Summary
miR-24, up-regulated during terminal differentiation of multiple lineages, inhibits cell cycle progression. Antagonizing miR-24 restores post-mitotic cell proliferation and enhances fibroblast proliferation, while over-expressing miR-24 increases the G1 compartment. The 248 mRNAs down-regulated upon miR-24 over-expression are highly enriched for DNA repair and cell cycle regulatory genes that form a direct interaction network with prominent nodes at genes that enhance (MYC, E2F2, CCNB1, CDC2) or inhibit (p27Kip1, VHL) cell cycle progression. miR-24 directly regulates MYC and E2F2 and some genes they transactivate. Enhanced proliferation from antagonizing miR-24 is abrogated by knocking down E2F2, but not MYC, and cell proliferation, inhibited by miR-24 over-expression, is rescued by miR-24-insensitive E2F2. Therefore, E2F2 is a critical miR-24 target. The E2F2 3′UTR lacks a predicted miR-24 recognition element. In fact, miR-24 regulates expression of E2F2, MYC, AURKB, CCNA2, CDC2, CDK4 and FEN1 by recognizing seedless, but highly complementary, sequences.
Introduction
microRNAs (miRNA) regulate key steps of cell differentiation and development by suppressing gene expression in a sequence-specific manner (Bartel, 2009). In mammals, the active strand miRNA sequence (typically ∼22 base pairs) is partially complementary to binding sites in the 3′UTR of genes, often with full complementarity to 7 or 8 nucleotides in the “seed region” (residues 2-9) of the miRNA. Gene suppression in mammals is thought to occur primarily by inhibiting translation (Olsen and Ambros, 1999). However, miRNAs in mammals also cause mRNA decay (Chang et al., 2007; Lim et al., 2005; Johnson et al., 2007); recent reports (Baek et al., 2008; Selbach et al., 2008) suggest that reduced protein is frequently associated with decreased mRNA.
miR-24 is consistently up-regulated during terminal differentiation of hematopoietic cell lines into a variety of lineages (Lal et al., 2009). miR-24 is also up-regulated during thymic development to naïve CD8 T cells (Neilson et al., 2007) and during muscle and neuronal cell differentiation (Sun et al., 2008; Fukuda et al., 2005). miR-24 is encoded with miR-23 and miR-27 in 2 duplicated gene clusters. One cluster (miR-23b, miR-27b, miR-24-1) is within a chromosome 9 EST and the other (miR-23a, miR-27a, miR-24-2) is in a chromosome 19 intergenic region. Both miR-24 genes are processed to the same active strand. Disruption or changes in expression of both sites have been linked to CLL prognosis (Calin et al., 2005). Because miR-24 is up-regulated in diverse cell types during terminal differentiation, we sought to identify its function and the target genes it regulates.
Common approaches to identify miRNA target genes are (1) bioinformatic algorithms that predict potential target genes that contain conserved 3′UTR sequences complementary to a seed region at the 5′-end of the miRNA active strand (Doench and Sharp, 2004; Lewis et al., 2005), (2) analysis of mRNAs that are down-regulated when a miRNA is over-expressed (Chang et al., 2007; Johnson et al., 2007; Lim et al., 2005), and (3) identifying mRNAs enriched in co-immunoprecipitates with tagged Argonaute or GW182 proteins in cells over-expressing the miRNA (Easow et al., 2007; Zhang et al., 2007). The bioinformatic approach is hampered by the fact that the existing algorithms have a high margin of error (most predicted genes are not real targets and some key targets, such as RAS for let-7, are not predicted (Johnson et al., 2005)). The utility of the biochemical approach involving Argonaute proteins for genome-wide target identification of miRNAs is still unclear since Argonaute over-expression globally increases miRNA levels, perhaps obscuring the effect of an individual over-expressed miRNA (Diederichs and Haber, 2007). Since miRNA-mediated mRNA degradation and protein down-regulation often occur together (Baek et al., 2008), identifying the mRNAs which decrease when a miRNA is over-expressed, might identify many of its targets. Although some bona fide miR-24 targets that are primarily regulated by translation will be missed by this approach and other down-regulated genes may not be directly regulated, this strategy has been successfully used to identify targets of some mammalian miRNAs, including miR-124 and miR-1 (Lim et al., 2005), miR-34a (Chang et al., 2007) and let-7 (Johnson et al., 2007). We therefore applied this approach to identify the genes regulated by miR-24 in HepG2 cells that express low levels of miR-24 and combined it with bioinformatics to uncover miR-24 regulated pathways. We find that miR-24 regulates a network of genes that control cell cycle progression and DNA repair (Lal et al., 2009). Over-expressing miR-24 increases the G1 population and reduces DNA replication, while antagonizing miR-24 increases cell proliferation, which can be rescued by knocking down E2F2, suggesting that E2F2 is a key miR-24 target gene. MYC and other genes important in cell cycle regulation that are transcriptionally regulated by MYC and E2Fs (AURKB, BRCA1, CCNA2, CDC2, CDK4, FEN1) are also direct miR-24 targets by luciferase assay. Of note, E2F2 and most of these genes lack 3′UTR miR-24 seed match sequences. However, miR-24 regulates these genes by base-pairing to “seedless” 3′UTR MREs with extensive base-pairing elsewhere in the sequence.
Results
miR-24 is up-regulated during hematopoietic differentiation
To understand the role of miRNAs during terminal differentiation, miRNA expression was analyzed by microarray in 2 human leukemia cell lines - K562 cells differentiated to megakaryocytes using 12-O-tetradecanoylphorbol-13-acetate (TPA) or to erythrocytes with hemin, and HL60 cells differentiated to macrophages using TPA or to monocytes using vitamin D3. miR-24 was one of only 6 miRNAs that was consistently up-regulated in all 4 systems of terminal differentiation (Lal et al., 2009). The other uniformly up-regulated miRNAs were 3 other members of the miR-24 clusters (miR-23a, miR-23b, miR-27a), miR-22 and miR-125a. miR-24 was the most up-regulated of these miRNAs. We therefore focused on miR-24, which we hypothesized might regulate terminal differentiation in multiple cell lineages. qRT-PCR confirmed the induction of miR-24 during differentiation of these hematopoietic cells (Fig. 1A) with the highest up-regulation in K562 cells treated with TPA. The mature miR-24 transcript increased 2- to 8-fold during differentiation into megakaryocytes, erythrocytes, macrophages, monocytes and granulocytes. Expression of the chromosome 19 miR-24 cluster primary transcript, encoding miR-23a, miR-27a and miR-24, increased in both cell lines within 6 h of TPA treatment, peaked at ∼12 hr and remained elevated for at least 2 d (Suppl. Fig.1A,B), suggesting that the observed increase in mature miR-24 was due to increased transcription. Up-regulation of the Dicer-cleaved mature miRNA was slightly delayed, becoming significant at 12-16 hr (Suppl. Fig. 1C,D). Mature miR-24 levels remained elevated for as long as was measured (4 d).
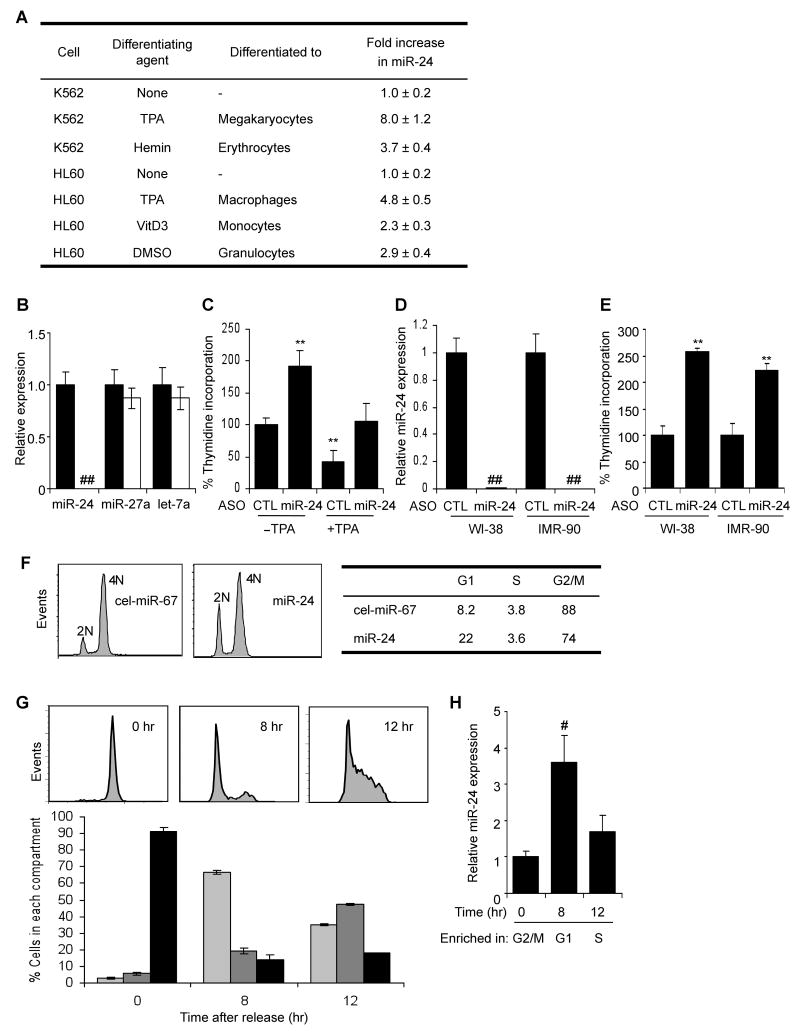
Fig. 1.
miR-24 is up-regulated during hematopoietic cell differentiation and inhibits cell proliferation (A) miR-24 expression, measured using qRT-PCR relative to untreated cells, increases in K562 cells differentiated to megakaryocytes or erythrocytes and HL60 cells differentiated to macrophages, monocytes and granulocytes. Differentiation in all experiments was verified by cell surface phenotype. Mature miR-24 levels were determined by qRT-PCR and normalized to U6. (B) miR-24 knockdown in K562 cells specifically decreases miR-24, assayed by qRT-PCR in cells transfected with miR-24 ASO (white) relative to control ASO (black). Expression relative to U6 snRNA is normalized to control cells. (C) miR-24 knockdown with ASO increases K562 cell proliferation measured by thymidine uptake, both in the presence and absence of TPA. The decline in proliferation with TPA is completely restored by antagonizing miR-24. (D, E) Knocking down miR-24 in WI-38 or IMR-90 cells also significantly increases proliferation as measured by thymidine incorporation 48 hr post-transfection. miR-24 knockdown by ASO measured by qRT-PCR is normalized to U6 snRNA. Error bars in (B-E) represent standard deviation from 3 experiments (**, p<0.01; ##, p<0.001). (F) miR-24 over-expression increases the G1 compartment in HepG2 cells. HepG2 cells transfected with miR-24 or control mimic for 48 hr were stained with propidium iodide and analyzed by flow cytometry. Representative analysis of 3 independent experiments is shown. (G) K562 cells, synchronized in G2/M by nocodazole and then released, were analyzed by flow cytometry. A representative experiment is shown in the top panel and the mean (±SD) percentage of cells in each phase of the cell cycle (from 3 independent experiments) below (G1, light grey; S, dark grey; G2/M, black). (H) qRT-PCR analysis of miR-24 (normalized to U6 snRNA) from the partially synchronized K562 cells in (G) shows that miR-24 is most highly expressed in G1 and declines as cells progress to S and G2/M phase. (#, p<0.005).
miR-24 inhibits cellular proliferation by increasing the G1 compartment
Since cessation of cell proliferation is a hallmark of terminal differentiation, we first examined whether proliferation is altered by either inhibiting or enhancing miR-24 function by transfecting cells with miR-24 2′-OMe antisense oligonucleotide (ASO) or miRNA mimics, respectively. When K562 cells were transfected with miR-24 ASO, miR-24 was dramatically and specifically reduced by qRT-PCR 36 hr later (Fig. 1B). DNA replication, measured by thymidine incorporation, doubled in cells transfected with miR-24 ASO compared to cells transfected with control ASO (Fig. 1C). When K562 cells were differentiated with TPA for 4 hr, thymidine incorporation declined by 60%. However, in cells transfected with miR-24 ASO and treated with TPA, thymidine uptake was indistinguishable from that of the control ASO-transfected, but TPA untreated, cells (Fig. 1C). Therefore, miR-24 ASO fully restored proliferation to differentiating K562 cells. To examine whether miR-24 also inhibits cell proliferation in nontransformed cells, we next antagonized miR-24 in early passage WI-38 and IMR-90 normal diploid fibroblasts. Antagonizing miR-24 in WI-38 and IMR-90 cells dramatically reduced miR-24 (Fig. 1D) and increased thymidine uptake >2-fold 48 hr after transfection (Fig. 1E). Conversely, over-expressing miR-24 in HepG2 cells synchronized with nocodazole, which typically leads to mitotic arrest and only ∼8% of cells in G1, increased G1 cells 3-fold (22%; miR-24 vs cel-miR-67, p<0.001) (Fig. 1F).
We next analyzed how miR-24 expression changes during normal cell cycle progression using K562 cells released at various times from nocodazole treatment, which synchronized them in G2/M (Bar-Joseph et al., 2008; O'Donnell et al., 2005) (Fig. 1G,H). Before release, 90% of cells were in G2/M; 8 hr later 65% were in G1 and 12 hr after removing nocodazole, 45% were in S phase. miR-24 was low in G2/M, increased >3-fold by 8 hr when most cells were in G1 and then declined by 12 hr as cells progressed into S phase. These results suggest that miR-24 is most highly expressed in G1. Taken together with our finding that cells transduced with miR-24 mimics accumulate in G1, these results suggest that miR-24 regulates cell cycle progression mostly by blocking or delaying the G1/S transition.
Most genes down-regulated by miR-24 contain a miR-24 seed in their 3′UTR
We next sought to identify miR-24-regulated targets and cellular pathways by comparing mRNA microarrays of cells transfected with miR-24 or control miRNA (cel-miR-67) mimic. Transfecting HepG2 cells, which have low endogenous miR-24 levels (Fig. 2A), with a miR-24 mimic increased miR-24 expression ∼80-fold compared with control cells (Fig. 2B). Total RNA, isolated from duplicate miRNA-transfected samples 48 hr later, was amplified, labeled and hybridized to Illumina mRNA microarrays. 248 mRNAs were down-regulated at least 2-fold by miR-24 over-expression (Z-ratio>1.5) (Suppl. Table 1). We validated the microarray data by performing qRT-PCR amplification for 9 randomly chosen down-regulated genes that spanned the range of significantly down-regulated genes (Z-ratios, 1.6-6.1), of which 6 (CNDP2, TOP1, PER2, MBD6, H2AFX, STX16) were predicted miR-24 targets by TargetScan 4.2 and 3 (UBD, BCL2L12, ZNF317) were not (Fig. 2C). H2AFX was previously shown to be directly regulated by miR-24 (Lal et al., 2009). All 9 genes were significantly down-regulated, and the extent of down-regulation correlated well with their reduced expression in the microarray, validating the quality of the microarray data.
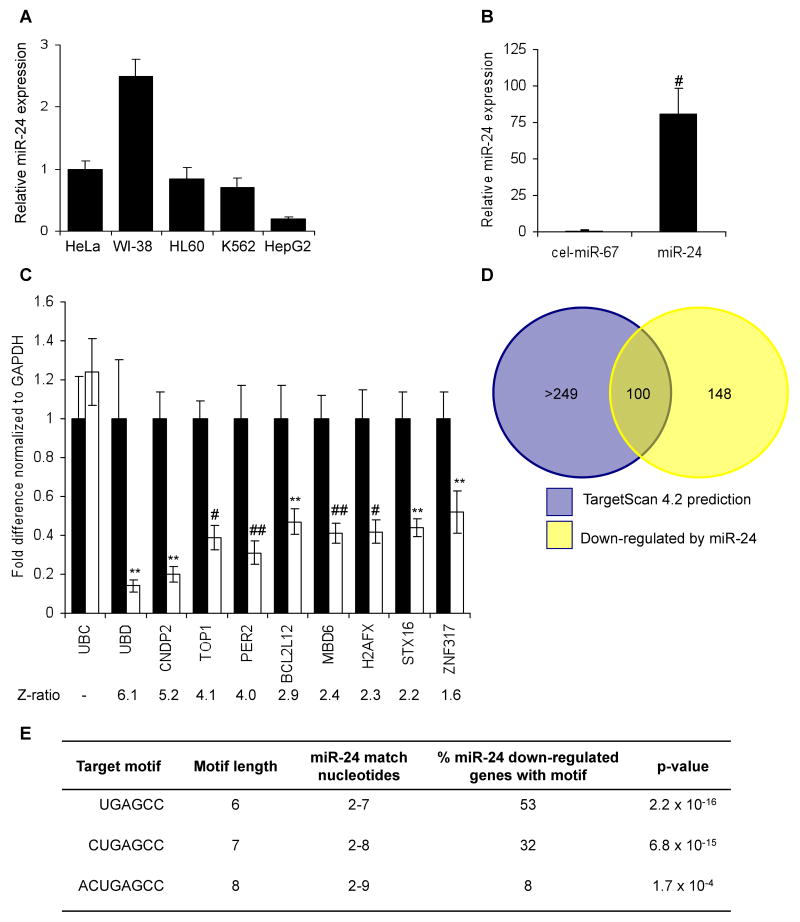
Fig. 2.
mRNA down-regulation after miR-24 over-expression (A) HepG2 cells express low levels of miR-24, assayed by qRT-PCR analysis normalized to U6, compared to HeLa, WI-38, HL60 and K562 cells. (B) Effective increase in miR-24 in HepG2 cells 48 hr after transfection with miR-24 mimic compared to cel-miR-67-transfected control cells. #, p<0.005. (C) Genes identified by microarray as down-regulated by miR-24 over-expression were confirmed to be down-regulated by qRT-PCR normalized to GAPDH. UBC is a housekeeping gene. Cells were transfected with cel-miR-67 (black) or miR-24 mimic (white). **, p<0.01; #, p<0.005, ##, p<0.001. The down-regulated genes are graphed in order of their down-regulation on the microarray; the Z-ratio of the microarray analysis is shown below. Error bars represent SD from 3 independent experiments (A, B and C). (D) Venn diagram of genes down-regulated by miR-24 in HepG2 cells and genes predicted to be regulated by miR-24 using TargetScan 4.2. Of the 100 predicted genes, whose mRNA is also significantly down-regulated, only 20 have conserved predicted miR-24 recognition sites. TargetScan 4.2 predicts 349 conserved miR-24 targets and many more that are not conserved. (E) Sites complementary to the miR-24 seed are enriched in the 3′UTR of down-regulated transcripts. The table shows the frequency of perfect hexamer (positions 2-7), heptamer (positions 2-8) and octamer (positions 2-9) miR-24 3′UTR seeds in the down-regulated genes.
To determine what fraction of the down-regulated genes are likely direct targets of miR-24, we used two approaches. First, we compared our experimental list of down-regulated transcripts with TargetScan predictions. 100 down-regulated genes were also predicted by TargetScan 4.2 (Lewis et al., 2003) (Fig. 2D, Suppl. Table 1). Amongst these 100 targets, however, only 20 have predicted miR-24 3′UTR miRNA recognition sites (MRE) conserved in human, mouse, rat and dog. Second, we examined the frequency of a 3′UTR sequence perfectly complementary to the miR-24 seed (hexamer (positions 2-7), heptamer (positions 2-8) and octamer (positions 2-9)). The down-regulated genes were highly enriched for miR-24 seed matches (Fig. 2E, Suppl. Table 1). Just over half of the 219 miR-24 down-regulated transcripts that have an annotated 3′UTR contain a 3′UTR complementary hexamer sequence (53%, p=2×10-16 relative to the background frequency of the seed in the known transcriptome), 32% have a heptamer match (p=7×10-15) and 8% have an octamer seed match (p=0.0002). This significant enrichment of predicted miR-24 target genes and the high frequency of genes containing perfect seed matches suggest that a substantial proportion of the down-regulated genes may be direct miR-24 targets.
Cell cycle and DNA repair genes are regulated by miR-24
We next performed a gene ontology (GO) analysis (GeneGo, Inc) to identify cellular pathways enriched in the set of genes down-regulated after ectopic miR-24 expression. This analysis did not yield a tightly focused set of functions for miR-24 (Suppl. Table 2). Therefore, we next looked at whether the 100 down-regulated genes, which were also predicted miR-24 targets, are enriched for specific biological processes. A functional enrichment and network analysis revealed statistically significant enrichment for 49 processes, many of which are overlapping (Fig. 3A; Suppl. Table 3). The top 3 most enriched GO processes involve DNA repair (DNA damage checkpoint, double strand break repair by homologous recombination, and recombinational repair; each enriched with significance of p=0.0001). We previously found that miR-24 interferes with the DNA damage response in terminally differentiated hematopoietic cells, predominantly by reducing expression of the histone variant H2AFX, which recruits and retains DNA repair factors at double-strand breaks (Lal et al., 2009). In addition, multiple GO processes involved in cell cycle regulation were also highly enriched (regulation of cell cycle, p=0.0002; DNA integrity checkpoint p=0.0003; cell cycle arrest, p=0.0007; cell cycle, p=0.001; DNA recombination, p=0.001). This was not surprising based on the effect of miR-24 on cell cycle progression. When networks were developed to identify known directly interacting proteins from these over-represented biological processes, there was one cluster of 6 genes centered around MYC (c-myc) and three other small clusters involving 2 or 3 genes (Suppl. Fig. 2A); the other 87 genes in the data set lack any previously annotated direct interactions. Because the TargetScan algorithm might miss some important miRNA-regulated genes, we also constructed a direct interaction network from the 248 down-regulated mRNAs. The direct interaction network constructed from all significantly down-regulated mRNAs was a highly interactive set of 68 interacting genes, many of which are important in cell cycle regulation. The major connected network of miR-24 down-regulated genes is shown in Fig. 3B; there were also some smaller networks (Suppl. Fig. 2B). Key nodes of the major network are MYC (22 interactions), E2F2 (6), VHL (6), CDC2 (6), CCNB1 (5), and CDKN1B (5). The MYC and E2F2 transcription factors play a central role in regulating G1/S transition and progression through S. They inhibit cell differentiation and apoptosis and promote cellular transformation (Bracken et al., 2004; Lebofsky and Walter, 2007). MYC regulates the transcription of other genes in the network, including E2F2, CDKN1B, CCNB1, CDC2, CDCA7, and RRM2. E2F2 also regulates the transcription of other genes in the network, including CDC2, MYC, RRM2 and the minichromosome maintenance proteins MCM4 and MCM10 that are essential for initiating DNA replication. These analyses support our experimental findings that miR-24 regulates cell cycle progression and DNA repair.
Fig. 3.
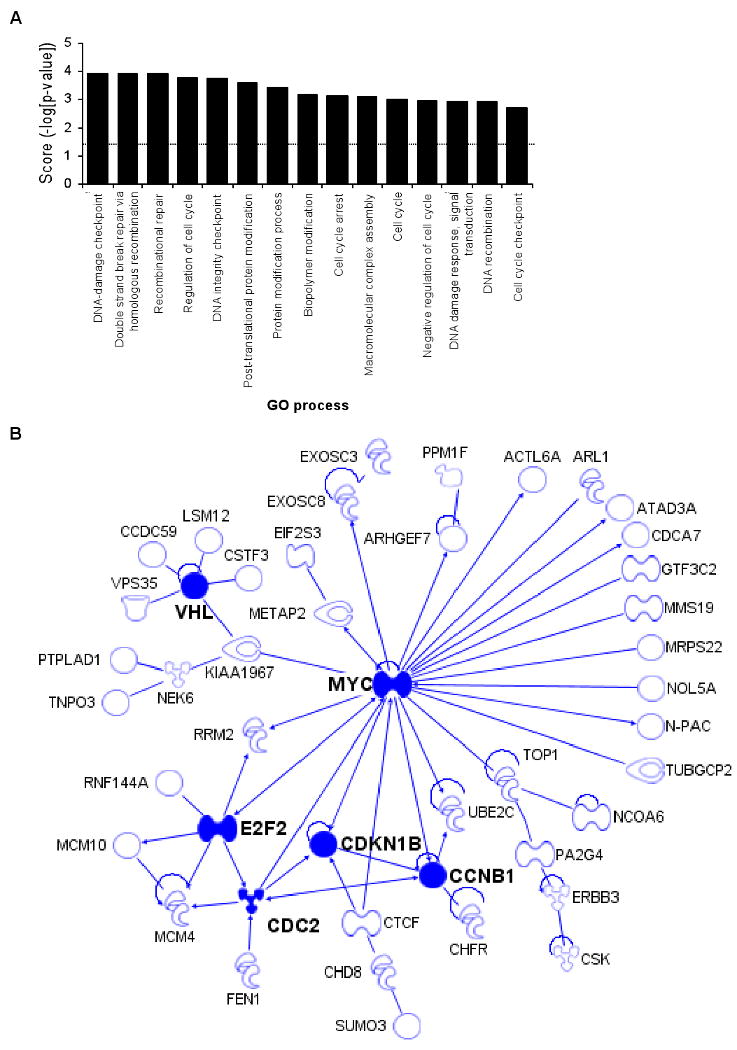
Bioinformatic analysis of miR-24 down-regulated genes suggests that miR-24 regulates cell cycle progression and DNA repair (A) Top 15 over-represented cellular processes for the 100 overlapping genes in Fig. 2D. Over-represented processes are sorted by Score (-log [p-value]). A highly positive score suggests that the subnetwork is highly saturated with genes identified experimentally and doesn't have many nodes not experimentally identified. The complete list of 100 genes in the overlap and further statistical analysis is provided in Suppl. Table 1 and 3. The dotted line represents the statistically significant limit. (B) Major direct interaction network of the 248 genes significantly down-regulated after transfection of HepG2 cells with miR-24 mimics. Nodes with at least 5 interactions (including auto-regulation) are highlighted (symbols: transcription factor,  ; enzyme,
; enzyme,  ; kinase,
; kinase,  ; ligand-dependent nuclear receptor,
; ligand-dependent nuclear receptor,  ; transporter,
; transporter,  ; phosphatase,
; phosphatase, 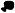 ; peptidase,
; peptidase,  ; translation regulator,
; translation regulator, 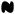 ; other, ●).
; other, ●).
miR-24 regulates MYC by binding to its 3′UTR
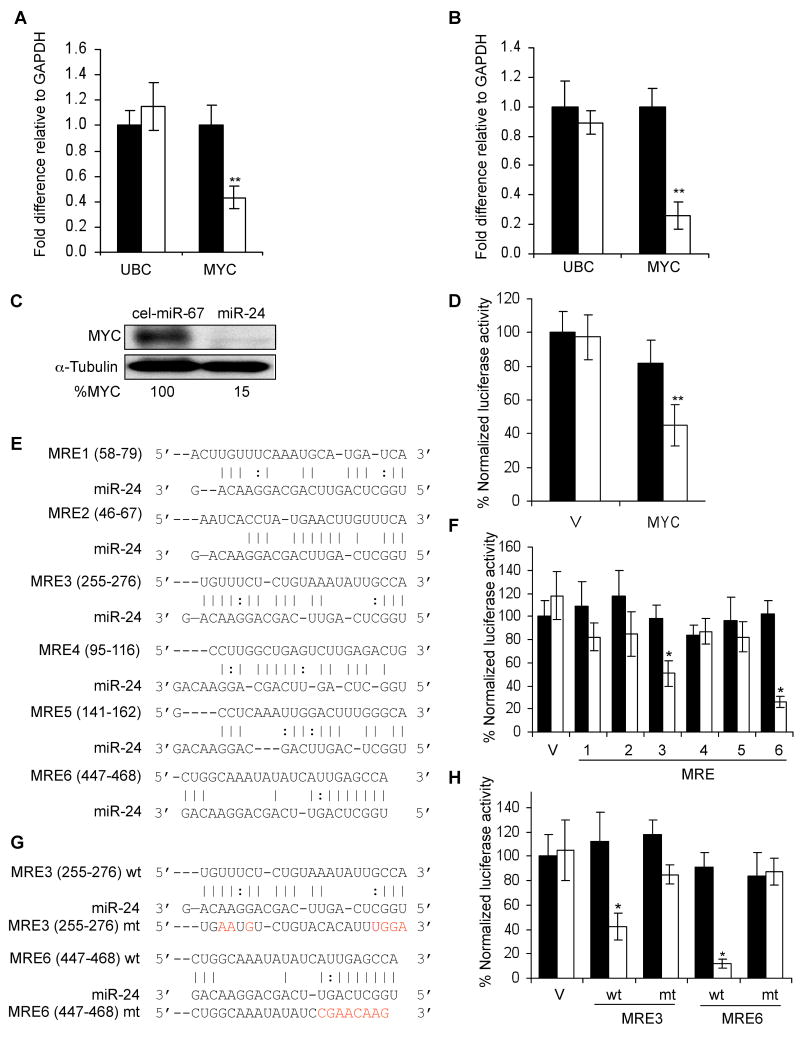
To determine whether miR-24 directly regulates the down-regulated genes, we began with MYC, since it is a key node of the interaction network, plays an important role in cell cycle progression and is a predicted miR-24 target. We transfected HepG2 and K562 cells with miR-24 mimics and 48 hr later, measured MYC mRNA by qRT-PCR. miR-24 over-expression decreased MYC mRNA by ∼2-4 fold relative to GAPDH in HepG2 and K562 cells (Fig. 4A,B). UBC mRNA, a control gene, did not change significantly. MYC protein also decreased substantially (85%) (Fig. 4C). To investigate whether reduced MYC expression was directly mediated by miR-24, we measured changes after miR-24 co-transfection in luciferase activity from a MYC 3′UTR reporter. miR-24 reduced luciferase activity 2.2-fold (Fig. 4D). We next sought to identify miR-24 MREs in the MYC 3′UTR. TargetScan 4.2 predicts a single MRE containing a poorly conserved 7-mer exact seed match at positions 462-468 (although the recent TargetScan 5.0 algorithm does not list MYC as a miR-24 target), while rna22, an algorithm that does not require a seed match (Miranda et al., 2006), identifies 6 potential miR-24 MREs in the 488 nt MYC 3′UTR, including the TargetScan 4.2-predicted MRE (MRE6) (Fig. 4E). miR-24 over-expression specifically and significantly reduced luciferase activity by 1.9- and 3.9-fold for MRE3 and MRE6, respectively, but the other MREs had no significant effect on luciferase activity (Fig. 4F). MRE3 has no seed, but has extensive complementarity with the 3′end of miR-24. Point mutations of MRE3 and MRE6 that disrupted miR-24 binding restored luciferase activity (Fig. 4G,H). These findings suggest that miR-24 binds to 2 partially complementary sites (MRE3 and MRE6) in the MYC 3′UTR.
Fig. 4.
miR-24 regulates MYC expression. miR-24 over-expression in HepG2 (A) or K562 cells (B) decreases MYC mRNA, analyzed by qRT-PCR and normalized to GAPDH (black, cel-miR-67; white, miR-24). (C) MYC protein is decreased in K562 cells upon miR-24 over-expression. Densitometry was used to quantify protein; α-tubulin served as loading control. (D) miR-24 targets the MYC 3′UTR in a luciferase reporter assay. HepG2 cells were transfected with control miRNA (black) or miR-24 (white) mimic for 48 hr and then with MYC 3′UTR-luciferase reporter (MYC) or vector (V) for 24 hr. (E) Predicted binding sites in the MYC 3′UTR for miR-24 (MRE1-6) by rna22. The numbers in parenthesis correspond to the position in the MYC 3′UTR. Perfect matches are indicated by a line; G:U pairs by :. (F-H) miR-24 regulates MRE3 and MRE6 by luciferase reporter assay. HepG2 cells were co-transfected with cel-miR-67 (black) or miR-24 (white) mimics and luciferase reporters containing the wild type (wt) MRE1-6 in (E,F) or mutated (mt) MRE3 and MRE6 in (G,H) or vector (V). Luciferase activity was measured 48 hr after transfection. In (G), red letters denote point mutations that disrupt base pairing. Mean ± SD, normalized to vector control, of 3 independent experiments is shown. *, p<0.05; **, p< 0.01.
miR-24 down-modulates E2F2
We next examined the effect of miR-24 on E2F2, since E2F2 is down-regulated by miR-24 over-expression by microarray, is a key node in the gene interaction network (Fig. 3B), and plays a crucial role in regulating progression through G1, where miR-24-over-expressing cells pile up (Polager and Ginsberg, 2006). qRT-PCR analysis confirmed that E2F2 mRNA was significantly down-regulated by over-expressing miR-24 (Fig. 5A). In addition, the related E2F family members, E2F1 and E2F3, were also significantly decreased, although these two genes were not identified by the less sensitive microarray analysis. E2F1 and E2F3 down-regulation may be secondary to E2F2 down-regulation since the E2F-family of transcription factors regulate each other (Bracken et al., 2004; Vernell et al., 2003) or may be mediated by MYC, since MYC and E2F1 have been shown to transactivate each other (Fernandez et al., 2003). As expected, E2F2 protein (Fig. 5B) was also substantially reduced (9-fold).
Fig. 5.
miR-24 over-expression downregulates E2Fs and some E2F-regulated genes (A) miR-24 significantly reduces mRNA levels of E2F1, E2F2 and E2F3 and of some E2F-target genes (RRM2, CHEK1, CCNA2, FEN1, MCM4, MCM10, CDC2, AURKB), but not BRCA1 and PCNA. CDK4, a key MYC target gene, is also down-regulated. HepG2 cells were transfected with miR-24 (white) or control mimics (black) for 48 hrs, and E2F and their target mRNAs were measured by qRT-PCR. Data normalized to GAPDH are expressed relative to control mimic-transfected cells. UBC is a control housekeeping gene. The Z-ratios refer to the significantly down-regulated mRNAs in the mRNA microarray in miR-24-over-expressing cells. (B) Protein expression of miR-24 target genes is substantially reduced in miR-24 mimic-transfected K562 cells 72 hr after transfection, relative to control miRNA-transfected cells. Densitometry was used to quantify protein relative to α-tubulin. HuR and α-tubulin are loading controls. (C) miR-24 levels significantly increased when K562 cells were transfected with 10 or 50 nM miR-24 mimics as measured by qRT-PCR analysis. Expression relative to U6 snRNA is depicted normalized to control cells. (D) A 4-fold increase in miR-24, obtained by transfecting 10 nM miR-24 mimic, reduces target proteins. Cell lysates of K562 cells, obtained 72 hr after transfection with indicated concentrations of control or miR-24 mimics, were analyzed by immunoblot. α-tubulin and HuR are loading controls. Error bars represent mean ± SD from 3 independent experiments. *, p<0.05, **, p<0.01; #, p<0.005.
miR-24 down-regulates multiple E2F- and MYC-regulated genes
The E2F transcription factors activate the transcription of many genes essential for DNA replication, cell cycle progression and DNA repair. If miR-24 over-expression down-regulates E2F2, E2F2 target gene mRNAs would also be expected to decline after ectopic miR-24 expression. The effect of ectopic miR-24 expression in HepG2 cells on transcripts of 10 E2F targets important for cell cycle progression and DNA repair (AURKB, BRCA1, CCNA2, CDC2, CHEK1, FEN1, PCNA, RRM2, MCM4, MCM10) was analyzed 48 hr later. Eight of the 10 transcripts, with the exception of BRCA1 and PCNA, were significantly reduced (>40%) (Fig. 5A). A subset of these genes (CDC2, MCM4, MCM10, RRM2 and FEN1) was also significantly down-regulated by miR-24 over-expression by microarray (Suppl. Table 1). mRNA microarray may not be sensitive enough to identify some genes whose expression is suppressed, either directly or indirectly, by a miRNA. Protein levels of E2F2 and all 7 miR-24 target genes examined (AURKB, BRCA1, CCNA2, CDC2, CHEK1, FEN1, PCNA), quantified by densitometry of immunoblots, decreased by at least 2-fold (Fig. 5B). A possible explanation for the lack of correlation between the mRNA and protein levels of BRCA1 and PCNA is that mRNA levels were measured 48 hr after transfection of one cell type (HepG2) while protein levels were assayed 72 hr post-transfection in K562 cells. In fact, BRCA1, but not PCNA, mRNA was reduced by ∼2-fold when K562 cells were transfected with miR-24 for 3 d (Suppl. Fig. 5).
Since miR-24 over-expression reduced MYC protein by 85% (Fig. 4C), MYC target genes should also be down-regulated. Consistent with this hypothesis, 11 known MYC-regulated genes (ATAD3A, ACTL6A, ARHGEF7, CCNB1, CDCA7, EXOSC8, E2F2, METAP2, N-PAC, RRM2 and UBE2C) were significantly down-regulated in miR-24 over-expressing HepG2 cells by mRNA microarray (Suppl. Table 1). Among MYC-regulated genes, CDK4 is an important mediator of MYC's effects on cellular proliferation (Hermeking et al., 2000). Although CDK4 mRNA was not significantly altered by microarray, CDK4 mRNA declined ∼4-fold after ectopic expression of miR-24 in HepG2 cells by more sensitive qRT-PCR assay (Fig. 5A) and CDK4 protein became undetectable (Fig. 5B). Therefore, miR-24 over-expression decreases the levels of many genes important in cell cycle progression.
In these experiments, we over-expressed miR-24 ∼80-fold above the level in undifferentiated K562 cells, whereas the physiological increase after TPA treatment of K562 cells is only 8-fold. To determine whether these genes are regulated by a physiological increase in miR-24, these experiments were repeated by transfecting K562 cells with varying miR-24 mimic concentrations (2-50 nM). Transfection of 2 nM miR-24 did not significantly alter miR-24, while 10 and 50 nM miR-24 increased miR-24 levels by 4- and 28-fold, respectively (Fig 5C). E2F2, MYC and 3 of 4 other E2F2-regulated mRNAs (AURKB, CCNA2, H2AX, but not PCNA) (Suppl. Fig. 5) and protein levels of all 7 genes tested (E2F2, AURKB, CHEK1, CCNA2, CDK4, MYC and PCNA) were all significantly reduced by a 4-fold increase in miR-24 (Fig. 5D). Therefore the genes identified by mRNA microarray down-regulated after ectopic miR-24 expression, are likely physiologically relevant direct and/or indirect miR-24 targets.
E2F2 and some E2F-target genes are directly regulated by “seedless” 3′UTR MREs
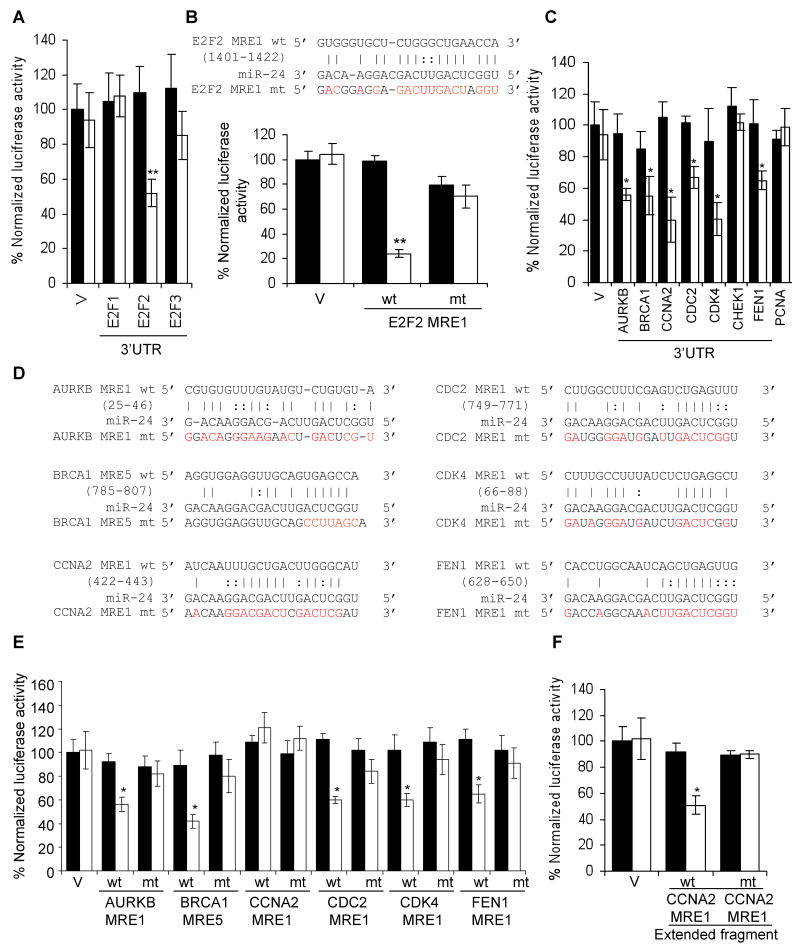
None of the 3 E2F paralogs (E2F1, E2F2 and E2F3), is a predicted target of miR-24 and their 3′UTRs do not contain a miR-24 seed match sequence. We nonetheless tested whether the E2F 3′UTRs might be directly regulated by miR-24 by luciferase assay. The E2F2, but not E2F1 or E2F3, 3′UTR significantly repressed luciferase activity in a miR-24-dependent manner (Fig. 6A) suggesting that E2F2 is a direct miR-24 target. rna22 identified 5 candidate E2F2 3′UTR miR-24 MREs (Fig. 6B, Suppl. Fig. 3). miR-24 significantly suppressed luciferase activity of a reporter gene containing the E2F2 MRE1. E2F2 MRE1 does not have a seed match in the 3′UTR, even if G:U wobbles are allowed, but has extensive complementarity to miR-24 elsewhere. Point mutations that disrupt base pairing between miR-24 and E2F2 MRE1 rescued luciferase expression, verifying that miR-24 specifically recognizes the E2F2 MRE1.
Fig. 6.
E2F2 and multiple E2F-target genes are direct targets of miR-24, recognized by “seedless” MREs. (A) miR-24 silences the expression of luciferase genes engineered with the 3′UTR of E2F2, but not with E2F1 or E2F3 3′UTRs, suggesting that E2F2 is a direct miR-24 target but E2F1 and E2F3 are down-regulated indirectly. Luciferase assays were performed in HepG2 cells over-expressing miR-24 (white) or control mimics (black). (B) miR-24 down-regulates luciferase activity of a reporter gene containing wild-type (wt) E2F2 MRE1. Mutations in the miR-24 pairing residues (mt) rescue luciferase expression (sequences and luciferase assays for candidate E2F2 wt 3′UTR MREs are shown in Suppl. Fig. 3). (C) miR-24 targets the 3′UTR of E2F-regulated genes (AURKB, BRCA1, CCNA2, CDC2, FEN1) and CDK4, a MYC-regulated gene. CHEK1 and PCNA 3′UTRs are not regulated by miR-24. HepG2 cells were co-transfected with a luciferase reporter containing the 3′UTR of the indicated gene and control miRNA (black) or synthetic miR-24 (white) for 48 hr. Expression of the unmodified luciferase vector (V) is unchanged by miR-24. (D) Predicted binding sites in the 3′UTR of genes whose 3′UTR was repressed by miR-24 in (C) and binding site mutations tested (indicated in red). (E) Expression of reporter genes containing wild-type (wt) AURKB MRE1, BRCA1 MRE5, CDC2 MRE1, CDK4 MRE1 and FEN1 MRE1 is significantly reduced upon co-transfection of HepG2 cells with miR-24 mimics (white) and not the control mimic (black). Mutations in the miR-24 pairing residues (mt) rescue luciferase expression (sequences and luciferase assays for all tested wt MREs for these genes in Suppl. Fig. 6,7). The CCNA2 MRE1 is not regulated. (F) However, miR-24 regulates a 181 nt region containing the CCNA2 MRE1 in the luciferase vector. Mutations in the binding residues of CCNA2 MRE1 within the extended sequence restore luciferase activity. Error bars in A, C and E represent mean ± SD from 3 independent experiments. **, p<0.01; *, p<0.05.
To verify further that MYC and E2F2 are direct targets of miR-24, we also looked at changes in expression of luciferase reporter genes 24 hr after transduction of HepG2 cells with miR-24 mimic. At this early time, thymidine uptake of HepG2 cells does not significantly change (Suppl. Fig. 4A), but ectopic miR-24 still suppresses luciferase reporters encoding MYC MRE3 or MRE6 or E2F2 MRE1 (Suppl. Fig. 4B). The identification of “seedless” E2F2 and MYC MREs confirms previous studies showing that MREs lacking a seed with good downstream complementarity can contribute to miRNA gene regulation (Didiano and Hobert, 2006, 2008; Vella et al., 2004).
Since “seedless” MREs contributed to the regulation of E2F2 and MYC by miR-24, we next investigated whether some of the E2F2 and MYC regulated genes, whose transcripts declined in response to miR-24, might also be direct miR-24 targets even though they might lack a predicted MRE. We selected 8 genes (AURKB, BRCA1, CCNA2, CHEK1, CDC2, CDK4, FEN1, PCNA) that play important roles in cell cycle progression and cloned their entire 3′UTRs into the luciferase reporter. The 3′UTR of 6 of 8 genes (AURKB, BRCA1, CCNA2, CDC2, CDK4, FEN1, but not CHEK1 or PCNA) was significantly repressed by miR-24, suggesting that these genes may be direct targets (Fig. 6C). To confirm that these genes are direct miR-24 targets, we next sought to identify the miR-24 MREs that regulate their expression. Among the 6 genes whose 3′UTR was specifically repressed by miR-24, BRCA1 is the only gene that is predicted by TargetScan. The BRCA1 3′UTR contains a nonconserved perfect 7-mer seed match sequence, which functions as a miR-24 MRE by luciferase assay (Fig. 6D,E). To identify potential miR-24 MREs in these E2F2 and MYC-regulated genes, we used the rna22 or PITA algorithms, which allow G:U wobbles or seed mismatches. These algorithms identified 1 candidate MRE for AURKB; 5 for BRCA1 (which included the TargetScan BRCA1 site); and 3 sites each for CCNA2, CDC2, CDK4 and FEN1 (Suppl. Fig. 6). miR-24 significantly repressed luciferase activity of one MRE for 5 of 6 of these reporter genes (AURKB MRE1, BRCA1 MRE5, CDC2 MRE1, CDK4 MRE1, FEN1 MRE1) (Fig. 6D,E; Suppl. Fig. 7). Although CCNA2 MRE1 appeared to be inactive, a longer fragment (181 nucleotides) from the CCNA2 3′UTR (that included only CCNA2 MRE1) significantly repressed luciferase expression in a miR-24-dependent manner when cloned into the luciferase vector 3′UTR (Fig. 6F). Point mutations that disrupt base-pairing between miR-24 and the 5 minimal MREs and the CCNA2 MRE within the extended sequence rescued luciferase expression, verifying that these MREs are regulated by miR-24 (Fig. 6D-F). Therefore we have identified, and verified by mutation, 7 seedless miR-24 MREs in genes important in cell cycle progression.
E2F2 down-regulation is key to miR-24's inhibition of cell proliferation
Because both MYC and E2F2 are important cell cycle progression regulators, we next examined their contributions to the increased cellular proliferation from antagonizing miR-24 by knocking down MYC and/or E2F2 in K562 cells co-transfected with miR-24 ASO (Fig. 7A; Suppl. Fig. 8). Introducing miR-24 ASO into K562 cells doubled thymidine incorporation (as in Fig. 1C). E2F2 knockdown completely abrogated the proliferative effect of miR-24 ASO, but MYC knockdown had no significant effect. Moreover, E2F2 down-regulation by miR-24 is physiologically relevant. When K562 cells were terminally differentiated to megakaryocytes with TPA, the decrease in E2F2 mRNA and protein was completely blocked by inhibiting miR-24 (Fig. 7B,C). Conversely, ectopic expression of miR-24 insensitive E2F2 lacking the 3′UTR restored proliferation to miR-24-treated K562 cells (Fig. 7D,E). Therefore the miR-24 antiproliferative effect is largely mediated by its down-regulation of E2F2.
Fig. 7.
E2F2 is a key miR-24 target gene. (A) Increased cell proliferation from antagonizing miR-24 in K562 cells is blocked by siRNA-mediated knockdown of E2F2, but not MYC. Knockdown is shown by immunoblot (Suppl. Fig. 5). K562 cells were co-transfected with or without miR-24 ASO plus control siRNA or siRNAs targeting E2F2 and/or MYC. The rate of cellular proliferation was determined 72 hr later by thymidine incorporation. Error bars represent mean ± SD from 3 independent experiments. (B,C) Down-regulation of E2F2 mRNA (B) and protein (C) during TPA-mediated differentiation of K562 cells to megakaryocytes is mediated by miR-24 and can be completely inhibited by antagonizing miR-24. K562 cells were transfected with miR-24 or a control (CTL) ASO for 72 hr and then treated with TPA for 6 hr. mRNA was assessed by qRT-PCR normalized to GAPDH and normalized to control cells transfected with CTL ASO. E2F2 protein was quantified by densitometry and normalized to α-tubulin. (D,E) Transfection of K562 cells with a miR-24 mimic reduces cell proliferation, which can be rescued by expressing miR-24-insensitive E2F2, lacking the 3′UTR. K562 cells were co-transfected with a vector expressing HA-tagged E2F2 or GFP and miR-24 or cel-miR-67 (CTL) mimics for 72 hr before measuring thymidine uptake. (E) Immunoblot probed for HA tag. Error bars represent mean ± SD from 3 independent experiments. *, p<0.05, **, p<0.01; #, p<0.005 (F) Model of the miR-24, miR-17∼92, MYC, and E2F network of cell cycle regulators. Here we show that miR-24 directly suppresses expression of MYC and E2F2 (and indirectly suppresses E2F1 and E2F3) and thereby regulates the G1/S transition. Expression of the opposing miRNAs encoded by the miR-17∼92 and miR-106b∼25 clusters that promote cell proliferation is transcriptionally activated by the same transcription factors that miR-24 suppresses (O'Donnell et al., 2005; Petrocca et al., 2008). Therefore miR-24 would be predicted to reduce expression of the proliferation-promoting miRNA clusters indirectly. These miRNAs also knockdown the E2F genes, but probably to fine tune their proliferative effect. MYC may also suppress miR-24 transcription (Gao et al., 2009).
Antagonizing miR-24 elevated MYC protein levels in untreated K562 cells and the down-regulation of MYC mRNA in TPA-treated K562 cells could be partially rescued by antagonizing miR-24 (Suppl. Fig. 9A,B). However, antagonizing miR-24 did not restore MYC protein to differentiating cells, suggesting that although miR-24 suppresses MYC expression, down-regulation of MYC protein during post-mitotic differentiation is also controlled by miR-24-independent changes in protein stability. This may help explain why MYC siRNAs had no significant effect on proliferation of cells transduced with miR-24 ASO (Fig. 7A).
Discussion
miR-24 and its clustered miRNAs are amongst only a handful of miRNAs consistently up-regulated during hematopoietic cell terminal differentiation. Here we show that miR-24 suppresses expression of several key genes that regulate cell cycle progression. Over-expressing miR-24 increases the percentage of cells in the G1 phase, while antagonizing it causes differentiating cells to keep proliferating. The anti-proliferative effect of miR-24 is not restricted to tumor cells (HepG2 and K562 cells) but also occurs in human diploid fibroblasts.
miRNAs can regulate expression of hundreds of genes. Genome-wide analysis of miRNA target genes has been assessed following miRNA over-expression or knockdown for only a handful of miRNAs (Chang et al., 2007; Johnson et al., 2007; Lim et al., 2005). Using this approach for miR-24 enabled us to identify 248 candidate genes that might be either directly or indirectly regulated by miR-24. Of these down-regulated genes, 40% are predicted miR-24-regulated genes by TargetScan and 53% have a 3′UTR hexamer sequence complementary to the miR-24 seed, suggesting that a large proportion of miR-24 down-regulated genes may be direct targets.
To make sense of the set of 248 genes down-regulated by miR-24 over-expression, we used bioinformatics to identify over-represented processes and direct interacting protein networks within this gene set. This type of analysis, which surprisingly has not been applied to understanding miRNA regulation, led to the hypothesis that miR-24 might regulate cell cycle progression during post-mitotic differentiation by targeting MYC and/or E2F2, since they constituted nodes of the major interaction network of the down-regulated gene set. Both MYC and E2F2 are directly regulated by miR-24, but neither of these genes is a predicted miR-24 target. MYC, which has a 3′UTR hexamer seed sequence, is regulated both by a seed-containing MRE and a noncanonical “seedless” MRE. E2F2 lacks any miR-24 seed match. However, E2F2 turned out to be the key gene for miR-24 inhibition of the cell cycle, since over-expressing miR-24-insensitive E2F2 completely restored proliferation.
The GO analysis of miR-24 down-regulated genes also suggested that miR-24 might regulate DNA repair. We recently verified this prediction by showing that over-expression of miR-24 enhances sensitivity to DNA damage (Lal et al., 2009). The key miR-24 target for this biological effect is H2AFX, which has 2 seed-bearing predicted MREs.
An unbiased analysis, which did not filter out genes whose 3′UTR lack seed binding sites, was critical for enabling us to identify E2F2 as the key miR-24 target gene for cell cycle regulation. In addition to MYC and E2F2, we found 5 other miR-24 down-regulated genes whose 3′UTR was inhibited by miR-24 through “seedless” MREs. These genes (AURKB, CCNA2, CDK4, CDC2, FEN1) are also transcriptionally regulated by E2Fs or MYC and play crucial roles in cell cycle progression. Our results suggest that in addition to genes containing miR-24 perfect seed matches, “seedless” MREs are also important. Indeed seedless MREs are critical for miR-24 function since the anti-proliferative effect of miR-24 can be recapitulated by silencing or obliterated by over-expressing the “seedless” E2F2 gene. However, the importance of recognition of “seedless” vs seed-bearing MREs could vary between miRNAs. An assessment of this question could be determined by experimental testing of a large set of randomly chosen genes, whose protein or mRNA is down-regulated by miRNA over-expression or increased by miRNA inhibition. In addition to “seedless” 3′UTR MREs, we previously identified a coding region miR-24 MRE in p16INK4A (Lal et al., 2008). Other recent studies also identified coding region MREs (Duursma et al., 2008; Tay et al., 2008). Taken together, these results suggest that target gene identification might be improved by not disregarding noncanonical MREs.
miR-24 directly regulates not only critical nodes of the interactome of cell cycle regulatory genes, but also genes downstream of these nodes. This multitiered gene regulation may guarantee that cell cycle arrest is not easily evaded. In fact we have preliminary data suggesting that miR-24 may directly regulate many additional periodic genes, including others that lack a canonical seed-bearing MRE. Because expression of many of these genes is suppressed in nondividing cells, we were careful to show that miR-24 mediated gene suppression occurs before miR-24 transduced cells have stopped dividing (Suppl. Fig. 4), so their down-regulation is a cause, not consequence, of cell cycle arrest.
MYC and E2F regulate progression through G1. Regulating the transition to S phase may be the major site of miR-24 action, since miR-24-treated cells accumulate in G1. Since MYC and E2F2 promote each other's transcription, miR-24 may prevent the reciprocal activation of these genes by regulating both of them. The dramatic down-regulation of both proteins in miR-24 over-expressing cells could therefore be a combined effect of post-transcriptional and transcriptional regulation. Other miR-24 targets that are also transcriptionally regulated by MYC or E2F2 are implicated in controlling progression through G1, the G1/S checkpoint, S, and G2/M. For example, over-expressing miR-24 down-regulated mRNA and protein of E2F-regulated CCNA2 and CDC2. CCNA2 binds to and activates CDC2, thereby promoting G1/S and G2/M transition. Although the mRNA for CDK6, which regulates G1 to S transition, was not significantly changed in our microarrays, we previously showed that CDK6 is directly regulated by miR-24 (Lal et al., 2008). CDK6 may be an example of a target gene regulated primarily by translational inhibition. p16INK4A is another direct target of miR-24 that is translationally regulated by miR-24 and therefore not down-regulated in the microarrays (Lal et al., 2008). In addition to genes, which act at the G1/S transition, cells transfected with miR-24 mimics also have decreased expression of genes that principally act at other phases of the cell cycle. Important genes required for DNA replication in S phase were also down-regulated by miR-24, including MCM4 and MCM10 in the pre-replication complex; RRM2, a ribonucleotide reductase that catalyses deoxyribonucleotide synthesis from ribonucleotides; PCNA, which forms a moving platform to recruit replication enzymes to the replication fork, and FEN1, a flap endonuclease involved in rejoining Okasaki fragments. Other down-regulated genes act principally to facilitate mitosis, including AURKB and CCNB1. Therefore, miR-24 may put the breaks on cell division at multiple steps in cell cycle progression.
Silencing the genes mentioned above would be expected to inhibit cell division. However, suppressing other miR-24 down-regulated genes would promote cell cycle progression, especially in the context of DNA damage. These genes include CHEK1, which participates in the G2/M checkpoint and is activated by ATR in response to unresolved DNA damage, and BRCA1, which is in a surveillance complex that activates double-strand break repair. Prominent in the down-regulated gene interaction network are the cyclin D inhibitor CDKN1B (p27KIP1) and VHL, a tumor suppressor protein. In addition, p16INK4A, a CDK inhibitor, is a validated direct miR-24 target (Lal et al., 2008). Thus, the role of miR-24 in regulating the cell cycle may be complex. If cells are unable to exit G1 and replicate their DNA, it may be economical to suppress the inhibitory genes that guard the genome from propagating damaged DNA. However, in some contexts, depending on the transcripts expressed in a particular cell, miR-24 might actually promote cell proliferation by suppressing these cell cycle inhibitory genes. In fact, inhibiting miR-24 decreases proliferation of A549 lung cancer cells, but has the opposite effect on HeLa cells (Cheng et al., 2005).
miR-24 is most highly expressed in G1. This is consistent with our finding that miR-24 regulates the G1/S transition. The E2F family of transcription factors regulates progression through this checkpoint. It therefore makes sense that miR-24 acts in large part by directly targeting E2F2 (and thereby indirectly suppressing E2F1 and E2F3 expression). The pattern of miR-24 expression is consistent with the known cell cycle variation of E2F family members (Sears et al., 1997). When miR-24 is high in G1, E2F1 and E2F2 are low; the E2F family begins to be expressed in late G1 and peaks in S phase when miR-24 is turned down. The E2Fs continue to be expressed in G2 and M (where they also have important functions) when miR-24 levels remain low.
miR-24 is not the only miRNA that regulates the cell cycle and targets the E2F family (Fig. 7F). For instance, the miR-17∼92 cluster directly down-regulates the E2F family (O'Donnell et al., 2005; Petrocca et al., 2008). However, unlike miR-24, whose expression varies inversely with E2F expression, miR-17∼92 appears to be expressed uniformly except in quiescent cells. Moreover, E2F2 down-regulation should antagonize the dominant effect of these miRNAs to promote cell proliferation. Thus E2F down-regulation is likely not a defining effect of miR-17∼92, but rather a secondary effect that fine tunes its major proliferative effect. Since miR-24 suppresses MYC and E2F expression and both MYC and the E2F family activate miR-17∼92 and miR-106b∼25 transcription (O'Donnell et al., 2005; Petrocca et al., 2008), miR-24 also likely inhibits proliferation by indirectly suppressing transcription of these cell cycle-promoting miRNAs. A recent paper also suggests another layer of complexity to the miR-24, miR-17∼92, MYC and E2F network (Gao et al., 2009). MYC suppresses the transcription of miR-23b, which is encoded with miR-24. Although one recent study suggests that miR-24 and miR-23b are independently transcribed (Sun et al., 2009), MYC might also regulate miR-24 transcription. It is worth noting that E2F1 mRNA and protein do not correlate during the cell cycle, consistent with post-transcriptional regulation by miRNAs (O'Donnell et al., 2005). Since E2F2 mRNA has kinetics similar to E2F1 (Sears et al., 1997), miRNA-dependent regulation of translation may be operating, possibly for all E2Fs that promote G1/S transition.
The integrated effect of miR-24 on a highly interacting set of key genes acts as a switch to stop cell division, rather than as a fine-tuning rheostat. It will be interesting to understand how expression of these two miR-24 gene clusters is regulated and to understand the function of the clustered miRNAs (miR-23 and miR-27). The only other miRNAs consistently upregulated during terminal differentiation are miR-22 and miR-125a (a mammalian ortholog of lin-4) (Lal et al., 2009). There are suggestions in the literature that these genes might also regulate important pathways of cell differentiation (Choong et al., 2007; Wu and Belasco, 2005).
miR-24 directly regulates both cell proliferation and DNA repair. Enhancing miR-24 function in cancer cells by introducing miR-24 mimics might be an attractive therapeutic, since potentially it could block dysregulated cell proliferation and also sensitize cancer cells to DNA damage from chemo- and radiotherapy.
Experimental procedures (also see Suppl. Materials)
GO analysis
The GO project provides structured controlled vocabularies, or ontologies, to describe genes relative to their biological processes. Each biological process consists of a series of events achieved by one or more molecular functions. The ontologies are stored in directed acyclic graphs where each node represents a biological process and each subsequent node corresponds to a more specialized term. Over-represented GO biological processes were determined using a MetaCore tool (GeneGo Inc., St Joseph, MI, http://www.genego.com), which utilizes the hypergeometric distribution to calculate the p-value for genes showing enrichment in a biological process. The value is equivalent to the probability of a subset of genes from a specific experiment (i.e., miR-24 over-expression) to arise by chance given the total number of genes associated with the biological process.
Network Analysis
We developed a graphical representation of the molecular relationships between proteins from the 248 genes down-regulated in miR-24 over-expressing cells. Network analysis was performed using the MetaCore Analytical Suite and visualization was performed using Ingenuity Pathways Analysis (Ingenuity Inc. www.ingenuity.com). Proteins are represented as nodes, and the biological relationship between two nodes is represented as an edge. Functional and pathway analysis was based on support for direct interaction edges in the networks from at least 1 reference from the literature, a textbook, or from canonical information stored in the network generation software database - manually curated for experimentally verified human protein-protein interactions and protein-DNA interactions.
Supplementary Material
Acknowledgments
This work was supported in part by NIH AI070302 and a GSK-IDI Alliance grant (JL), by the NIA-IRP, NIH (KGB, MG), the Harry Oppenheimer Memorial Trust (WH), a GSK-IDI Alliance fellowship (FN) and the Harvard Center for AIDS Research (NY). We thank N. Dyson (Harvard Medical School) for the HA-E2F2 expression plasmid, Ray McGovern for programming support and Lieberman laboratory members for useful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Joseph Z, Siegfried Z, Brandeis M, Brors B, Lu Y, Eils R, Dynlacht BD, Simon I. Genome-wide transcriptional analysis of the human cell cycle identifies genes differentially regulated in normal and cancer cells. Proc Natl Acad Sci U S A. 2008;105:955–960. doi: 10.1073/pnas.0704723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken AP, Ciro M, Cocito A, Helin K. E2F target genes: unraveling the biology. Trends Biochem Sci. 2004;29:409–417. doi: 10.1016/j.tibs.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choong ML, Yang HH, McNiece I. MicroRNA expression profiling during human cord blood-derived CD34 cell erythropoiesis. Exp Hematol. 2007;35:551–564. doi: 10.1016/j.exphem.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Didiano D, Hobert O. Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat Struct Mol Biol. 2006;13:849–851. doi: 10.1038/nsmb1138. [DOI] [PubMed] [Google Scholar]
- Didiano D, Hobert O. Molecular architecture of a miRNA-regulated 3′ UTR. RNA. 2008;14:1297–1317. doi: 10.1261/rna.1082708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederichs S, Haber DA. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131:1097–1108. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duursma AM, Kedde M, Schrier M, le Sage C, Agami R. miR-148 targets human DNMT3b protein coding region. RNA. 2008;14:872–877. doi: 10.1261/rna.972008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easow G, Teleman AA, Cohen SM. Isolation of microRNA targets by miRNP immunopurification. RNA. 2007;13:1198–1204. doi: 10.1261/rna.563707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez PC, Frank SR, Wang L, Schroeder M, Liu S, Greene J, Cocito A, Amati B. Genomic targets of the human c-Myc protein. Genes Dev. 2003;17:1115–1129. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda Y, Kawasaki H, Taira K. Exploration of human miRNA target genes in neuronal differentiation. Nucleic Acids Symp Ser (Oxf) 2005:341–342. doi: 10.1093/nass/49.1.341. [DOI] [PubMed] [Google Scholar]
- Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, Dang CV. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermeking H, Rago C, Schuhmacher M, Quing Li, Barrett JF, Obaya AJ, O'Connell BC, Mateyak MK, Tam W, Kohlhuber F, et al. Identification of CDK4 as a target of c-MYC. Proc Natl Acad Sci U S A. 2000;97:2229–2234. doi: 10.1073/pnas.050586197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- Lal A, Kim HH, Abdelmohsen K, Kuwano Y, Pullmann R, Jr, Srikantan S, Subrahmanyam R, Martindale JL, Yang X, Ahmed F, et al. p16(INK4a) translation suppressed by miR-24. PLoS ONE. 2008;3:e1864. doi: 10.1371/journal.pone.0001864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal A, Pan Y, Navarro F, Dykxhoorn D, Moreau L, Meire E, Bentwich Z, Lieberman J, Chowdhury D. miR-24-mediated down-regulation of H2AX suppresses DNA repair in terminally differentiated blood cells. Nature Struct Mol Biol. 2009;16:492–498. doi: 10.1038/nsmb.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebofsky R, Walter JC. New Myc-anisms for DNA replication and tumorigenesis? Cancer Cell. 2007;12:102–103. doi: 10.1016/j.ccr.2007.07.013. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- Neilson JR, Zheng GX, Burge CB, Sharp PA. Dynamic regulation of miRNA expression in ordered stages of cellular development. Genes Dev. 2007;21:578–589. doi: 10.1101/gad.1522907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, Iliopoulos D, Pilozzi E, Liu CG, Negrini M, et al. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272–286. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- Polager S, Ginsberg D. E2F - at the crossroads of life and death. Trends Cell Biol. 2008;18:528–535. doi: 10.1016/j.tcb.2008.08.003. [DOI] [PubMed] [Google Scholar]
- Sears R, Ohtani K, Nevins JR. Identification of positively and negatively acting elements regulating expression of the E2F2 gene in response to cell growth signals. Mol Cell Biol. 1997;17:5227–5235. doi: 10.1128/mcb.17.9.5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Sun Q, Zhang Y, Yang G, Chen X, Cao G, Wang J, Sun Y, Zhang P, Fan M, Shao N, Yang X. Transforming growth factor-beta-regulated miR-24 promotes skeletal muscle differentiation. Nucleic Acids Res. 2008;36:2690–2699. doi: 10.1093/nar/gkn032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Wang J, Pan Q, Yu Y, Zhang Y, Wan Y, Wang J, Li X, Hong A. Characterization of function and regulation of miR-24-1 and miR-31. Biochem Biophys Res Commun. 2009;380:660–665. doi: 10.1016/j.bbrc.2009.01.161. [DOI] [PubMed] [Google Scholar]
- Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- Vella MC, Choi EY, Lin SY, Reinert K, Slack FJ. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′UTR. Genes Dev. 2004;18:132–137. doi: 10.1101/gad.1165404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernell R, Helin K, Muller H. Identification of target genes of the p16INK4A-pRB-E2F pathway. J Biol Chem. 2003;278:46124–46137. doi: 10.1074/jbc.M304930200. [DOI] [PubMed] [Google Scholar]
- Wu L, Belasco JG. Micro-RNA regulation of the mammalian lin-28 gene during neuronal differentiation of embryonal carcinoma cells. Mol Cell Biol. 2005;25:9198–9208. doi: 10.1128/MCB.25.21.9198-9208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ding L, Cheung TH, Dong MQ, Chen J, Sewell AK, Liu X, Yates JR, 3rd, Han M. Systematic identification of C. elegans miRISC proteins, miRNAs, and mRNA targets by their interactions with GW182 proteins AIN-1 and AIN-2. Mol Cell. 2007;28:598–613. doi: 10.1016/j.molcel.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.