Abstract
The effect of dextran nanoparticles on the conformation and compaction of single DNA molecules confined in a nanochannel was investigated with fluorescence microscopy. It was observed that the DNA molecules elongate and eventually condense into a compact form with increasing volume fraction of the crowding agent. Under crowded conditions, the channel diameter is effectively reduced, which is interpreted in terms of depletion in DNA segment density in the interfacial region next to the channel wall. Confinement in a nanochannel also facilitates compaction with a neutral crowding agent at low ionic strength. The threshold volume fraction for condensation is proportional to the size of the nanoparticle, due to depletion induced attraction between DNA segments. We found that the effect of crowding is not only related to the colligative properties of the agent and that confinement is also important. It is the interplay between anisotropic confinement and osmotic pressure which gives the elongated conformation and the possibility for condensation at low ionic strength.
Keywords: depletion, dextran, fluorescence, nanofluidics, nanoparticles
A substantial fraction of the total volume of biological media is occupied by macromolecules, which do not directly participate in biochemical reactions. Nevertheless, it is now well established that these background species have an important effect on molecular transport, reaction rates, and chemical equilibrium (1). The steric repulsion between impenetrable macromolecules in a crowded medium is a major factor in determining the thermodynamic activities of the reactants. Crowding by an inert osmotic agent can also affect macromolecular structure. A well known example is the transition of DNA to a compact form (condensation) in the presence of overthreshold concentrations of simple neutral polymers and simple salts (2, 3, 4). It has been proposed that macromolecular crowding is the basis for phase separation in the cytoplasm (5) and condensation of DNA into the nucleoid of bacterial cells (6). The latter hypothesis is supported by the observation that DNA can be condensed by cytoplasmic extracts from Escherichia coli at extract concentrations corresponding to about ½ the cellular concentration (7). Besides background species, the cytoplasm of most eukaryotic cells contains stationary elements such as fiber lattices and membranes. These structures affect macromolecular conformation through confinement in 1D or 2D. Accordingly, macromolecular crowding and confinement are intimately related and deserve an integrated approach to understand their modes of operation and how they couple.
DNA condensation can be assisted and directed by a surface. In surface directed condensation, DNA is first adsorbed onto an interface, after which it is condensed with an agent. Examples that have been reported in the literature are the condensation of single molecules into rods and toroidal structures with protamine or ethanol (8, 9). Single DNA molecules can be confined and visualized with fluorescence microscopy in quasi 1D nanochannels. The extension in the longitudinal direction of these channels has been measured as a function of channel diameter (10, 11) and ionic strength of the supporting medium (12, 13). Other studies have focused on the dynamics to elucidate the variation in the extension due to thermal fluctuation (14) or the response of the molecular motion to entropic, electrophoretic, and frictional forces (15). To the best of our knowledge, the effect of macromolecular crowding on the configurational properties of a single DNA molecule confined in a nanochannel has not been reported before. Here, we report the effect of the generic crowding agent dextran on the conformation and condensation of DNA confined in long, straight, and rectangular nanochannels with a depth of 300 nm and a width in the range 150–300 nm. Due to the cross-sectional diameter of a few hundred nm, the elongated DNA molecules remain coiled (16). The advantage of such a configuration is that the data can be interpreted using well established polymer theory, including the effects of the local bending rigidity (persistence length) and interaction of spatially close segments which are separated over a long distance along the contour (excluded volume or self-avoidance).
Dextran is a neutral branched polysaccharide made of glucose monomers. The dextran molecules used in the present study have a radius of gyration Rg in the range 2.6–17 nm. They behave as spherical nanoparticles, which readily dissolve in water. Because of their inert behavior, dextran is often used as crowding agent to mimic the intracellular crowded environment in vitro (1). Our experiments are done using nanofluidic devices made of poly(dimethyl siloxane) (PDMS). We have used a lithography process with proton beam writing to fabricate a nanopatterned stamp (17, 18). The stamp was subsequently replicated in PDMS, followed by curing and sealing with a glass slide (19). The advantage of this method is that around 100 chips can be replicated using a single stamp, which allows the use of a fresh chip for every experiment. To visualize the DNA molecules with fluorescence microscopy, it is necessary to stain them with a dye. The intercalation ratio was 23 base-pairs per YOYO-1 molecule. For such a low level of intercalation, the distortion of the secondary DNA structure is minimal; the contour length has increased from 57 to 60 μm and the DNA charge is reduced by a factor 42/46 (20, 21). Furthermore, there is no appreciable effect on the bending rigidity, as inferred from previously reported measurements of the extension of DNA in nanochannels with different concentrations of dye (13). T4-DNA (166 kbp) molecules were brought into the nanochannels with an electric field and their extensions were measured in buffer of various ionic strength and in the presence of various volume fractions of dextran. We have also monitored the condensation of DNA, including its time evolution, for overthreshold values of the dextran volume fraction. For reference, the effect of dextran on the size of DNA in the bulk phase was measured with dark field microscopy.
Results
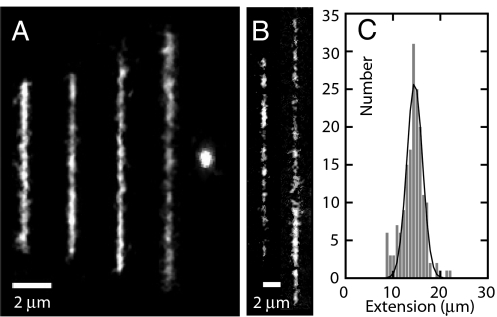
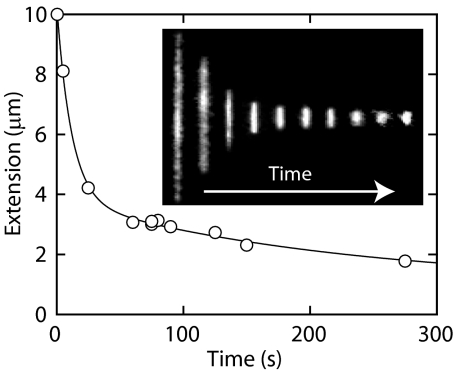
Montages of images of single T4-DNA molecules confined in nanochannels are shown in Fig. 1. With increasing dextran concentration but constant ionic strength and channel diameter, the DNA molecules elongate. For the smallest channel diameter, lowest ionic strength, and highest volume fraction of dextran, the maximum value of the extension is 29 μm, i.e., about half the contour length. For overthreshold volume fractions of dextran, condensation into a compact form is observed. Compacted DNA molecules are visible as bright fluorescence spots and can easily be discerned. The images refer to well-equilibrated structures. After the electric field has been removed, the nonequilibrium extension is determined by the balance of electrophoretic, entropic, and frictional forces. Without dextran or for subthreshold volume fractions of dextran, the molecules elongate and relax to their equilibrium state within 60 s. Besides elongation or compaction, we did not observe an obvious dependence of the kinetics on the concentration of dextran. It is possible to witness the condensation process in situ and in real time. The process is illustrated by the time lapse series of images in Fig. 2, captured after the electric field has been switched off. It is seen that condensation occurs at 2 different time scales. The first time scale is 10 s, over which the molecule condenses into a structure with an extension of a few μm. This rapid decay is related to the elastic response of the coil to the osmotic compaction force, because its time constant corresponds with the molecular relaxation time (15). At a longer time scale of a few minutes, the condensed structure becomes even more compacted with a final extension of around 1 μm. The longer time scale is possibly due to the progressive rearrangement of the DNA segments into a more orderly configuration.
Fig. 1.
(A) Montage of fluorescence images of T4-DNA in 300 × 300 nm2 channels and 1×T buffer (2.9 mM TrisCl, 7.1 mM Tris, pH 8.5). The molecules are crowded by dextran (Rg = 6.9 nm) with volume fraction ϕ = 0, 4.2 × 10−4, 4.2 × 10−3, 4.2 × 10−2, and 6.3 × 10−2 from left to right. (B) As in panel (A), but in 150 × 300 nm2 channels, 1/10×T, and ϕ = 4.2 × 10−4 (Left) and 4.2 × 10−2 (Right). (C) Distribution in extension of a population of 170 molecules in 300 × 300 nm2 channels, 1×T, and ϕ = 4.2 × 10−2. A Gaussian fit gives R∥ = 15 ± 2 μm.
Fig. 2.
Time lapse series showing condensation after switching off the electric field. The solid curve represents a biexponential fit with fast and slow time constants of 12 and 220 s, respectively. The DNA molecule is in a 300 × 300 nm2 channel, in 1×T buffer, and crowded by dextran (Rg = 6.9 nm) with ϕ = 6.3 × 10−2.
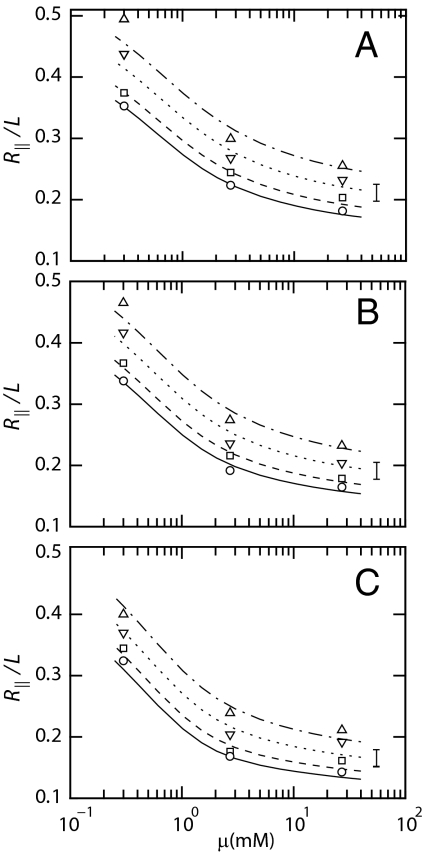
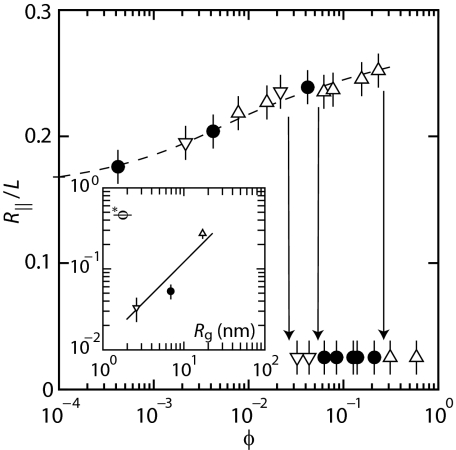
We first focus on the elongation of the DNA molecules as a function of the ionic strength and size and density of the nanoparticles. For this purpose, we have measured the extension of T4-DNA confined in channels with 3 different cross-sections: 300 × 300, 200 × 300, and 150 × 300 nm2. For each experimental condition, i.e., ionic strength, channel diameter, dextran volume fraction, and molecular weight, we have measured around 200 molecules. The fluctuation induced distribution in extension is close to Gaussian (14). An example of such a distribution is also shown in Fig. 1. DNA fragments can easily be discerned, because their extensions fall below the values pertaining to the intact molecules. For the cut-off, we have used the mean value minus 2 times the standard deviation. Resolution-broadening can be neglected because the optical resolution is one order of magnitude smaller than the variance. The mean relative extensions, i.e., the mean extensions divided by the contour length, are set out in Fig. 3 as a function of the ionic strength. The buffer also contains dextran with Rg = 6.9 nm (data obtained without dextran are also shown in Fig. 3). The DNA molecules take a more extended conformation with decreasing ionic strength, irrespective the presence of dextran. With increasing volume fraction of dextran, the DNA molecules elongate. There is, however, no qualitative change in the dependencies of the extension on the ionic strength and channel diameter, i.e., in the presence of dextran the relative extension is uniformly shifted toward higher values. We also measured the extension of T4-DNA in 300 × 300 nm2 channels in 1×T buffer and using dextran with Rg = 2.6, 6.9, and 17 nm. As shown in Fig. 4, the elongations induced by dextran of different sizes collapse to a single master curve if they are set out against the volume fraction. Note that the relative extensions are typically in the range 0.1–0.4, which implies that the DNA molecules remain coiled.
Fig. 3.
Relative extension R∥/L of T4-DNA in 150 × 300 (A), 200 × 300 (B), and 300 × 300 (C) nm2 channels versus the ionic strength μ of the buffer (1/10, 1, and 10×T composed of 0.27/0.73, 2.9/7.1, and 31/69 mM TrisCl/Tris, respectively, pH 8.5). The dextran (Rg = 6.9 nm) volume fractions are ϕ = 0 (○), 4.2 × 10−4 (□), 4.2 × 10−3 (▿), and 4.2 × 10−2 (△). The curves represent a fit of the blob model with C = 1.46 (solid), 1.53 (dashed), 1.66 (dotted), and 1.80 (dashed-dotted). The bars denote the typical width of the distribution in extension.
Fig. 4.
Relative extension R∥/L of T4-DNA in 300 × 300 nm2 channels and 1×T buffer versus the volume fraction ϕ of dextran. The dextran Rg = 2.6 (▿), 6.9 (●), and 17 ( ) nm. The dashed curve is drawn as an aid to the eye and the arrows denote the condensation thresholds. The inset shows the critical volume fraction ϕ* pertaining to condensation versus Rg. The solid line denotes ϕ*∝Rg.
) nm. The dashed curve is drawn as an aid to the eye and the arrows denote the condensation thresholds. The inset shows the critical volume fraction ϕ* pertaining to condensation versus Rg. The solid line denotes ϕ*∝Rg.
For overthreshold volume fractions of dextran, the DNA molecules condense into a compact form. This condensation is facilitated by the confinement inside the nanochannel, because we did not observe condensation in the feeding microchannels and/or the reservoirs of the chip. As shown in the inset of Fig. 4, the critical volume fraction for condensation is approximately proportional to the size of the particles ϕ*∝Rg. The condensation of DNA for overthreshold densities of a condensing agent including neutral polymer is well known (3). Here, we report condensation inside nanochannels in Tris buffer with an ionic strength of around 3 mM. This ionic strength is markedly lower than the one used in polymer and salt induced (psi) condensation of DNA in the bulk phase (2). For psi-condensation, it is generally necessary to increase the ionic strength to a value exceeding 100 mM to suppress electrostatic repulsion between segments of the negatively charged DNA molecule. As we will see shortly, condensation by dextran is no different in this respect. Condensed DNA usually has an ordered morphology, in which the segments of the molecule are arranged in a hexagonal fashion (22). Inside a nanochannel, the morphology of the condensed molecules is probably also hexagonal. This could, however, not be confirmed because of difficulties associated with molecular imaging of enclosed molecules.
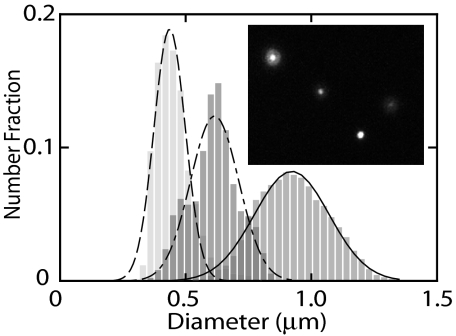
We have investigated how the size of the DNA molecule in the bulk phase changes with the addition of dextran and salt with dark field microscopy. An example of an image of single DNA molecules is shown in Fig. 5. We have measured the diameter by taking an isotropic average, despite the fact that the molecules are slightly anisotropic at high dextran and salt concentration. The distribution in diameter is also shown in Fig. 5. With a high volume fraction of dextran (ϕ = 0.3, Rg = 6.9 nm), but low ionic strength (1×T), the mean diameter is reduced from 0.9 to 0.6 μm due to the depletion of dextran from the interior of the coil. In the nanochannel, these experimental conditions result in a condensed rather than elongated form of the DNA molecules. In the bulk phase, the molecules are, however, not condensed. This is shown by the progressive decrease in number of molecules with a diameter around 0.6 μm and a concomitant increase in molecules with a diameter around 0.4 μm, once the buffer contains NaCl with a concentration increasing from 50 to 250 mM. We have checked that in the presence of 250 mM NaCl, an increase in dextran volume fraction from ϕ = 0.3 to 0.5 does not give a further reduction in size. The molecules with a diameter close to the optical resolution of around 0.4 μm are condensed. Furthermore, the coexistence of condensed and uncondensed molecules agrees with a first order phase transition, as previously reported for T4-DNA crowded by poly(ethylene glycol) (4).
Fig. 5.
Distribution in diameter of T4-DNA molecules in the bulk phase. The solvents are 1×T buffer (Right), 1×T with dextran (Middle), and 1×T with dextran and 250 mM NaCl (Left). Gaussian fits give diameters 0.45 ± 0.06, 0.62 ± 0.10, and 0.92 ± 0.15 μm from Left to Right. The inset shows a dark field image of single DNA coils in Tris buffer with dextran, but without NaCl. The dextran Rg = 6.9 and ϕ = 0.3.
The behavior of DNA confined in a nanochannel has 2 unique aspects. First, for subthreshold volume fractions of dextran the molecules elongate in the longitudinal direction. In the bulk phase, we observed an isotropic contraction resulting in a smaller coil size. Second, in the channel, the DNA condenses into a compact structure for overthreshold volume fractions of dextran at an ionic strength of a few mM. In the bulk phase, it is necessary to increase the salt concentration to around 100 mM to get a significant fraction of condensed DNA.
Discussion
Neutral dextran nanoparticles with a size around 10 nm easily penetrate the DNA coil. Dextran is not known to complex on DNA. Accordingly, both the elongation and the collapse of DNA should be related to entropic effects. This is also supported by the fact that the presence of dextran has no qualitative effect on the dependencies of the extension on the channel diameter and ionic strength (Fig. 3). We can hence assume that there is no change in DNA bending rigidity induced by dextran. The depletion interaction between DNA segments which are spatially close, but separated over far distances along the contour, is attractive and is expected to reduce the swelling of the chain. A reduced swelling would result in a contracted rather than elongated chain conformation with respect to the dextran-free state, which is not in agreement with our data for DNA inside a nanochannel (in the bulk phase the chain is contracted). Furthermore, self-avoidance effects on the extension of confined DNA are expected to be moderate, due to the small DNA correlation length on the order of the diameter of the nanochannel (13). Accordingly, in the analysis of the extension, we have also ignored the effect of dextran on the excluded volume interaction between the segments of the DNA molecule.
In de Gennes scaling approach, a DNA molecule inside the nanochannel is considered a sequence of linearly packed subcoils (blobs) of uniform size (16, 23). The size of the blobs is given by their radius of gyration Rb, which is on the order of the diameter D of the channel. The exact relationship between Rb and D is unknown, but we assume D = C Rb, with C being an adjustable parameter. If the contour length stored in a single blob is given by Lb, the total number of blobs in the aligned sequence of blobs is L/Lb. The extension is then given by R‖ = C RbL/Lb with second moment 〈δR‖2〉 = C2Rb2L/Lb. The blob size Rb and contour length Lb depend on the persistence length Lp and effective diameter Deff of the DNA duplex, both modified by electrostatic interaction. Following the procedure as described in ref. 13, we have calculated Rb and Lb as a function of the ionic strength. In the calculation, we have used a bare persistence length of 45 nm and a DNA charge and contour length corresponding with a level of intercalation of 23 base pairs per dye molecule. The theoretical extension was fitted to the data by optimization of C. To account for the electrostatic interaction of DNA with the channel wall, we have used a reduced channel diameter by subtracting 2 times the electrostatic screening length from the bare diameter. Furthermore, for asymmetric cross-sections we have taken the square root of the depth times the width as effective diameter.
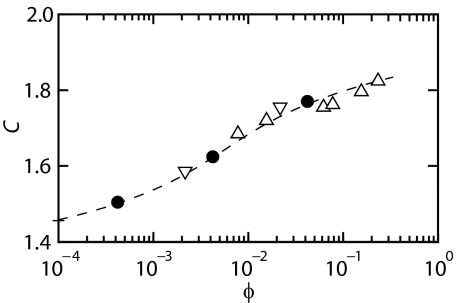
The data in Fig. 3 are well reproduced with the optimized values of C listed in the corresponding legend, irrespective channel diameter and ionic strength. The values of C pertaining to the data in Fig. 4, i.e., for T4-DNA in 300 × 300 nm2 channels, 1×T buffer, and with Rg = 2.6, 6.9, and 17 nm dextran, are shown in Fig. 6. The variation in extension due to thermal fluctuation also agrees with the theoretical prediction (13). With increasing volume fraction, C increases from 1.46 pertaining to the dextran-free state to 1.82 for the most congested state. This increase in C corresponds with a decrease in effective channel diameter from 205 to 165 nm. We inferred the 40 nm decrease in diameter in an indirect way from the extension in the longitudinal direction. Direct molecular imaging of such a decrease is, however, difficult, if not impossible, due to problems associated with the optical resolution and the sealing of the channels. If the DNA segments do not exert a full excluded volume effect, C would be 9% larger at most (13). Note that for subthreshold volume fractions of the crowding agent, the DNA molecules remain self-avoiding and are always elongated with respect to the dextran-free state. The increase in C implies that the blob size decreases according to Rb = D/C with increased volume fraction of dextran. The diameter of the nanochannel is hence effectively reduced by the crowding agent.
Fig. 6.
Confinement constant C for T4-DNA in 300 × 300 nm2 channels and 1×T buffer versus the volume fraction ϕ of dextran. The symbols and curve are as in Fig. 4.
Dressing of the walls by dextran is not a plausible explanation for the effective reduction in channel diameter. With fluorescence microscopy of rhodamine labeled dextran, we did not observe sticking of dextran to PDMS after flushing the chip with buffer. Furthermore, a single adsorption layer of dextran should have a thickness on the order of Rg (2.6–17 nm), whereas we observed a maximum reduction in channel diameter by 40 nm, irrespective particle size. It should also be noted that for each set of experimental conditions a fresh chip was used, so that accumulation of dextran at the wall from different experiments is excluded. A qualitative explanation can be proposed based on the existence of an interfacial layer next to the channel wall in which the DNA segment density is depleted. This effect results in a blob size smaller than the channel diameter and a value of C exceeding unity (without dextran C = 1.46). The thickness of the interfacial region is on the order of the DNA persistence length, i.e., 50 to 150 nm depending on ionic strength (13). The nanoparticles are also excluded from the wall, but to a smaller spatial extent due to their size of around 10 nm. As a result of the hard core repulsion between the nanoparticles and DNA, the nanoparticle density inside the blob is depleted. The concomitant difference in density between the interfacial and inner regions of the channel causes an osmotic pressure in the transverse, inward direction. Due to the anisotropy in pressure, the DNA molecule elongates at the cost of elastic energy. Based on these arguments, we will derive a scaling theory describing the effect of a neutral crowding agent on the chain statistics inside a nanochannel. This theory will be used for a qualitative derivation of the critical blob size that would balance the inward osmotic pressure created by crowding.
For a flexible chain of N segments with length Lp in a wide channel (Lp≪D), the scaling relation of the free energy of confinement is given by Fconf/kT≃N(Lp/Rb) 5/3. Furthermore, the chain has extension R‖≃L(Lp/Rb)2/3, volume V≃Rb2R‖, and segment density ρs≃Lp−3(Lp/Rb)4/3 (16, 23). The stretching pressure follows from the derivative of the free energy of confinement with respect to the volume of the chain and reads Πconf≃kT/Rb3. We can also derive a scaling relation for the work of insertion of a small particle into the coil, following a procedure originally proposed by de Gennes (24). The work should be on the order of thermal energy and follow a scaling law in the ratio of the particle and blob size, i.e., w/kT≃(Rg/Rb)4/3. The exponent 4/3 has been chosen for a linear dependence of the work on the segment density, so that w/kT≃Rg4/3Lp5/3 ρs. Here, we have assumed that the DNA segments exert a full excluded volume effect. It has been argued that the statistics of the segments next to the nanoparticle is ideal, although the whole chain is expanded by volume interactions (25). For a chain under theta conditions, the work of insertion is proportional to RgLp2. Notice that the repulsive depletion interaction depends on the size of the particle as well as the persistence length. The DNA molecule is immersed in a medium with nanoparticle density ρ0. From the balance in chemical potentials, the difference in particle density between the interior and surrounding medium is to leading order given by Δρ≃ρ0(Rg/Rb)4/3 (Rg≪Rb). With a typical experimental ratio Rg/Rb≃0.02, the particle density is depleted by a few percent. The osmotic pressure Πosmo = kTΔρ should balance the stretching pressure Πosmo = Πconf, from which follows the scaling relation for the blob size Rb≃ρ0−3/5Rg−4/5. For a typical nanoparticle density ρ0≃10−5 nm−3 and size Rg≃5 nm, the inward osmotic pressure is sufficient to reduce the blob size to a value in the range 100–300 nm. The scaling theory predicts, however, a too steep dependence of the blob size on the particle density and does not correctly predict the observed dependency on the particle volume fraction. The collapse of the extension versus volume fraction to a single master curve suggests that the depletion interaction is proportional to Rg3 rather than Rg4/3Lp5/3. In view of the many uncertain parameters describing the intricate interactions between DNA, nanoparticles, and the wall, we have, however, refrained from further elaboration of the theoretical model.
The condensation at high volume fraction of dextran is due to depletion induced attraction between DNA segments. Each segment is surrounded by a cylindrical volume in which the nanoparticles cannot penetrate for steric reasons. The diameter of this volume is approximately equal to the sum of the diameter of the DNA duplex and the radius of gyration of the particles: DDNA+Rg with DDNA = 2 nm. For 2 parallel cylinders with their center lines of mass separated by a distance r with DDNA<r<DDNA+Rg, there is an attractive force due to the exclusion of the particles from the overlap region. Based on an Asakura-Oosawa type of treatment, the interaction energy per unit length is given by the cross-sectional area of the overlap region times the osmotic pressure exerted by particles and takes the approximate form
with ρ the density of the particles inside the coil (16, 26, 27). The attractive depletion interaction is set off against the electrostatic repulsion, leading to a net potential depending on the relative strengths of the electrostatic and depletion interactions. Condensation occurs when the absolute value of the depletion interaction energy exceeds a certain critical value. The critical volume fraction is approximately proportional to the size of the particles ϕ*∝Rg, due to the fact that to leading order the cross-sectional area of the overlap region is proportional to Rg2. The theoretical critical volume fractions for condensation by small protein have been reported in the literature (27). Our results are in qualitative agreement with the predicted values of around 0.2. A quantitative analysis is, however, unfeasible, because the precise locations of the theoretical phase boundaries are very sensitive to the unknown particle interactions including solubility.
Conclusions
We have investigated the effects of the generic crowding agent dextran on the structure and condensation of single DNA molecules confined in a nanochannel. It was observed that with increasing volume fraction of the crowding agent, the DNA molecules progressively elongate with respect to the agent-free state. Eventually, for overthreshold volume fractions of the agent, the molecules condense into a compact structure. A crowding induced elongation is counter-intuitive, because the attractive depletion interaction between the segments of the DNA molecule is expected to result in a contracted rather than elongated chain conformation. We did not observe qualitative changes in the dependencies of the extension on the channel diameter and ionic strength, which indicates that the local chain statistics is not significantly modified. The elongation was analyzed with the blob model and it was found that the data could be reproduced by the adjustment of a factor which expresses the ratio of the channel diameter and the DNA correlation length. The channel diameter is effectively reduced under crowded conditions. A plausible explanation for this effect is based on depletion in DNA segment density next to the wall. As a result of the hard core repulsion between DNA segments and nanoparticles, the latter will be depleted from the interior of the coiled molecule. The concomitant osmotic pressure in the inward direction across the channel then results in a smaller blob size and an elongation of the DNA molecule at the cost of elastic energy. In the bulk phase, the pressure generated by the depletion of the nanoparticles is isotropic and gives a uniform reduction in coil size.
For overthreshold volume fractions of the crowding agent, the DNA molecules condense into a compact structure. The condensation takes place over at least 2 different time scales: a faster relaxation time of around 10 s and a longer one of several minutes. The existence of these 2 time scales can be rationalized in terms of a fast initial collapse into a globular state and subsequent slower progress into a more ordered morphology. We could, however, not obtain information about the morphology of condensed DNA, due to experimental difficulties associated with molecular imaging of DNA inside a sealed nanochannel. The crowding induced condensation of DNA confined in a nanochannel bears resemblance to polymer and salt induced condensation in the bulk phase. It should be noted, however, that the environmental condition is fundamentally different, in the sense that the latter condensation always requires a high concentration of salt. We have shown that condensation by dextran is no different in this respect. Confinement inside a nanochannel facilitates compaction at a relatively low ionic strength of a few mM. This observation supports the previously reported notion that (electrostatic) excluded volume effects on the chain statistics inside a nanochannel are moderate, due to the relatively short correlation length (blob size) of a few hundred nm imposed by the channel diameter (13). We also observed that the threshold volume fraction for condensation is approximately proportional to the size of the nanoparticles. This can be rationalized in terms of an attractive depletion interaction between almost parallel segments of the DNA molecule due to the exclusion of nanoparticles from the overlap region.
Our results have implications for biotechnology as well as biophysics. Nanofluidics provides a platform for single molecule studies of increasing importance. The fabrication of channels with a cross-sectional diameter less than 100 nm is still problematic, although recently progress has been made (28). Here, we have shown that the channel diameter can effectively be reduced in a simple way, i.e., by the addition of a crowding agent to the medium. From a biophysical point of view, crowding agents such as dextran are often used to mimic the intracellular environment and the nanochannel can be considered a model for the scaffolding of the cell. Our results show that the effect of crowding is not simply related to the colligative properties of the crowding agent, but the confinement also needs to be taken into account. It is the interplay of anisotropic confinement and osmotic pressure which gives the elongated rather than contracted conformation and the possibility for condensation at low ionic strength. Furthermore, we have seen that even the smallest investigated nanoparticles with a radius of gyration of 2.6 nm can be used to condense DNA inside a nanochannel. This size is comparable to the typical size of bacterial proteins, which indicates that osmotic compaction of the genome by nonbinding protein is a feasible mechanism for gene regulation.
Experimental Methods
Fabrication of the Nanofluidic Chip.
The nanofluidic device was made by replication in PDMS of a patterned master stamp (13). The stamp was fabricated in hydrogen silsesquioxane (HSQ) resist (Dow Corning) using a lithography processes with proton beam writing (17, 18). The 300 nm height of the positive channel structures on the stamp was measured with atomic force microscopy (Dimension 3000, Veeco). The stamp was replicated in PDMS followed by curing with a curing agent (Sylgard, Dow Corning) at 337 K for 24 h (19). Finally, the PDMS replica was sealed with a glass slide after both substrates were plasma oxidized (Harrick). The widths of the channels in the PDMS replica were measured with atomic force microscopy and the values agreed with those obtained from the scanning electron microscopy images of the HSQ master stamp. The nanochannels are rectangular-shaped and have a depth of 300 ± 5 nm and a width of 150 ± 5, 200 ± 5, and 300 ± 5 nm, respectively.
DNA Sample Preparation.
T4-DNA was purchased from Nippon Gene and used without further purification. YOYO-1 was purchased from Invitrogen. Water was deionized and purified by a Millipore system and had a conductivity less than 1 × 10−6 Ω−1cm−1. Samples were prepared by dialyzing solutions of T4-DNA against Tris/HCl (T) buffer in microdialyzers. The buffer concentrations were 1/10, 1, and 10×T (1×T is 10 mM Tris adjusted with HCl to pH 8.5, i.e., 2.9 mM TrisCl and 7.1 mM Tris). Dextran with molecular weights Mw of 5, 50, and 410 kg/mol was purchased from Sigma-Aldrich and dissolved in Tris buffer. Solutions of dextran and T4-DNA in Tris buffer were subsequently mixed in equal volumes and incubated for 24 h at 277 K. The final DNA concentration is 0.003 g/L. Before fluorescence microscopy, T4-DNA was stained with YOYO-1. No antiphoto bleaching agent was used. The ionic strength of the buffer was calculated with the Davies equation for estimating the activity coefficients of the ions and a dissociation constant pK = 8.08 for Tris. For dark field imaging, T4-DNA was dissolved in 1×T, various amounts of NaCl, and dextran to a DNA concentration of 0.01 g/L and incubated for at least 48 h at 277 K. The radii of gyration Rg of dextran were calculated using the empirical relation Rg = 0.066×Mw0.43 with Mw in g/mol and Rg in nm (29). The small value of the exponent is due to the branched nature of dextran.
Fluorescence Imaging.
The YOYO-1-stained DNA molecules dispersed in the relevant buffer/dextran solution were loaded into one of the 2 reservoirs connected by the nanochannels. The DNA molecules were subsequently driven into the channels by an electric field. For this purpose, 2 platinum electrodes were immersed in the reservoirs and connected to an electrophoresis power supply with a voltage in the range 10–30 V (Bio-Rad). Once the DNA molecules were localized inside the nanochannels, the electric field was switched off and the molecules were allowed to relax to their equilibrium state for at least 60 s. The stained DNA molecules were visualized with an Olympus IX71 inverted fluorescence microscope equipped with a 100 W mercury lamp, a UV filter set and a 100× oil immersion objective. The exposure time was controlled by a UV light shutter. Images were collected with a charge coupled device (CCD) camera (Olympus DP 70) and the extension of the DNA molecules inside the channels was measured with ImageJ software (http://rsb.info.nih.gov/ij/).
Dark Field Imaging.
A droplet of solution was deposited on a microscopy slide and sealed with a cover slip separated by a 0.12 mm spacer. The DNA molecules were imaged with a Nikon Eclipse 50i microscope with a CytoViva illuminator and a 100× oil immersion objective. Video was collected with a CCD camera (JVC TK-C921EG) connected via an analog-to-digital video converter (Canopus, ADVC55) to a computer. For each sample, around 10 min of video was analyzed with MATLAB and the sizes of the molecules were obtained with public domain tracking software (http://physics.georgetown.edu/matlab/).
Acknowledgments.
We thank Andrej Grimm for discussions. This research was supported by Grant R-144–000-177–112 from the Singapore Ministry of Education.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Zimmerman SB, Minton AP. Macromolecular crowding: Biochemical, biophysical and physiological consequences. Annu Rev Biophys Biomol Struct. 1993;22:27–65. doi: 10.1146/annurev.bb.22.060193.000331. [DOI] [PubMed] [Google Scholar]
- 2.Lerman LS. A transition to a compact form of DNA in polymer solutions. Proc Natl Acad Sci USA. 1971;68:1886–1890. doi: 10.1073/pnas.68.8.1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bloomfield VA. DNA condensation. Curr Opin Struc Biol. 1996;6:334–341. doi: 10.1016/s0959-440x(96)80052-2. [DOI] [PubMed] [Google Scholar]
- 4.Kojima M, Kubo K, Yoshikawa K. Elongation/compaction of giant DNA caused by depletion interaction with a flexible polymer. J Chem Phys. 2006;124 doi: 10.1063/1.2145752. 024902. [DOI] [PubMed] [Google Scholar]
- 5.Walter H, Brooks DE. Phase separation in cytoplasm, due to macromolecular crowding, is the basis for microcompartmentation. FEBS Lett. 1995;361:135–139. doi: 10.1016/0014-5793(95)00159-7. [DOI] [PubMed] [Google Scholar]
- 6.Zimmerman SB, Murphy LD. Macromolecular crowding and the mandatory condensation of DNA in bacteria. FEBS Lett. 1996;390:245–248. doi: 10.1016/0014-5793(96)00725-9. [DOI] [PubMed] [Google Scholar]
- 7.Murphy LD, Zimmerman SB. Condensation and cohesion of λ DNA in cell extracts and other media: Implications for the structure and function of DNA in prokaryotes. Biophys Chem. 1995;57:71–92. doi: 10.1016/0301-4622(95)00047-2. [DOI] [PubMed] [Google Scholar]
- 8.Allen MJ, Bradbury EM, Balhorn R. AFM analysis of DNA-protamine complexes bound to mica. Nucleic Acids Res. 1997;25:2221–2226. doi: 10.1093/nar/25.11.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang C, van der Maarel JRC. Surface-directed and ethanol-induced DNA condensation on mica. J Phys Chem B. 2008;112:3552–3557. doi: 10.1021/jp0776560. [DOI] [PubMed] [Google Scholar]
- 10.Reisner W, et al. Statics and dynamics of single DNA molecules confined in nanochannels. Phys Rev Lett. 2005;94:196101. doi: 10.1103/PhysRevLett.94.196101. [DOI] [PubMed] [Google Scholar]
- 11.Persson F, Utko P, Reisner W, Larsen NB, Kristensen A. Confinement spectroscopy: Probing single DNA molecules with tapered nanochannels. Nano Lett. 2009;9:1382–1385. doi: 10.1021/nl803030e. [DOI] [PubMed] [Google Scholar]
- 12.Reisner W, et al. Nanoconfinement-enhanced conformational response of single DNA molecules to changes in ionic environment. Phys Rev Lett. 2007;99 doi: 10.1103/PhysRevLett.99.058302. 058302. [DOI] [PubMed] [Google Scholar]
- 13.Zhang C, Zhang F, van Kan JA, van der Maarel JRC. Effects of electrostatic screening on the conformation of single DNA molecules confined in a nanochannel. J Chem Phys. 2008;128:225109. doi: 10.1063/1.2937441. [DOI] [PubMed] [Google Scholar]
- 14.Tegenfeldt JO, et al. The dynamics of genomic-length DNA molecules in 100-nm channels. Proc Natl Acad Sci USA. 2004;101:10979–10983. doi: 10.1073/pnas.0403849101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mannion JT, Reccius CH, Cross JD, Craighead HG. Conformational analysis of single DNA molecules undergoing entropically induced motion in nanochannels. Biophys J. 2006;90:4538–4545. doi: 10.1529/biophysj.105.074732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van der Maarel JRC. Introduction to Biopolymer Physics. Singapore: World Scientific; 2008. [Google Scholar]
- 17.van Kan JA, Bettiol AA, Watt F. Three-dimensional nanolithography using proton beam writing. Appl Phys Lett. 2003;83:1629–1631. [Google Scholar]
- 18.van Kan JA, Bettiol AA, Watt F. Proton beam writing of three-dimensional nanostructures in hydrogen silsesquioxane. Nano Lett. 2006;6:579–582. doi: 10.1021/nl052478c. [DOI] [PubMed] [Google Scholar]
- 19.Shao PG, van Kan JA, Ansari K, Bettiol AA, Watt F. Poly (dimethyl siloxane) micro/nanostructure replication using proton beam written masters. Nucl Instrum Methods Phys Res, Sect B. 2007;260:479–482. [Google Scholar]
- 20.Johansen F, Jacobsen JP. 1H NMR studies of bis-intercalation of a homodimeric oxazole yellow dye in DNA oligonucleotides. J Biomol Struct Dyn. 1998;16:205–222. doi: 10.1080/07391102.1998.10508240. [DOI] [PubMed] [Google Scholar]
- 21.Bakajin OB, et al. Electrohydrodynamic stretching of DNA in confined environments. Phys Rev Lett. 1998;80:2737–1740. [Google Scholar]
- 22.Hud NV, Vilfan ID. Toroidal DNA condensates: Unraveling the fine structure and the role of nucleation in determining size. Annu Rev Biophys Biomol Struc. 2005;34:295–318. doi: 10.1146/annurev.biophys.34.040204.144500. [DOI] [PubMed] [Google Scholar]
- 23.de Gennes P-G. Scaling Concepts in Polymer Physics. Ithaca, NY: Cornell Univ Press; 1979. [Google Scholar]
- 24.de Gennes P-G. Suspension colloïdales dans une solution de polymères. C. R. Acad. Sc. Paris. 1979;288 B:359–361. [Google Scholar]
- 25.Odijk T. Protein macromolecule interactions. Macromolecules. 1996;29:1842–1843. [Google Scholar]
- 26.Asakura S, Oosawa F. On interaction between two bodies immersed in a solution of macromolecules. J Chem Phys. 1955;22:1255–1256. [Google Scholar]
- 27.de Vries R. Depletion-induced instability in protein-DNA mixtures: Influence of protein charge and size. J Chem Phys. 2006;125 doi: 10.1063/1.2209683. 014905. [DOI] [PubMed] [Google Scholar]
- 28.Liang X, Morton KJ, Austin RH, Chou SY. Single sub-20 nm wide, centimeter-long nanofluidic channel fabricated by novel nanoimprint mold fabrication and direct imprinting. Nano Lett. 2007;7:3774–3780. doi: 10.1021/nl072253x. [DOI] [PubMed] [Google Scholar]
- 29.Senti FR, et al. Viscosity, sedimentation, and light-scattering properties of fraction of an acid-hydrolyzed dextran. J Polym Sci. 1955;17:527–546. [Google Scholar]