1-Olink-C1’ | 114.4 | 115.0 | 0.6 | |
| Φ | 58.1 | 60.3 | 2.2 | |
| Ψ | −97.1 | −95.9 | 1.2 | |
| C1-Olink | 1.428 | 1.419 | −0.009 | |
| C1’-Olink | 1.403 | 1.418 | 0.014 | |
| 5 | Oring-C1-Olink | 111.2 | 109.7 | −1.5 |
| C2-C1-Olink | 106.7 | 108.7 | 1.9 | |
| C1-Olink-C1’ | 112.7 | 113.2 | 0.6 | |
| Φ(=Ψ) | 68.6 | 73.1 | 4.4 | |
| C1-Olink | 1.425 | 1.417 | −0.008 | |
| 6 | Oring-C1-Olink | 107.5 | 109.4 | 1.9 |
| C2-C1-Olink | 108.1 | 106.9 | −1.3 | |
| C1-Olink-C1’ | 113.6 | 113.0 | −0.6 | |
| Φ(=Ψ) | −68.8 | −67.9 | 0.9 | |
| C1-Olink | 1.405 | 1.414 | 0.009 |
Valence angles and dihedral angles are in degrees, bond lengths are in Å.
1→2, 1→3, and 1→4 glycosidic linkages
Using two molecules of tetrahydropyran linked by a bridging ether as model compounds for the 1→2, 1→3, and 1→4 glycosidic linkages leads to a prohibitively large number of possibilities for all combinations of linkage type and linkage stereochemistry. Because the Oring atom in the second ring in these types of linkages is at least two atoms removed from Olink, a cyclohexane molecule was used to represent the second ring for the respective model compounds. For 1→2 linked pyranose disaccharides, the resultant model compounds, with all possibilities for stereochemistry at the C1 and C2’ positions, are compounds 7, 8, 9, and 10. Additionally, simply by renumbering the atoms on the cyclohexane ring, these four molecules correspond to all possible stereochemistries for 1→3 and 1→4 glycosidic linkage. Thus, energies and geometries of these four molecules were used to develop parameters for 1→2, 1→3, and 1→4 linked hexopyranose disaccharides. As with the 1→1 linkages, a single set of parameters was developed for all model compounds in the series.
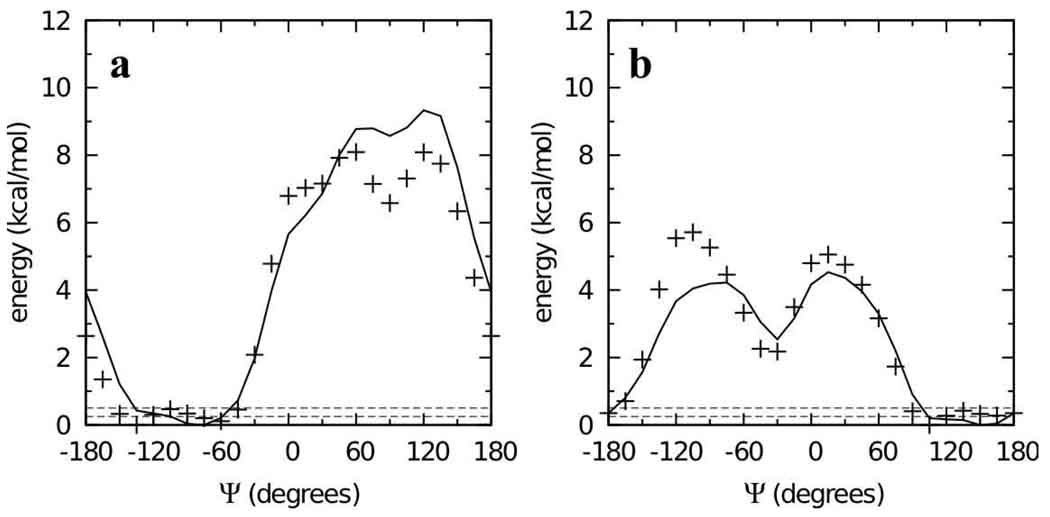
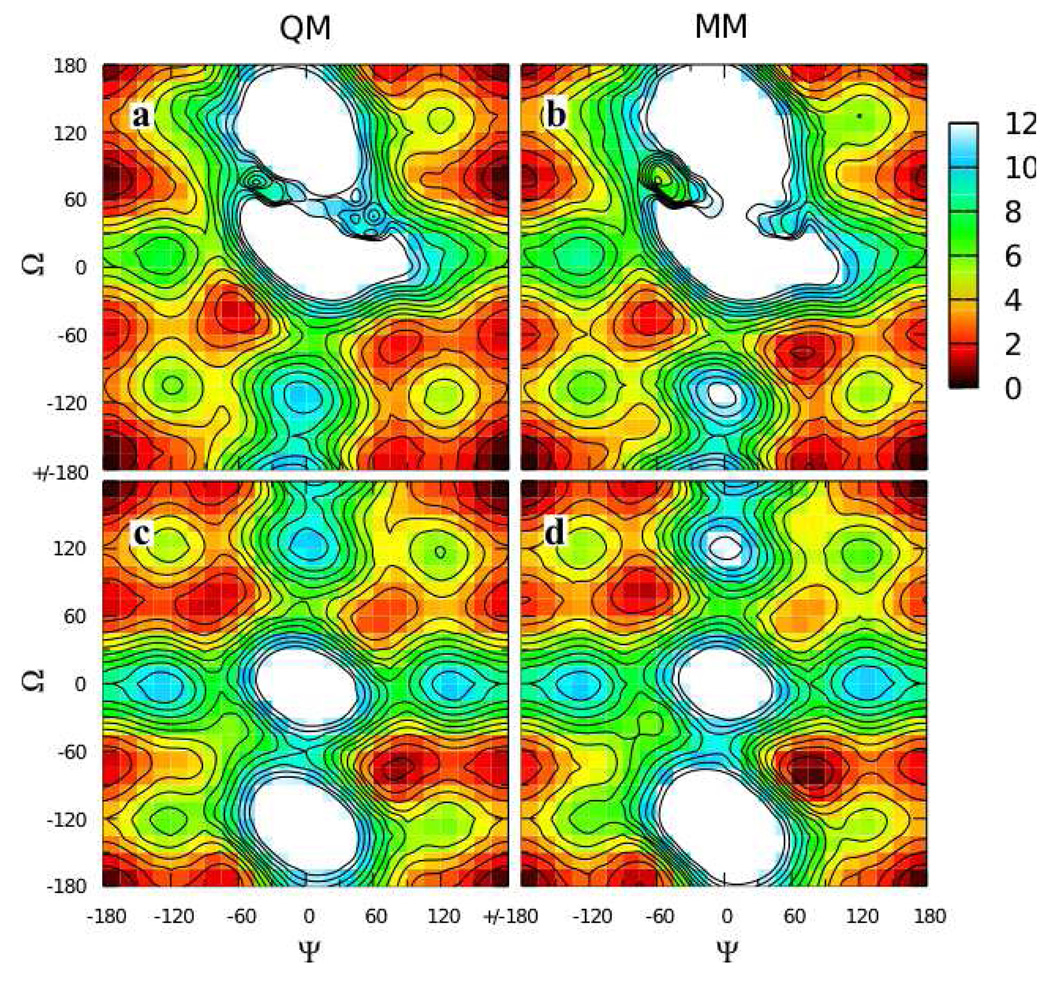
Parameters except for the Oring-C1-Olink-C2’, C2-C1-Olink-C2’, C1-Olink-C2’-C1’, and C1-Olink-C2’-C3’ dihedral parameters were transferred from the 1→1 linkages, and cyclohexane parameters were those previously developed in the context of hexopyranoses.19 Automated fitting of these four dihedrals, with the C1-Olink-C2’-C1’ and C1-Olink-C2’-C3’ parameters equivalenced (i.e. constrained to be the same),56 was undertaken by including in the fit all points having QM energies less than 12 kcal/mol relative to the respective global minima on the four two-dimensional MP2/cc-pVTZ// MP2/6-31G(d) scans. While the fits were generally good, two issues arose: systematic deviation of the MM C1-Olink and Olink-C2’ bond and C1-Olink-C2’ angle values from the QM data, and difficulty reproducing the global energy minima on the surfaces for 9 and 10 where the energy is nearly constant over a large range of Ψ for a value of Φ = −60° (Φ = Oring-C1-Olink-C2’, Ψ = C1-Olink-C2’-C1’).
To overcome these limitations, a special weighting scheme was applied to the dihedral fitting, and the relevant bond and angle parameters were optimized to target the QM data. With regard to dihedral fitting, the automated dihedral fitting algorithm was extended to allow separate RMS alignment of each of the four MM surfaces to its corresponding QM surface during the simultaneous fitting of all four surfaces, as described in the Methods. This allowed the fitting to overcome any discrepancies in the MM ΔE values between any pair of surfaces as compared to the QM ΔE. Additionally, points with an energy value within 0.25 kcal/mol of the global minimum for each surface were given a weight wi of 1000, compared to a weight of 1 for other points below the 12 kcal/mol cutoff.56 This emphasis on a few low-lying points was helpful in fitting the flat global minima for compounds 9 and 10 (Figure 6). There were sufficiently few points with this high weighting such that, combined with optimized bond and angle parameters, the full shapes of the energy surfaces were all well reproduced (Figure 7). Bond optimization entailed adjusting the equilibrium values of the C1-Olink and Olink-C2’ bonds to 1.395 and 1.435 Å, respectively. Additionally, the final optimized C1-Olink-C2’ parameters (force constant 50 kcal*mol−1*radian−2 and equilibrium value of 109.2°) gave impressive reproduction of the angle distortion as a function of Φ/Ψ (Figure 8).
Figure 6.
Φ = −60° slices from the two-dimensional Φ/Ψ potential energy surfaces for model compounds 9 (a) and 10 (b) in the QM (crosses) and MM using fit parameters (solid lines) representations. Dashed horizontal lines are at 0.25 and 0.50 kcal/mol.
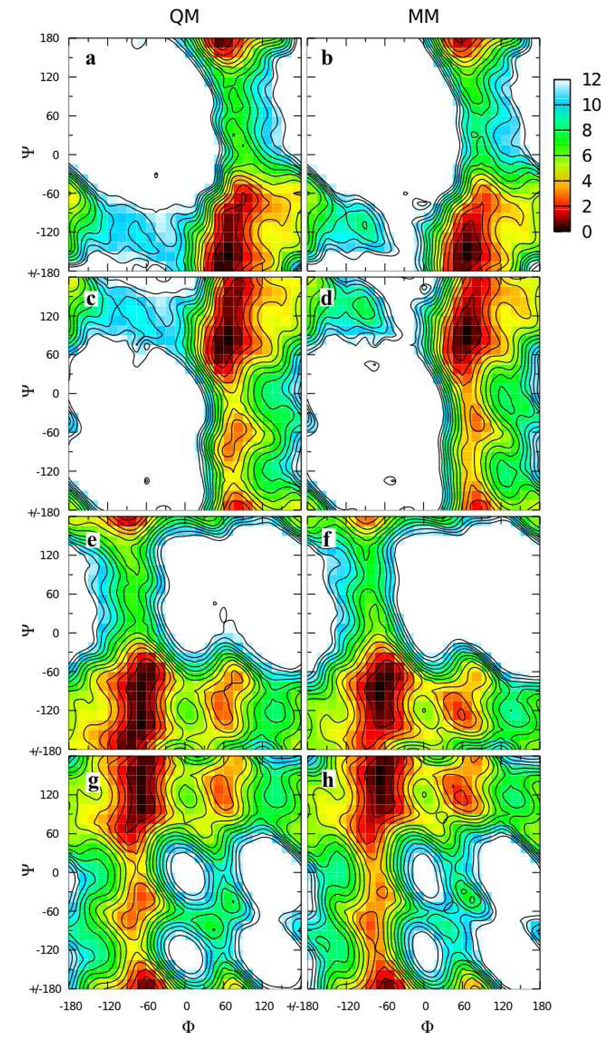
Figure 7.
Φ/Ψ potential energy surfaces for model compounds 7 (top row), 8 (second row), 9 (third row), and 10 (bottom row) in the QM (first column) and MM using fit parameters (last column) representations. Energies are in kcal/mol with contours every 1 kcal/mol.
Figure 8.
C1-Olink-C2’ valence angle as a function of Φ/Ψ in relaxed potential energy scans for model compounds 7 (top row), 8 (second row), 9 (third row), and 10 (bottom row) in the QM (first column) and MM using fit parameters (last column) representations. Angles are in degrees with contours every 2 degrees.
Intramolecular geometries and vibrational frequencies demonstrated the appropriateness of the final bonded parameters, and water pair interaction distances and energies confirmed the transferability of the Olink partial charge and Lennard-Jones nonbonded parameters from model compounds 4–6. Fully unconstrained MP2/6-31G(d) geometry optimizations done on the global minimum energy conformations from the two-dimensional Φ/Ψ scans showed little change in the value of the dihedral angles relative to the constrained values from the scans. Using the geometries from fully unconstrained QM optimizations, MM optimizations on compounds 7 and 8 gave structures in excellent agreement with the optimized QM structures (Table 6). However, for model compounds 9 and 10, the Ψ dihedral needed to be restrained to the QM value during the MM geometry optimization to prevent changes in this angle by +49.3° and +46.2° respectively. It is important to note that this simply reflects the very flat minimum energy well that these conformations exist in (Figure 6 and Figure 7e–h) and not a significant deficit in the force field. From Figure 6, it is apparent that Ψ values spanning 60° for 9 and 10 in the global minimum energy wells correspond to conformations within 0.5 kcal/mol of each other, with a majority of these being within 0.25 kcal/mol of each other. Because of the differences in the QM- versus unrestrained MM-optimized conformations for 9 and 10, vibrational frequencies for comparison were computed only for 7 and 8, and these had the same degree of accuracy as for model compounds 2–6 (Supplementary Material Figure S2).
Table 6.
Optimized geometries for model compounds 7–10 in the MP2/6-31G(d) (QM) and MM representations.
| model compound |
valence angle / dihedral angle / bond lengtha |
QM | MM | MM-QM |
|---|---|---|---|---|
| 7 | Oring-C1-Olink | 111.8 | 110.2 | −1.7 |
| C2-C1-Olink | 106.7 | 108.4 | 1.7 | |
| C1’-C2’-Olink | 106.3 | 107.2 | 0.9 | |
| C3’-C1’-Olink | 109.7 | 109.4 | −0.3 | |
| C1-Olink-C2’ | 113.7 | 114.0 | 0.3 | |
| Φ | 66.3 | 68.0 | 1.7 | |
| Ψ | −151.9 | −149.2 | 2.7 | |
| C1-Olink | 1.414 | 1.400 | −0.014 | |
| C2’-Olink | 1.446 | 1.441 | −0.004 | |
| 8 | Oring-C1-Olink | 111.9 | 110.2 | −1.7 |
| C2-C1-Olink | 106.8 | 108.5 | 1.8 | |
| C1’-C2’-Olink | 110.5 | 109.3 | −1.2 | |
| C3’-C1’-Olink | 107.1 | 107.3 | 0.2 | |
| C1-Olink-C2’ | 113.6 | 113.7 | 0.0 | |
| Φ | 66.3 | 67.6 | 1.3 | |
| Ψ | 88.2 | 91.5 | 3.3 | |
| C1-Olink | 1.414 | 1.401 | −0.013 | |
| C2’-Olink | 1.441 | 1.440 | −0.001 | |
| 9 | Oring-C1-Olink | 108.6 | 110.3 | 1.7 |
| C2-C1-Olink | 108.5 | 106.6 | −1.9 | |
| C1’-C2’-Olink | 106.5 | 107.6 | 1.1 | |
| C3’-C1’-Olink | 109.4 | 108.8 | −0.7 | |
| C1-Olink-C2’ | 114.4 | 114.1 | −0.3 | |
| Φ | −64.6 | −61.0 | 3.7 | |
| Ψ | −136.8 | b | ||
| C1-Olink | 1.391 | 1.398 | 0.007 | |
| C2’-Olink | 1.447 | 1.442 | −0.006 | |
| 10 | Oring-C1-Olink | 108.6 | 110.1 | 1.5 |
| C2-C1-Olink | 108.5 | 106.7 | −1.9 | |
| C1’-C2’-Olink | 110.0 | 108.8 | −1.1 | |
| C3’-C1’-Olink | 107.5 | 107.8 | 0.2 | |
| C1-Olink-C2’ | 114.2 | 113.7 | −0.5 | |
| Φ | −63.9 | −62.7 | 1.2 | |
| Ψ | 104.2 | b | ||
| C1-Olink | 1.391 | 1.398 | 0.008 | |
| C2’-Olink | 1.443 | 1.440 | −0.003 |
Valence angles and dihedral angles are in degrees, bond lengths are in Å.
Value restrained to the QM geometry during optimization.
Water pair interaction energies and distances for the QM and MM optimized conformations showed excellent agreement for 8 and 10, while hydrogen bonds were shorter and stronger in the MM representation relative to the QM for 7 and 9 (Table 7). Importantly, the hydrogen bonds for 8 and 10 are significantly shorter and more favorable in both representations than for 7 and 9, which have relatively long and weak hydrogen bonding interactions with the water molecule regardless of its orientation. Thus, the MM model is properly reproducing the relative hydrogen bond strengths for the different anomers. The disagreement for the weaker hydrogen bonding interactions may be ascribed to the inability of the water molecule to approach the Olink atom for 7 and 9 due to steric hinderance, and the MM model predicts a shorter interaction distance because it properly includes favorable dispersion interactions via the Lennard-Jones force field term whereas the Hartree-Fock level of theory used in the QM water pair interaction calculations does not model dispersion.
Table 7.
Water pair interaction energies and distances for model compounds 7–10.
| model compound |
water orientationa |
energy (kcal/mol) |
distance (Å) |
||||
|---|---|---|---|---|---|---|---|
| 1.16*HFb | MM | MM- QM |
HF-0.20b | MM | MM- QM |
||
| 7 | −180 | −1.19 | −2.48 | −1.29 | 3.10 | 2.84 | −0.25 |
| −90 | −1.28 | −2.60 | −1.33 | 2.96 | 2.81 | −0.15 | |
| 0 | −0.88 | −2.19 | −1.32 | 3.03 | 2.84 | −0.18 | |
| 90 | −1.24 | −2.27 | −1.03 | 2.99 | 2.84 | −0.15 | |
| average | −1.24 | −0.19 | |||||
| standard deviation |
0.14 | 0.05 | |||||
| 8 | −180 | −3.88 | −4.19 | −0.32 | 2.00 | 2.07 | 0.06 |
| −90 | −4.16 | −4.17 | −0.01 | 1.99 | 2.06 | 0.07 | |
| 0 | −3.27 | −3.49 | −0.21 | 2.24 | 2.10 | −0.14 | |
| 90 | −3.61 | −3.75 | −0.14 | 2.11 | 2.09 | −0.03 | |
| average | −0.17 | −0.01 | |||||
| standard deviation |
0.13 | 0.10 | |||||
| 9 | −180 | −1.87 | −2.98 | −1.11 | 2.82 | 2.64 | −0.17 |
| −90 | −2.13 | −3.26 | −1.13 | 2.60 | 2.62 | 0.02 | |
| 0 | −1.56 | −2.57 | −1.02 | 2.53 | 2.67 | 0.14 | |
| 90 | −1.97 | −2.45 | −0.49 | 2.67 | 2.67 | 0.00 | |
| average | −0.94 | −0.01 | |||||
| standard deviation |
0.30 | 0.13 | |||||
| 10 | −180 | −5.56 | −6.13 | −0.57 | 1.87 | 1.75 | −0.12 |
| −90 | −6.37 | −6.56 | −0.19 | 1.82 | 1.74 | −0.08 | |
| 0 | −5.46 | −5.42 | 0.04 | 1.87 | 1.77 | −0.10 | |
| 90 | −6.05 | −5.44 | 0.62 | 1.83 | 1.76 | −0.06 | |
| average | −0.02 | −0.09 | |||||
| standard deviation |
0.50 | 0.03 |
Dihedral angle (degrees) defined by C1-Olink…Owater-H2 where H2 is the non-interacting water hydrogen atom. In all cases, the water H1-Owater bond vector is in the C1-Olink-C1’ angle plane and along the C1-Olink-C1’ bisector.
HF target energies have been scaled by 1.16 and distances have been shortened by 0.20 Å.
As discussed at the beginning of this section, the use of two molecules of tetrahydropyran linked by a bridging ether as model compounds for all possible 1→2, 1→3, and 1→4 glycosidic linkages would have been prohibitive. To confirm the appropriateness of the dihedral parameters developed using 7–10, MM scans using these parameters were done on analogs of 7–10 (7O, 8O, 9O, 10O) in which cyclohexane was replaced by a second molecule of tetrahydropyran, and compared to QM data. The scans were done at fixed values of Φ, chosen based on the global minima on the Φ/Ψ surfaces for 7–10 (Figure 9). In all four cases, the MM scans capture the features of the QM scans, and for 8O, 9O, and 10O (Figure 9b–d), the global minimum is correctly captured. In the case of 7O (Figure 9a), the global minimum is at Ψ = −90° in the QM representation as compared to Ψ = −150° in the MM. More than anything else, this is a reflection of the flat well surrounding the global minimum, also seen for compound 7 (Figure 7a). In the case of 7O, energies in this well that ranges from Ψ = −180° to Ψ = −75° are within 1.5 kcal/mol of each other. The MM representation correctly captures the span of this wide well and also the adjacent peak at Ψ = +15°. Thus, the dihedral parameters developed on 7–10 appear also to be appropriate when the cyclohexane ring is replaced by a second tetrahydropyran ring.
Figure 9.
Energy as a function of Ψ for analogs of compounds 7–10 (7O, 8O, 9O, 10O), in which the cyclohexane ring has been replaced with a second tetrahydropyran ring. Φ = +60° for compounds 7O (a) and 8O (b), and Φ = −60° for compounds 9O (c) and 10O (d). QM energies are represented as crosses and MM energies (using dihedral parameters developed on compounds 7–10) as solid lines.
1→6 glycosidic linkages
Compared to 1→1, 1→2, 1→3, and 1→4 linkages, 1→6 glycosidic linkages have an additional dihedral degree of freedom, making full QM characterization of an analogous model compound too computationally expensive. The three dihedral degrees of freedom in a 1→6 linkage (Φ = Oring-C1-Olink-C6’, Ψ = C1-Olink-C6’-C5’, Ω = Olink-C6’-C5’-Oring’) were thus parametrized based on the results for 2 and 3 as models for the Φ dihedral angle, and 11 and 12 as models for the Ψ and Ω dihedral angles. A single set of dihedral parameters gave excellent results for the Ψ/Ω surfaces for both 11 and 12 (Figure 10). These dihedral parameters along with optimization of the Olink-C6’-C5’ angle geometry also gave excellent minimum-energy geometries (Table 8) and good agreement with vibrational frequencies (Supplementary Material Figure S3), completing the parameter development for all the model compounds.
Figure 10.
Ψ/Ω potential energy surfaces for model compounds 11 (top row) and 12 (bottom row) in the QM (first column) and MM using fit parameters (last column) representations. Energies are in kcal/mol with contours every 1 kcal/mol.
Table 8.
Optimized geometries for model compounds 11 and 12 in the MP2/6-31G(d) (QM) and MM representations.
| model compound |
valence angle / dihedral angle / bond lengtha |
QM | MM | MM-QM |
|---|---|---|---|---|
| 11 | C1-Olink-C6’ | 111.5 | 111.2 | −0.3 |
| Olink-C6’-C5’ | 107.1 | 107.6 | 0.5 | |
| C6’-C5’-Oring | 110.5 | 110.4 | −0.1 | |
| C6’-C5’-C4’ | 113.5 | 114.3 | 0.8 | |
| Ψ | 178.9 | 179.3 | 0.4 | |
| Ω | −170.8 | −169.7 | 1.1 | |
| Olink-C6’ | 1.420 | 1.424 | 0.004 | |
| C6’-C5’ | 1.525 | 1.510 | −0.015 | |
| 12 | C1-Olink-C6’ | 111.4 | 111.1 | −0.3 |
| Olink-C6’-C5’ | 107.6 | 107.8 | 0.2 | |
| C6’-C5’-Oring | 105.1 | 106.8 | 1.7 | |
| C6’-C5’-C4’ | 113.2 | 113.1 | −0.1 | |
| Ψ | 179.3 | 179.5 | 0.2 | |
| Ω | 177.7 | 177.8 | 0.1 | |
| Olink-C6’ | 1.418 | 1.424 | 0.006 | |
| C6’-C5’ | 1.515 | 1.504 | −0.011 |
Valence angles and dihedral angles are in degrees, bond lengths are in Å.
Further parameter optimization in full disaccharides
The parameters developed in the model compound analogs of disaccharides, along with existing parameters for hexopyranose monosaccharides,19 enabled the construction and simulation of disaccharides in the crystalline state (Table 9). Analysis of intramolecular geometries obtained from MD simulations of various disaccharide crystals showed several systematic differences in bond and valence angle geometries as determined by the model compound parameters, and these parameters were further optimized to correct for these systematic deviations. For C1-Olink-C1’ linked compounds, the model compound vs. full disaccharide parametrized equilibrium valence angle values were as follows: C2-C1-Olink 109.0° vs. 105.0°, and O5-C1-Olink 110.0° vs. 112.0°, with the model compound values followed by the full disaccharide values. For C1-Olink-Cn’ (n=2,3,4), the equilibrium bond and angle values were: Olink-Cn' 1.435 Å vs. 1.415 Å, C2-C1-Olink 109.0° vs. 105.0°, and O5-C1-Olink 110.0° vs. 112.0°. Of note is that the changes to the C2-C1-Olink and O5-C1-Olink angle parameters were the same for both C1-Olink-C1’ and C1-Olink-Cn’ (n=2,3,4) linked disaccharides relative to the model compounds. Perhaps it is not surprising that these two angle parameters required special attention; even in the case of model compounds 2 and 3, the changes in QM angle geometries going from the α- to the β-anomer resulted in MM errors having magnitudes of 1.5° to 1.8° but of differing signs depending on the anomer (Table 2), attesting to the challenge of developing general parameters involving the anomeric oxygen. The final set of parameters for all linkages between hexopyranoses is listed in Supplementary Material S4–S9.
Table 9.
Disaccharide crystals.
| CSD accession code |
common name | non-reducing end pyranose |
reducing end pyranose | linkage | linkage geometrya |
# of disacsb |
# of water moleculesb |
|---|---|---|---|---|---|---|---|
| DEKYEX | α,α-trehalose | α-glucose | α-glucose | α(1→1)α | aa | 2 | 0 |
| TREHAL01 | α,α-trehalose dihydrate |
α-glucose | α-glucose | α(1→1)α | aa | 4 | 8 |
| YOXFOG | α,α-allo-trehalose trihydrate |
α-allose | α-allose | α(1→1)α | aa | 2 | 6 |
| YOXFUM | α,α-galacto-trehalose | α-galactose | α-galactose | α(1→1)α | aa | 4 | 0 |
| TIQDUS | α,β-trehalose monohydrate |
α-glucose | β-glucose | α(1→1)β | ae | 2 | 2 |
| FABYOW10 | α-mannose | α-1-O-methyl-mannose | α(1→2) | aa | 2 | 0 | |
| RESMOR | α-mannose | β-1-O-methyl-glucose | α(1→2) | ae | 4 | 0 | |
| MOGLPR | methyl α-nigeroside | α-glucose | α-1-O-methyl-glucose | α(1→3) | ae | 2 | 0 |
| MALTOS11 | β-maltose monohydrate |
α-glucose | β-glucose | α(1→4) | ae | 2 | 2 |
| MMALTS | methyl β-maltoside monohydrate |
α-glucose | β-1-O-methyl-glucose | α(1→4) | ae | 4 | 4 |
| MELIBM10 | α-melibiose monohydrate |
α-glucose | α-glucose | α(1→6) | ae | 4 | 4 |
| SOPROS | α-sophorose monohydrate |
β-glucose | α-glucose | β(1→2) | ee | 4 | 4 |
| WAGBOV | methyl β- laminarabioside monohydrate |
β-glucose | β-1-O-methyl-glucose | β(1→3) | ee | 4 | 4 |
| BLACTO | β-lactose | β-galactose | β-glucose | β(1→4) | ee | 2 | 0 |
| CELLOB02 | β-cellobiose | β-glucose | β-glucose | β(1→4) | ee | 2 | 0 |
| GENTBS01 | β-gentiobiose | β-glucose | β-glucose | β(1→6) | ee | 4 | 0 |
Indicates the configuration at the ring carbons involved in the glycosidic linkage. For example, “ae” indicates that the glycosidic linkage ring carbon on the non-reducing end pyranose has the glycosidic linkage directed axially while the ring carbon on the reducing end pyranose has it directed equatorially.
Number of molecules in the unit cell used for MD simulation.
Validation
A number of molecular dynamics (MD) simulations were undertaken to validate the development of bonded and nonbonded parameters for glycosidic linkages, and included a variety of disaccharides both in the aqueous phase and the crystalline phase. Crystal simulations were used primarily to confirm the ability of the force field to reproduce intramolecular geometries, thereby testing bond, angle, and dihedral parameters, with additional analysis of crystal cell volumes and their relevance to the nonbonded parameters. Aqueous phase simulations served to test the conformational properties of the various glycosidic linkages as determined by the dihedral parameters, as well as the ability of the force field model to capture aqueous solution density as a function of concentration. The results of these extensive simulations compared favorably to the available experimental data, thereby serving to validate the force field parametrization.
Crystalline disaccharide intramolecular geometries and unit cell parameters
A comprehensive list of disaccharides representative of the parametrized linkages was simulated as infinite crystals using MD at constant temperature and pressure to test the ability of the force field to maintain intramolecular geometries and unit cell parameters. This list of 16 compounds, taken from the Cambridge Structural Database (CSD) of small molecules,61 included disaccharides containing not only glucose as a component hexopyranose monosaccharide but also allose, galactose, and mannose (Table 9). Additionally, a number of the disaccharides were O-methylated at the reducing end pyranose and/or contained water molecules in the crystal unit cell, both of which further tested the robustness of the force field. Finally, in all cases the simulated unit cell contained at least two independent disaccharide molecules, and a number of the simulations had four independent disaccharides. Overall, MD results of intramolecular geometries and unit cell parameters tabulated by averaging over snapshots from the trajectories, and over the independent disaccharide molecules in the case of intramolecular geometries, were quite consistent with the experimental values.
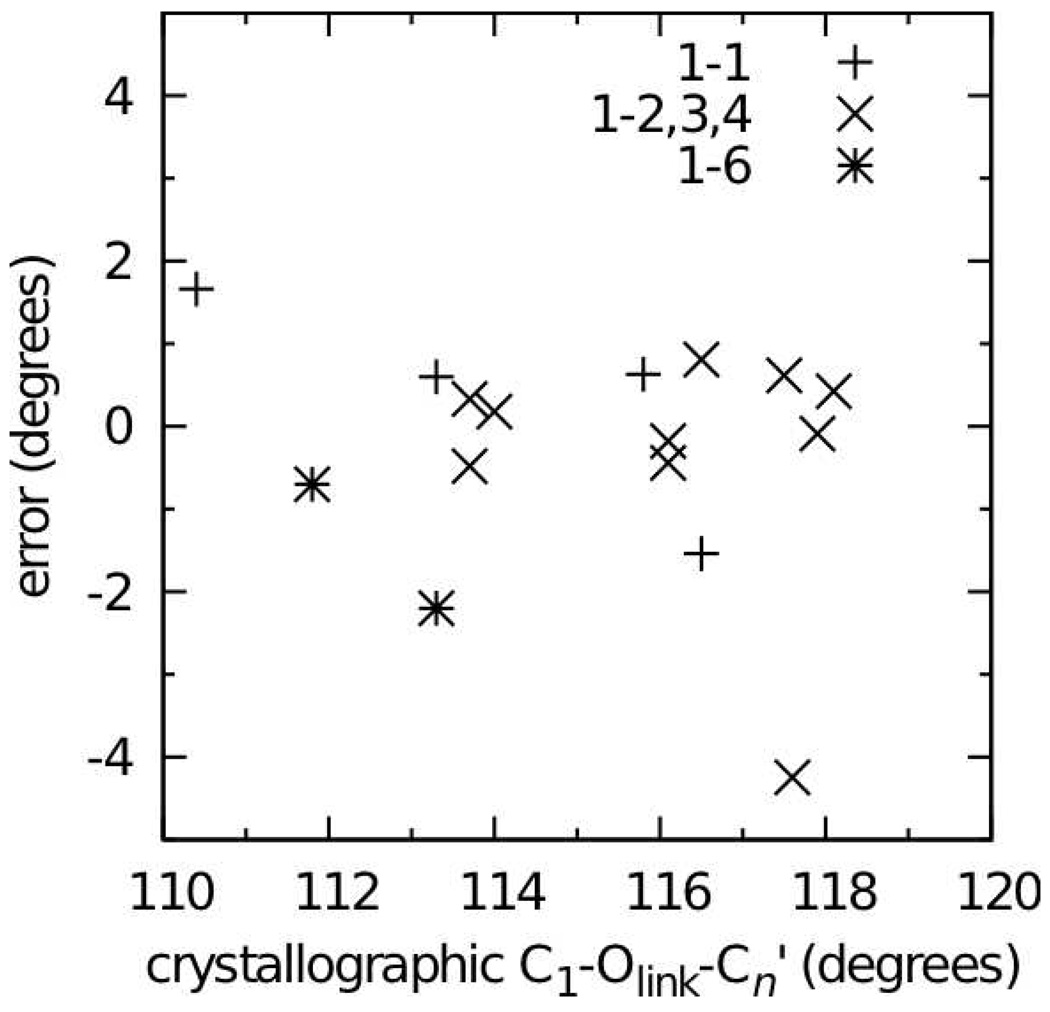
Errors in the disaccharide intramolecular geometries over the entire set of 16 crystals were impressively small for bonds, valence angles, and dihedral angles involving the glycosidic oxygen Olink (Table 10). Both bonds involving this atom had average errors no larger than 0.01 Å, and the spread in errors, as judged by the standard deviation in the errors, was also quite small, reflecting the fact that these bond lengths were well reproduced across the entire set of crystals. The situation was similar for valence angles, with all average errors for valence angles being less than 1.5° and having similarly small standard deviations in the errors. Of particular note is the C1-Olink-Cn’ valence angle with an average error of only −0.3°. Parameters for this angle were from careful parametrization aimed at reproducing not only the geometries of minimum-energy conformations of the disaccharide analog model compounds 4–10, but also the distortion of this angle with changing Φ/Ψ, including high-energy regions in the MP2/cc-pVTZ// MP2/6-31G(d) QM surfaces (Figure 4, Figure 5, Figure 7, and Figure 8). These parameters were directly used in the simulations of the full disaccharides and, along with C1-Olink-C6’ parameters transferred from linear ethers, yielded good reproduction of this glycosidic link angle across the full set of 1→1, 1→2, 1→3, 1→4, and 1→6 linked disaccharides (Figure 11). Finally, Φ, Ψ, and Ω dihedral angle values were well reproduced in the disaccharide crystal simulations using the dihedral parameters developed on model compounds 2–12 (Table 10).
Table 10.
Crystalline disaccharide internal geometries.a
| C1- Olink |
Olink- Cn’ |
C1-Olink- Cn’ |
Oring-C1- Olink |
C2-C1- Olink |
Olink-Cn’- Cn+1’ b |
Olink-Cn’- Xc |
Φ | Ψ | Ωd | |
|---|---|---|---|---|---|---|---|---|---|---|
| average error | 0.008 | 0.002 | −0.3 | 0.7 | 0.9 | 0.6 | 1.3 | − 0.8 |
− 3.9 |
− 3.1 |
| standard deviation of errors |
0.008 | 0.012 | 1.4 | 1.2 | 1.2 | 1.5 | 1.2 | 6.0 | 8.2 | n/a |
Data are from simulations of crystals listed in Table 9; MD data for a particular crystal were averaged over the independent disaccharide molecules in the crystal as well as over the MD trajectory; bonds are in Å and valence and dihedral angles are in degrees.
Data exclude MELIBM10 and GENTBS01 because these are 1→6 linked disaccharides.
X=Oring’ for 1→1 linked disaccharides and Cn-1’ for all others.
Data include only MELIBM10 and GENTBS01.
Figure 11.
C1-Olink-Cn’ valence angle errors in MD simulations of disaccharide crystals. Errors are calculated as the MD value (averaged over the simulation and over all independent disaccharides in the unit cell) minus the crystallographic value.
Disaccharide crystal unit cell geometries and volumes were also tabulated from the MD simulations of the systems in Table 9, and were in good accord with the experimental values (Table 11). The crystal unit cell length parameters A, B, and C were well maintained for 15 of the 16 simulated systems. In those systems where the crystal unit cell angle parameter β was not 90°, it was allowed to vary during the simulation, and in all of the 9 total systems for which this was the case, β stayed close to the crystallographic value. The outlier with regard to the crystal unit cell parameter data was α,α-allo-trehalose trihydrate (YOXFUG). This case is the most challenging in the entire set in that it is a trihydrate, with 6 molecules of water and 2 of the disaccharide in the unit cell. Furthermore, it is the only crystal in the set to have P1 symmetry, thus all three angular unit cell parameters α, β, and γ were allowed to vary unconstrained during the simulation, whereas all other crystals had at most one unconstrained angular parameter (β) (Table 11). It is important to note that unit cell volumes are systematically overestimated across the entire set of compounds, a trend that has been noted in prior parametrizations of cyclic19 and linear carbohydrates.20 As noted in these prior works, this likely is a reflection of the difficulty in applying nonbonded parameters developed from simulations of neat liquid alcohols19 to the highly-directional hydrogen bonding networks in crystals. Also as noted in these prior works, when used in aqueous simulations, the parameters give excellent reproduction of solution densities, as discussed next.
Table 11.
Crystalline disaccharide unit cell geometries and volumes
| A (Å) | B (Å) | C (Å) | β (degrees) a |
volume (Å3) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| expt | MD | % error |
expt | MD | % error |
expt | MD | % error |
expt | MD | % error |
expt | MD | % error |
|
| DEKYEX | 12.97 | 13.16 | 1.5 | 8.23 | 8.15 | −0.9 | 6.79 | 7.05 | 3.9 | 98.12 | 99.62 | 1.5 | 717.4 | 746.0 | 4.0 |
| TREHAL01 | 12.23 | 12.36 | 1.1 | 17.89 | 17.94 | 0.3 | 7.60 | 7.85 | 3.3 | 90.00 | 90.00 | 1661.7 | 1740.7 | 4.8 | |
| YOXFOG | 9.40 | 8.56 | −8.9 | 11.03 | 11.26 | 2.0 | 9.15 | 10.35 | 13.1 | 96.8 | 96.5 | −0.4 | 853.8 | 931.0 | 9.0 |
| YOXFUM | 11.07 | 11.13 | 0.5 | 18.28 | 18.91 | 3.4 | 6.87 | 6.85 | −0.2 | 90.00 | 90.00 | 1389.6 | 1442.0 | 3.8 | |
| TIQDUS | 9.77 | 10.05 | 2.8 | 8.58 | 8.13 | −5.2 | 10.10 | 10.50 | 4.0 | 111.68 | 109.39 | −2.1 | 786.0 | 808.1 | 2.8 |
| FABYOW10 | 8.09 | 8.17 | 1.0 | 9.78 | 9.88 | 1.0 | 9.98 | 10.16 | 1.8 | 104.58 | 103.15 | −1.4 | 763.0 | 797.5 | 4.5 |
| RESMOR | 9.38 | 9.32 | −0.6 | 10.65 | 10.76 | 1.0 | 15.83 | 16.57 | 4.7 | 90.00 | 90.00 | 1580.4 | 1661.9 | 5.2 | |
| MOGLPR | 6.59 | 6.90 | 4.6 | 13.24 | 13.73 | 3.7 | 9.45 | 9.23 | 2.3 | 108.00 | 101.40 | −6.1 | 784.1 | 853.2 | 8.8 |
| MALTOS11 | 4.87 | 5.06 | 3.9 | 15.08 | 15.30 | 1.4 | 10.70 | 10.76 | 0.5 | 97.07 | 97.80 | 0.8 | 779.2 | 823.6 | 5.7 |
| MMALTS | 22.64 | 24.22 | 7.0 | 4.84 | 4.90 | 1.3 | 17.32 | 17.01 | −1.8 | 117.3 | 120.7 | 2.9 | 1687.2 | 1736.9 | 2.9 |
| MELIBM10 | 8.90 | 9.08 | 2.0 | 10.89 | 11.03 | 1.2 | 15.87 | 16.03 | 1.1 | 90.00 | 90.00 | 1538.5 | 1604.8 | 4.3 | |
| SOPROS | 22.05 | 22.14 | 0.4 | 14.44 | 14.38 | −0.4 | 4.87 | 5.03 | 3.5 | 90.00 | 90.00 | 1548.5 | 1603.3 | 3.5 | |
| WAGBOV | 14.55 | 14.63 | 0.6 | 24.25 | 24.23 | −0.1 | 4.94 | 5.07 | 2.7 | 90.00 | 90.00 | 1742.2 | 1798.0 | 3.2 | |
| BLACTO | 10.84 | 11.03 | 1.8 | 13.35 | 13.55 | 1.5 | 4.95 | 5.06 | 2.2 | 91.31 | 86.43 | −5.3 | 716.6 | 752.5 | 5.0 |
| CELLOB02 | 10.97 | 11.07 | 0.9 | 13.05 | 13.35 | 2.3 | 5.09 | 5.14 | 0.9 | 90.83 | 91.22 | 0.4 | 728.8 | 758.6 | 4.1 |
| GENTBS01 | 8.87 | 9.01 | 1.5 | 22.85 | 23.31 | 2.0 | 7.20 | 7.22 | 0.3 | 90.00 | 90.00 | 1459.1 | 1516.6 | 3.9 | |
| average | 1.3 | 0.9 | 2.4 | −1.1 | 4.7 | ||||||||||
| standard deviation |
3.3 | 2.1 | 3.5 | 3.0 | 1.8 |
Constrained to 90° in the simulation if equal to 90° in the experimental crystal, otherwise allowed to vary independently during the simulation.
Solution densities
Aqueous solutions at various concentrations of different disaccharides were simulated under the experimental conditions of 298 K and 1 atm to calculate densities (Eqs. 5 and 6), which compared favorably with experimental values across all solutes. For all disaccharides and all densities, the calculated solution densities are in good agreement with the experimental solution densities, with all errors within 1.5% (Table 12). For the fifteen aqueous solutions that were simulated, the average total error in the densities is 0.85% and the average total absolute error is 1.10%. Moreover, the force field model is able to reproduce experimental results both for dilute and concentrated solutions (maltose). Specifically, for simulations of dilute aqueous trehalose, cellobiose, gentiobiose, and melibiose, ranging in concentration from 0.1 to 0.3 mol/kg, errors range from 0.87% to 1.48%. For simulations of concentrated maltose, with concentrations between 1.5 to 2.6 mol/kg, errors range from −0.26% to −1.07%. For the most concentrated system, 2.6 molal maltose, nearly one third of the atoms in the system belong to solute molecules. As a result, there is significant direct contact between solute molecules, and it is not entirely surprising that the density is somewhat overestimated given that in the limiting case in which there are no water molecules, i.e. crystalline disaccharides, densities are systematically overestimated. More important though is that for all the aqueous systems, errors in density lie within a relatively small window, and the results are quite favorable given the 25-fold difference in concentrations between the most dilute versus the most concentrated systems studied.
Table 12.
Comparison of the calculated and experimental densities for different molal concentrations of trehalose, cellobiose, gentiobiose and mellibiose in 1100 TIP3P waters at T = 298 K and P = 1 atm.
| molality (mol/kg) | Nsolute | Expt. | MD | % error |
|---|---|---|---|---|
| trehalose + water64 | ||||
| 0.106 | 2 | 1.011 | 1.024 | 1.29 |
| 0.159 | 3 | 1.018 | 1.029 | 1.08 |
| 0.210 | 4 | 1.024 | 1.034 | 0.98 |
| cellobiose+water64 | ||||
| 0.148 | 3 | 1.016 | 1.030 | 1.38 |
| 0.202 | 4 | 1.022 | 1.035 | 1.27 |
| 0.271 | 5 | 1.031 | 1.040 | 0.87 |
| gentiobiose+water64 | ||||
| 0.146 | 3 | 1.016 | 1.030 | 1.38 |
| 0.205 | 4 | 1.023 | 1.036 | 1.27 |
| 0.270 | 5 | 1.030 | 1.041 | 1.07 |
| melibiose+water64 | ||||
| 0.144 | 3 | 1.015 | 1.030 | 1.48 |
| 0.200 | 4 | 1.022 | 1.036 | 1.37 |
| 0.255 | 5 | 1.029 | 1.041 | 1.17 |
| maltose+water65 | ||||
| 1.465 | 29 | 1.144 | 1.141 | −0.26 |
| 1.717 | 34 | 1.163 | 1.156 | −0.60 |
| 2.626 | 52 | 1.216 | 1.203 | −1.07 |
| Average | 0.85 | |||
Disaccharide conformational properties in aqueous solution
Karplus-type equations have previously been developed for both vicinal 13C-1H and 13C-13C spin-couplings to relate the observed three-bond coupling constant values 3 J to dihedral angle values in the glycosidic linkage (see Methods).57–59 Such equations provide a means of comparing the conformational properties of the glycosidic linkage in MD simulations of disaccharides in water to NMR observables. Consequently, they can be used to help validate the dihedral parameters developed using gas-phase MP2/cc-pVTZ//MP2/6-31G(d) QM data from model compounds when applied to full disaccharides in aqueous solution.
The selection of disaccharides for MD J-coupling studies was based on the availability of experimental data as well as the desire to test all the different dihedral parameters. As described above, separate glycosidic dihedral parameters were developed for 1→1 linkages (model compounds 4–6; “Category 1”), for 1→2, 1→3, and 1→4 linkages (model compounds 7–10; “Category 2”), and for 1→6 linkages (model compounds 2–3 and 11–12; “Category 3”). Accordingly, disaccharides were chosen that fell into each of these three categories. α,α-trehalose was used to test Category 1 linkage parameters, maltose and cellobiose for Category 2 linkage parameters, and melibiose and gentiobiose for Category 3 parameters. In addition to the availability of recent NMR J-coupling data for these molecules,62,63 the tested Category 2 and 3 disaccharides are representative of both α and β linkages, which is important given that the same dihedral parameters are used for a given type of linkage regardless of the chiralities at the ring carbon atoms involved in the linkage. While Category 2 encompasses 1→2 and 1→3 linkages as well as 1→4 linkages, the 1→4 linked molecules maltose and cellobiose were chosen to represent this category because of their biological significance. Although the set of disaccharides does cover all of the dihedral parameters developed for glycosidic linkages based on the conformational properties of model compounds 2–12, it is worth noting that the configuration at the carbon on the reducing-end monosaccharide involved in the glycosidic linkage in all of these disaccharides is equatorial, reflecting a lack of available experimental data.
The referenced experimental glycosidic 3 JCOCH coupling constants for trehalose, maltose, cellobiose, melibiose, and gentiobiose span the range of 2.36 to 4.75 Hz (Table 13). Values for 3 JCOCH calculated from the MD simulations are representative of this range and correlate well with the experimental data. Interestingly, depending on whether Eqn. 7 or Eqn. 8 was used to convert the MD conformational data to 3 JCOCH, the calculated values for a given dihedral in a given simulation differ by as much as 0.85 Hz (cellobiose 3 J(C1OlinkCn’Hn’)). Furthermore, the variability of the calculated values can lead to underestimation when using one equation and overestimation when using the other, as is the case for both 3 J(H1C1OlinkCn’) and 3 J(C1OlinkCn’Hn’) for maltose. Readers interested in further detail are directed to Ref. 61, Table 1, which summarizes both older and more recent experimental and computed 3 JCOCH values, and shows that for e.g. maltose, experimental data can vary by as much as 2.2 Hz and computed data by 2.4 Hz depending on the source. In the case of 3 JCOCC values, the experimental data are limited to cellobiose and gentiobiose (Table 14). The reference data range from ≤0.5 up to 3.0 Hz, and both the low and the high 3 JCOCC are faithfully reproduced by the simulations. The overall closeness of the calculated data to the experimental data across all compounds, especially taking into account the inherent uncertainties introduced by using empirical equations to convert observed MD conformational data to NMR 3 J values, suggests the force field parameters are properly reproducing the true aqueous solution conformational behavior of this variety of glycosidic linkages.
Table 13.
3JCOCH coupling constants (Hz).
| disaccharide | linkage | 3J (H1C1OlinkCn’) | 3J (C1OlinkCn’Hn’) | ||||
|---|---|---|---|---|---|---|---|
| Expt.62,a | MD Eqn. 7 |
MD Eqn. 8 |
Expt.62,a | MD Eqn. 7 | MD Eqn. 8 | ||
| trehalose | α(1→1)α | 3.19 (0.05) | 2.54 | 2.72 | |||
| maltose | α(1→4) | 4.12 (0.2) | 3.87 | 4.44 | 4.68 (0.1) | 4.35 | 5.08 |
| cellobiose | β(1→4) | 4.08 (0.04) | 3.15 | 3.51 | 4.75 (0.25) | 4.83 | 5.69 |
| melibiose | α(1→6) | 3.1 (0.4) | 2.39 | 2.52 | 2.9 (0.4) | 1.78/2.04b | 1.75/2.10 b |
| gentiobiose | β(1→6) | 2.68 (0.08) | 2.50 | 2.67 | 2.36 (0.06) | 1.87/2.01 b | 1.88/2.05 b |
Experimental errors are in parentheses.
Data are for both protons on C6’.
Table 14.
3JCOCC coupling constants (Hz).
| disaccharide | linkage |
3J C2C1OlinkCn’) |
3J (C1OlinkCn’Cn- 1’) |
3J (C1OlinkCn’Cn+1’) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Expt.63 | MD Eqn. 9 |
MD Eqn. 10 |
Expt.63 | MD Eqn. 9 |
MD Eqn. 10 |
Expt.63 | MD Eqn. 9 |
MD Eqn. 10 |
||
| trehalose | α(1→1)α | 3.40 | 3.43 | n/a | n/a | |||||
| maltose | α(1→4) | 2.57 | 2.62 | 0.54 | 0.57 | 2.18 | 2.23 | |||
| cellobiose | β(1→4) | 3.0 | 3.04 | 3.08 | ≤0.5 | 0.70 | 0.76 | 2.0 | 1.63 | 1.70 |
| melibiose | α(1→6) | 3.42 | 3.45 | 3.35 | 3.37 | n/a | n/a | |||
| gentiobiose | β(1→6) | 3.0 | 3.40 | 3.42 | 2.6 | 3.30 | 3.33 | n/a | n/a | n/a |
Conclusions
The present work extends the developing CHARMM all-atom additive force field for carbohydrates19,20 to hexopyranose glycosides. The newly-developed and validated parameters allow for the modeling of all possible 1→1, 1→2, 1→3, 1→4, and 1→6 linkages between hexopyranoses, as well as O-methylation at the C1 position. The parameter development protocol and the force field functional form are consistent not only with recent CHARMM carbohydrate parametrizations,19,20 but also with the CHARMM all-atom additive force fields for proteins, nucleic acids, and lipids; the parameter set is therefore an extension of the CHARMM all-atom additive biomolecular force field. Given the comprehensive coverage of glycosidic linkages and the compatability with other CHARMM biomolecular force fields, the presented force field is expected be of utility for the simulation of linear, cyclic, and branched hexopyranose glycosides in solution, including in heterogeneous systems that include proteins, lipids, and/or nucleic acids.
The present work has focused on the development of a highly-optimized force field for the glycosidic linkages between hexopyranoses, with validation focusing on disaccharide crystalline intramolecular geometries and unit cell parameters, solution densities, and conformational properties in aqueous solution. Preliminary data on dynamic properties such as diffusion coefficients and NMR relaxation rates show promise with regard to experimental results, and a thorough analysis of simulated dynamic properties and existing experimental data is under preparation.
Supplementary Material
Acknowledgements
This work was supported by NIH GM070855 (ADM) and F32CA119771 (OG). The authors acknowledge computer time and resources from the National Cancer Institute Advanced Biomedical Computing Center, Department of Defense High Performance Computing, and the Pittsburgh Supercomputing Center.
Footnotes
Supplementary Material
Supplementary material includes three figures and force field parameters. This information is available free of charge via the Internet at http://pubs.acs.org. The force field topologies and parameters in CHARMM readable format may be obtained from the web site of ADM at http://mackerell.umaryland.edu.
References
- 1.Rao VSR, Qasba PK, Balaji PV, Chandrasekaran R. Conformation of Carbohydrates. Amsterdam: Harwood Academic Publishers; 1998. [Google Scholar]
- 2.Melberg S, Rasmussen K. Carbohydr. Res. 1979;69:27. [Google Scholar]
- 3.Ha SN, Madsen LJ, Brady JW. Biopolymers. 1988;27:1927. doi: 10.1002/bip.360271207. [DOI] [PubMed] [Google Scholar]
- 4.Homans SW. Biochemistry. 1990;29:9110. doi: 10.1021/bi00491a003. [DOI] [PubMed] [Google Scholar]
- 5.Grootenhuis PDJ, Haasnoot CAG. Mol. Simul. 1993;10:75. [Google Scholar]
- 6.Glennon TM, Zheng YJ, Legrand SM, Shutzberg BA, Merz KM. J. Comput. Chem. 1994;15:1019. [Google Scholar]
- 7.Woods RJ, Dwek RA, Edge CJ, Fraserreid B. J. Phys. Chem. 1995;99:3832. [Google Scholar]
- 8.Kouwijzer M, Grootenhuis PDJ. J. Phys. Chem. 1995;99:13426. [Google Scholar]
- 9.Reiling S, Schlenkrich M, Brickmann J. J. Comput. Chem. 1996;17:450. [Google Scholar]
- 10.Ott KH, Meyer B. J. Comput. Chem. 1996;17:1068. [Google Scholar]
- 11.Senderowitz H, Still WC. J Org Chem. 1997;62:1427. [Google Scholar]
- 12.Momany FA, Willett JL. Carbohydr. Res. 2000;326:194. doi: 10.1016/s0008-6215(00)00042-2. [DOI] [PubMed] [Google Scholar]
- 13.Eklund R, Widmalm G. Carbohydr. Res. 2003;338:393. doi: 10.1016/s0008-6215(02)00503-7. [DOI] [PubMed] [Google Scholar]
- 14.Lii JH, Chen KH, Allinger NL. J. Comput. Chem. 2003;24:1504. doi: 10.1002/jcc.10271. [DOI] [PubMed] [Google Scholar]
- 15.Vergoten G, Mazur I, Lagant P, Michalski JC, Zanetta JP. Biochimie. 2003;85:65. doi: 10.1016/s0300-9084(03)00052-x. [DOI] [PubMed] [Google Scholar]
- 16.Lins RD, Hunenberger PH. J. Comput. Chem. 2005;26:1400. doi: 10.1002/jcc.20275. [DOI] [PubMed] [Google Scholar]
- 17.Lii JH, Chen KH, Johnson GP, French AD, Allinger NL. Carbohydr. Res. 2005;340:853. doi: 10.1016/j.carres.2005.01.032. [DOI] [PubMed] [Google Scholar]
- 18.Kirschner KN, Yongye AB, Tschampel SM, González-Outeiriño J, Daniels CR, Foley BL, Woods RJ. J. Comput. Chem. 2008;29:622. doi: 10.1002/jcc.20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guvench O, Greene SN, Kamath G, Brady JW, Venable RM, Pastor RW, MacKerell AD., Jr J. Comput. Chem. 2008;29:2543. doi: 10.1002/jcc.21004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hatcher ER, Guvench O, MacKerell AD., Jr J. Chem. Theory. Comput. 2009;5:1315. doi: 10.1021/ct9000608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacKerell AD, Jr, Brooks B, Brooks CL, III, Nilsson L, Roux B, Won Y, Karplus M. CHARMM: The energy function and its paramerization with an overview of the program. In: Schleyer PvR, Allinger NL, Clark T, Gasteiger J, Kollman PA, Schaefer HF, III, Schreiner PR., editors. Encyclopedia of Computational Chemistry. Vol. 1. Chichester: John Wiley & Sons; 1998. p. 271. [Google Scholar]
- 22.MacKerell AD., Jr J. Comput. Chem. 2004;25:1584. doi: 10.1002/jcc.20082. [DOI] [PubMed] [Google Scholar]
- 23.Guvench O, MacKerell AD., Jr . Comparison of protein force fields for molecular dynamics simulations. In: Kukol A, editor. Molecular Modeling of Proteins. New Jersey: Humana Press, Inc.; 2008. p. 63. [DOI] [PubMed] [Google Scholar]
- 24.MacKerell AD, Jr, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiórkiewicz-Kuczera J, Yin D, Karplus M. J. Phys. Chem. B. 1998;102:3586. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 25.MacKerell AD, Jr, Feig M, Brooks CL., III J. Comput. Chem. 2004;25:1400. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 26.MacKerell AD, Jr, Wiórkiewicz-Kuczera J, Karplus M. J. Am. Chem. Soc. 1995;117:11946. [Google Scholar]
- 27.Foloppe N, MacKerell AD., Jr J. Comput. Chem. 2000;21:86. [Google Scholar]
- 28.MacKerell AD, Jr, Banavali NK. J. Comput. Chem. 2000;21:105. [Google Scholar]
- 29.Schlenkrich M, Brinkman J, MacKerell AD, Jr, Karplus M. An empirical potential energy function for phospholipids: criteria for parameter optimization and applications. In: Merz KM, Roux B, editors. Membrane Structure and Dynamics. Boston: Birkhauser; 1996. p. 31. [Google Scholar]
- 30.Feller SE, Yin DX, Pastor RW, MacKerell AD., Jr Biophys. J. 1997;73:2269. doi: 10.1016/S0006-3495(97)78259-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yin DX, MacKerell AD., Jr J. Comput. Chem. 1998;19:334. [Google Scholar]
- 32.Feller SE, MacKerell AD., Jr J. Phys. Chem. B. 2000;104:7510. [Google Scholar]
- 33.Feller SE, Gawrisch K, MacKerell AD., Jr J. Am. Chem. Soc. 2002;124:318. doi: 10.1021/ja0118340. [DOI] [PubMed] [Google Scholar]
- 34.Vorobyov I, Anisimov VM, Greene S, Venable RM, Moser A, Pastor RW, MacKerell AD., Jr J. Chem. Theory. Comput. 2007;3:1120. doi: 10.1021/ct600350s. [DOI] [PubMed] [Google Scholar]
- 35.Guvench O, MacKerell AD., Jr J. Phys. Chem. A. 2006;110:9934. doi: 10.1021/jp0623241. [DOI] [PubMed] [Google Scholar]
- 36.Woodcock HL, Moran D, Pastor RW, MacKerell AD, Jr, Brooks BR. Biophys. J. 2007;93:1. doi: 10.1529/biophysj.106.099986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. J. Comput. Chem. 1983;4:187. [Google Scholar]
- 38.Brooks BR, Brooks CL, III, MacKerell AD, Jr, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. J. Comput. Chem. 2009;30:1545. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Allen MP, Tildesley DJ. Computer Simulation of Liquids. Oxford: Oxford University Press; 1987. [Google Scholar]
- 40.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J. Chem. Phys. 1983;79:926. [Google Scholar]
- 41.Durell SR, Brooks BR, Ben-Naim A. J. Phys. Chem. 1994;98:2198. [Google Scholar]
- 42.Ryckaert JP, Ciccotti G, Berendsen HJC. J. Comput. Phys. 1977;23:327. [Google Scholar]
- 43.Steinbach PJ, Brooks BR. J. Comput. Chem. 1994;15:667. [Google Scholar]
- 44.Darden T, York D, Pedersen L. J. Chem. Phys. 1993;98:10089. [Google Scholar]
- 45.Hockney RW. The potential calculation and some applications. In: Alder B, Fernbach S, Rotenberg M, editors. Methods in Computational Physics. Vol. 9. New York: Academic Press; 1970. p. 136. [Google Scholar]
- 46.Nosé S. Mol. Phys. 1984;52:255. [Google Scholar]
- 47.Hoover WG. Phys. Rev. A. 1985;31:1695. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 48.Feller SE, Zhang YH, Pastor RW, Brooks BR. J. Chem. Phys. 1995;103:4613. [Google Scholar]
- 49.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA, Vreven T, Jr, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda K, Kitao O, Nakai H, Klene M, Li TW, Knox JE, Hratchian HP, Cross JB, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA. Gaussian 03. Revision B.04 ed. Pittsburgh, PA: Gaussian, Inc.; 2003. [Google Scholar]
- 50.Møller C, Plesset MS. Phys. Rev. 1934;46:618. [Google Scholar]
- 51.Hariharan PC, Pople JA. Theor. Chim. Acta. 1973;28:213. [Google Scholar]
- 52.Scott AP, Radom L. J. Phys. Chem. 1996;100:16502. [Google Scholar]
- 53.Pulay P, Fogarasi G, Pang F, Boggs JE. J. Am. Chem. Soc. 1979;101:2550. [Google Scholar]
- 54.Dunning TH. J. Chem. Phys. 1989;90:1007. [Google Scholar]
- 55.MacKerell AD, Jr, Karplus M. J. Phys. Chem. 1991;95:10559. [Google Scholar]
- 56.Guvench O, MacKerell AD., Jr J. Mol. Model. 2008;14:667. doi: 10.1007/s00894-008-0305-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tvaroska I, Hricovíni M, Petráková E. Carbohydr. Res. 1989;189:359. [Google Scholar]
- 58.Cloran F, Carmichael I, Serianni AS. J. Am. Chem. Soc. 1999;121:9843. doi: 10.1021/ja002525x. [DOI] [PubMed] [Google Scholar]
- 59.Bose B, Zhao S, Stenutz R, Cloran F, Bondo PB, Bondo G, Hertz B, Carmichael I, Serianni AS. J. Am. Chem. Soc. 1998;120:11158. [Google Scholar]
- 60.Capon B. Chem. Rev. 1969;69:407. [Google Scholar]
- 61.Allen FH. Acta Crystallogr. Sect. B-Struct. Sci. 2002;58:380. doi: 10.1107/s0108768102003890. [DOI] [PubMed] [Google Scholar]
- 62.Cheetham NWH, Dasgupta P, Ball GE. Carbohydr. Res. 2003;338:955. doi: 10.1016/s0008-6215(03)00069-7. [DOI] [PubMed] [Google Scholar]
- 63.Olsson U, Serianni AS, Stenutz R. J. Phys. Chem. B. 2008;112:4447. doi: 10.1021/jp710977k. [DOI] [PubMed] [Google Scholar]
- 64.Galema SA, Hoiland H. J. Phys. Chem. 1991;95:5321. [Google Scholar]
- 65.Lourdin D, Colonna P, Ring SG. Carbohydr. Res. 2003;338:2883. doi: 10.1016/j.carres.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 66.Humphrey W, Dalke A, Schulten K. J. Mol. Graph. 1996;14:33. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.