Abstract
The mitochondrial genomes of flowering plants possess a promiscuous proclivity for taking up sequences from the chloroplast genome. All characterized chloroplast integrants exist apart from native mitochondrial genes, and only a few, involving chloroplast tRNA genes that have functionally supplanted their mitochondrial counterparts, appear to be of functional consequence. We developed a novel computational approach to search for homologous recombination (gene conversion) in a large number of sequences and applied it to 22 mitochondrial and chloroplast gene pairs, which last shared common ancestry some 2 billion years ago. We found evidence of recurrent conversion of short patches of mitochondrial genes by chloroplast homologs during angiosperm evolution, but no evidence of gene conversion in the opposite direction. All 9 putative conversion events involve the atp1/atpA gene encoding the alpha subunit of ATP synthase, which is unusually well conserved between the 2 organelles and the only shared gene that is widely sequenced across plant mitochondria. Moreover, all conversions were limited to the 2 regions of greatest nucleotide and amino acid conservation of atp1/atpA. These observations probably reflect constraints operating on both the occurrence and fixation of recombination between ancient homologs. These findings indicate that recombination between anciently related sequences is more frequent than previously appreciated and creates functional mitochondrial genes of chimeric origin. These results also have implications for the widespread use of mitochondrial atp1 in phylogeny reconstruction.
Keywords: gene conversion, gene transfer, recombination
Back in the dark ages of organelle “genomics,” well before the term was even coined and genome sequencing became routine, it came as a shock to discover through Southern blot analysis that mitochondrial genomes of flowering plants are rich in sequences captured from the chloroplast genome (1, 2). Indeed, this discovery led Ellis (3) to coin the very term “promiscuous DNA.” Each of the few dozen diverse angiosperm mitochondrial genomes that have been thoroughly examined, by either Southern blots or genome sequencing, contains numerous chloroplast-derived sequences (e.g., ref. 4). For reasons that are only partially clear, transfer of foreign DNA (including mitochondrial sequences) in the opposite direction, i.e., into chloroplast genomes, occurs far more rarely (4, 5).
The functional impact of chloroplast-to-mitochondrial promiscuity is thought to be limited virtually entirely to the occasional functional replacement of a native mitochondrial tRNA gene (or a nuclear gene encoding a mitochondrially imported tRNA) by a captured chloroplast tRNA gene (6). Aside from tRNA gene replacements, the only suspected functional impact of plant interorganellar promiscuity is the apparent recruitment of a chloroplast-derived promoter sequence by a native mitochondrial gene in rice (7). All of the many chloroplast protein genes known to be captured by mitochondrial genomes are thought to have had no impact on mitochondrial gene function, with their only fate being to decay as pseudogenes.
This study seeks to address the following question: Are chimeric mitochondrial genes of bicompartmental origin ever created via recombination/gene conversion between homologous chloroplast and mitochondrial genes? Two well-established features and 2 recent sets of discoveries have prompted us to search carefully for this potential impact of DNA promiscuity on mitochondrial gene evolution. First, fully half of the 40 mitochondrial protein genes present in the common ancestor of angiosperms have homologs present in chloroplast genomes. Second, the generally low rates of point mutation (8, 9) and the relatively high degree of sequence conservation of chloroplast and plant mitochondrial genes increase the probability that these genes might at least occasionally recombine with one another. Third, although the frequency of homologous recombination correlates positively and tightly with DNA sequence similarity and decreases sharply with the level of relatedness between donor and recipient (10, 11), several cases have nonetheless been reported in recent years of recombination between sequences that are distantly related in time and/or sequence (5, 12–15). Finally, 2 cases are now known of plant mitochondrial housekeeping genes that have a clear history of chimerism, through recombination between native mitochondrial genes and foreign copies acquired via horizontal gene transfer from other, distantly related angiosperms (16, 17).
To search comprehensively for evidence of recombination between chloroplast and mitochondrial genes, we developed a novel computational approach to examine a large number of sequences. We found evidence of repeated recombination events during angiosperm evolution between the 2 members of the best-conserved and most widely sequenced pair of mitochondrial/chloroplast protein gene homologs.
Results
We searched for evidence of interorganellar recombination for all 20 protein genes with homologs present in both plant chloroplast and mitochondrial genomes (Table 1), plus the small and large subunit rRNA genes. The high-throughput test for recombination developed for this study (see Methods) detected, at the P < 0.001 significance level, 9 putative conversion events in which short segments of a chloroplast gene replaced cognate regions of the endogenous mitochondrial homolog (Table 2; see supporting information (SI) Table S1 for a list of all potential events detected with P < 0.05). No putative conversions were detected in the opposite direction, i.e., from mitochondrial to chloroplast genes (the best hits in this direction were over an order-of-magnitude higher than the P < 0.001 significance threshold used in this study). All 9 cases involve the same gene pair, atp1 and atpA, encoding the alpha subunit of ATP synthase (the “coupling factor”) of chloroplasts and mitochondria, respectively. The large number (529) of mitochondrial atp1 sequences analyzed requires corrections for multiple tests. Applying the conventional, Bonferroni correction, P-values for atp1/atpA gene conversion remain significant in 6 of the 9 putatively converted groups (Table 2). However, the Bonferroni procedure is known to be overly conservative when the number of tests is large, because it ignores dependencies among the data (18). Simulations were therefore conducted using the consensus of angiosperm atp1 genes to obtain a more reasonably adjusted P-value. The 5% critical value of the simulated P-values was considered as the adjusted P-value and equals 2.20 × 10−04 (Fig. S1). This P-value is larger than the P-value of 9.45 × 10−05 from the Bonferroni correction, and, consequently, 2 additional recombinant segments were considered significant (Table 2).
Table 1.
Homologous protein and rRNA genes present in some or all angiosperm chloroplast (cp) and mitochondrial (mt) genomes
| mt genesa |
cp homologsb |
Alignment length | Identity |
Gaps |
No. mt segmencesd | |||
|---|---|---|---|---|---|---|---|---|
| Name | Length | Name | Length | No. | %c | %c | ||
| Protein | ||||||||
| atp1 | 507 | atpA | 507 | 519 | 286 | 56.2 | 2.7 | 529 |
| atp6 | 385 | atpI | 249 | 397 | 64 | 16.2 | 40.1 | 38 |
| atp9 | 85 | atpH | 81 | 89 | 21 | 23.6 | 1.1 | 58 |
| nad1 | 325 | ndhA | 360 | 367 | 134 | 36.4 | 6.0 | 35 |
| nad2 | 499 | ndhB | 512 | 526 | 159 | 30.6 | 6.5 | 26 |
| nad3 | 118 | ndhC | 120 | 121 | 41 | 33.9 | 0.8 | 47 |
| nad4 | 495 | ndhD | 500 | 511 | 140 | 27.7 | 4.3 | 31 |
| nad5 | 669 | ndhF | 746 | 779 | 227 | 29.4 | 17.3 | 33 |
| nad6 | 205 | ndhG | 176 | 218 | 50 | 22.9 | 6.0 | 28 |
| nad7 | 394 | ndhH | 393 | 403 | 148 | 37.0 | 4.0 | 21 |
| nad9 | 190 | ndhJ | 158 | 196 | 36 | 20.6 | 11.7 | 31 |
| rpl2 | 349 | rpl2 | 274 | 377 | 68 | 18.8 | 30.8 | 16 |
| rpl16 | 179 | rpl16 | 135 | 179 | 61 | 34.1 | 24.6 | 27 |
| rps2 | 531 | rps2 | 236 | 544 | 57 | 10.5 | 53.7 | 88 |
| rps3 | 556 | rps3 | 218 | 557 | 65 | 11.9 | 58.9 | 29 |
| rps4 | 362 | rps4 | 201 | 370 | 57 | 17.3 | 37.0 | 24 |
| rps7 | 148 | rps7 | 155 | 163 | 46 | 29.1 | 11.0 | 25 |
| rps12 | 125 | rps12 | 123 | 125 | 72 | 57.5 | 0.0 | 46 |
| rps14 | 118 | rps14 | 100 | 119 | 39 | 38.2 | 2.5 | 31 |
| rps19 | 94 | rps19 | 92 | 97 | 31 | 33.7 | 3.1 | 78 |
| DNA | ||||||||
| atp1 | 1521 | atpA | 1521 | 1557 | 882 | 57.8 | 2.7 | 529 |
| rrn18S | 1935 | rrn16S | 1491 | 1995 | 1061 | 53.3 | 28.3 | 66 |
| rrn26S | 2568 | rrn23S | 2810 | 3186 | 1504 | 47.5 | 30.6 | 36 |
aMost of the genes used for these analyses are from the Arabidopsis mitochondrial genome (NC_001284), while the rps14 and rps19 sequences are from the Vitis genome (NC_012119) and the rps2 and nad3 sequences are from the Zea NB genome (NC_007982).
bFrom the Arabidopsis chloroplast genome (NC_000932).
cEnd gaps were excluded when calculating the percentages of identical and gapped sites. Consequently, these percentages differ from those calculated by dividing the number of identical sites by the alignment length. Because this table does, however, include all internal gaps, it presents a very different view of sequence conservation than do the sequence plots in Fig. 1, Fig. S3, and Fig. S4, for which all gaps are excluded.
dThe number of homologs was determined by a TBLASTN search with an E-value < 10−20 and are limited to angiosperm mitochondrial sequences.
Table 2.
Segments in mitochondrial atp1 genes derived by conversion with chloroplast atpA sequences at P < 0.001
| Speciesa | Chloroplast-derived segments |
||
|---|---|---|---|
| Start | End | P-valueb,c | |
| Lamialesd | 1110 | 1141 | |
| Streptocarpus holstii | 5.35 × 10−07 | ||
| Catalpa bignonioides | 6.87 × 10−09 | ||
| Empetrum nigrum | 1128 | 1149 | 9.49 × 10−05 |
| Rhododendron impeditum | 1104 | 1141 | 2.50 × 10−04 |
| (Vaccinium arboreum | 1128 | 1141 | 6.60 × 10−03)e |
| (Vaccinium uliginosumi | 1128 | 1141 | 8.86 × 10−03)e |
| Chimaphila umbellata | 1128 | 1141 | 9.81 × 10−04 |
| Clethra arborea | 1119 | 1141 | 3.30 × 10−07 |
| Clethra barbinervis | 1119 | 1141 | 3.15 × 10−07 |
| Passiflora suberosa | 1128 | 1141 | 1.01 × 10−04 |
| Cynomorium coccineum | 1119 | 1149 | 1.66 × 10−05 |
| Citrus sp. | 1110 | 1141 | 3.98 × 10−04 |
| Apodanthes caseariae | 942 | 1020 | 1.27 × 10−10 |
| Ranunculus sp. | 957 | 970 | 3.58 × 10−06 |
| Myrtus communis | 1008 | 1029 | 1.24 × 10−05 |
aSpecies order is shown as in Fig. 2.
bNumbers shaded in gray are significant at P < 0.05 after Bonferroni correction.
cNumbers in bold are significant at P < 0.05 as per simulations (Fig. S1).
dOf the 27 species of Lamiales examined, only those with the largest and smallest P-values are shown.
eThese are included because they belong to a clade that otherwise meets the P < 0.001 criterion (Fig. 2A).
Six of the 9 putative gene conversion events involve largely overlapping subsets of a 40-nucleotide (NT) region that is centered in the region of greatest atp1/atpA conservation (Table 2 and Figs. 1 and 2A). As shown in Fig. 2A, all members of the asterid order Lamiales except the 2 basal lineages (represented here by Syringa, Jasminum, and Jovellana) possess a 32-NT segment in mitochondrial atp1 that is identical (in most Lamiales) to the same region of the angiosperm chloroplast atpA consensus sequence, but different (again, in most Lamiales) at fully 8 positions from the angiosperm (Fig. 2A) and asterid (Fig. S2) mitochondrial atp1 consensus. We infer that early in Lamiales' evolution, a region of chloroplast atpA of between 32 and 44 NT in length replaced the homologous region of mitochondrial atp1. Chloroplast/mitochondrial conversion events involving a subset of (or in 1 case, a potential overlap with) the Lamiales' conversion region appear to have occurred separately in 5 other groups of angiosperms (Fig. 2A, Table 2).
Fig. 1.
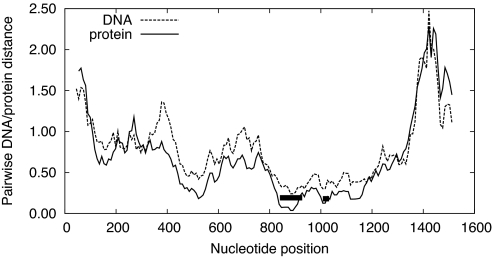
Amino acid and nucleotide conservation between mitochondrial atp1 and chloroplast atpA of Arabidopsis thaliana. The plots used sliding windows of 30 aa or 90 NT, slid 3 aa or 9 NT at a time. The y axis corresponds to the estimated number of substitutions/changes per site, with DNA distance measured using the F84 matrix and protein distance using the JTT matrix. The regions involved in gene conversion are marked by bars, with the first corresponding to nucleotide positions 942-1029 (Fig. 2B) and the second to positions 1110–1149 (Fig. 2A). Note that these plots are based solely on the alignment of the 2 Arabidopsis sequences; some coordinates will differ as compared to the multiple sequence alignments.
Fig. 2.
Chloroplast-like sequences embedded in plant mitochondrial atp1 genes. The NT coordinates shown correspond to those in the whole-gene alignments of Figs. S2 and S7. Filled circles on the trees indicate the timing of putative gene conversion events. Regions that are or are not significant as per simulations are shaded in dark or light gray, respectively (Table 2 and Fig. S1). See SI Text Sources of the Topologies Shown in Fig. 2 for sources of the tree topologies shown.
About 150 NT upstream of this conversion region (Fig. 2A) is a region that appears to have experienced 3 overlapping chloroplast-to-mitochondrial conversion events (Table 2, Fig. 2B). Two of the mitochondrial atp1 conversion tracts are identical to the angiosperm atpA chloroplast consensus: Ranunculus and Myrtus atp1 each contain all 7 and 6 chloroplast-specific NTs within conversion tracts of minimum length 14 and 22 NT, respectively (Fig. 2B). Apodanthes atp1 contains the longest region of putative chloroplast origin found in this study. This 79-NT tract possesses 16 of 20 NTs specific to the angiosperm chloroplast consensus sequence (Fig. 2B; see SI Text Analysis of Chloroplast Donor Sequences) and encompasses or overlaps both the Ranunculus and Myrtus conversion tracts.
Discussion
Advantages and Limitations of the Methodology.
Most existing programs for detecting gene conversion/recombination perform exhaustive searches on all possible sequence combinations, comparing 3 sequences at a time. When the number of sequences is large, it becomes extremely computationally difficult to conduct calculations for all possible combinations of gene triplets. For instance, for the 616 angiosperm atp1/atpA sequences analyzed (529 atp1 genes and 87 atpA genes), the number of possible triplets is (3616) ≈ 3.9 × 107. In this study, to reduce the number of comparisons and thus the computational burden, we fixed 2 of the 3 sequences in each comparison by using the angiosperm chloroplast and mitochondrial consensus sequences. The rationale for this approach is that because most plant mitochondrial lineages have extremely low substitution rates (8, 9), because chloroplast lineages also generally have low substitution rates (ref. 8; but not as low as those of plant mitochondria), and because mitochondria and chloroplasts diverged so anciently (ca. 2 billion years ago), chloroplast regions embedded within angiosperm mitochondrial genes should be notably different in sequence from cognate regions of other angiosperm mitochondrial genes but highly similar to those of angiosperm chloroplast genes.
The method used in this study (Table 2) is more sensitive than the GENECONV program (Table S2) for detecting recombination. However, the use of consensus sequences, while computationally advantageous, carries a risk (see SI Text Analysis of Chloroplast Donor Sequences) that recombination events involving chloroplast regions that differ significantly from the chloroplast consensus sequence will be missed (whereas such events would be detected in analyses using the actual donor sequence or sequences closely related to it). By the same token, the consensus approach also poses a (probably smaller) risk of yielding false positives (this can be effectively ruled out for the 4 conversion cases for which chloroplast atpA sequences are available for closely related species (see SI Text Analysis of Chloroplast Donor Sequences). A general problem for all recombination detection programs is a lack of sensitivity for detecting events involving highly conserved regions with few informative sites. For these reasons, and also considering the relatively stringent significance threshold (P < 0.001) used for our recombination search, the number of interorganellar recombinations detected in this study should probably be considered a lower bound on the actual number of such events. In particular, some of the 15 lowest-scoring but nonetheless not-unreasonable candidate cases for chloroplast/mitochondrial recombination listed in Table S1 (those with P < 0.05 but >0.001) may well be bona fide cases. Such cases would be considerably strengthened if much denser taxon sampling were to reveal an abrupt phylogenetic divide in the sequence (chloroplast vs. mitochondrial) of the region in question.
Mechanisms and Historical Patterns of Conversion and Gene Transfer.
Various alternative explanations that could in theory account for these findings—i.e., mitochondrial retroprocessing, experimental artefacts, and intense, directional purifying selection—can be ruled out as discussed in SI Text Alternative Explanations for the Findings of This Study. We are therefore confident that all 9 “chloroplast-like” segments that are embedded in plant mitochondrial atp1 genes did indeed ultimately arise from gene conversion events between a “visiting” chloroplast atpA gene and a resident mitochondrial atp1 gene. Given the extensive and sometimes long-term incorporation of chloroplast sequences within angiosperm mitochondrial genomes (1, 2, 4), it seems likely that most if not all of these conversions occurred by intramitochondrial recombination, probably occurring well after the integration of chloroplast atpA genes in various mitochondrial genomes [the only sequenced mitochondrial genome from the 9 converted lineages (Digitalis from the Lamiales; J. P. Mower and J. D. Palmer, unpublished results) lacks chloroplast atpA]. However, it is difficult if not impossible to rule out more direct events, whereby a transiently introduced, nonintegrated chloroplast atpA sequence converted directly with its resident mitochondrial homolog.
The extensive overlap if not positional identity (see Passiflora and the Empetrum-Vaccinium clade) among many of the chloroplast conversion tracts raises the possibility of a nonindependent history of some of these conversion tracts. Because Passiflora and Empetrum et al. are distantly related angiosperms, with many phylogenetically intervening lineages that lack this tract (far more than shown in Fig. 2A), they must have obtained it by separate events. However, whether these 2 separate events were fully independent of one another is entirely another matter, especially in light of the relatively frequent horizontal transfer of genes from one mitochondrial genome to another in angiosperm evolution (16, 17, 19). Either 2 independent chloroplast/mitochondrial conversion events occurred in Passiflora and Empetrum et al., or a single such conversion event was followed by the spread of the resulting recombinant mitochondrial atp1 gene into a second mitochondrial lineage by mitochondrial-to-mitochondrial horizontal transfer. As discussed in SI Text Possible Spread of the Chloroplast Conversion Tract in Parasitic Angiosperms via Mitochondrial Horizontal Gene Transfer, mitochondrial horizontal transfer should also be considered for the nonphotosynthetic parasites Apodanthes and Cynomorium, which could have acquired their conversion tracts from photosynthetic donors, such as their host plants. In any event, regardless of the relative balance between these 2 types of transfer, both processes involve recombination/conversion between native and foreign sequences, of greater (chloroplast/mitochondrial) or lesser (mitochondrial/mitochondrial) divergence.
Why Are All Chloroplast/Mitochondrial Conversions in atp1?
At least 3 factors could contribute to this very biased pattern. First, and most importantly, atp1 is (with but 1 exception) by far the most conserved of the 20 protein genes examined, and sequence conservation is crucial to both the occurrence and fixation of gene conversion events. Table 1 shows that only atp1 (56.2%) and rps12 (57.5%) have greater than 38% amino acid conservation relative to their chloroplast homologs among the 20 protein genes examined. atp1 is 4 times longer than rps12. Importantly, atp1 also contains several short stretches that are substantially better conserved than any regions in rps12 (or any of the 18 shared protein genes; Fig. S3), and all 9 conversion tracts are restricted to the 2 regions of highest amino acid and nucleotide conservation in atp1/atpA (Fig. 1). These findings fit well with observations in other systems that the frequency of homologous recombination correlates positively and strongly with the degree of sequence conservation and is particularly associated with the presence of shared blocks of high similarity (10, 11). A high level of sequence conservation is important in 2 respects: First, it both promotes the physical occurrence of gene recombination/conversion and also introduces foreign sequences with relatively few differences such that the resulting chimeric gene is more likely to be fixed. Second, atp1 is by far the most widely sequenced of the 22 mitochondrial genes examined in this study, with 529 angiosperm sequences being available for atp1 compared to only 16–88 (mean = 37, median = 31) for the other 21 genes (Table 1). The third factor was described in the preceding section, i.e., a relatively small number of actual chloroplast/mitochondrial conversions (“founder events”) could have ramified into a larger number of apparent conversions via subsequent mitochondrial-specific horizontal transfer.
The small and large subunit rRNA genes are better conserved at the nucleotide level than is atp1 as viewed in plots in which all gaps are excluded (Fig. S4). Why, then, were no conversion events detected between chloroplast and mitochondrial rRNA genes? First, as already noted, these genes have been much less widely sequenced (66 and 36 times) in angiosperm mitochondria than has atp1 (519 times). Second, what Fig. S4 does not show is that there are many gaps in the rRNA alignments, whereas the atp1/atpA alignment is virtually free of gaps, and gaps will reduce the odds of successful recombination between otherwise well-conserved sequences. Third, the best conserved regions in rRNA genes are for the most part involved in secondary base pairing of the rRNAs. Given both the distribution of conservation across rRNA genes, such that most physically successful conversions are likely to be short, and also the often disparate location of paired elements in the primary sequence of the gene, most conversions will affect only one-half of a stem region and therefore be so destabilizing of secondary structure as to be grossly deleterious. Finally, the redundancy of the genetic code means that many nucleotide substitutions in atp1/atpA are silent, and indeed this is particularly important in the 2 regions of recurrent recombination (Figs. 1 and 2; also see below), whereas rRNA lacks such genetic buffering. Nonetheless, at least 1 case of a successful, fixed recombination between anciently divergent rRNA genes has been reported (in cyanobacteria; ref. 13), and we would not be surprised if greatly expanded sequencing of mitochondrial rRNA genes does not reveal the occasional chloroplast conversion event.
Functionality of Chloroplast/Mitochondrial Chimeric Genes.
For the following combination of reasons, we think that all 9 sets of chimeric, mitochondrially located (see SI Text Mitochondrial Provenance and Copy Number of Chimeric atp1 Genes) atp1 genes that contain short conversion tracts of chloroplast origin are probably functional. First, all of these chimeric genes have intact ORFs insofar as sequenced and show no evidence of being pseudogenes. Second, there is no reason to suspect that any of these are cryptic, mitochondrial pseudogenes (i.e., pseudogenes that have not yet sustained mutations that readily identify them as such), with the functional copy of atp1 having been transferred to the nucleus. This is because, in sharp distinction to the frequent functional transfer of many other mitochondrial genes in plants, there is no evidence that atp1 has ever been functionally transferred in plants, despite survey of hundreds of diverse angiosperms (9, 20). Third, and related to the preceding point, most if not all of these chimeric genes are probably the only copy of atp1 present in the mitochondrial genome (see SI Text Mitochondrial Provenance and Copy Number of Chimeric atp1 Genes). Fourth, approximately all 13–16 or so (see SI Text Analysis of Chloroplast Donor Sequences on ambiguities for Apodanthes) putatively postrecombination substitutions within the recombinant regions are synonymous, consistent with the regions (and genes) still being under functional constraint. Fifth, the chloroplast-derived segments within the atp1 genes are remarkably similar to the corresponding native mitochondrial sequences at the amino acid level, introducing but a single amino acid replacement among all 9 cases (Fig. S5). Finally, chimeric atp1 transcripts are well represented in EST libraries available for several relevant taxa (see SI Text Functionality of Chimeric atp1 Genes—Evidence of Transcription and Fig. S6). Thus, there is little reason to think that the introduction of these chloroplast regions has created significantly maladapted mitochondrial atp1 genes. This study therefore presents unique evidence for the occurrence of gene conversion, creating functional chimeric genes, across the ca. 2-billion-year divide between chloroplast and mitochondrial genes. It thereby establishes a unique role for chloroplast-to-mitochondrial transfer in modifying the mitochondrial proteome of plants.
Gene Conversion and Phylogenetic Inference.
atp1 is 1 of 3 mitochondrial genes that has been widely sequenced and used to help reconstruct angiosperm phylogeny (the other 2, cox1 and matR, lack chloroplast homologs). It is therefore important to consider the potential phylogenetic repercussions of recurrent chloroplast conversion of atp1. On the one hand, gene conversion events, when properly recognized as such, represent so-called “rare genomic changes,” which have the potential to serve as individual phylogenetic characters of notable significance (21). On the other hand, when unrecognized (as has been the case heretofore), these “single characters” will be improperly overweighted in sequence-based phylogenetic analyses; the more NT differences they introduce, the greater the overweighting. In conjunction with the recurrent, homoplasious nature of the atp1/atpA conversions and the very low substitution rates in most plant mitochondrial genomes, overweighting because of unrecognized conversion could cause serious phylogenetic error (although it does not seem to with respect to the currently available data; analyses not shown).
General Implications for Recombination.
This study adds significantly to the number of known cases (5, 12–15) of gene conversion between distantly related sequences. The frequency of gene conversion is so well established to be strongly inversely proportional to sequence divergence (10, 11) that it is hardly surprising that studies of gene conversion in nuclear genomes have focused on recently duplicated genes (11). Nonetheless, our findings recommend that increased attention be given to the possibility of rarer, but potentially evolutionarily significant conversion among the hundreds to thousands of anciently arisen gene families that inhabit many nuclear genomes.
Finally, our findings further emphasize the importance of recombination, and the diversity of ways in which it is manifest, in the evolution and function of angiosperm mitochondrial genomes. The importance of recombination first became apparent 25 years ago at the whole-genome level, in the context of generating multipartite genomes (22, 23). Since then, recombination has been shown to create chimeric functional genes in 3 very different contexts: intramitochondrial recombination to generate functionally novel, chimeric genes involved in cytoplasmic male sterility (24); recombination between native and foreign mitochondrial genes to create hybrid forms of canonical mitochondrial genes (16, 17); and, now, recombination between anciently divergent chloroplast and mitochondrial genes. One wonders what recombinational tricks plant mitochondrial genomes might still have up their sleeves.
Methods
Sequence similarity searches were performed with TBLASTN against the nonredundant GenBank database using the Arabidopsis atp1 protein sequence as the query sequence. All significant hits were required to have an expect-value <10−20 and to match the query sequence by over 70% of its length. A total of 529 mitochondrial atp1 sequences were retained for analysis. Chloroplast atpA sequences of angiosperms were extracted only when their complete chloroplast genomes were available (87 in total).
DNA sequences were extracted from GenBank files and translated into protein sequences. These were aligned using MUSCLE (25), and nucleotide alignments were created by replacing each amino acid with its corresponding codon using in-house PERL scripts. Phylogenies were constructed using PhyML (26) with a GTR + Γ + I model. Parameters estimated from the 529 atp1 and 87 atpA sequences were used in further simulation studies.
The core calculation for detecting potential recombination/conversion tracts was conducted using a method modified from the Recombination Detection Program (27). In brief, this program (the source code for which is available upon request of the first author, with a user-friendly version in preparation as a separate publication) compares 3 sequences at a time by examining only informative sites. The probability of observing 1 recombination follows a binomial distribution:
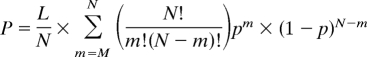 |
where L is the length of informative sites, N is the length of the putative recombinant segment, and M and p are the number and proportion, respectively, of nucleotides shared between the putative recombinant sequences. We calculated the probability of recombination by comparing each mitochondrial gene sequence analyzed with the 2 fixed sequences for that gene (see Discussion). Segments with a significantly higher similarity to the chloroplast than the mitochondrial consensus were deemed to be recombinant.
We first adjusted for multiple comparisons using the Bonferroni correction (P < 0.05/k, k is the number of tests = the number of atp1 genes = 529), and P-values would have to be smaller than 8.38 × 10−5 to be considered significant at the 5% level. Second, as a less conservative adjustment, we performed simulations to estimate the distribution of the calculated P-values. In brief, sequences were simulated on the basis of the phylogeny of the 529 atp1 and 87 atpA genes with parameters estimated by PhyML using the Seq-Gen program (28). Simulated sequences were then analyzed for recombination using the above procedure. Five hundred iterations were conducted, and the smallest P-value for recombination in each iteration was recorded. The 5% quantile of the P-value distribution was used to determine significance.
Supplementary Material
Acknowledgments.
We thank Andy Alverson and Brian Golding for comments on the manuscript; Todd Barkman and Claude dePamphilis for supplying the previously undeposited Citrus atp1 sequence; Todd Barkman for providing other unpublished data and for very helpful discussion; and the 2 reviewers for exceptionally helpful and insightful comments. This work was supported by National Institutes of Health research grant RO1-GM-70612 (to J.D.P) and by the METACyt Initiative of Indiana University, funded in part through a major grant from the Lilly Endowment, Inc.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908766106/DCSupplemental.
References
- 1.Stern DB, Lonsdale DM. Mitochondrial and chloroplast genomes of maize have a 12-kilobase DNA sequence in common. Nature. 1982;299:698–702. doi: 10.1038/299698a0. [DOI] [PubMed] [Google Scholar]
- 2.Stern DB, Palmer JD. Extensive and widespread homologies between mitochondrial DNA and chloroplast DNA in plants. Proc Natl Acad Sci USA. 1984;81:1946–1950. doi: 10.1073/pnas.81.7.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ellis J. Promiscuous DNA: Chloroplast genes inside plant mitochondria. Nature. 1982;299:678–679. doi: 10.1038/299678a0. [DOI] [PubMed] [Google Scholar]
- 4.Goremykin VV, Salamini F, Velasco R, Viola R. mtDNA of Vitis vinifera and the issue of rampant horizontal gene transfer. Mol Biol Evol. 2008;26:99–110. doi: 10.1093/molbev/msn226. [DOI] [PubMed] [Google Scholar]
- 5.Rice DW, Palmer JD. An exceptional horizontal gene transfer in plastids: Gene replacement by a distant bacterial paralog and evidence that haptophyte and cryptophyte plastids are sisters. BMC Biol. 2006;4:31. doi: 10.1186/1741-7007-4-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miyata S, Nakazono M, Hirai A. Transcription of plastid-derived tRNA genes in rice mitochondria. Curr Genet. 1998;34:216–220. doi: 10.1007/s002940050389. [DOI] [PubMed] [Google Scholar]
- 7.Nakazono M, Nishiwaki S, Tsutsumi N, Hirai A. A chloroplast-derived sequence is utilized as a source of promoter sequences for the gene for subunit 9 of NADH dehydrogenase (nad9) in rice mitochondria. Mol Gen Genet. 1996;252:371–378. doi: 10.1007/BF02173001. [DOI] [PubMed] [Google Scholar]
- 8.Wolfe KH, Li WH, Sharp PM. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci USA. 1987;84:9054–9058. doi: 10.1073/pnas.84.24.9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mower JP, Touzet P, Gummow JS, Delph LF, Palmer JD. Extensive variation in synonymous substitution rates in mitochondrial genes of seed plants. BMC Evol Biol. 2007;7:135. doi: 10.1186/1471-2148-7-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Majewski J, Cohan FM. DNA sequence similarity requirements for interspecific recombination in Bacillus. Genetics. 1999;153:1525–1533. doi: 10.1093/genetics/153.4.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen JM, Cooper DN, Chuzhanova N, Ferec C, Patrinos GP. Gene conversion: Mechanisms, evolution and human disease. Nat Rev Genet. 2007;6:762–777. doi: 10.1038/nrg2193. [DOI] [PubMed] [Google Scholar]
- 12.Archibald JM, Roger AJ. Gene conversion and the evolution of euryarchaeal chaperonins: A maximum likelihood-based method for detecting conflicting phylogenetic signals. J Mol Evol. 2002;55:232–245. doi: 10.1007/s00239-002-2321-5. [DOI] [PubMed] [Google Scholar]
- 13.Miller SR, et al. Discovery of a free-living chlorophyll d-producing cyanobacterium with a hybrid proteobacterial/cyanobacterial small-subunit rRNA gene. Proc Natl Acad Sci USA. 2005;102:850–855. doi: 10.1073/pnas.0405667102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inagaki Y, Susko E, Roger AJ. Recombination between elongation factor 1α genes from distantly related archaeal lineages. Proc Natl Acad Sci USA. 2006;103:4528–4533. doi: 10.1073/pnas.0600744103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keeling PJ, Palmer JD. Lateral transfer at the gene and subgenic levels in the evolution of eukaryotic enolase. Proc Natl Acad Sci USA. 2001;98:10745–10750. doi: 10.1073/pnas.191337098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bergthorsson U, Adams KL, Thomason B, Palmer JD. Widespread horizontal transfer of mitochondrial genes in flowering plants. Nature. 2003;424:197–201. doi: 10.1038/nature01743. [DOI] [PubMed] [Google Scholar]
- 17.Barkman TJ, et al. Mitochondrial DNA suggests at least 11 origins of parasitism in angiosperms and reveals genomic chimerism in parasitic plants. BMC Evol Biol. 2007;7:248. doi: 10.1186/1471-2148-7-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakagawa S. A farewell to Bonferroni: The problems of low statistical power and publication bias. Behav Ecol. 2004;15:1044–1045. [Google Scholar]
- 19.Richardson AO, Palmer JD. Horizontal gene transfer in plants. J Exp Bot. 2007;58:1–9. doi: 10.1093/jxb/erl148. [DOI] [PubMed] [Google Scholar]
- 20.Adams KL, Qiu YL, Stoutemyer M, Palmer JD. Punctuated evolution of mitochondrial gene content: High and variable rates of mitochondrial gene loss and transfer to the nucleus during angiosperm evolution. Proc Natl Acad Sci USA. 2002;99:9905–9912. doi: 10.1073/pnas.042694899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rokas A, Holland PW. Rare genomic changes as a tool for phylogenetics. Trends Ecol Evol. 2000;15:454–459. doi: 10.1016/s0169-5347(00)01967-4. [DOI] [PubMed] [Google Scholar]
- 22.Palmer JD, Shields CR. Tripartite structure of the Brassica campestris mitochondrial genome. Nature. 1984;307:437–440. [Google Scholar]
- 23.Lonsdale DM, Hodge TP, Fauron CMR. The physical map and organization of the mitochondrial genome from the fertile cytoplasm of maize. Nucleic Acids Res. 1984;12:9249–9261. doi: 10.1093/nar/12.24.9249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newton KJ, Gabay-Laughnan S, De Paepe R. Mitochondrial mutations in plants. In: Day DA, Millar AH, Whelan J, editors. Plant Mitochondria: From Genome to Function. Dordrecht, The Netherlands: Kluwer; 2004. pp. 121–142. [Google Scholar]
- 25.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 27.Martin D, Rybicki E. RDP: Detection of recombination amongst aligned sequences. Bioinformatics. 2000;16:562–563. doi: 10.1093/bioinformatics/16.6.562. [DOI] [PubMed] [Google Scholar]
- 28.Rambaut A, Grassly NC. Seq-Gen: an application for the Monte Carlo simulation of DNA sequence evolution along phylogenetic trees. Comput Appl Biosci. 1997;13:235–238. doi: 10.1093/bioinformatics/13.3.235. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.