Abstract
The superficial zone (SZ) of articular cartilage is critical in maintaining tissue function and homeostasis and represents the site of the earliest changes in osteoarthritis. Mechanisms that regulate the unique phenotype of SZ chondrocytes and maintain SZ integrity are unknown. We recently demonstrated that expression of the chromatin protein high mobility group box (HMGB) protein 2 is restricted to the SZ in articular cartilage suggesting a transcriptional regulation involving HMGB2 in SZ. Here, we show that an interaction between HMGB2 and the Wnt/β-catenin pathway regulates the maintenance of the SZ. We found that the Wnt/β-catenin pathway is active specifically in the SZ in normal mouse knee joints and colocalizes with HMGB2. Both Wnt signaling and HMGB2 expression decrease with aging in mouse joints. Our molecular studies show that HMGB2 enhances the binding of Lef-1 to its target sequence and potentiates transcriptional activation of the Lef-1-β-catenin complex. The HMG domain within HMGB2 is crucial for interaction with Lef-1, suggesting that both HMGB2 and HMGB1 may be involved in this function. Furthermore, conditional deletion of β-catenin in cultured mouse chondrocytes induced apoptosis. These findings define a pathway where protein interactions of HMGB2 and Lef-1 enhance Wnt signaling and promote SZ chondrocyte survival. Loss of the HMGB2-Wnt signaling interaction is a new mechanism in aging-related cartilage pathology.
Keywords: aging, osteoarthritis, apoptosis, superficial zone
Articular cartilage is a tissue that provides biomechanical properties that allow near frictionless joint movement and dispersion of mechanical loads. Cartilage is composed of a single cell lineage but differences in the organization, phenotype and function of cells in the various layers of cartilage have been recognized (1–4). The superficial zone (SZ) is the most unique. SZ cells produce lubricin, also termed proteoglycan-4 (PRG4) or superficial zone protein (SZP), an important joint lubricant (5–7), and are more responsive to stimulation by catabolic cytokines such as IL-1 (8). Recent studies also suggest that the SZ contains cells that express mesenchymal stem cell markers (9–11).
Articular cartilage is among the tissues that undergo profound aging-related changes and aging represents the major risk factor for osteoarthritis (OA), the most prevalent joint disease (12). Aging-related changes in cartilage include reduced cellularity, increased apoptosis and altered cellular responses to growth factors, cytokines and mechanical stress (13–15). Cartilage changes in aging and OA begin in the SZ and once the SZ is disrupted this is followed by progressive erosion of the remaining cartilage layers (16).
To address mechanisms that maintain the unique phenotype of SZ cells we performed gene expression analyses and observed that expression of the chromatin protein HMGB2 is restricted to the SZ (17). Joint aging in humans and mice leads to loss of HMGB2 expression and this is correlated with the onset of OA-like changes. Mice deficient in Hmgb2 develop early onset and more severe OA, and this is associated with a reduction in cartilage cellularity attributable to increased cell death (17).
Wnt proteins are secreted factors that regulate cell proliferation and differentiation during early stages of chondrogenesis (18, 19). Overexpression of β-catenin in prechondrogenic cells inhibits overt chondrocytic differentiation (20) and overexpression in chick limb buds accelerates hypertrophic differentiation (21). In contrast, inhibition of β-catenin signaling by overexpression of Frzb-1, dominant negative Wnt receptors, results in delayed maturation (22). Homozygous deletion of β-catenin is embryonic lethal but conditional deletion in cartilage was associated with delayed chondrocyte hypertrophy and reduced chondrocyte proliferation in growth plates (23). Conditional mutant mice deficient in Wnt/β-catenin signaling displayed a defective flat cell layer normally abutting the synovial cavity and markedly reduced levels of PRG4/SZP (24). This supports the importance of Wnt signaling in skeletal development and early stages of chondrocyte differentiation.
Recent studies indicate that Wnt signaling has a role in adult articular cartilage. Increased Wnt signaling due to loss of sFRPS function represents a risk factor for OA (25). Similarly, overexpression of β-catenin in chondrocytes stimulates the expression of matrix degradation enzymes (26). However, Wnt signaling also contributes to differentiation and maintenance of articular cartilage chondrocytes. Inhibition of β-catenin signaling by transgenic overexpression of its intracellular antagonist ICAT results in progressive SZ degradation and development of OA (27). These studies suggest that the precise temporal and spatial activation of Wnt signaling in articular cartilage determines its homeostatic versus pathogenic effects.
Taken together, these reports on Wnt/β-catenin and our observations on HMGB2 suggest possible interactions in the maintenance of the SZ in adult cartilage. Here, we define a molecular mechanism by which HMGB2 and β-catenin regulate cartilage SZ integrity.
Results
β-Catenin Signaling Is Activated in the SZ of Articular Cartilage and Decreases with Aging.
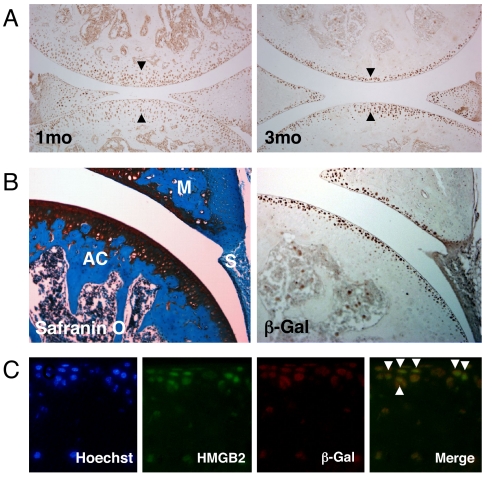
β-catenin is an important regulator of chondrocyte maturation in growth plate and its expression and function during skeletal development have been characterized (19, 21, 28). To analyze β-catenin in adult cartilage we used the TOPGAL transgenic mouse model where the β-galactosidase gene is under the control of a LEF/TCF and β-catenin inducible promoter and allows direct detection of cells and tissues with active Wnt signaling (29). Wnt/β-catenin signaling has been reported to be active at early stages during joint formation and to remain active and prominent at later stages in small and large joints (24). In 1-month-old TOPGAL mice, we detected β-galactosidase activity in all zones of articular cartilage. At 3 months with joint maturation it became more restricted to the cartilage surface (Fig. 1A). At this stage, β-galactosidase protein was expressed in the SZ of articular cartilage in meniscus but not in synovium (Fig. 1B). Because this pattern is similar to that of HMGB2 (17), we performed double immunofluorescence assay, and verified that most SZ cells express both HMGB2 and β-galactosidase (Fig. 1C).
Fig. 1.
Active Wnt signaling and correlation with HMGB2 expression in the articular cartilage SZ. (A) Immunohistochemistry was performed with β-galactosidase antibody on knee joint sections from 1 and 3-month-old TOPGAL mice. Between 1 and 3 months of age the β-galactosidase positive cells become more restricted to the superficial cell layers in articular cartilage. (B) β-galactosidase (β-Gal) positive cells are found in articular cartilage and meniscus, whereas synovium is negative. Safranin O staining of the adjacent section. AC, articular cartilage; M, meniscus; S, synovium. X100. (C) HMGB2 and β-galactosidase expression by immunofluorescence assay. Colocalization of HMGB2 and β-galactosidase (β-Gal) positive cells is found in the SZ in articular cartilage at 3 months of age (arrowheads). Hoechst dye 33258 was used to stain nuclei. (Magnification: ×400.)
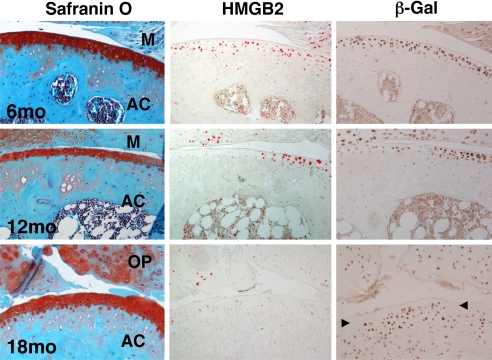
Articular cartilage in C57BL/6J mice undergoes aging-related changes that are similar to osteoarthritis joint pathology (30), and this was also observed in TOPGAL mice on CD1 background (Fig. 2). At 6 months of age articular cartilage had normal appearance, and HMGB2 and β-galactosidase positive cells were present in the superficial cell layers. At 12 months of age there was a reduction in cartilage thickness and cellularity and surface irregularities were prominent in the central weight-bearing areas of the tibial plateau. At 12 months HMGB2 and β-galactosidase were both absent in the SZ in the weight bearing areas, and HMGB2 was completely absent in all regions of articular cartilage by 18 months (Fig. 2). In contrast, β-galactosidase was enhanced in the mid and deep zone, in calcified cartilage, subchondral bone and in osteophytes at 18 months. These findings demonstrate a correlated aging-related loss of HMGB2 and β-catenin signaling in the SZ of articular cartilage and this is associated with OA-like pathology.
Fig. 2.
HMGB2 and β-galactosidase expression during aging in TOPGAL mice. Safranin-O stained sections of joints from TOPGAL mice show normal cartilage at 6 months, reduced thickness and cellularity at 12 and 18 months. HMGB2 and β-galactosidase (β-Gal) are detected by immunohistochemistry at 6 months in the articular cartilage surface. At 12 months both are absent in the weight bearing areas, and HMGB2 is completely absent in the articular cartilage by 18 months. In contrast, at 18 months β-galactosidase becomes detectable in all other zones of articular cartilage except for the SZ (arrowheads). AC, articular cartilage; M, meniscus; OP, osteophyte. X100.
Functional Interactions of β-Catenin and HMGB2.
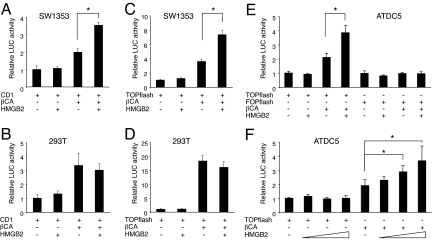
The in vivo colocalization of HMGB2 and Wnt/β-catenin activity (Fig. 1) and the correlation of their loss in OA-like pathology suggest interaction of HMGB2 and Wnt/β-catenin in the SZ. To study this in detail, we performed luciferase-reporter assays. Using cyclin D1 promoter (−962CD1) (31), β-catenin transfection caused the expected increase in luciferase activity in both SW1353 chondrosarcoma cells and 293T kidney epithelial cells (Fig. 3 A and B). Transfection of HMGB2 (32) did not change luciferase activity but cotransfection of HMGB2 and β-catenin resulted in synergistic enhancement in SW1353 chondrosarcoma cells (Fig. 3A); this synergy was not observed in 293T kidney epithelial cells (Fig. 3B). Similar differences between cell types were obtained using the TOPflash promoter, which contains multiple repeats of the β-catenin-TCF/LEF consensus sequences (33) (Fig. 3 C and D). The synergistic activity of HMGB2 and β-catenin was also seen in chondrogenic ATDC5 cells. Transfection of the FOPflash promoter with a mutated Lef-1 binding site showed no activity (Fig. 3E) but the activity of TOPflash promoter was enhanced by HMGB2 in a dose-dependent manner under β-catenin transfection (Fig. 3F). These experiments demonstrate synergistic interaction of β-catenin and HMGB2 in enhancing Lef-1 responsive promoters, specifically in chondrogenic cell types.
Fig. 3.
Synergy of HMGB2 and β-catenin. HMGB2 and β-catenin (βCA) were cotransfected with the cyclin D1 (CD1) (A and B) and TOPflash reporter genes (C–F) in SW1353 cells (A and C) and 293T cells (B and D), and luciferase assay was performed after 24 h. In SW1353 cells, HMGB2 enhances luciferase activity when cotransfected with β-catenin (A and C), whereas this synergistic effect is not seen in 293T cells (B and D). This synergistic effect is also found with TOPflash in a dose-dependent manner (F), but not with FOPflash reporter genes in ATDC5 cells (E) (*, P < 0.05).
Physical Interactions of HMGB2 and Lef-1.
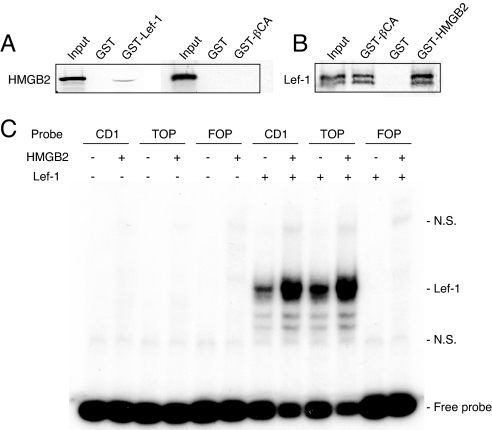
To examine molecular interactions between β-catenin, Lef-1 and HMGB2, GST-pull down assays were performed using bacterially expressed GST-HMGB2, β-catenin and Lef-1 and in vitro-translated HMGB2, β-catenin and Lef-1 (34). We observed that in vitro-translated HMGB2 bound GST-Lef-1, but not GST-β-catenin (Fig. 4A). The results from the reverse experiment showed that in vitro-translated Lef-1 interacted with GST-HMGB2 and GST-β-catenin (Fig. 4B). When in vitro-translated β-catenin protein was incubated with GST-HMGB2 and GST-Lef-1, only GST-Lef-1 but not GST-HMGB2 was pulled down (Fig. S1). This indicates a specific interaction between HMGB2 and Lef-1, leading to enhanced transcriptional activation of the Lef-1-β-catenin complex.
Fig. 4.
GST-pull down assay for β-catenin (βCA), Lef-1 and HMGB2. (A) In vitro-translated HMGB2 interacts with GST-Lef-1, but not with GST-β-catenin. (B) In vitro-translated Lef-1 interacts with both GST-β-catenin and GST-HMGB2. (C) DNA binding interactions of HMGB2 and Lef-1 by EMSA. Using nuclear extracts of SW1353 cells transfected with Lef-1, binding of Lef-1 with both cyclin D1 (CD1) and TOP probes was detected. This was enhanced by the addition of HMGB2 protein (1 μg). In contrast, no binding was detected on the FOP probe. N.S., nonspecific.
Interaction Domains of HMGB2 and Lef-1.
To define the Lef-1 interaction domain within HMGB2, in vitro GST pull-down assays were performed using GST-Lef-1 and HMGB2 deletion mutants (A-box, B-box, acidic tail) as shown in Fig. S2A. The results demonstrated that none of these 3 HMGB2 mutants interacts with Lef-1 (Fig. S2B). Then we constructed A-box and B-box domains with linker regions and found that both bind Lef-1 (Fig. S2B). Delta box, which contains linker and acidic tail but not A-box or B-box, did not interact with Lef-1. These results indicate that A-box or B-box together with the linker region are required for HMGB2 binding to Lef-1.
Next we determined the domains in Lef-1 that are required for interaction with HMGB2. GST pull-down assays were performed with GST-HMGB2 and Lef-1 deletion mutant plasmids (FL, ΔN113, ΔN295, ΔN113-ΔC102) (35). The results showed that GST-HMGB2 could pull down Lef-1 FL (Fig. 4B), Lef-1 ΔN113 and Lef-1 ΔN295 but not Lef-1 ΔN113-ΔC102 (Fig. S2C), indicating that the HMG domain in Lef-1 is responsible for the physical interaction with HMGB2.
DNA Binding Interactions of HMGB2 and Lef-1.
To understand how the HMGB2-Lef-1 interaction contributes to Wnt/β-catenin activity, we examined whether the interaction affects Lef-1 DNA binding. Gel shift assays were performed to determine binding specificity and interactions of HMGB2 and Lef-1. We prepared oligonucleotide probes with Lef-1 binding sites (cyclin D1 and TOP) and a probe with a mutated Lef-1 binding site (FOP) as described in ref. 36. Using nuclear extracts of Lef-1 transfected SW1353 chondrosarcoma cells, we detected binding of Lef-1 to both cyclin D1 and TOP probes, and this binding was enhanced by the addition of purified HMGB2 (Fig. 4C). In contrast, no binding was detected on the FOP probe. HMGB2 did not interact with cyclin D1 and TOP probes without overexpressed Lef-1.
c-Jun is a target gene for the β-catenin-Tcf/Lef transcriptional complex (37), and Wnt signaling induces c-Jun expression in chondrocytes (38). We also detected binding of Lef-1 to c-Jun probes in SW1353 cells in the presence of Lef-1, and this binding was potentiated by the addition of HMGB2 protein (Fig. S3). These results suggest that interaction between HMGB2 and Lef-1 enhances DNA binding affinity of Lef-1, to enhance Wnt/β-catenin signaling.
HMGB2 and Wnt/β-Catenin Target Gene Expression.
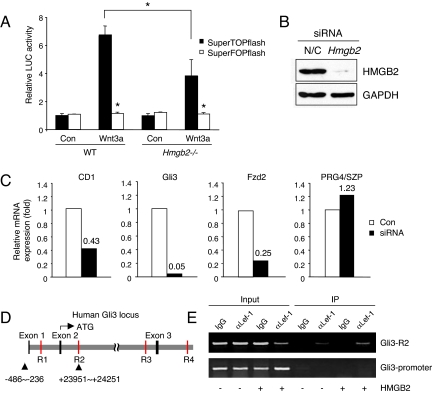
To examine whether the interaction between HMGB2 and Lef-1 potentiates Wnt/β-catenin signaling activity, we examined Wnt/β-catenin signaling in WT and Hmgb2−/− chondrocytes using SuperTOPflash, which contains 8 TCF/LEF binding sites and the corresponding negative control vector SuperFOPflash (39). We did not detect a difference in luciferase activity between two groups when the cells were unstimulated; however, in response to stimulation with recombinant Wnt3a luciferase activity was increased. Importantly, this activation was significantly lower in Hmgb2−/− chondrocytes than in WT chondrocytes (Fig. 5A).
Fig. 5.
Wnt/β-catenin signaling in WT and Hmgb2−/− chondrocytes. (A) SuperTOPflash or SuperFOPflash reporter genes were transfected into murine WT or Hmgb2−/− chondrocytes. Upon addition of recombinant Wnt3a, stronger luciferase activity was found in WT chondrocytes compared with Hmgb2−/− chondrocytes. SuperFOPflash vector was used as negative control (*, P < 0.01). HMGB2 siRNA reduces Wnt/β-catenin target genes in articular chondrocytes. (B) Murine chondrocytes were transfected by oligo-siRNA negative control (N/C) or HMGB2 and cultured for 48 h, followed by measurement of HMGB2 by Western blot analysis. HMGB2 siRNA specifically and efficiently down-regulated protein levels of HMGB2. (C) mRNA levels of Wnt/β-catenin target genes were measured by real-time PCR. The expression levels of cyclin D1 (CD1), Gli3 and Fzd2 but not PRG4/SZP were reduced in chondrocytes with HMGB2 siRNA. (D) Schematic representation of the human GLI3 locus. Gray indicates the Gli3 gene, and Tcf/Lef-binding sites as putative enhancers are depicted in red (R1–4). (E) Chromatin immunoprecipitation assay shows that HMGB2 potentiates binding affinity of Lef-1 on human Gli3 enhancer within R2 (Upper). Sequences not predicted to contain Tcf/Lef-1 binding sites upstream of the Exon 1 were not pulled down (Lower). IP, immunoprecipitation.
To further examine this, we analyzed levels of Wnt/β-catenin targets genes. HMGB2 was reduced by siRNA in immature murine articular chondrocytes, which strongly express endogenous HMGB2 (17) (Fig. 5B). Quantitative PCR shows that HMGB2 siRNA reduced cyclin D1 mRNA expression. Additional Wnt/β-catenin target genes, Gli3 and Frizzled 2 (Fzd2), which are expressed in articular cartilage (26, 40), were also reduced by HMGB2 siRNA, whereas PRG4/SZP that is expressed in murine Hmgb2−/− chondrocytes and WT chondrocytes was unaffected (17) (Fig. 5C).
To verify that HMGB2 does bind to targets of Lef1/β-catenin genes, we used chromatin immunoprecipitation assay. We observed that HMGB2 can facilitate binding affinity of Lef-1 to the human Gli3 enhancer (R2) (Fig. 5 D and E), which shows strong activity among highly conserved noncoding DNA regions that contained Tcf/Lef binding sequence within human Gli3 locus (R1–4) (41). These results further support our notion that HMGB2 potentiates Wnt/β-catenin activity.
Loss of β-Catenin Signaling Results in Chondrocyte Apoptosis.
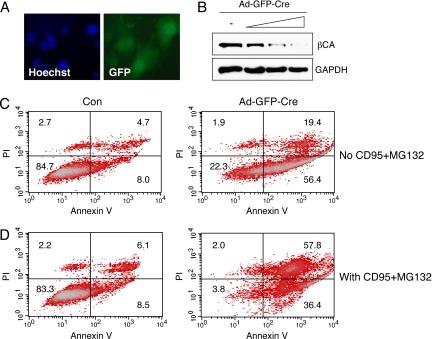
To further test the functional significance of β-catenin signaling we conditionally inactivated β-catenin. Chondrocytes were isolated from knee and hip cartilage of β-catenin floxed mice (Ctnnb1flox/flox), infected with adenovirus-GFP-Cre and cultured for 72 h. Immunofluorescence analysis of GFP demonstrated effective adenoviral transduction (Fig. 6A). β-catenin protein levels were reduced with increasing amounts of adenovirus-GFP-Cre in Ctnnb1flox/flox chondrocytes (Fig. 6B). Next, chondrocytes from Ctnnb1flox/flox mice with or without adenovirus-GFP-Cre infection were analyzed by flow cytometry for viability and the apoptosis marker Annexin V. Upon adenovirus-GFP-Cre infection there was a significant increase in apoptotic cells (Fig. 6C) without addition of apoptosis inducers. When the chondrocytes were stimulated with anti-Fas antibody CD95 and proteasome inhibitor MG132 (17), a higher percentage of apoptotic chondrocytes was found after adenovirus-GFP-Cre infection compared with control (Fig. 6D). Thus, β-catenin signaling promotes chondrocyte survival under basal conditions and in response to apoptosis inducers.
Fig. 6.
Conditional inactivation of β-catenin and chondrocyte survival. βCAflox/flox chondrocytes were infected with GFP-Cre adenovirus and cultured for 72 h. (A) Immunofluorescence analysis shows GFP expression in infected cells. (B) Western blot shows reduction in β-catenin protein levels with increasing amounts of GFP-Cre adenovirus (Ad-GFP-Cre). (C and D) FACS analysis for annexin V and propidium iodide (PI) staining. GFP-Cre adenovirus infected cultures showed a higher percentage of apoptotic chondrocytes compared with noninfected (Con) cells in the absence or presence of anti-Fas antibody CD95 (1 μg/mL) and MG132 (20 μM).
Discussion
Understanding mechanisms that control articular cartilage formation and maintenance is of significance to cartilage tissue engineering and the prevention and treatment of diseases affecting articular cartilage. In regards to cartilage tissue engineering a major unmet challenge is the generation of a tissue that recapitulates the zonal organization of normal cartilage. In regards to joint diseases, the major current deficit is in the lack of therapies for OA, the most prevalent form of arthritis. The initial lesions in OA are at the articular surface and once the SZ of cartilage is disrupted, the chronic cartilage remodeling and degradation process is initiated.
To begin elucidating molecular mechanisms that govern the SZ phenotype we showed that the chromatin protein HMGB2 is uniquely expressed in the SZ (17). Aging in humans and mice is associated with a loss of HMGB2 expression, which correlates with OA-like cartilage changes and mice with Hmgb2 deletion show early onset and more severe OA (17). This observation presented a starting point to further characterize the signaling network in which HMGB2 operates to control SZ cell survival and function.
The Wnt/β-catenin pathway presented a candidate based on a series of recent observations. Most notably, loss of β-catenin signaling leads to OA-like pathology (27). The first observations in this study addressed β-catenin activation patterns in articular cartilage. Wnt/β-catenin signaling is active at multiple embryonic stages of joint formation (24). Postnatally, we observed remarkable similarities between localization of HMGB2 and β-catenin. HMGB2 expression and β-catenin activation were found in all zones of articular cartilage in newborn mice. With joint maturation both became more restricted to the SZ and both showed an aging-related loss in the SZ. Although HMGB2 eventually was completely absent, β-catenin was activated in the other cartilage zones.
To determine molecular mechanisms related to these similarities in expression patterns we analyzed functional and physical interactions. Our EMSA data showed that HMGB2 does not directly bind to regulatory DNA elements but it augments DNA binding of Lef-1. HMGB2 does not alter the electrophoretic mobility of Lef-1 complexed with oligonucleotides, suggesting that HMGB2 dissociates from the complex after having provided its architectural activity (42). Similar results were observed for HMGB1, which increased the affinity of p53 complexes with oligonucleotides (43).
Transfection of HMGB2 did not activate β-catenin responsive promoters but cotransfection of HMGB2 and β-catenin did result in synergistic activation of Lef-1 responsive promoters. This synergy was seen in two chondrogenic cell types, including SW1353 chondrosarcoma cells and ATDC5 prechondrogenic cells but not in lineages such as kidney epithelial cells, suggesting other lineage specific factors mediate this interaction or the difference in both HMGB2 and HMGB1 between chondrogenic cells and 293T cells is responsible (Fig. S4).
Physical interaction studies showed there is no direct binding of HMGB2 and β-catenin. However, HMGB2 binds to Lef-1 and the complex that contains HMGB2, β-catenin, Lef-1 and probably other components leads to enhanced expression of genes containing Lef-1 binding sites. Mapping of interaction domains revealed that the HMG domain in Lef-1 is required for HMGB2 binding. The HMG domain is also responsible for interaction with Notch intracellular domain (44). Notch1 is expressed in developing articular cartilage surface (45) in a pattern similar to HMGB2 (17), indicating that Notch might be involved in Lef1-HMGB2 complex formation in temporally and spatially specific patterns during cartilage formation. HMGB2 has been reported to interact with steroid receptors (46), p53 and p73 (47). Stros et al. reported that B-box within human HMGB1 required the TKKKFKD motif that is included in the linker for interaction with p73, whereas A-box itself can bind p73 (47). It has also been shown that A-box, which contains the linker region within HMGB1, can interact with p53 (48). Our results demonstrate that the A-box or B-box within HMGB2 contribute to binding with Lef-1 only when the linker is present, because A-box, B-box and Δbox deletion mutants did not bind with Lef-1. HMGB1 can also interact with Lef-1 (Fig. S5). Considering that HMGB1 is ubiquitously expressed in the nuclei throughout normal articular cartilage (Fig. S6), we cannot exclude the possibility that HMGB1 and HMGB2 may function cooperatively as coactivators for Wnt/β-catenin signaling in the SZ (17).
The findings on interactions between HMGB2 and the Wnt signaling pathway are similar to a report demonstrating that HMG-17 was responsive to Wnt/β-catenin signaling. HMG-17 forms a chromatin complex with PITX2 to repress PITX2 transcriptional activity. This complex is inactive and switched to an active transcriptional complex through the interaction of β-catenin with PITX2 (49).
To determine functional consequences of the HMGB2/Lef-1 interaction in a cellular context, we analyzed cell survival and expression of representative Lef-1 target genes. The conditional deletion of β-catenin by Cre adenovirus infection of chondrocytes from β-catenin floxed mice increased basal and in vitro induced apoptosis. This observation is consistent with our earlier findings that Hmgb2 deficient cells are more susceptible to CD95/Fas mediated apoptosis (17). Inhibition of Wnt proteins promotes programmed cell death in different types of cancer cells (50, 51). In human OA cartilage, FrzB-2 is highly expressed and is associated with chondrocyte apoptosis (52). In Col2a1-ICAT-transgenic mice in which β-catenin signaling is selectively blocked in chondrocytes, apoptosis is increased (27).
Our results also show that promoters with Lef-1 binding sites were less responsive to Wnt3a treatment in Hmgb2−/− chondrocytes compared with WT chondrocytes. Then we analyzed cyclin D1, Gli3 and Fzd2, three representative and well characterized Lef-1 target genes in cartilage (26, 40, 53). The expression levels of these genes were reduced by HMGB2 siRNA in chondrocytes. Thus, it is possible that reduction of these three genes at least in part explains the increased apoptosis seen in both the Hmgb2 deficient mice (17) and in chondrocytes with deficient β-catenin (27).
In conclusion, this study demonstrates similar expression and activation patterns of HMGB2 and β-catenin in articular cartilage and that a loss of these pathways in the SZ of articular cartilage may lead to altered gene expression, cell death and OA-like changes.
Materials and Methods
Mice.
The β-catenin floxed mice (β-cateninflox/flox) with loxP sites in introns 1 and 6 of the β-catenin gene (6.129-Ctnnb1tmKem/KnwJ line) and TOPGAL mice (Tg(Fos-lacZ)34Efu/J line) (29) were purchased from the Jackson Laboratory. Mice were used according to protocols approved by the Institutional Animal Care and Use Committee at The Scripps Research Institute.
Plasmid Construction.
The HMGB2 deletion constructs were prepared by PCR amplification of full-length murine HMGB2 cDNA and cloned into pcDNA3-flag vector after the mapping of A-box and B-box within human HMGB2 (54). pGEX-HMGB2 and pGEX-HMGB1 were constructed by subcloning of murine HMGB2 or human HMGB1 into pGEX (Promega), respectively. pGEX-β-catenin was provided by X. He (Harvard Medical School, Boston) and pGEX-Lef-1 by M.R. Stallcup (University of Southern California, Los Angeles).
GST Pull-Down Assay.
The wild-type and deletion mutants of HMGB2 and Lef-1 were in vitro transcribed/translated with the TNT reticulocyte lysate kit (Promega) in the presence of [35S]methionine. GST-null, GST-HMGB2, GST-Lef-1, or GST-β-catenin proteins were produced in E. coli and purified, and then incubated overnight at 4 °C rotating with the 35S-met-labeled proteins in PC100+βME buffer (55). After extensive washes, we added SDS loading buffer to the beads, boiled them, and separated the supernatant on SDS/PAGE gels. As a positive control, the amount of 35S-met-labeled protein loaded was 20% of the input. The gels were dried and then exposed to X-ray film.
Electrophoretic Mobility-Shift Assay (EMSA).
Preparation of DNA for EMSA with 32P-labeled duplex oligonucleotide probes for CD1, CD1TOP and CD1FOP was described earlier (36). We also generated synthetic duplex oligonucleotides encompassing regions evolutionarily conserved in the c-Jun promoter (37). The binding reaction contained 40,000 cpm of 32P-labeled DNA that was incubated with nuclear extracts from SW1353 cells with or without transfection of HA-tagged Lef-1 expression vector (gift from P.K. Vogt, The Scripps Research Institute) in the presence of purified calf thymus HMGB2 protein (Shino-Test), following Gel Shift Assay Systems Protocol (Promega). DNA-protein complexes were electrophoresed in 6% DNA retardation gel (Invitrogen) and visualized by autoradiography.
Quantitative PCR.
Total RNA was extracted and oligo(dT)-primed cDNA was prepared from 500 ng of total RNA by using SuperScript III (Invitrogen). The resulting cDNAs were analyzed by using the SYBR green system for quantitative analysis of specific transcripts as described in ref. 56. All mRNA expression data were normalized to GAPDH expression in the same sample. The primers used in real-time PCR are listed in SI Text.
Apoptosis Induction and Analysis in Vitro.
Chondrocytes were prepared from 5-day-old β-catenin floxed mice (β-cateninflox/flox) as described in ref. 57. The cells were plated in 6-well plates at semiconfluence, infected with adenovirus expressing both Cre recombinase and green fluorescent protein (GFP) (Adv-Cre-GFP) and cultured for 72 h. We used an E1/E3-deleted, replication-incompetent, serotype 5 adenovirus-expressing Cre recombinase and GFP under control of the cytomegalovirus (CMV) promoter. Medium was changed to DMEM/F12 with 0.5% FBS and chondrocytes were stimulated with NA/LE hamster anti-mouse CD95 antibody (BD PharMingen) and proteasome inhibitor MG132 (Sigma) for 12 h, which induces apoptosis in articular chondrocytes (58). Cells were incubated with FITC-labeled annexin V (BD PharMingen) or propidium iodide (Sigma) and analyzed on a BD FACSCalibur as described in ref. 17.
Statistical Analysis.
Results are expressed as mean ± standard deviation. Statistical comparison between genotypes or treatment groups was performed with a two-tailed Student's t test. P values <0.05 were considered significant.
Supplementary Material
Acknowledgments.
We thank Lilo Creighton, Jean Valbracht and Diana Brinson for technical support and Marco E. Bianchi (San Raffaele University, Milan, Italy) for helpful discussions. This work was supported by National Institutes of Health Grants AG007996 and AG033409 (to M.L.), the Arthritis National Research Foundation and Japan Orthopaedics and Traumatology Foundation, Inc. No. 179 (to N.T.), and the Sam and Rose Stein Endowment Fund (M.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0904414106/DCSupplemental.
References
- 1.Korver GH, van de Stadt RJ, van Kampen GP, van der Korst JK. Composition of proteoglycans synthesized in different layers of cultured anatomically intact articular cartilage. Matrix. 1990;10:394–401. doi: 10.1016/s0934-8832(11)80147-2. [DOI] [PubMed] [Google Scholar]
- 2.Hunziker EB, Quinn TM, Hauselmann HJ. Quantitative structural organization of normal adult human articular cartilage. Osteoarthritis Cartilage. 2002;10:564–572. doi: 10.1053/joca.2002.0814. [DOI] [PubMed] [Google Scholar]
- 3.Darling EM, Hu JC, Athanasiou KA. Zonal and topographical differences in articular cartilage gene expression. J Orthop Res. 2004;22:1182–1187. doi: 10.1016/j.orthres.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 4.Aydelotte MB, Kuettner KE. Differences between sub-populations of cultured bovine articular chondrocytes. I. Morphology and cartilage matrix production. Connect Tissue Res. 1988;18:205–222. doi: 10.3109/03008208809016808. [DOI] [PubMed] [Google Scholar]
- 5.Flannery CR, et al. Articular cartilage superficial zone protein (SZP) is homologous to megakaryocyte stimulating factor precursor and Is a multifunctional proteoglycan with potential growth-promoting, cytoprotective, and lubricating properties in cartilage metabolism. Biochem Biophys Res Commun. 1999;254:535–541. doi: 10.1006/bbrc.1998.0104. [DOI] [PubMed] [Google Scholar]
- 6.Rhee DK, et al. The secreted glycoprotein lubricin protects cartilage surfaces and inhibits synovial cell overgrowth. J Clin Invest. 2005;115:622–631. doi: 10.1172/JCI200522263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schumacher BL, Hughes CE, Kuettner KE, Caterson B, Aydelotte MB. Immunodetection and partial cDNA sequence of the proteoglycan, superficial zone protein, synthesized by cells lining synovial joints. J Orthop Res. 1999;17:110–120. doi: 10.1002/jor.1100170117. [DOI] [PubMed] [Google Scholar]
- 8.Hauselmann HJ, Stefanovic-Racic M, Michel BA, Evans CH. Differences in nitric oxide production by superficial and deep human articular chondrocytes: Implications for proteoglycan turnover in inflammatory joint diseases. J Immunol. 1998;160:1444–1448. [PubMed] [Google Scholar]
- 9.Hiraoka K, Grogan S, Olee T, Lotz M. Mesenchymal progenitor cells in adult human articular cartilage. Biorheology. 2006;43(3–4):447–454. [PubMed] [Google Scholar]
- 10.Alsalameh S, Amin R, Gemba T, Lotz M. Identification of mesenchymal progenitor cells in normal and osteoarthritic human articular cartilage. Arthritis Rheum. 2004;50:1522–1532. doi: 10.1002/art.20269. [DOI] [PubMed] [Google Scholar]
- 11.Dowthwaite GP, et al. The surface of articular cartilage contains a progenitor cell population. J Cell Sci. 2004;117(Pt 6):889–897. doi: 10.1242/jcs.00912. [DOI] [PubMed] [Google Scholar]
- 12.Bierma-Zeinstra SM, Koes BW. Risk factors and prognostic factors of hip and knee osteoarthritis. Nat Clin Pract Rheumatol. 2007;3:78–85. doi: 10.1038/ncprheum0423. [DOI] [PubMed] [Google Scholar]
- 13.Goldring MB. The role of the chondrocyte in osteoarthritis. Arthritis Rheum. 2000;43:1916–1926. doi: 10.1002/1529-0131(200009)43:9<1916::AID-ANR2>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 14.Kuhn K, D'Lima DD, Hashimoto S, Lotz M. Cell death in cartilage. Osteoarthritis Cartilage. 2004;12:1–16. doi: 10.1016/j.joca.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 15.Abramson SB. Inflammation in osteoarthritis. J Rheumatol Suppl. 2004;70:70–76. [PubMed] [Google Scholar]
- 16.Poole AR, Guilak F, Abramson S. Etiopathogenesis of osteoarthritis. In: Moskowitz R, editor. Osteoarthritis. Philadelphia: Wolters Kluwer; 2007. [Google Scholar]
- 17.Taniguchi N, et al. Aging-related loss of the chromatin protein HMGB2 in articular cartilage is linked to reduced cellularity and osteoarthritis. Proc Natl Acad Sci USA. 2009;106:1181–1186. doi: 10.1073/pnas.0806062106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson ML, Kamel MA. The Wnt signaling pathway and bone metabolism. Curr Opin Rheumatol. 2007;19:376–382. doi: 10.1097/BOR.0b013e32816e06f9. [DOI] [PubMed] [Google Scholar]
- 19.Goldring SR, Goldring MB. Eating bone or adding it: The Wnt pathway decides. Nat Med. 2007;13:133–134. doi: 10.1038/nm0207-133. [DOI] [PubMed] [Google Scholar]
- 20.Ryu JH, Chun JS. Opposing roles of WNT-5A and WNT-11 in interleukin-1β regulation of type II collagen expression in articular chondrocytes. J Biol Chem. 2006;281:22039–22047. doi: 10.1074/jbc.M601804200. [DOI] [PubMed] [Google Scholar]
- 21.Hartmann C, Tabin CJ. Dual roles of Wnt signaling during chondrogenesis in the chicken limb. Development. 2000;127:3141–3159. doi: 10.1242/dev.127.14.3141. [DOI] [PubMed] [Google Scholar]
- 22.Enomoto-Iwamoto M, et al. The Wnt antagonist Frzb-1 regulates chondrocyte maturation and long bone development during limb skeletogenesis. Dev Biol. 2002;251:142–156. doi: 10.1006/dbio.2002.0802. [DOI] [PubMed] [Google Scholar]
- 23.Day TF, Guo X, Garrett-Beal L, Yang Y. Wnt/β-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev Cell. 2005;8:739–750. doi: 10.1016/j.devcel.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 24.Koyama E, et al. A distinct cohort of progenitor cells participates in synovial joint and articular cartilage formation during mouse limb skeletogenesis. Dev Biol. 2008;316:62–73. doi: 10.1016/j.ydbio.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loughlin J, et al. Functional variants within the secreted frizzled-related protein 3 gene are associated with hip osteoarthritis in females. Proc Natl Acad Sci USA. 2004;101:9757–9762. doi: 10.1073/pnas.0403456101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tamamura Y, et al. Developmental regulation of Wnt/β-catenin signals is required for growth plate assembly, cartilage integrity, and endochondral ossification. J Biol Chem. 2005;280:19185–19195. doi: 10.1074/jbc.M414275200. [DOI] [PubMed] [Google Scholar]
- 27.Zhu M, et al. Inhibition of β-catenin signaling in articular chondrocytes results in articular cartilage destruction. Arthritis Rheum. 2008;58:2053–2064. doi: 10.1002/art.23614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo X, et al. Wnt/β-catenin signaling is sufficient and necessary for synovial joint formation. Genes Dev. 2004;18:2404–2417. doi: 10.1101/gad.1230704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DasGupta R, Fuchs E. Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development. 1999;126:4557–4568. doi: 10.1242/dev.126.20.4557. [DOI] [PubMed] [Google Scholar]
- 30.Stanescu R, Knyszynski A, Muriel MP, Stanescu V. Early lesions of the articular surface in a strain of mice with very high incidence of spontaneous osteoarthritic-like lesions. J Rheumatol. 1993;20:102–110. [PubMed] [Google Scholar]
- 31.Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 32.Ronfani L, et al. Reduced fertility and spermatogenesis defects in mice lacking chromosomal protein Hmgb2. Development. 2001;128:1265–1273. doi: 10.1242/dev.128.8.1265. [DOI] [PubMed] [Google Scholar]
- 33.Korinek V, et al. Constitutive transcriptional activation by a β-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 34.Kawakami Y, et al. Transcriptional coactivator PGC-1α regulates chondrogenesis via association with Sox9. Proc Natl Acad Sci USA. 2005;102:2414–2419. doi: 10.1073/pnas.0407510102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Diamond E, Amen M, Hu Q, Espinoza HM, Amendt BA. Functional interactions between Dlx2 and lymphoid enhancer factor regulate Msx2. Nucleic Acids Res. 2006;34:5951–5965. doi: 10.1093/nar/gkl689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shtutman M, et al. The cyclin D1 gene is a target of the β-catenin/LEF-1 pathway. Proc Natl Acad Sci USA. 1999;96:5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mann B, et al. Target genes of β-catenin-T cell-factor/lymphoid-enhancer-factor signaling in human colorectal carcinomas. Proc Natl Acad Sci USA. 1999;96:1603–1608. doi: 10.1073/pnas.96.4.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hwang SG, Yu SS, Lee SW, Chun JS. Wnt-3a regulates chondrocyte differentiation via c-Jun/AP-1 pathway. FEBS Lett. 2005;579:4837–4842. doi: 10.1016/j.febslet.2005.07.067. [DOI] [PubMed] [Google Scholar]
- 39.Veeman MT, Slusarski DC, Kaykas A, Louie SH, Moon RT. Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Curr Biol. 2003;13:680–685. doi: 10.1016/s0960-9822(03)00240-9. [DOI] [PubMed] [Google Scholar]
- 40.Robinson JA, et al. Wnt/β-catenin signaling is a normal physiological response to mechanical loading in bone. J Biol Chem. 2006;281:31720–31728. doi: 10.1074/jbc.M602308200. [DOI] [PubMed] [Google Scholar]
- 41.Alvarez-Medina R, Cayuso J, Okubo T, Takada S, Marti E. Wnt canonical pathway restricts graded Shh/Gli patterning activity through the regulation of Gli3 expression. Development. 2008;135:237–247. doi: 10.1242/dev.012054. [DOI] [PubMed] [Google Scholar]
- 42.Agresti A, Bianchi ME. HMGB proteins and gene expression. Curr Opin Genet Dev. 2003;13:170–178. doi: 10.1016/s0959-437x(03)00023-6. [DOI] [PubMed] [Google Scholar]
- 43.Jayaraman L, et al. High mobility group protein-1 (HMG-1) is a unique activator of p53. Genes Dev. 1998;12:462–472. doi: 10.1101/gad.12.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ross DA, Kadesch T. The notch intracellular domain can function as a coactivator for LEF-1. Mol Cell Biol. 2001;21:7537–7544. doi: 10.1128/MCB.21.22.7537-7544.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hayes AJ, Dowthwaite GP, Webster SV, Archer CW. The distribution of Notch receptors and their ligands during articular cartilage development. J Anat. 2003;202:495–502. doi: 10.1046/j.1469-7580.2003.00185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boonyaratanakornkit V, et al. High-mobility group chromatin proteins 1 and 2 functionally interact with steroid hormone receptors to enhance their DNA binding in vitro and transcriptional activity in mammalian cells. Mol Cell Biol. 1998;18:4471–4487. doi: 10.1128/mcb.18.8.4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stros M, Ozaki T, Bacikova A, Kageyama H, Nakagawara A. HMGB1 and HMGB2 cell-specifically down-regulate the p53- and p73-dependent sequence-specific transactivation from the human Bax gene promoter. J Biol Chem. 2002;277:7157–7164. doi: 10.1074/jbc.M110233200. [DOI] [PubMed] [Google Scholar]
- 48.Imamura T, et al. Interaction with p53 enhances binding of cisplatin-modified DNA by high mobility group 1 protein. J Biol Chem. 2001;276:7534–7540. doi: 10.1074/jbc.M008143200. [DOI] [PubMed] [Google Scholar]
- 49.Amen M, et al. Chromatin-associated HMG-17 is a major regulator of homeodomain transcription factor activity modulated by Wnt/β-catenin signaling. Nucleic Acids Res. 2008;36:462–476. doi: 10.1093/nar/gkm1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He B, et al. Blockade of Wnt-1 signaling induces apoptosis in human colorectal cancer cells containing downstream mutations. Oncogene. 2005;24:3054–3058. doi: 10.1038/sj.onc.1208511. [DOI] [PubMed] [Google Scholar]
- 51.You L, et al. An anti-Wnt-2 monoclonal antibody induces apoptosis in malignant melanoma cells and inhibits tumor growth. Cancer Res. 2004;64:5385–5389. doi: 10.1158/0008-5472.CAN-04-1227. [DOI] [PubMed] [Google Scholar]
- 52.James IE, et al. FrzB-2: A human secreted frizzled-related protein with a potential role in chondrocyte apoptosis. Osteoarthritis Cartilage. 2000;8:452–463. doi: 10.1053/joca.1999.0321. [DOI] [PubMed] [Google Scholar]
- 53.Akiyama H, et al. Interactions between Sox9 and β-catenin control chondrocyte differentiation. Genes Dev. 2004;18:1072–1087. doi: 10.1101/gad.1171104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vaccari T, Beltrame M, Ferrari S, Bianchi ME. Hmg4, a new member of the Hmg1/2 gene family. Genomics. 1998;49:247–252. doi: 10.1006/geno.1998.5214. [DOI] [PubMed] [Google Scholar]
- 55.Tsuda M, Takahashi S, Takahashi Y, Asahara H. Transcriptional co-activators CREB-binding protein and p300 regulate chondrocyte-specific gene expression via association with Sox9. J Biol Chem. 2003;278:27224–27229. doi: 10.1074/jbc.M303471200. [DOI] [PubMed] [Google Scholar]
- 56.Taniguchi N, et al. Stage-specific secretion of HMGB1 in cartilage regulates endochondral ossification. Mol Cell Biol. 2007;27:5650–5663. doi: 10.1128/MCB.00130-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Salvat C, Pigenet A, Humbert L, Berenbaum F, Thirion S. Immature murine articular chondrocytes in primary culture: A new tool for investigating cartilage. Osteoarthritis Cartilage. 2005;13:243–249. doi: 10.1016/j.joca.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 58.Kuhn K, Lotz M. Regulation of CD95 (Fas/APO-1)-induced apoptosis in human chondrocytes. Arthritis Rheum. 2001;44:1644–1653. doi: 10.1002/1529-0131(200107)44:7<1644::AID-ART287>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.