Abstract
Dynamic post-translational modifications (PTMs) regulate and diversify protein properties and cellular behaviors. Real-time monitoring of these modifications has been made possible with biosensors based on fluorescent proteins (FPs) and fluorescence resonance energy transfer (FRET), which can provide spatiotemporal information of PTMs with little perturbation to the cellular environment. In this review, we highlight available fluorescent biosensors applicable to detect PTMs in living cells and how they have shed light on biological questions that have been difficult to address otherwise. In addition, we also provide discussions about various engineering strategies for overcoming potential challenges associated with the development and application of such biosensors.
Introduction
Protein post-translational modifications (PTMs) are the chemical modifications that occur on specific amino acid(s) of a protein after it is being translated, including covalent attachment of phosphate, alkyl groups, carbohydrates, polypeptides, and others. A few well-known examples of PTMs are phosphorylation, methylation, glycosylation and ubiquitination. These modifications serve the important purpose of increasing a protein's functional diversity by altering its basic physical and chemical properties such as its structure, activity, stability, cellular localization and interaction profile [1]. In fact, the very same protein can carry out distinct functions depending on its specific modifications [2]. As a result, a limited number of genes can be regulated in such a way to generate seemingly far more complex cellular behaviors.
Much of our knowledge about PTMs has come from studies employing analytical methods such as in vitro biochemical assays, immunofluorescence microscopy and mass spectrometry [3,4]. While these conventional methods have provided important information regarding the identifications and regulations of various PTMs, in general, they lack the ability to track real-time dynamics of PTMs in single living cells. However, it is believed that such dynamic regulation is essential to their biological functions [5]. Therefore, it is important to develop methods and tools that offer such capability. In recent years, direct visualization of a variety of PTM events in living cells has been made possible by the development of fluorescent biosensors based on fluorescent proteins (FPs) and fluorescence resonance energy transfer (FRET). These genetically encoded biosensors have enabled us to better understand the dynamic regulation of protein behaviors by various PTMs in their native context, the living cell.
FRET-based biosensors for post-translational modification dynamics
FRET is the nonradiative transfer of energy from an excited donor fluorophore to an acceptor fluorophore in its proximity [6]. For FRET to occur, the emission spectrum of the donor fluorophore should overlap with the excitation spectrum of the acceptor fluorophore and the two fluorophores must be in close proximity (<10 nm). Importantly, FRET efficiency is highly sensitive to the distance and/or orientation between the donor and acceptor fluorophores [7]. For FRET-based biosensors discussed here, two FPs are chosen so that the emission spectrum of the donor FP sufficiently overlaps with the excitation spectrum of the acceptor FP, for example cyan FP (CFP) and yellow FP (YFP). Furthermore, all these biosensors have been designed to achieve a FRET change via either a conformational change (for intramolecular or unimolecular biosensors) or a distance change (for intermolecular or bimolecular biosensors) [7,8•]. The change in FRET is then used as a readout for the change in PTM dynamics. While there are many types of PTMs, currently available biosensors (Table 1) are limited to only a few, including phosphorylation/dephosphorylation, methylation, ubiquitination, and glycosylation.
Table 1.
A list of biosensors that detect dynamics of different PTMs.
| Currently available biosensors to detect dynamics of different PTMs | ||
|---|---|---|
| Biosensor name | References | |
| Glycosylation | ||
| β-O-GlcNAcylation | 17 | |
| Methylation | ||
| Histone K9/K27 methylation | K9/K27 reporter | 16 |
| Phosphorylation (kinase/phosphatase activity reporters) | ||
| Protein Kinase A (PKA) | AKAR | 9• |
| Protein Kinase B (PKB/Akt) | BKAR, AktAR | 9•,10 |
| Protein Kinase C (PKC) | CKAR | 9•,27 |
| Protein Kinase D (PKD) | DKAR | 9• |
| Extracellular Signal Regulated Kinase (ERK) | ERKUS, EKAR | 9•,12 |
| Crk II adaptor protein | Picchu | 9• |
| Src, EGFR | 9• | |
| Src | 11 | |
| Aurora B Kinase | 14•• | |
| Histone H3 phosphorylation | 13 | |
| Phosphorylation | Phocus | 9• |
| Calcineurin activity reporter | CaNAR | 15 |
| Ubiquitination | ||
| β-Arrestin ubiquitination | 19 | |
Protein phosphorylation is the covalent attachment of a phosphate group, for example at the serine amino acid residue, to a target protein. This modification is carried out by a specialized family of enzymes known as protein kinases, which are involved in every aspect of signal transduction. Therefore, understanding of the spatiotemporal regulation of protein kinases in living cells holds the key to deciphering the logic and language of signal transduction. Toward this end, FRET-based kinase activity reporters for various serine/threonine and tyrosine kinases have been developed for monitoring their phosphorylation dynamics in living cells [9•-14••]. The modular design of a typical kinase activity reporter consists of two FPs (Figure 1a), a target substrate for the specific kinase of interest and a phopho-amino acid binding domain that serves as an interaction partner of the phopho-substrate to induce a conformational change upon substrate phosphorylation. On the other hand, dephosphorylartion is carried out by another family of enzymes known as protein phosphatases that often work alongside protein kinases to control the phosphorylation state of their substrates in a cellular context. Recently, a FRET-based biosensor has been developed to specifically track the activity of the Ca2+/calmodulin-dependent serine/threonine phosphatase, calcineurin, in living cells [15]. This reporter design utilizes a phosphatase activity-dependent molecular switch based on a specific substrate of calcineurin, NFAT, sandwiched between CFP and YFP, and should be generally applicable to other protein phosphatases.
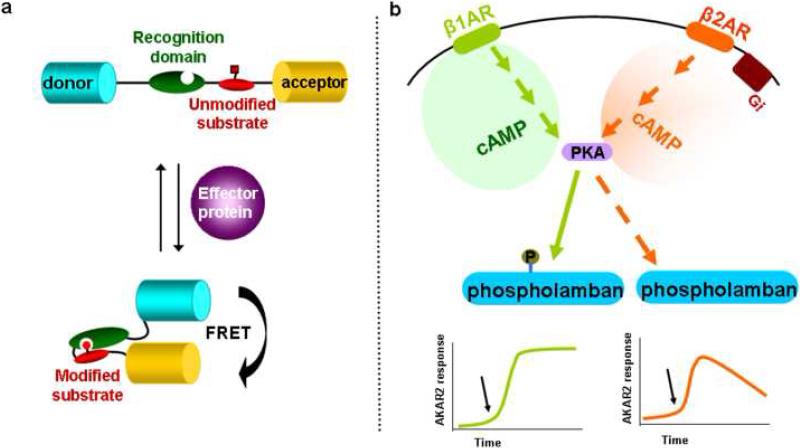
Figure 1. General biosensor design and its application in living cells.
(a) A schematic construction of unimolecular FRET-based biosensors for post-translational modification. The modular components are a FRET-donor and acceptor pair, a substrate domain that is subject to modification and a recognition domain that can recognize the particular post-translational modification. PTM dynamics can be measured by changes in FRET. (b) Temporal dynamics of PKA phosphorylation observed by PKA activity reporter, AKAR2. Black arrow indicates the activation of respective receptors.
Protein methylation is the covalent attachment of a methyl group, for example at the lysine amino acid residue of a histone protein, to a target protein. In particular, methylation of the N-terminal tails of histones is involved in transcriptional regulation and can be quite dynamic. To monitor the dynamics of histone methylation in living cells, a couple of FRET-based fluorescent reporters have been constructed using residues 1−13 (K9 methylation) or 24−35 (K27 methylation) of histone H3 as the substrate and HP1 chromodomain or Polycomb chromodomain as the methylation recognition domain for the K9 and K27 methylation, respectively [16]. These reporters take advantage of the same modular design employed by the FRET-based kinase activity reporters since, like phosphorylation, methylation occurs on specific substrates and is recognized by specific binding domains.
O-GlcNAcylation, a type of protein glycosylation by which the β-N-acetylglucosamine (O-GlcNAc) attaches to the serine/threonine residue of a target protein, is a dynamic modification that plays an important role in signal transduction. To visualize the O-GlcNAcylation dynamics in living cells, a FRET-based biosensor has been developed using a substrate for O-GlcNAc transferase derived from casein kinase II and an O-GlcNAc-binding domain, GafD [17]. On the other hand, O-GlcNAcylation has been shown to compete for the same modification sites with protein kinases and thereby negatively regulate kinase signaling [18]. As such, biosensors like this should provide unique opportunities to dissect the dynamic interplay between O-GlcNAcylation and phosphorylation in living cells.
Additionally, a biosensor for ubiquitination, a modification that attaches a small protein called ubiquitin to a target protein, has been developed based on bioluminescence resonance energy transfer (BRET), a process similar to FRET but using bioluminescence instead [19].
Biological insights
Importantly, applications of these fluorescent biosensors have enabled us to gain biological insights into the spatiotemporal regulation of various PTMs in single living cells. Highlighted in the following paragraphs are a few recent examples.
In cardiac myocytes, cAMP/PKA signaling via β-adrenergic receptors (β-ARs) represents a key mechanism for controlling cardiac contraction under neurohormonal stimulation. Upon activation by cAMP, PKA phosphorylation of multiple proteins targets, including phospholamban and activated receptors, lead to enhancement of myocyte contraction as well as desensitization of activated β-ARs [20, 21]. Given the many protein targets involved in this process and their distinct functions as well as subcellular confinement, it is important to understand how signaling specificity from PKA to its respective substrates is achieved. Using A-kinase activity reporter (AKAR), which can dynamically monitor the level of PKA substrate phosphorylation by providing a corresponding FRET signal [22], it was found that in both wild-type and β2-AR-KO cardiac myocytes, activation of β-ARs lead to sustained PKA activation that results in an increase in phospholamban phosphorylation and thus enhanced myocyte contraction [23••]. In contrast, transient cAMP accumulation has been reported under similar conditions [24,25]. Furthermore, selective stimulation of β2-AR induces transient PKA activation that is sufficient to phosphorylate activated receptors on plasma membrane but not phospholamban, which is on the sarcoplasmic reticulum, suggesting spatiotemporal control of PKA activation can lead to differential substrate phosphorylation (Figure 1b). Together, these findings have provided answers to two important outstanding questions with regard to β-AR signaling in cardiac myocytes: 1. Although it is well-accepted that β-AR-stimulated cAMP signaling is responsible for the enhancement of myocyte contraction, there exists an apparent lack of temporal correlation between the transient cAMP accumulation and the more sustained contraction responses; 2. While stimulation of β2-AR induces significant cellular cAMP accumulation, it has little effect on the myocyte contraction responses.
Aurora B kinase is a key mitotic regulator that plays multiple roles during cell division, including ensuring proper chromosome attachment to the mitotic spindle [26]. However, little has been known about the connection between its sptiotemporal regulation and its diverse functional roles. In a recent study, aurora B signaling patterns in living cells has been quantitatively analyzed using a FRET-based aurora B kinase activity reporter [14••]. It was found that reporters targeted to different subcellular locations showed site-dependent aurora B activity kinetics during anaphase. In addition, the kinetics of the chromosome-targeted reporters was similar to that of chromosome segregation, suggesting a correlation between aurora B activity and chromosome position. Indeed, further analysis of reporter signal with regard to centromere position at time points early in anaphase revealed a striking spatial phosphorylation gradient with the highest phosphorylation level at the spindle midzone. This phosphorylation gradient due to midzone activation of aurora B has been proposed to provide important spatial information for mitotic events.
Ubiquitination is the PTM that enables rapid protein degradation. It is a highly dynamic and regulated process that also plays a prominent role in a variety of other cellular signaling processes [27]. Thus, being able to visualize the real-time dynamics of ubiquitination should lead to better understanding of how ubiquitination regulates various cellular functions. To study the ubiquitination dynamics of β-arrestin, an important regulator of G-protein coupled receptor (GPCR) signaling, a BRET-based ubiquitination biosensor has been developed. This bimolecular biosensor consists of a luciferasetethered ubiquitin substrate, β-arrestin, and a GFP-tethered ubiquitin, where luciferase and GFP serve as energy donor and energy acceptor, respectively [19]. Using this biosensor, it was shown in living cells that activation of β2-AR produces a transient β-arrestin ubiquitination, whereas selective activation of another GPCR, V2-vasopressin receptor, leads to a sustained β-arrestin ubiquitination. In addition, parallel tracking of β-arrestin recruitment to the activated GPCRs using a similar BRET-based approach revealed a tight correlation between the β-arrestin ubiquitination and its recruitment to the activated receptors. Together, these findings suggest that the distinct kinetic changes of β-arrestin ubiquitination with regard to GPCR signaling are indeed determined by its interaction profiles with the receptors [28].
Discussions
Most PTMs involved in cellular signaling are very dynamic and the signals are often encoded in relative changes in the PTM state of their target substrates. Therefore, it is imperative that PTM biosensors have sufficient dynamic range to track the physiologically relevant changes of PTM dynamics which can be relatively small. Furthermore, improved dynamic range should make high-throughput monitoring of PTM dynamics in living cells possible and enables efficient screening for specific small molecule regulators of PTMs [29]. In turn, these potential regulators should help elucidate the molecular mechanisms underlying the dynamic regulation of various PTMs. Moreover, these regulators can be potentially useful for correcting dysregulated PTM signaling. Several practical strategies have been successfully applied to the improvement of various existing biosensors [22,30]. Among these, the most successful one to date is the change of FPs used as the FRET pair in the biosensor, including using FPs with improved spectral properties for FRET and circular permutated FPs, since FRET efficiency depends on the spectral properties of the donor and acceptor fluorophores and is highly sensitive to the relative orientation between them.
Not only PTMs are dynamic, often they are also associated with spatially defined signaling patterns. To be able to visualize such spatially regulated signaling, genetically encoded biosensors can be targeted to subcellular regions of interest via specific amino acid targeting sequences. For example, while the chromosome-targeted aurora B kinase activity reporters revealed a midzone phosphorylation gradient during anaphase, the untargeted version of the reporter failed to do so [14••]. In another example, targeting C-kinase activity reporter (CKAR) to the plasma membrane where protein kinase C (PKC) is activated, revealed oscillatory phosphorylation in HeLa cells in response to histamine [31]. In contrast, untargeted CKAR not only did not detect phosphorylation oscillation, it actually did not show discernable signal at all under the same condition.
In addition, PTMs do not happen in isolation in a cellular context. Rather, a single signaling process may involve multiple PTMs [32]. Furthermore, multiple PTMs, for example, O-GlcNAcylation and phosphorylation, can compete for the same modification site on a particular protein substrate [18,33]. Thus, it is advantageous to study the dynamics of related PTMs in parallel, allowing for precise spatiotemporal correlation of these PTMs in the same cellular context. Toward this end, recent advances in FP engineering have afforded us with a rainbow of FPs [34•], making it possible to employ spectrally distinct FRET pairs for parallel tracking of multiple events in a single living cell [35•]. A recent progress in multiparameter imaging is the development of a single-excitation dual-FRET method that allows precise parallel tracking of fast signal dynamics in cells undergoing significant morphological changes [36].
So far, only a limited number of PTMs can be dynamically tracked with fluorescent biosensors. This is, in part, due to the lack of specific information about many PTMs, including substrate specificity, localization pattern, interaction profile and regulation. Such information can be obtained from traditional methods such as in vitro biochemical assays, immunofluorescence microscopy and mass spectrometry. However, to accelerate the development process, alternative methods and tools are needed to yield information more relevant to the development of biosensors for particular PTMs. For example, FP-based fragment complementation assay can provide information about protein-protein interactions in living cells [37], thereby facilitating simultaneous selection of a specific target substrate and a corresponding recognition domain: two important modular components in a FRET-based biosensor for detecting PTM dynamics. Similarly, FRET- or BRET-based assays can also be developed to provide such information. In a recent study, selective recognition of acetylated histones by bromodomain proteins in living cells has been determined employing a FRET-based flow cytometry method [38]. Specifically, cells expressing different combinations of FP-tagged (CFP and YFP, respectively) bromodomain and histone proteins were analyzed for FRET signals between CFP and YFP in order to determine the specific interactions. This may be an important step toward the development of a biosensor for dynamic histone acetylation.
Conclusion
In conclusion, FRET-based fluorescent biosensors have served as important tools in real-time tracking of spatiotemporal dynamics of PTMs in living cells. These genetically encoded biosensors offer precise molecular targeting, as well as high spatiotemporal resolution in living cells. In addition, the very same advantages can be extended to more complex living systems such as tissues and whole organisms. Therefore, continuing developments of biosensor for other PTMs along with better engineering strategies to allow for improved dynamic range and simultaneous monitoring should lead to better understanding of the complex molecular mechanisms underlying these dynamic signaling processes in living systems.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wold F. In Vivo Chemical modification of proteins (Post-translational modification). Ann Rev Biochem. 1981;50:783–814. doi: 10.1146/annurev.bi.50.070181.004031. [DOI] [PubMed] [Google Scholar]
- 2.Walsh CT, Garneau-Tsodikova S, Gatto GJ. Protein posttranslational modifications: The chemistry of proteome diversifications. Angew Chem Int Ed Engl. 2005;44:7342–7372. doi: 10.1002/anie.200501023. [DOI] [PubMed] [Google Scholar]
- 3.Hoffman MD, Sniatynski MJ, Kast J. Current approaches for global post-translational modification discovery and mass spectrometric analysis. Anal Chim Acta. 2008;627:50–61. doi: 10.1016/j.aca.2008.03.032. [DOI] [PubMed] [Google Scholar]
- 4.Reinders J, Sickmann A. Modificomics: Posttranslational modifications beyond protein phosphorylation and glycosylation. Biomol Eng. 2007;24:169–177. doi: 10.1016/j.bioeng.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 5.Seet BT, Dikic I, Zhou M, Pawson T. Reading protein modifications with interaction domains. Nat Rev Mol Cell Biol. 2006;7:473–483. doi: 10.1038/nrm1960. [DOI] [PubMed] [Google Scholar]
- 6.Lakowicz JR. Principles of Fluorescence Spectroscopy. Springer; 2006. Energy transfer. pp. 443–472. [Google Scholar]
- 7.Ananthanarayanan B, Ni Q, Zhang J. Molecular Sensors Based on Fluorescence Resonance Energy Transfer to Visualize Cellular Dynamics. Methods Cell Biol. 2008;89:37–57. doi: 10.1016/S0091-679X(08)00602-X. [DOI] [PubMed] [Google Scholar]
- •8.VanEngelenburg SB, Palmer AE. Fluorescent biosensors of protein functions. Curr Opin Chem Biol. 2008;12:60–65. doi: 10.1016/j.cbpa.2008.01.020. [This recent review covers available genetically encoded fluorescent biosensors based on various biosensor platforms as well as applications of these biosensors in understanding localized signaling in living cells and in high throughput screening.] [DOI] [PubMed] [Google Scholar]
- •9.Zhang J, Allen MD. FRET-based biosensors for protein kinases: illuminating the kinome. Mol BioSyst. 2007;3:759–765. doi: 10.1039/b706628g. [This is a recent review covering the genetically-encodable, FRET-based kinase biosensors.] [DOI] [PubMed] [Google Scholar]
- 10.Gao X, Zhang J. Spatiotemporal analysis of differential Akt regulation in plasma membrane microdomains. Mol Biol Cell. 2008;19:4366–4373. doi: 10.1091/mbc.E08-05-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ouyang M, Sun J, Chien S, Wang Y. Determination of hierarchical relationship of Src and Rac at subcellular locations with FRET biosensors. Proc Natl Acad Sci USA. 2008;105:14353–14358. doi: 10.1073/pnas.0807537105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harvey CD, Ehrhardt AG, Cellurale C, Zhong H, Yasuda R, Davis RJ, Svoboda K. A genetically encoded fluorescent sensor of ERK activity. Proc natl Acad Sci. 2008;105:19264–19269. doi: 10.1073/pnas.0804598105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin CW, Ting AY. A genetically encoded fluorescent reporter of histone phosphorylation in living cells. Angew Chem Int Ed Engl. 2004;43:2940–2943. doi: 10.1002/anie.200353375. [DOI] [PubMed] [Google Scholar]
- ••14.Fuller BG, Lampson MA, Foley EA, Rosasco-Nitcher S, Le KV, Tobelmann P, Brautigan DL, Stukenberg PT, Kapoor TM. Midzone activation of aurora B in anaphase produces an intracellular phosphorylation gradient. Nature. 2008;453:1132–1136. doi: 10.1038/nature06923. [This paper describes an interesting observation of an anaphase phosphorylation gradient at the spindle midzone using a FRET-based aurora B kinase activity reporter. This phosphorylation gradient is proposed to provide important spatial cues for various mitotic events.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Newman RH, Zhang J. Visualization of phosphatase activity in living cells with a FRET-based calcineurin activity sensor. Mol Biosyst. 2008;4:496–501. doi: 10.1039/b720034j. [DOI] [PubMed] [Google Scholar]
- 16.Lin CW, Jao CY, Ting AY. Genetically encoded fluorescent reporters of histone methylation in living cell. J Am Chem Soc. 2004;126:5982–5983. doi: 10.1021/ja038854h. [DOI] [PubMed] [Google Scholar]
- 17.Carrillo LD, Krishnamoorthy L, Mahal LK. A cellular FRET-based sensor for β-O-GlcNAc, a dynamic carbohydrate modification involved in signaling. J Am Chem Soc. 2006;128:14768–14769. doi: 10.1021/ja065835+. [DOI] [PubMed] [Google Scholar]
- 18.Vosseller K, Wells L, Lane MD, Hart GW. Elevated nucleocytoplasmic glycosylation by O-GlcNAc results in insulin resistance associated with defects in Akt activation in 3T3-L1 adipocytes. Proc Natl Acad Sci USA. 2002;99:5313–5318. doi: 10.1073/pnas.072072399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perroy J, Pontier S, Charest PG, Aubry M, Bouvier M. Real-time monitoring of ubiquitination in living cells by BRET. Nat Methods. 2004;1:203–208. doi: 10.1038/nmeth722. [DOI] [PubMed] [Google Scholar]
- 20.Xiang Y, Kobilka BK. Myocyte Adrenoceptor Signaling Pathways. Science. 2003;300:1530–1532. doi: 10.1126/science.1079206. [DOI] [PubMed] [Google Scholar]
- 21.Lefkowitz RJ. Seven transmembrane receptors: something old, something new. Acta Physiologica. 2007;190:9–19. doi: 10.1111/j.1365-201X.2007.01693.x. [DOI] [PubMed] [Google Scholar]
- 22.Zhang J, Hupfeld CJ, Taylor SS, Olefsky JM, Tsien RY. Insulin disrupts beta-adrenergic signalling to protein kinase A in adipocytes. Nature. 2005;437:569–573. doi: 10.1038/nature04140. [DOI] [PubMed] [Google Scholar]
- ••23.Soto D, Arcangelis VD, Zhang J, Xiang Y. Dynamic protein kinase A activities induced by beta-adrenoceptors dictate signaling propagation for substrate phosphorylation and myocyte contraction. Circ Res. 2009;104:770–779. doi: 10.1161/CIRCRESAHA.108.187880. [Using an activity reporter for PKA, differential regulation of PKA activities by different β-AR subtypes has been visualized in living cardiac myocytes. Findings from this study have shed light on distinct biological consequences due to spatiotemporal regulation of cAMP/PKA signaling in cardiac myocytes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiang Y, Naro F, Zoudilova M, Jin S-C, Conti M, Kobilka B. Phosphodiesterase 4D is required for β2 adrenoceptor subtype-specific signaling in cardiac myocytes. Proc Natl Acad Sci USA. 2005;102:909–914. doi: 10.1073/pnas.0405263102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Violin JD, DiPilato LM, Yildirim N, Elston TC, Zhang J, Lefkowitz RJ. {beta}2-Adrenergic receptor signaling and desensitization elucidated by quantitative modeling of real time cAMP dynamics. J Biol Chem. 2008;283:2949–2961. doi: 10.1074/jbc.M707009200. [DOI] [PubMed] [Google Scholar]
- 26.Kelly AE, Funabiki H. Correcting aberrant kinetochore microtubule attachments: an Aurora B-centric view. Curr Opin Cell Biol. 2009;21:51–58. doi: 10.1016/j.ceb.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 28.Shenoy SK, Lefkowitz RJ. Trafficking patterns of beta-arrestin and G protein-coupled receptors determined by the kinetics of beta-arrestin deubiquitination. J Biol Chem. 2003;278:14498–14506. doi: 10.1074/jbc.M209626200. [DOI] [PubMed] [Google Scholar]
- 29.Allen MD, DiPilato LM, Rahdar M, Ren YR, Chong C, Liu JO, Zhang J. Reading dynamic kinase activity in living cells for high-throughput screening. ACS Chem Biol. 2006;1:371–376. doi: 10.1021/cb600202f. [DOI] [PubMed] [Google Scholar]
- 30.Nagai T, Yamada S, Tominaga T, Ichikawa M, Miyawaki A. Expanded dynamic range of fluorescent indicators for Ca2+ by circularly permuted yellow fluorescent proteins. Proc Natl Acad Sci USA. 2004;101:10554–10559. doi: 10.1073/pnas.0400417101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Violin JD, Zhang J, Tsien RY, Newton AC. A genetically encoded fluorescent reporter reveals oscillatory phosphorylation by protein kinase C. J Cell Biol. 2003;161:899–909. doi: 10.1083/jcb.200302125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang X. Multisite protein modification and intramolecular signaling. Oncogene. 2004;24:1653–1662. doi: 10.1038/sj.onc.1208173. [DOI] [PubMed] [Google Scholar]
- 33.Liu F, Iqbal K, Grundke-Iqbal I, Hart GW, Gong C. O-GlcNAcylation regulates phosphorylation of tau: A mechanism involved in Alzheimer's disease. Proc Natl Acad Sci USA. 2004;101:10804–10809. doi: 10.1073/pnas.0400348101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •34.Shaner NC, Patterson GH, Davidson MW. Advances in fluorescent protein technology. J Cell Sci. 2007;120:4247–4260. doi: 10.1242/jcs.005801. [This chronologic review provides detailed information about different FP variants and highlights emerging technologies in engineering optical tools using FPs.] [DOI] [PubMed] [Google Scholar]
- •35.Carlson HJ, Campbell RE. Genetically encoded FRET-based biosensors for multiparameter fluorescence imaging. Curr Opin Biotechnol. 2009;20:19–27. doi: 10.1016/j.copbio.2009.01.003. [This is a comprehensive review of multiparameter imaging using different FRET pairs.] [DOI] [PubMed] [Google Scholar]
- 36.Nino Y, Hotta K, Oka K. Simultaneous live cell imaging using dual FRET sensors with a single excitation light. Plos ONE. 2009 doi: 10.1371/journal.pone.0006036. doi:10.1371/journal.pone.0006036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fang D, Kerppola TK. Ubiquitin-mediated fluorescence complementation reveals that Jun ubiquitinated by Itch/AIP4 is localized to lysosomes. Proc Natl Acad Sci. 2004;101:14782–14787. doi: 10.1073/pnas.0404445101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kanno T, Kanno Y, Siegel RM, Jang MK, Lenardo MJ, Ozato K. Selective recognition of acetylated histones by bromodomain proteins visualized in living cells. Mol Cell. 2004;13:33–43. doi: 10.1016/s1097-2765(03)00482-9. [DOI] [PubMed] [Google Scholar]