Abstract
We describe the role of PSF protein and VL30–1 RNA, a mouse retroelement noncoding RNA, in the reversible regulation of proto-oncogene transcription, cell proliferation, and tumorigenesis in mice. The experiments involved increasing expression of PSF or VL30–1 RNA in NIH/3T3 fibroblast cells and B16F10 melanoma cells by transfecting the respective coding genes under control of a strong promoter or decreasing expression by transfecting a shRNA construct that causes degradation of PSF mRNA or VL30–1 RNA. The results are as follows: (i) PSF binds to the proto-oncogene Rab23, repressing transcription, and VL30–1 RNA binds and releases PSF from Rab23, activating transcription; (ii) increasing expression of PSF or decreasing expression of VL30–1 RNA suppresses cell proliferation in culture and tumorigenesis in mice; and (iii) decreasing expression of PSF or increasing expression of VL30–1 RNA promotes cell proliferation in culture and tumorigenesis in mice. These results indicate that PSF is a major tumor-suppressor protein and VL30–1 RNA is a major tumor-promoter RNA in mice. Although VL30–1 RNA can integrate into the cell genome, tumor promotion by VL30–1 RNA involves a trans effect rather than a cis effect on gene transcription. Expression of VL30–1 RNA is 5- to 8-fold higher in mouse tumor lines than in mouse fibroblast or myoblast lines, whereas expression of PSF mRNA does not decrease in the tumor lines, suggesting that tumorigenesis is driven by an increase of VL30–1 RNA rather than a decrease of PSF. A similar regulatory mechanism functions in human cells, except that human PSF-binding RNAs replace VL30–1 RNA, which is not encoded in the human genome. We propose that PSF protein and PSF-binding RNAs have a central role in the reversible regulation of mammalian cell proliferation and tumorigenesis and that increasing PSF expression or decreasing PSF-binding RNA expression in tumor cells is a potential therapeutic strategy for cancer.
Keywords: cancer therapy, Rab23, regulatory RNA, tumor suppression
Preceding studies have described a reversible mechanism controlling gene transcription that involves PSF protein (1) and VL30–1 RNA, a member of the VL30 family of mouse retroelement noncoding RNAs (2, 3). PSF contains a DNA-binding domain (DBD) that binds and represses transcription of genes that have a PSF-binding site (4–6) and 2 RNA-binding domains (RBDs) that bind VL30–1 RNA, forming a PSF-RNA complex that dissociates from a gene and activates transcription (5–7). Increasing expression of PSF in a human tumor cell suppressed tumorigenesis (6), and ectopic expression of VL30–1 RNA in a human tumor cell promoted metastasis (3). VL30 RNAs are expressed in virtually all tissues of adult mice (8) and are associated with Ras-mediated transformation of mouse fibroblast cells (9). Here, we extend our studies of the function of PSF and VL30–1 RNA to the regulation of proto-oncogene transcription, cell proliferation, and tumorigenesis in mice. The results indicate that PSF is a major tumor-suppressor protein and VL30–1 RNA is a major tumor-promoter RNA in mice.
Results
Expression of PSF and VL30–1 RNA in NIH/3T3 and B16F10 Cell Lines.
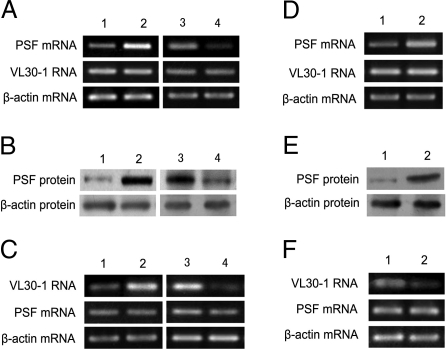
NIH/3T3 and B16F10 cells were transfected with a transgene encoding PSF (NIH/3T3-PSF↑ and B16F10-PSF↑ lines) or VL30–1 RNA (NIH/3T3-VL30↑ line) or with a plasmid encoding a shRNA that causes degradation of PSF mRNA (NIH/3T3-PSF↓ line) or VL30–1 RNA (NIH/3T3-VL30↓ and B16F10-VL30↓ lines). The control lines were transfected with an empty plasmid pcDNA3.1(+) (NIH/3T3-pcDNA3.1 and B16F10-pcDNA3.1 cell lines) or a plasmid encoding sh-Luciferase (NIH/3T3-shLuc and B16F10-shLuc lines). The NIH/3T3 and B16F10 cell lines were assayed for expression of PSF mRNA and VL30–1 RNA (Fig. 1 and Table 1) and PSF protein (Fig. 1).
Fig. 1.
Expression of PSF and VL30–1 RNA in NIH/3T3 and B16F10 cell lines. (A and B) 1, NIH/3T3-pcDNA3.1 cells; 2, NIH/3T3-PSF↑ cells; 3, NIH/3T3-shLuc cells; 4, NIH/3T3-PSF↓ cells. (C) 1, NIH/3T3-pcDNA3.1 cells; 2, NIH/3T3-VL30↑ cells; 3, NIH/3T3-shLuc cells; 4, NIH/3T3-VL30↓ cells. (D and E) 1, B16F10-pcDNA3.1 cells; 2, B16F10-PSF↑ cells. (F) 1, B16F10-shLuc cells; 2, B16F10-VL30↓ cells. (A, C, D, and F) Assay of PSF mRNA and VL30–1 RNA by semiquantitative RT-PCR. (B and E) PSF protein assay by Western blot.
Table 1.
Expression of PSF mRNA and VL30–1 RNA in NIH/3T3 and B16F10 cell lines
| Cell line | Transgene | PSF mRNA | VL30–1 RNA |
|---|---|---|---|
| NIH/3T3-pcDNA3.1 | None | ++ | ++ |
| NIH/3T3-shLuc | sh-Luciferase | ++ | ++ |
| NIH/3T3-PSF↑ | PSF | +++ | ++ |
| NIH/3T3-PSF↓ | shPSF | + | ++ |
| NIH/3T3-VL30↑ | VL30–1 RNA | ++ | +++ |
| NIH/3T3-VL30↓ | shVL30–1 RNA | ++ | + |
| B16F10-pcDNA3.1 | None | ++ | +++ |
| B16F10-shLuc | sh-Luciferase | ++ | +++ |
| B16F10-PSF↑ | PSF | +++ | +++ |
| B16F10-VL30↓ | shVL30–1 RNA | ++ | + |
The scale +/++/+++ indicates relative levels of expression.
Binding of PSF to the Regulatory DNAs of Mouse Genes.
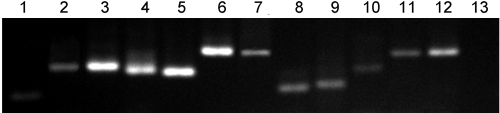
Chromatin fragments from NIH/3T3 cells were coimmunoprecipitated with anti-PSF antibody, and the DNAs in the fragments were tested for hybridization to a mouse gene-promoter chip (NimbleGen; Roche) that contains 385,000 regulatory DNAs from the mouse genome. A total of 57 DNA fragments were identified by chip assay, of which 12 mapped to characterized mouse coding genes [supporting information (SI) Tables S1 and S2]. The 12 DNAs bound to PSF in NIH/3T3 and B16F10 cells, as shown by a ChIP assay (Fig. 2); the P450scc gene had been identified as a PSF-binding gene in earlier studies (4, 6). We chose the Rab23 gene, a member the RAS gene family (10, 11), to analyze the role of PSF and VL30–1 RNA in the regulation of proto-oncogene transcription in mice.
Fig. 2.
Identification of mouse genes that bind PSF. An anti-PSF antibody was used to immunoprecipitate PSF from NIH/3T3 cells, and 12 genes identified by ChIP-analysis (Tables S1 and S2) were tested by semi-quantitative PCR for immunoprecipitation with PSF. 1, Bex2; 2, Ifnz; 3, Kars; 4, Nsfl1c; 5, P450scc; 6, Pik3c3; 7, Rab23; 8, Sema6a; 9, SfiI; 10, Srms; 11, Svep1; 12, Tnfrsf8; 13, β-actin (standard).
Binding of PSF to Rab23 Regulatory DNA and the Effect on Transcription of the Rab23 Gene in NIH/3T3 and B16F10 Cell Lines.
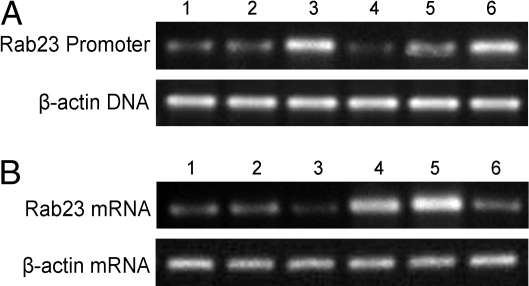
(i) The binding assays were done by ChIP in NIH/3T3 and B16F10 lines expressing high or low levels of PSF (Table 1). Binding of Rab23 DNA to PSF is higher in NIH/3T3-PSF↑ and B16F10-PSF↑ cells and lower in NIH/3T3-PSF↓ cells than in NIH/3T3 or B16F10 control cells (Fig. 3A). (ii) Transcription of Rab23 was analyzed in the same cell lines by RT-PCR. Transcription is lower in NIH/3T3-PSF↑ and B16F10-PSF↑ cells and higher in NIH/3T3-PSF↓ cells than in NIH/3T3 or B16F10 control cells (Fig. 3B). The results indicate that PSF binds and represses transcription of the Rab23 gene in all the NIH/3T3 and B16F10 cell lines.
Fig. 3.
Effect of PSF binding to Rab23 regulatory DNA on transcription of Rab23. 1, NIH/3T3-pcDNA3.1 cells; 2, NIH/3T3-shLuc cells; 3, NIH/3T3-PSF↑ cells; 4, NIH/3T3-PSF↓ cells; 5, B16F10-pcDNA3.1 cells; 6, B16F10-PSF↑ cells. (A) PSF binding assay. Anti-PSF antibody was added to the cell extracts, and the amount of Rab23 regulatory DNA in the immunoprecipitates was determined by semi-quantitative PCR. Upper shows the amount of Rab23 regulatory DNA, and Lower shows the amount of β-actin DNA in the cells before immunoprecipitation, which was used to normalized the total DNA in the samples. (B) Rab23 transcription assay. The amount of Rab23 mRNA was determined by semi-quantitative RT-PCR. Upper shows the amount of Rab23 mRNA, and Lower shows the amount of β-actin mRNA, which was used to normalize the total RNA in the samples.
Effect of PSF on the Regulation of Cell Proliferation and Tumorigenesis in NIH/3T3 and B16F10 Cell Lines.
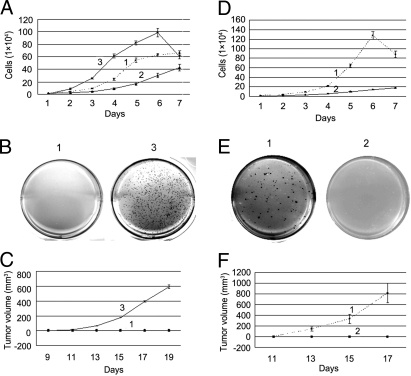
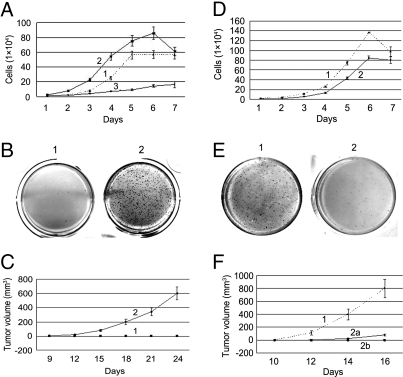
(i) Cell proliferation in culture was faster in NIH/3T3-PSF↓ cells and slower in NIH/3T3-PSF↑ cells than in NIH/3T3 control cells, and it was inhibited by contact in NIH/3T3 control cells but not in NIH/3T3-PSF↓ cells (Fig. 4A). (ii) NIH/3T3-PSF↓ cells but not NIH/3T3-shLuc cells formed colonies in agar and tumors in mice (Fig. 4 B and C). (iii) Cell proliferation in culture was faster in B16F10-pcDNA3.1 cells than in B16F10-PSF↑ cells. (Fig. 4D). (iv) B16F10-pcDNA3.1 cells but not B16F10-PSF↑ cells formed colonies in agar and tumors in mice (Fig. 4 E and F). The results indicate that PSF suppresses cell proliferation and tumorigenesis in the NIH/3T3 and B16F10 cell lines.
Fig. 4.
Effect of PSF on cell proliferation and tumorigenesis in NIH/3T3 and B16F10 cell lines. (A–C) 1, NIH/3T3 control cells (NIH/3T3-pcDNA3.1 and NIH/3T3-shLuc); 2, NIH/3T3-PSF↑ cells; 3, NIH/3T3-PSF↓ cells. (D–F) 1, B16F10-pcDNA3.1 cells; 2, B16F10-PSF↑ cells. (A and D) Rates of cell proliferation. The error bars for each point show the variation in 3 experiments. (B and E) Colony formation in soft agar. (C and F) Tumor formation following injection of the cells into two sites in the necks of 3 nude mice (C) or 3 C57BL/6J mice (F). The error bars for each point show the variation at 6 sites.
Binding of VL30–1 RNA to PSF and the Effect on Binding of PSF to Rab23 DNA and Transcription of the Rab23 Gene.
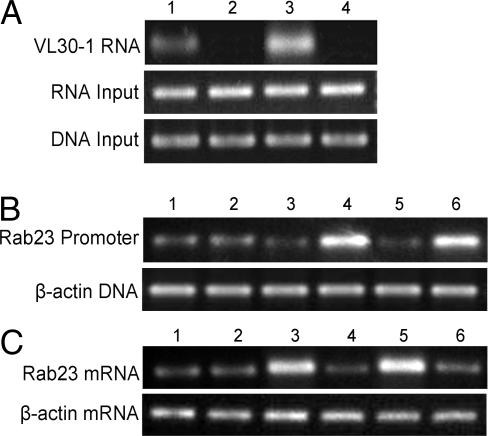
(i) The PSF proteins in NIH/3T3-pcDNA3.1 and NIH/3T3-VL30↑ cells were immunoprecipitated with an anti-PSF antibody, and the amount of VL30–1 RNA coprecipitated with PSF was assayed by RNA immunoprecipitation (RIP) assay. The amount of VL30–1 RNA is higher in NIH/3T3-VL30↑ cells than in NIH/3T3-pcDNA3.1 cells (Fig. 5A), consistent with the expression level of VL30–1 RNA in the 2 cell lines (Fig. 1 and Table 1). It was shown in earlier studies that VL30–1 RNA binds selectively to the RBDs but not to the DBD of the PSF molecule (6). (ii) The PSF proteins in NIH/3T3 and B16F10 cell lines expressing high or low levels of VL30 RNA-1 were immunoprecipitated with an anti-PSF antibody, and the amount of Rab23 DNA coprecipitated with PSF was assayed by ChIP. The amount of Rab23 DNA is lower in NIH/3T3-VL30↑ cells and higher in NIH/3T3-VL30↓ and B16F10-VL30↓ cells than in NIH/3T3 and B16F10 control cells (Fig. 5B). (iii) Rab23 transcription was analyzed using RT-PCR in the same cell lines as above. Transcription was higher in NIH/3T3-VL30↑ cells and lower in NIH/3T3-VL30↓ and B16F10-VL30↓ cells than in NIH/3T3 or B16F10 control cells (Fig. 5C). The results indicate that VL30–1 RNA binds to PSF, reducing the binding of PSF to Rab23 DNA and increasing the transcription of Rab23 mRNA. The mechanism probably involves the release of PSF from Rab23 DNA by VL30–1 RNA, as reported for the human proto-oncogene GAGE6 (9).
Fig. 5.
Binding of VL30–1 RNA to PSF and the effect on binding of PSF to Rab23 regulatory DNA and transcription of the Rab23 gene. (A) Binding of VL30–1 RNA to PSF protein by RIP assay. 1 and 2, NIH/3T3-pcDNA3.1 cells; 3 and 4, NIH/3T3-VL30 RNA↑ cells. PSF antibody was added to cell extracts in 1 and 3 but not in 2 and 4. DNA and RNA inputs of β-actin were used to normalize the amount of RIP products. (B and C) 1, NIH/3T3-pcDNA3.1 cells; 2, NIH/3T3-shLuc cells; 3, NIH/3T3-VL30↑ cells; 4, NIH/3T3-VL30↓ cells; 5, B16F10-shLuc cells; 6, B16F10-VL30↓ cells. (B) ChIP assay for binding of PST to Rab23 regulatory DNA. Upper shows the amount of Rab23 promotor DNA, and Lower shows the amount of β-actin DNA used to normalize the total DNA in the samples. (C) Semi-quantitative RT-PCR assays for transcription of the Rab23 gene. Upper shows the amount of Rab23 mRNA, and Lower shows the amount of β-actin mRNA used to normalize the total RNA in the samples.
Effect of VL30–1 RNA on the Regulation of Cell Proliferation and Tumorigenesis in NIH/3T3 and B16 F10 Cell Lines.
(i) Cell proliferation is faster in NIH/3T3-VL30↑ cells and slower in NIH/3T3-VL30↓ cells than in NIH/3T3 control cells, and it is inhibited by contact in NIH/3T3 control cells but not in NIH/3T3-VL30↑ cells (Fig. 6A). (ii) Colony formation in agar and tumor formation in mice are higher in NIH/3T3-VL30↑ cells than in NIH/3T3-pcDNA3.1 cells (Fig. 6 B and C). (iii) Cell division is faster in B16F10-shLuc cells than in B16-VL30↓ cells, and it is not inhibited by contact in B16F10-shLuc cells (Fig. 6D). (iv) Colony formation in agar and tumor formation in mice are higher in B16F10-shLuc cells than in B16-VL30↓ cells (Fig. 6 E and F). The results indicate that VL30–1 RNA promotes cell proliferation and tumorigenesis in the NIH/3T3 and B16F10 cell lines.
Fig. 6.
Effect of VL30–1 RNA on cell proliferation and tumorigenesis in NIH/3T3 and B16 F10 cell lines. (A–C) 1, NIH/3T3 control cells (NIH/3T3-pcDNA3.1 and NIH/3T3-shLuc); 2, NIH/3T3-VL30↑ cells; 3, NIH/3T3-VL30↓ cells. (D–F) 1, B16F10-shLuc cells; 2, B16F10-VL30↓ cells. (A and D) Rates of cell proliferation. The error bars for each point show the variation in 3 experiments. (B and E) Colony formation in soft agar. (C and F) Tumor formation following injection of the cells into two sites in the necks of 3 nude mice (C) or 3 C57BL/6J mice (F). The error bars for each point show the variation at 6 sites. Curve 2a in F shows the tumors size at 3 sites, and curve 2b shows the other 3 sites that had no visible tumor.
Expression of PSF mRNA and VL30–1 RNA in Differentiated Cells and Tumor Cells.
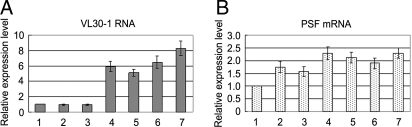
The purpose of this experiment was to determine whether tumorigenesis in mouse tumor lines is driven by decreased expression of PSF, increased expression of VL30–1 RNA, or both. The expression levels of PSF mRNA and VL30–1 RNA were assayed by real-time RT-PCR in differentiated fibroblast and myoblast lines and 4 tumor lines. The level of VL30–1 RNA is 5 to 8 times higher in the tumor lines than in the fibroblast and myoblast lines, whereas the level of PSF mRNA does not decrease in the tumor lines (Fig. 7 A and B). The results suggest that increased expression of VL30–1 RNA, rather than decreased expression of PSF protein, drives tumorigenesis in mouse tumor cells.
Fig. 7.
Expression of VL30–1 RNA and PSF mRNA in mouse fibroblast cells, myoblast cells, and tumor cells. Cell lines: 1, primary fibroblast; 2, NIH/3T3 fibroblast; 3, C2C12 myoblast; 4, TC-1 lung tumor; 5, Y-1 adrenal tumor; 6, EMT6 mammary tumor; 7, B16F10 melanoma tumor. RNA expression levels were assayed by real-time RT-PCR and normalized to the level in primary fibroblasts.
Discussion
Previous studies showed that PSF binds and represses transcription of multiple genes in a human tumor cell line, mostly proto-oncogenes that drive the cell cycle, and VL30–1 RNA binds and releases PSF from a proto-oncogene and activates transcription (5–7). Here, we analyzed the regulatory functions of PSF and VL30–1 RNA in mouse NIH/3T3 fibroblast and B16F10 melanoma cell lines. The experiments involved transfecting a coding gene for PSF or VL30–1 RNA to increase expression (PSF↑ and VL30↑ cell lines) or a shRNA construct that causes degradation of PSF mRNA or VL30–1 RNA to decrease expression (PSF↓ and VL30↓ cell lines). The cell lines were compared for binding of PSF to the proto-oncogene Rab23, transcription of Rab23, rate of cell proliferation in culture, and tumorigenesis by colony formation in agar and tumor formation in mice (Table 2).
Table 2.
Regulatory functions of PSF and VL30-1 RNA in NIH/3T3 abd B16F10 cell lines
| Cell line | Rab23 DNA binding to PSF | Rab23 transcription | Cell proliferation | Tumorigenesis |
|---|---|---|---|---|
| NIH/3T3-pcDNA3.1 | ++ | ++ | ++ | 0 |
| NIH/3T3-shLuc | ++ | ++ | ++ | 0 |
| NIH/3T3-PSF↑ | +++ | + | + | Not tested |
| NIH/3T3-PSF↓ | + | +++ | +++ | +++ |
| NIH/3T3-VL30↑ | + | +++ | +++ | +++ |
| NIH/3T3-VL30↓ | +++ | + | + | Not tested |
| B16F10-pcDNA3.1 | + | +++ | +++ | +++ |
| B16F10-shLuc | + | +++ | +++ | +++ |
| B16F10-PSF↑ | +++ | + | + | 0 |
| B16F10-VL30↓ | +++ | + | ++ | 0 to + |
The scale 0/+/++/+++ indicates relative levels of activity.
The results indicate that PSF is a tumor-suppressor protein and VL30–1 RNA is a tumor-promoter RNA in mice. All cell lines express PSF and VL30–1 RNA; a high molar ratio of PSF/VL30–1 RNA suppresses and a low molar ratio promotes proto-oncogene transcription, cell proliferation, and tumorigenesis. The tumor-promoter function of VL30–1 RNA involves binding to PSF (Fig. 5A) and forming a PSF-RNA complex that dissociates from a repressed proto-oncogene (4, 5, 9). PSF and VL30–1 RNA appear to have a primary role in tumorigenesis, because a decrease of PSF expression or increase of VL30–1 RNA expression transforms NIH/3T3 cells, whereas an increase of PSF expression or decrease of VL30–1 RNA expression suppresses tumorigenesis of B16F10 cells. Although VL30–1 RNA can integrate into the cell genome (3, 12), the tumor-promoter function of VL30–1 RNA involves a trans effect rather than a cis effect on gene transcription. A similar regulatory mechanism functions in human cells with human PSF-binding RNAs replacing VL30–1 RNA (13), which is not encoded in the human genome.
To determine whether tumorigenesis in mice is associated with decreased expression of PSF, increased expression of VL30–1 RNA, or both, we compared expression of PSF mRNA and VL30–1 RNA in 4 tumor lines with fibroblast and myoblast lines (Fig. 7). Expression of VL30–1 RNA increased 5- to 8-fold in the tumor lines compared with the fibroblast and myoblast lines, whereas expression of PSF mRNA did not decrease in the tumor lines, suggesting that tumorigenesis is driven by an increase in VL30–1 RNA expression rather than a decrease in PSF expression. Tumorigenesis also can result from mutations affecting the synthesis or function of PSF, as indicated by the human cervical tumor line HeLa, which has a mutation in the PSF gene that deleted the coding region for the DBD of PSF and was the probable cause of the HeLa tumor (5).
Tumorigenesis can progress via multiple pathways controlled by proto-oncogenes (14). The results reported here and in other studies (5–7, 12) suggest that tumorigenic pathways are initiated and driven by increased expression of PSF-binding RNAs, which reverse repression of proto-oncogenes by PSF. The regulation of tumorigenesis by PSF and PSF-binding RNAs identifies 2 previously undescribed markers for tumor cells: a reduced level of proto-oncogene promoters bound to PSF and an elevated level of PSF-binding RNAs. The former can be assayed by ChIP, and the latter can be assayed by RT-PCR. Both markers could be the basis of therapeutic procedures for cancer that increase the amount of PSF or deplete the amount of PSF-binding RNAs in tumor cells.
Materials and Methods
Cell Lines.
The mouse fibroblast line NIH/3T3, myoblast line C2C12, melanoma line B16F10, and mammary carcinoma line EMT6 were cultured in DMEM; the mouse lung tumor line TC-1 was cultured in RPMI 1640; and the mouse adrenal line Y-1 was cultured in F12 medium. Mouse primary fibroblast cells were isolated from the dorsal skin of 15-day C57BL/6J embryos. All cells were grown as attached monolayers in the indicated medium supplemented with 10% (vol/vol) FBS, penicillin, and streptomycin in a CO2 incubator at 37 °C.
Plasmids Encoding a shRNA.
A shRNA target site in PSF mRNA or VL30–1 RNA was identified, and complementary sense and antisense oligonucleotides encoding a shRNA with a 19-mer stem derived from the target site were synthesized as follows: (i) for PSF mRNA, the plasmid was called pGenesil-shPSF: 5′-GATCCGTTAAGGCGCATGGAGGAACTTCAAGAGAGTTCCTCCATGCGCCTTAATTTTTTA-3′ and 5′-AGCTTAAAAAATTAAGGCGCATGGAGGAACTCTCTTGAAGTTCCTCCATGCGCCTTAACG-3′ and (ii) for VL30–1 RNA, the plasmid was called pGenesil-shVL30: 5′-GATCCGcagaaaaactgctttcactTTCAAGAGAAGTGAAAGCAGTTTTTCTGTTTTTTA-3′ and 5′-AGCTTAAAAAAcagaaaaactgctttcactTCTCTTGAAAGTGAAAGCAGTTTTTCTGCG-3′.
The sense oligonucleotide contained the BamH1 3′ residues (5′-gatcc-3′), followed by the 19-nucleotide target sequence, a 9-nucleotide loop (5′-ttcaagaga-3′), a 19-nucleotide reverse sequence complementary to the target sequence, a TTTTTT tract to terminate transcription, and a HindIII 5′ residue. The antisense oligonucleotide was complementary to the sense oligonucleotide except for the upstream HindIII 3′ residue (5′-agctt-3′) and the downstream BamH1 5′ residues -g. To facilitate transcription by mouse U6 promoter, an additional guanine was added to the target sequence that began with a pyrimidine. The sense and antisense oligonucleotides were annealed and inserted into the BamH1/HindIII sites of the plasmid pGenesil-1 (Genesil Company). The control plasmid pGenesil-shLuc targeted the luciferase mRNA sequence GTAGCGCGGTGTATTATAC.
Stably Transfected Cell Lines.
(i) The NIH/3T3-VL30↑ cell line was constructed by transfecting NIH/3T3 cells with the plasmid pcDNA-VL30 encoding the 319-bp VL30–1 RNA fragment containing the PSF-binding sequences (5). (ii) The NIH/3T3-PSF↑ or B16F10-PSF↑ cell line was constructed by transfecting NIH/3T3 or B16F10 cells with plasmid pcDNA-PSF (5). (iii) The NIH/3T3-VL30↓ or B16F10-VL30↓ cell line was constructed by transfecting NIH/3T3 or B16F10 cells with plasmid pGenesil-shVL30. (iv) The NIH/3T3-PSF↓ cell line was constructed by transfecting NIH/3T3 cells with the plasmid pGenesil-shPSF. (v) NIH/3T3-pcDNA3.1 or B16F10-pcDNA3.1, the control cell line for VL30↑ or PSF↑, was constructed by transfecting NIH/3T3 or B16F10 cells with the empty plasmid pcDNA-3.1(+). (vi) NIH/3T3-shLuc or B16F10-shLuc, the control cell line for VL30↓ or PSF↓, was constructed by transfecting NIH/3T3 or B16F10 cells with the plasmid pGenesil-shLuc. The transfection reagent Lipofectamine 2000 (Invitrogen) was used for all transfections. Two days after transfection, transfected cells were selected with 800 μg/mL G418 for 10 days and maintained with 500 μg/mL G418. Cell clones derived from a single cell were isolated, and the expression levels of target genes were analyzed by semiquantitative RT-PCR.
Semiquantitative RT-PCR.
Cells were cultured to approximately 80% confluence in 6-well plates, washed with ice-cold PBS, and lysed in 1 mL of TRIzol (Invitrogen), and RNA was isolated according to the manufacturer's protocol. cDNA was synthesized from 1 μg of RNA in a 20-μL reaction volume containing 4 μL of 5× reaction buffer, 1 μL of dNTP mix (10 mM each), 1 μL of random hexamers (10 μM), 2 μL of 0.1 M DTT, 0.5 μL of RNaseOUT Recombinant Ribonuclease Inhibitor (40 U/μL; Invitrogen), 0.5 μL of M-MLV reverse transcriptase (200 U/μL; Invitrogen), and H2O. β-actin mRNA was used to normalize the amounts of RNA in the samples. The forward and reverse PCR primers were as follows: β-actin: 5′-CTCCTCCCTGGAGAAGAGCTA-3′ and 5′-CCTTCTGCATCCTGTCGGCAA-3′, PSF: 5′-CAAGATCTGATGAGACGCCAGGAA-3′ and 5′-CCATCTCACGTTGGCGAATCATCA-3′, VL30: 5′-GAGAGCAGCCAGCGGGTCACAGT-3′ and 5′-GGCGCTCTTGGCCGGCTACTCG-3′, and Rab23: 5′-TTCTTAGTGGCGGGCACAGAGC-3′ and 5′-GAGCATCCGCCCCACTTCACTC-3′. The PCR conditions were 95 °C for 1 min and suitable cycles at 95 °C for 10 sec, 64 °C for 30 sec, and 72 °C for 20 sec, followed by 72 °C for 2 min.
Real-Time Quantitative RT-PCR.
One μg RNA sample was reverse-transcribed into cDNA in a 20-μL reaction volume as described previously, and the PCR assay was performed with 1 μL of the cDNA using 2× TaKaRa SYBR Green Premix (TaKaRa) on a BioRad iCycler. All reactions were done in a-20 μL reaction volume in triplicate. The specificity of amplification was verified by melt-curve analysis and agarose gel electrophoresis, and the data were collected using BioRad iQ5 software. The relative amount of target gene mRNA was normalized to a β-actin mRNA standard. For the VL30–1 RNA assay, the primers for VL30–1 RNA and β-actin mRNA were the same as for semiquantitative RT-PCR. The PCR conditions were 1 min at 95 °C, followed by 40 cycles of PCR at 95 °C for 10 sec, 64 °C for 30 sec, and 72 °C for 20 sec. For the PSF mRNA assay, the primers were: PSF forward 5′-TGGCAGTGTACCTTGTGCCACCGAAT-3′ and reverse 5′-AGGGGGGGCACCATTTGTAGAAAGCA-3′ and β-actin forward 5′-CCGCATCCTCTTCCTCCCTGGAGAA-3′ and reverse 5′-GCCTGGGTACATGGTGGTACCACCA-3′. The PCR conditions were 1 min at 95 °C, followed by 40 cycles at 95 °C for 10 sec and 68 °C for 50 sec.
Cell Proliferation.
Cells were grown in 12-well plates at 37 °C as attached monolayers in DMEM with 10% (vol/vol) FBS. Samples were recovered from 3 wells each day by treatment with trypsin, and the number of viable cells was counted. Each point is the mean ± SE (n = 3).
Soft-Agar Colony Assay.
Cells were suspended in 1 mL of 0.3% melted agar in DMEM with 10% (vol/vol) FBS and plated in 6-well plates containing a solidified layer of 0.6% agar in the same medium. After a suitable period of incubation, 0.5 mg of nitro blue tetrazolium in 400 μL of PBS was added to the dishes; 4 h later, the colonies were scanned with an Epson 4990 Scanner.
Tumor Formation in Mice.
Cells were detached from the culture plates by a brief incubation with 2 mM EDTA in PBS, suspended in DMEM/10% (vol/vol) FBS, washed, and resuspended in PBS. The cells were inoculated s.c. at 2 sites on the neck as follows: 2 × 105 cells from the NIH/3T3 lines were inoculated into 3 female 6-week-old nude mice, and 4 × 104 cells from the B16F10 lines were inoculated into 3 C57BL/6J mice. The length (L) and width (W) of the resulting tumors were measured every 2 or 3 days, and tumor volumes were calculated by the formula: volume = L × W2/2.
ChIP and RIP Assays.
Cells were cultured in 100-mm plates to ≈70–80% confluence, reversibly cross-linked with formaldehyde, and immunoprecipitated with an anti-PSF polyclonal antibody (from James Patton, Vanderbilt University, Nashville, TN). (i) The ChIP assays were done according to the protocol provided with the ChIP assay kit (Upstate Biotechnology). The DNA fragments in PSF-DNA complexes were analyzed by semiquantitative PCR. PCR primers of the precipitated DNAs were designed based on the NimbleGen's promoter database (Roche) and University of California Santa Cruz mouse genome data MM8 (Table S2). The PCR parameters were 95 °C for 1 min and 22 cycles at 95 °C for 10 sec, 64 °C or 68 °C for 30 sec, and 72 °C for 20 sec. The PCR product was diluted 10-fold, and 2 μL of template was used for the next round of the PCR assay according to the same parameters as above. (ii) The RIP assays were done as described (15). VL30–1 RNA in PSF-RNA complexes was analyzed by extracting the RNA with TRIzol reagent and assaying by RT-PCR.
ChIP-Chip Assay.
The ChIP-chip system (NimbleGen; Roche), which combines the chromatin immunoprecipitation (ChIP) and promoter microarray (chip) assays, was used to identify the genes associated with the PSF-binding DNAs. Following the NimbleGen ChIP-chip protocol, the ChIP products and input control were purified and amplified using ligation-mediated (LM) PCR. The LM-PCR products were purified using the Qiaquick PCR Purification kit (Qiagen) and sent to the NimbleGen for the chip assay.
Western Blot Analysis.
The cellular proteins were fractionated by 8% (w/v) SDS/PAGE and electronically transferred to a PVDF membrane at 100 V for 1 h. The membrane was rinsed with PBS-tween (PBST), blocked for 2 h in PBST containing 5% (w/v) powdered dried milk, and incubated with primary antibody for 1 h. The membrane was washed with PBST and incubated for 1 h with secondary antibody conjugated to HRP. Blots were visualized using an ECL detection kit (Amersham Pharmacia Biotech) and exposed to x-ray films. The primary antibody for PSF was mouse anti-PSF (Sigma), and the secondary antibody was HRP-conjugated anti-mouse IgG; the primary antibody for β-actin was rabbit anti-β-actin, and the secondary antibody was HRP-conjugated anti-rabbit IgG.
Supplementary Material
Acknowledgments.
This work was supported by the National Natural Science Foundation of China (Grants 30700459 and 90919005), the Ph.D. Programs Foundation of the Ministry of Education of China (Grant 20070610170), and the Scientific Research Foundation for Returned Scholars, Ministry of Education of China (Grant 20071108–18-9).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0909022106/DCSupplemental.
References
- 1.Patton JG, Porro EB, Galceran J, Tempst P, Nadal-Ginard B. Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes Dev. 1993;7:393–406. doi: 10.1101/gad.7.3.393. [DOI] [PubMed] [Google Scholar]
- 2.French NS, Norton JD. Structure and functional properties of mouse VL30 retrotransposons. Biochim Biophys Acta. 1997;1352:33–47. doi: 10.1016/s0167-4781(97)00009-2. [DOI] [PubMed] [Google Scholar]
- 3.Song X, et al. Retroviral-mediated transmission of a mouse VL30 RNA to human melanoma cells promotes metastasis in an immunodeficient mouse model. Proc Natl Acad Sci USA. 2002;99:6269–6273. doi: 10.1073/pnas.092112199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Urban RJ, Bodenburg Y. PTB-associated splicing factor regulates growth factor-stimulated gene expression in mammalian cells. Am J Physiol. 2002;283:E794–E798. doi: 10.1152/ajpendo.00174.2002. [DOI] [PubMed] [Google Scholar]
- 5.Song X, Sui A, Garen A. Binding of mouse VL30 retrotransposon RNA to PSF protein induces genes repressed by PSF: Effects on steroidogenesis and oncogenesis. Proc Natl Acad Sci USA. 2004;101:621–626. doi: 10.1073/pnas.0307794100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song X, Sun Y, Garen A. Roles of PSF protein and VL30 RNA in reversible gene regulation. Proc Natl Acad Sci USA. 2005;102:12189–12193. doi: 10.1073/pnas.0505179102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garen A, Song X. Regulatory roles of tumor-suppressor proteins and noncoding RNAs in cancer and normal cell functions. Int J Cancer. 2008;122:1687–1689. doi: 10.1002/ijc.23285. [DOI] [PubMed] [Google Scholar]
- 8.Harrigan MT, Baughman G, Campbell N, Bourgeois S. Isolation and characterization of glucocorticoid and cyclic AMP-induced genes in T lymphocytes. Mol Cell Biol. 1989;9:438–446. doi: 10.1128/mcb.9.8.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Owen RD, Bortner DM, Ostrowski MC. ras oncogene activation of a VL30 transcriptional element is linked to transformation. Mol Cell Biol. 1990;10:1–9. doi: 10.1128/mcb.10.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bos JL. ras oncogenes in human cancer: A review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- 11.Eggenschwiler JT, Espinoza E, Anderson KV. Rab23 is an essential negative regulator of the mouse Sonic hedgehog signalling pathway. Nature. 2001;412:194–198. doi: 10.1038/35084089. [DOI] [PubMed] [Google Scholar]
- 12.Noutsopoulos D, Vartholomatos G, Kolaitis N, Tzavaras T. SV40 large T antigen up regulates the retrotransposition frequency of viral-like 30 elements. J Mol Biol. 2006;301:450–461. doi: 10.1016/j.jmb.2006.06.030. [DOI] [PubMed] [Google Scholar]
- 13.Li L, et al. Role of human noncoding RNAs in the control of tumorigenesis. Proc Natl Acad Sci USA. 2009;106:12956–12961. doi: 10.1073/pnas.0906005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frank SA. Dynamics of Cancer. Princeton: Princeton Univ Press; 2007. [PubMed] [Google Scholar]
- 15.Sun BK, Deaton AM, Lee JT. A transient heterochromatic state in Xist preempts X-inactivation choice without RNA stabilization. Mol Cell. 2006;21:617–628. doi: 10.1016/j.molcel.2006.01.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.