Abstract
Inositol requiring enzyme-1 (IRE1), a protein located on the endoplasmic reticulum (ER) membrane, is highly conserved from yeast to humans. This protein is activated during ER stress and induces cellular adaptive responses to the stress. In mice, IRE1α inactivation results in widespread developmental defects, leading to embryonic death after 12.5 days of gestation. However, the cause of this embryonic lethality is not fully understood. Here, by using in vivo imaging analysis and conventional knockout mice, respectively, we showed that IRE1α was activated predominantly in the placenta and that loss of IRE1α led to reduction in vascular endothelial growth factor-A and severe dysfunction of the labyrinth in the placenta, a highly developed tissue of blood vessels. We also used a conditional knockout strategy to demonstrate that IRE1α-deficient embryos supplied with functionally normal placentas can be born alive. Fetal liver hypoplasia thought to be responsible for the embryonic lethality of IRE1α-null mice was virtually absent in rescued IRE1α-null pups. These findings reveal that IRE1α plays an essential function in extraembryonic tissues and highlight the relationship of physiological ER stress and angiogenesis in the placenta during pregnancy in mammals.
The endoplasmic reticulum (ER) plays a central role in the synthesis and modification of secretory and membrane proteins in all eukaryotic cells. Under normal conditions, these proteins are correctly folded and assembled in the ER. However, when cells are exposed to disturbed environments, such as overproduction of ER proteins, viral infection, and glucose deprivation, these proteins accumulate as unfolded or misfolded forms in the ER lumen and, consequently, cause ER stress. To maintain cellular homeostasis, cells induce some adaptive responses to ER stress. One is the unfolded protein response (UPR), which up-regulates the transcription of various genes to increase the protein-folding and protein-degradation activity in the ER.
Inositol requiring enzyme-1 (IRE1) is an ER-located type I transmembrane protein with a kinase domain and RNase domain in the cytosolic region and has a unique function of relieving ER stress in cells. When the amino-terminal luminal region senses perturbations in the ER environment, via trans-autophosphorylation and activation of its RNase domain, IRE1 induces unconventional splicing of mRNA coding a specific transcription factor for activating the UPR (1–6). IRE1 is highly conserved from yeast to humans, and two IRE1 paralogues have been reported in mammals: IRE1α and IRE1β (7–9). IRE1α induces unconventional splicing of XBP1 mRNA under ER stress conditions, which results in the removal of a 26-nucleotide intron from XBP1 mRNA and the translational frame shift of XBP1 mRNA (5, 10). The spliced XBP1 mRNA is translated into a functional transcription factor to up-regulate gene expression for ER quality control. IRE1 is also reported to activate proapoptotic JNK signaling under ER stress conditions (11). Other than IRE1, two ER-located transmembrane proteins, PERK and ATF6, play important roles in the mammalian UPR (12, 13). On sensing ER stress, PERK induces the UPR via trans-autophosphorylation, phosphorylation of eIF2α, and translational activation of a specific transcription factor, ATF4 (14). On the other hand, ATF6 translocates into the Golgi body and is cleaved by Site-1 and Site-2 proteases to induce the UPR under ER stress conditions (15, 16). It has also been shown by in vitro analysis using mouse embryonic fibroblasts (MEFs) from knockout animals that PERK and ATF6 as well as IRE1α are required for the full activation of the UPR (14, 17–19).
PERK, ATF6, and IRE1 have common features as UPR inducers. Nevertheless, there are several differences among their functions in vivo. The following results have so far been obtained from analyses using mice. PERK is highly activated in the pancreas and is essential for the viability of exocrine and endocrine pancreatic cells (20). ATF6α and ATF6β are expressed ubiquitously, and double knockout of ATF6α and ATF6β in mammals causes embryonic lethality in the early developmental stage (by 8.5 days of gestation), although a single knockout of each gene does not cause developmental abnormality (18, 19). IRE1α is also known to be expressed ubiquitously in fetal and adult mice and to be essential for mammalian developmental processes (7, 11). Therefore, IRE1α inactivation results in widespread developmental defects, leading to embryonic death after 12.5 days of gestation in mice (21). However, the cause of this embryonic lethality is not fully understood. In contrast, it is known that yeast and nematodes have only one IRE1 gene in their genome, and that inactivation of this gene is not lethal to these organisms under normal conditions (1, 22). These lines of evidence suggest that IRE1α has a unique function in mammalian developmental processes, but it has been hitherto unclear in which tissues and how IRE1α functions during embryogenesis.
Results
IRE1α Is Activated Predominantly in the Placenta.
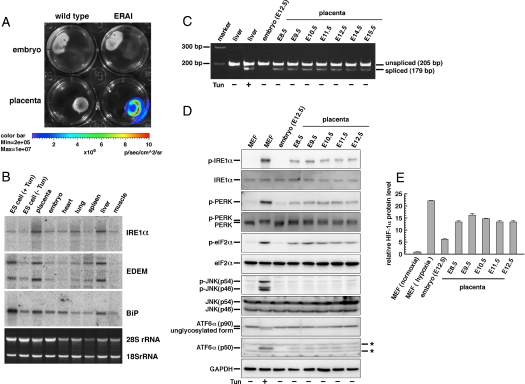
We previously reported the ER stress-activated indicator (ERAI) mouse, a model system for monitoring ER stress in vivo using GFP (23). This mouse can also be used as a tool for visualizing IRE1α activity in vivo. Here, another type of ERAI transgenic mouse was generated by using the luciferase gene instead of GFP (Fig. S1). By using ERAI-LUC mice, we examined luminescence signals in embryos at various developmental stages to detect where IRE1α activity exists. Interestingly, intense luminescence was detected from the placenta, but that from the embryo proper was very weak (Fig. 1A). We also examined the expression level of the endogenous IRE1α gene and two other genes, BiP and EDEM, which are up-regulated in an IRE1α-dependent manner, in various tissues in wild-type mice. Northern blot analysis and quantitative PCR analysis revealed that the placenta expressed not only IRE1α but also BiP and EDEM at high levels. (Fig. 1B and Fig. S2A). By RT-PCR analysis and quantitative PCR analysis of endogenous XBP1, we clearly detected the spliced form of XBP1 mRNA and calculated the ratio of spliced XBP1 mRNA level to total XBP1 mRNA level in the placenta at all developmental stages from embryonic days 8.5 (E8.5) to E15.5 (Fig. 1C and Fig. S2B). In this connection, we also examined endogenous XBP1 splicing level in various tissues by quantitative PCR analysis (Fig. S2C). Moreover, we detected the activated (phosphorylated) form of IRE1α in the placenta by Western blot analysis, although the signals were very weak compared with those in the positive control (Fig. 1D). In contrast, the spliced form of XBP1 mRNA and the activated form of IRE1α were slightly or hardly detected in the embryo proper (Fig. 1 C and D and Fig. S2B). Moreover, we examined the level of activation of other ER stress-responsive molecules by Western blot analysis. The activated (phosphorylated) forms of PERK and eIF2α and the activated (cleaved) form of ATF6α were detected in the placenta but were slightly or hardly detected in the embryo proper. On the other hand, the activated (phosphorylated) form of JNK was hardly detected either in the placenta or the embryo proper (Fig. 1D). These results indicate that IRE1α is activated predominantly in the placenta and suggest that IRE1α activation is possibly dependent on the ER stress produced in the placenta during mouse embryogenesis.
Fig. 1.
Activity and expression of IRE1α in the placenta. (A) Bioluminescence imaging of embryos and placentas of wild-type and ERAI-LUC mice (E14.5). (B) Northern blot analysis of IRE1α, EDEM, and BiP in the embryo (E11.5), placenta (E11.5), and various tissues of the adult wild-type mouse. +Tun and −Tun indicate treatment with and without 2 μg/mL tunicamycin (ER stressor) for 6 h, respectively. ES cells were used as control samples. Ethidium bromide staining (Bottom) shows the RNA loading control. (C) RT-PCR analysis of XBP1 in embryos and placentas. +Tun indicates i.p. injection with tunicamycin (500 ng/g body weight) 16 h before tissue collection. Liver tissues were used as control samples. (D) Western blot analysis of IRE1α, PERK, eIF2α, JNK, and ATF6α in embryos and placentas. p- indicates the phosphorylated form of each protein. ATF6α (p50) is the cleaved form. +Tun indicates treatment with 2.5 μg/mL tunicamycin for 6 h. MEFs were used as control samples. GAPDH was used as an internal standard. *, Nonspecific signals. (E) ELISA of HIF-1α in embryos and placentas. Normoxia and hypoxia indicate culture under 20% O2 or 1% O2 for 6 h, respectively. MEFs were used as control samples. Error bar indicates SEM (n = 3).
Loss of IRE1α Affects Not Only the Embryo Proper but Also the Placenta.
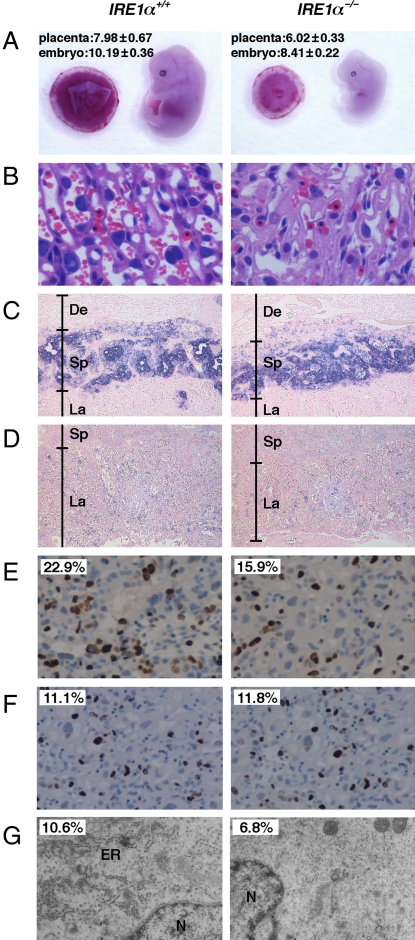
Considering that IRE1α is activated predominantly in the placenta, we hypothesized that IRE1α disruption might be detrimental to placental function. To test this hypothesis, we generated an IRE1α KO mouse and assessed the placental phenotype. Our IRE1α KO mouse was generated as shown in Fig. S3. A comparison of the gross morphology of IRE1α+/+ and IRE1α−/− littermates showed that IRE1α−/− mice were smaller than IRE1α+/+ mice with respect to the size of the placenta as well as that of the embryo proper (Fig. 2A). Thus, IRE1α disruption affects not only the embryo proper but also the placenta.
Fig. 2.
Placental dysplasia in IRE1α−/− mice. (A) Gross morphology of E13.5 IRE1α+/+ and IRE1α−/− embryos and placentas. Each number indicates the body length of embryo and the diameter of placenta (millimeters) as mean ± SD (n = 16). (B) H&E-stained E13.5 placental sections showing a region of the labyrinth layer in the placenta. (C and D) Cell lineage marker analysis of E13.5 placental sections by using in situ hybridization. Tpbp (C) and Tfeb (D) were used as a spongiotrophoblast cell-specific and a labyrinth cell-specific marker, respectively. De, deciduas; Sp, spongiotrophoblast; La, labyrinth. Sections were counterstained with eosin. (E) BrdU immunohistochemically stained E13.5 placental sections. Sections were counterstained with hematoxylin. Number indicates the percentage of BrdU-positive cells. (F) TUNEL assays for E13.5 placental sections. Sections were counterstained with hematoxylin. Number indicates the percentage of TUNEL-positive cells. (G) Electron micrographs of E13.5 placental sections. N, nucleus. Number indicates the percentage of the ER luminal space compared with the cytoplasmic space. Each space was measured from 10 photographs with Image Gauge version 4.21 software (Fuji film). (Original magnification: A, 1×; B, 20×; C and D, 5×; E and F, 10×; and G, 4,000×.)
Loss of IRE1α Leads to Severe Dysfunction of the Placenta.
At E13.5, three layers—the labyrinth, spongiotrophoblasts, and deciduas—are easily discernible in the placenta. To assess phenotypes in the placental tissues of IRE1α−/− mice, we analyzed H&E-stained sections obtained from wild-type and IRE1α-deficient placentas. The labyrinth is the site of oxygen and nutrient exchange between the mother and the fetus and is a highly developed tissue of blood vessels. The IRE1α mutant labyrinth layer was severely disrupted relative to the normal labyrinth, with a reduction in the internal space of fetal and maternal blood vessels (Fig. 2B). In contrast, the spongiotrophoblast and decidua layers of IRE1α mutant mice appeared normal (data not shown). In addition, we performed cell marker analysis of the spongiotrophoblast and labyrinth layers by using an in situ hybridization method. Tpbp (24) and Tfeb (25), spongiotrophoblast layer-specific and labyrinth layer-specific markers, respectively, were expressed in the correct spatial pattern in both wild-type and IRE1α mutant placentas (Fig. 2 C and D). The result of this cell marker analysis suggests that the histological abnormality of the IRE1α mutant placenta is not caused by misdifferentiation of the labyrinth layer cells.
The reduction in the blood space in the IRE1α-deficient labyrinth suggests that there would be restricted nutrient and/or oxygen exchange between the mother and the developing embryos. To directly assess placental transport, we measured the accumulation of essential fatty acids (EFAs) in the fetuses and placentas of the IRE1α mutant and wild-type E13.5 embryos by gas chromatographic analysis (26, 27). EFAs are obtained from the diet, and thus fetal EFA content is a direct reflection of placental transport capacity. As shown in Table S1, IRE1α-deficient fetuses, relative to wild-type fetuses, had significantly lower amounts of the three EFAs (linoleic acid, arachidonic acids, and docosahexaenoic acid) but had normal levels of non-EFAs (palmitic acid and stearic acid). Because the fatty acid composition of the placenta was not affected by the loss of IRE1α (Table S1), the lower level of EFAs in IRE1α−/− fetuses suggests a malfunctioning placenta.
Loss of IRE1α Affects Expression Level of VEGF-A in the Placenta.
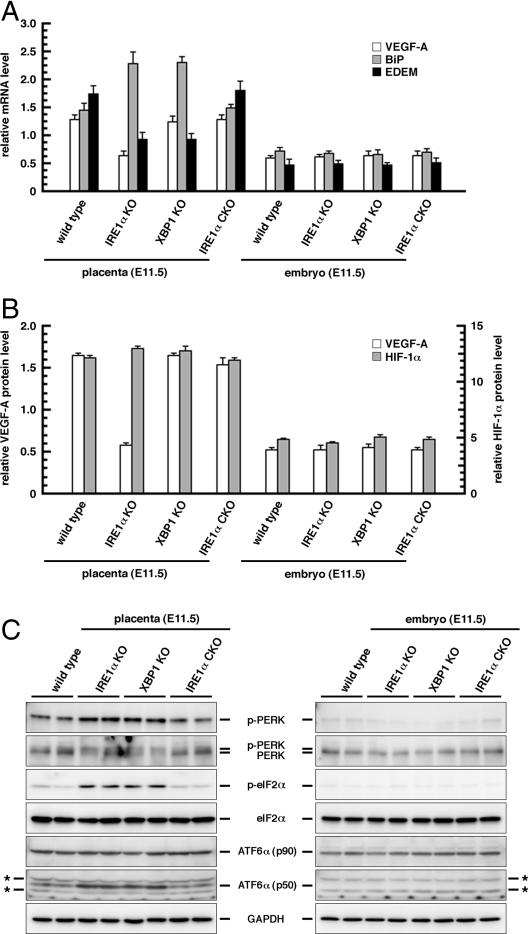
VEGF-A is essential for development of the labyrinth in the placenta, and VEGF-A expression is elevated by ER stress (28, 29). Based on these lines of evidence and phenotypes in the IRE1α-deficient placenta described above, we hypothesized that IRE1α performs a function for VEGF-A expression in the placenta. To test this hypothesis, we compared the expression level of VEGF-A by quantitative PCR analysis and ELISA. As expected, IRE1α-deficient placentas had half the level of VEGF-A compared with wild-type placentas (Fig. 3 A and B). This result indicates that VEGF-A expression in the placenta is at least partially dependent on IRE1α. Moreover, we generated an XBP1 KO mouse and assessed the phenotype. Our XBP1 KO mouse was generated as shown in Fig. S4. Unlike IRE1α-deficient mice, XBP1-deficient placentas had the same level of VEGF-A compared with wild-type placentas. Data shown in Fig. S4 also revealed that the morphological and histological phenotypes in the placenta of XBP1-deficient mice were mild compared with those of IRE1α-deficient mice. On the other hand, both IRE1α-deficient embryos and XBP1-deficient embryos had the same level of VEGF-A as that in wild-type embryos (Fig. 3 A and B). HIF-1 is a transcription factor that activates VEGF-A expression under ischemic stress (30, 31) and is essential for angiogenesis in the labyrinth of the placenta (32, 33). Generally, the transcriptional activity of HIF-1 is dependent on the level of HIF-1α protein. Here, we examined the level of HIF-1α protein by ELISA. HIF-1α protein in both mutant placentas was expressed at the same level as that of the wild-type placenta. Also, HIF-1α protein in the embryo was expressed at the same level among each genotype (Fig. 3B). We also demonstrated that the level of HIF-1α protein was 2- or 3-fold higher in the placenta than in the embryo proper (Fig. 1E). These results indicate that the effect of VEGF-A on the IRE1α mutant is prominent in the placenta, that HIF-1α was not decreased in the IRE1α mutant placenta, and that VEGF-A expression in the placenta is partially dependent on IRE1α but not on XBP1.
Fig. 3.
Expression or activation level of VEGF-A, HIF-1α, and ER stress-responsive molecules in the placenta (E11.5) and embryo (E11.5). (A) Quantitative PCR analysis of VEGF-A, BiP, and EDEM. Error bar indicates SEM (n = 3). (B) ELISA of VEGF-A and HIF-1α. Error bar indicates SEM (n = 3). (C) Western blot analysis of PERK, eIF2α, and ATF6α. p- indicates the phosphorylated form of each protein. ATF6α (p50) is the cleaved form. GAPDH was used as an internal standard. The genotype of IRE1α CKO is Mox2+/Cre; IRE1αΔNeo/ΔR. Each sample used here was derived from the embryonic part (labyrinth and spongiotrophoblasts) of each placenta. *, Nonspecific signals.
Loss of IRE1α Affects Proliferation, ER Development, and ER Stress Level in Trophoblasts of the Placenta.
The effects of IRE1α mutant in the placenta may not be restricted to the angiogenic phenotypes described above. We therefore examined UPR induction, cell growth level, ER structure, and apoptosis level in IRE1α−/− trophoblasts. BrdU incorporation and TUNEL assays showed that IRE1α−/− trophoblasts had a lower BrdU index than that in IRE1α+/+ controls, although the IRE1α−/− trophoblasts had a level of apoptosis similar to that in the IRE1α+/+ controls (Fig. 2 E and F). These results suggest that the small placentas of IRE1α KO mice may be due to reduced cell proliferation of trophoblasts but not their accelerated apoptosis. Observation by transmission electron microscopy also revealed poor ER development in IRE1α−/− trophoblasts (Fig. 2G and Fig. S5). Quantitative PCR analysis demonstrated that the BiP expression level of the IRE1α−/− and XBP1−/− placentas was 1.5-fold higher than that of the wild-type placenta, and that the EDEM expression level of the IRE1α−/− and XBP1−/− placentas was half that of the wild-type placenta (Fig. 3A). Western blot analysis also revealed that PERK, eIF2α, and ATF6 were more activated in the IRE1α−/− and XBP1−/− placentas than in the wild-type placenta (Fig. 3C). On the other hand, the activated form of JNK was hardly detected in the placenta or in the embryo proper in wild-type, IRE1α−/−, and XBP1−/− mice (data not shown). These data suggest that ER stress in the placenta may be enhanced by disruption of IRE1α and XBP1, but this is not proapoptotic. BiP is known to be regulated not only by the IRE1α/XBP1 pathway but also by the PERK and ATF6 pathways in UPR induction. BiP expression may be activated by PERK and ATF6 in IRE1α and XBP1 KO mouse placentas. On the other hand, EDEM expression may be reduced in those KO mouse placentas because EDEM is regulated only by the IRE1α/XBP1 pathway.
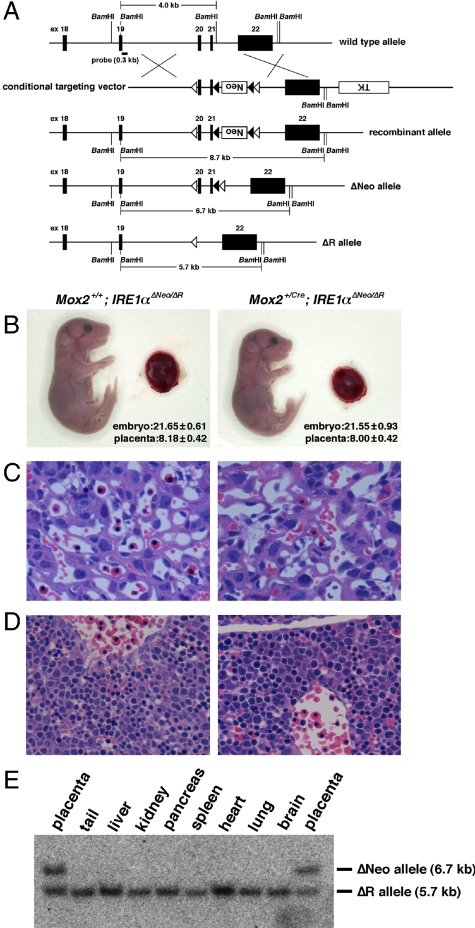
Extraembryonic Function of IRE1α Rescues Lethality of IRE1α-Deficient Embryos.
Considering the phenotype in IRE1α−/− embryos, we hypothesized that a defective IRE1α−/− placenta may cause embryonic lethality. To explore this possibility, we used IRE1α conditional KO mice and Mox2+/Cre transgenic mice (34) to reconstitute IRE1α-deficient embryos with functionally normal placentas. Previous studies have shown that the Cre recombinase gene under the control of the endogenous Mox2 promoter (Mox2+/Cre) is efficiently expressed and functionally active in all cells of the embryo proper but is not expressed at all in the trophoblasts or extraembryonic endoderm lineages (27, 34). IRE1α conditional KO mice were generated as shown in Fig. 4A, and basic characterization of these mice was performed as shown in Fig. S6. By breeding Mox2+/Cre mice harboring one ΔR allele (Mox2+/Cre; IRE1α+/ΔR) with IRE1αΔNeo/ΔNeo mice (Fig. S7), we were able to generate viable IRE1α conditional KO mice (Mox2+/Cre; IRE1αΔNeo/ΔR) which, interestingly, were born at near-Mendelian ratios (28 of 129 offspring; a frequency of 21.7% compared with the expected 25%). The IRE1α conditional KO mice rescued by IRE1α expression in the placenta showed no placental abnormalities or liver hypoplasia, both of which are the hallmark phenotypes of IRE1α−/− mice (Fig. 4 B–D). Also, the expression of VEGF-A and HIF-1α and the activation of ER stress-responsive molecules of the conditional KO mice were at the same levels as those of the wild-type mice both in the placental and embryonic tissues (Fig. 3). The results of the Southern blot analysis demonstrated that although both ΔNeo and ΔR alleles could be readily detected in the placentas of the conditional KO mice, the ΔNeo allele could not be detected in other tissues of these mice (Fig. 4E). This finding indicates that, as expected, the Cre-dependent deletion of exons 20–21 from the floxed IRE1α allele (IRE1αΔNeo to IRE1αΔR) occurred without any leakage in any tissues other than the extraembryonic tissues. Therefore, the results of this genetic rescue experiment suggest that IRE1α has a critical function in extraembryonic cells that are vital for fetal viability.
Fig. 4.
IRE1α-deficient mice with normal placentas as reconstituted by using conditional knockout approaches. (A) Schematic diagram showing the IRE1α conditional knockout strategy. Neo and TK indicate expression units of the neomycin resistance gene and thymidine kinase gene for positive and negative selections, respectively. Closed and open arrowheads indicate FRT and loxP elements, respectively. (B) Gross morphology of E18.5 Mox2+/+; IRE1αΔNeo/ΔR and Mox2+/Cre; IRE1αΔNeo/ΔR embryos and placentas. Each number indicates the body length of embryo and the diameter of placenta (millimeters) as mean ± SD (n = 10). (C) H&E-stained E13.5 placental sections showing a region of the labyrinth layer in the placenta. (D) H&E-stained E13.5 liver sections. (Original magnifications: B, 1×; C, 20×; and D, 40×.) (E) Southern blot analysis of various tissues from Mox2+/Cre; IRE1αΔNeo/ΔR mice.
Discussion
Previous studies have established the essential role of IRE1α during embryogenesis (11). Here, we addressed the question, “Why is IRE1α necessary for embryogenesis?” We initially found that IRE1α is activated predominantly in the placenta during mouse embryogenesis (Fig. 1). Then, we observed that the loss of IRE1α reduced VEGF-A level and aggravated ER stress in the placenta, and that the labyrinth structure was markedly altered in IRE1α-deficient placentas, leading to defective oxygen/nutrient exchange between the mother and the fetus (Figs. 2 and 3 and Table S1). Our genetic rescue analysis also indicated that functional defects in IRE1α−/− placental cells led to embryonic lethality in IRE1α−/− mice (Fig. 4). Finally, we demonstrated that IRE1α plays a critical role in the placenta that is vital for fetal viability.
Why is IRE1α activated in the placenta? We found that other ER stress-responsive molecules were activated at a higher level in the placenta than in the embryo. We also observed that the activation levels of those molecules in the placenta were lower than those in cells treated with an ER stressor (Fig. 1 B and D). Moreover, JNK activation was hardly detected in the placenta (Fig. 1D). Apoptosis level in the IRE1α-deficient placenta was very low and was similar to that in the wild-type placenta (Fig. 2F), although severe ER stress is known to induce apoptosis, and IRE1α is believed to be necessary for mitigating ER stress. Considering these findings, we suggest that the placenta is possibly under a mild ER stress condition, which might partially activate IRE1α. However, we still do not have a clear answer as to why the placenta is under this ER stress condition. As described in the Introduction, ER stress is caused by glucose deprivation and overproduction of ER proteins, which are also the likely causes of placental ER stress. Moreover, some molecules for hypoxic stress response (e.g., HIF-1β, B-Raf, and PHD2) are involved in angiogenesis in the labyrinth of the placenta (32, 33, 35, 36). Here, we further revealed that HIF-1α protein was expressed at a 2- or 3-fold higher level in the placenta than in the embryo proper. Therefore, the relationship between ER stress in the placenta and the production of secretory and membrane proteins, glucose deprivation, and hypoxia is interesting and needs to be investigated in the future. What type of placental cell is exposed to ER stress? The answer to this question may provide important clues for understanding the cause of ER stress in the placenta. In this study, however, we have not yet completely answered this question because in the placental sections, we detected neither ERAI-LUC signals nor specific immunohistochemical signals of ER stress-responsive molecules (data not shown). Furthermore, we also need to develop novel and additional sensitive methods for detecting ER stress in tissue sections.
Interestingly, why do IRE1α KO mice die at the embryonic stage? The study of conditional KO mice showed that embryonic lethality of IRE1α KO mice was caused by the loss of IRE1α in the placenta (Fig. 4). However, we still do not have a clear answer to this question: What effects of an IRE1α-deficient placenta cause embryonic lethality of IRE1α KO mice? Here, we showed that the primary effects of IRE1α-deficient placenta include VEGF-A reduction, labyrinth dysfunction, UPR induction, reduced proliferation of trophoblasts, and poor ER development in trophoblasts (Figs. 2 E and G and 3, and Table S1). Mature placental trophoblasts with a normally developed ER produce many secretory proteins, such as placental lactogens and growth factors, as well as VEGF-A (37–40). On the other hand, immature, IRE1α-deficient trophoblasts with a poorly developed ER might reduce the production and/or secretion of these placental proteins. In fact, this study showed VEGF-A reduction in the IRE1α-deficient placenta. A previous study of VEGF-A gene-targeting mice showed that VEGF-A+/− mice had angiogenic abnormality in the labyrinth of the placenta and other tissues, and they died at the embryonic stage, indicating that the tight dose-dependent regulation by VEGF-A was critical for normal angiogenesis in the labyrinth of the placenta and embryonic development and viability (28). Therefore, a detailed investigation into the relationship between VEGF-A reduction and embryonic lethality of IRE1α KO is needed, although the embryonic lethality of these mice might be caused by a vicious spiral or negative interaction of the multiple primary effects of IRE1α-deficient placenta described above.
How is VEGF-A gene expression regulated in the placenta? Generally, IRE1α regulates the expression of the UPR target gene via the XBP1. However, the loss of XBP1 showed no effect on the expression level of VEGF-A in the placenta (Fig. 3 A and B). Another research group reported that XBP1 KO mice died at the embryonic stage (41), and that XBP1 KO mice rescued with an XBP1 transgene specifically expressed in the liver were born at near-Mendelian ratios but died soon after birth because of pancreatic dysfunction (42). Considering these lines of evidence, XBP1 possibly plays an essential role in the liver and pancreas but not in the placenta. Therefore, we need to clarify how other molecule(s) function as downstream target(s) of IRE1α for VEGF-A expression in the placenta. As described above, HIF-1 is a transcription factor that activates VEGF-A expression (30, 31) and is essential for angiogenesis in the labyrinth of the placenta (32, 33). On the other hand, the up-regulation of VEGF-A in response to glucose deprivation was reported to be independent of HIF-1 (43, 44). These reports and our results suggest that IRE1α may play a role in VEGF-A expression in the placenta through the XBP1- and HIF1-independent pathways. However, we have not yet determined the downstream target(s) of IRE1α in VEGF-A expression. Considering that the up-regulation of VEGF-A in response to glucose deprivation was also reported to be due to an increase in mRNA stability (45), it is possible that the downstream target(s) of IRE1α in VEGF-A expression might be molecule(s) regulating mRNA stability. In addition, we need to consider not only XBP1-independent IRE1α function but also IRE1α-independent XBP1 function. Presently, not one report exists on XBP1 splicing via any mechanism other than that induced by IRE1α. However, genetic rescue experiments of IRE1α and XBP1 KO mice demonstrated that IRE1α-independent XBP1 was essential for fetal liver function. This suggests that the unspliced form of XBP1 may have an important function in the fetal liver for mouse embryonic viability.
Regarding ER stress (responsive molecules), in vivo function is an important topic. To date, it has been reported that IRE1α is required for B-cell differentiation (21) and insulin biosynthesis (46), and that it is involved in diabetes (47) and some neurodegenerative diseases (48, 49). Recently, some researchers have also focused on the relationship between IRE1α and cancer, and they reported the angiogenic function of IRE1α via the up-regulation of VEGF-A in tumors (50). Here, we found a relationship between the function of IRE1α and the development of the mouse placenta. However, until now, little has been known regarding the relationship between ER stress and pregnancy. ER stress and IRE1α might also be involved in other physiological phenomena and diseases. The IRE1α conditional KO mice and ERAI-LUC transgenic mice developed here will undoubtedly be important tools for research on these involvements in the future.
Materials and Methods
Gene Constructs.
For ERAI-LUC transgenic mice, pCAX-F-XBP1ΔDBD-LUC was constructed as described previously (51). For IRE1α conventional KO mice, pKOV1-IRE1α-1 was constructed by the insertion of the 5′ and 3′ IRE1α homology arms into the KpnI/XhoI and EcoRI sites, respectively, of pKOV1. The 5′ and 3′ IRE1α homology arms were produced by PCR using specific primers and mouse genomic DNA as a template. For XBP1 conventional KO mice, pKOV1-XBP1-1 was constructed by the insertion of the 5′ and 3′ XBP1 homology arms into the KpnI/XhoI and BamHI sites, respectively, of pKOV1. The 5′ and 3′ XBP1 homology arms were produced by PCR. For IRE1α conditional KO mice, pKOV2-IRE1αCKO-1 was constructed by the insertion of the 5′ CKO homology arm, conditional targeting region, and 3′ CKO homology arm into the SalI/NotI, HindIII, and BamHI sites, respectively, of pKOV2. The 5′ CKO homology arm, conditional targeting region, and 3′ CKO homology arm were produced by PCR. The primers are shown in Table S2.
Mouse Strain and Genotyping.
Mox2+/Cre (34) and ROSA26Flp/Flp (52) transgenic mice were obtained from the Jackson Laboratory and were maintained on a mixed (C57BL/6 × 129/SvJae) background. ERAI-LUC mice were generated as described previously (23) and were maintained on a C57BL/6 background. IRE1α conventional KO mice, XBP1 conventional KO mice, and IRE1α conditional KO mice were generated as described previously (53, 54) and were maintained on a mixed (C57BL/6 × 129/SvE) background. Experimental protocols involving animals were approved by the Animal Studies Committees at RIKEN and the Nara Institute of Science and Technology.
Northern Blot Analysis, RT-PCR, and Quantitative PCR.
Northern blot analysis was performed as described previously (23). To detect ERAI-LUC, IRE1α, EDEM, BiP, XBP1, and VEGF-A, the following regions were used as probes: luciferase, 1–1,650 nt of the coding region; IRE1α, 1,235–2,048 nt or 2,529–2,721 nt; EDEM, 3–752 nt; BiP, 3–682 nt; XBP1, 207–584 nt; and VEGF-A, 1–645 nt. RT-PCR was also performed as described previously (23). Quantitative PCR analysis of each transcript was performed by using TaqMan probe and 7900HT (Applied Biosystems) in accordance with the manufacturer's instructions. The GAPDH transcript was used as an internal control in the analysis. The results of quantitative PCR analysis were expressed as mean ± SEM from triplicate experiments using RNA isolated from three independent tissues and cell cultures. Each probe/primer set was purchased from Applied Biosystems.
Western Blot Analysis.
Western blot analysis was performed as described previously (23). The following antibodies were used for Western blot analysis: anti-eIF2α polyclonal antibody (Cell Signaling Technology), anti-phospho-eIF2α (Ser-51) polyclonal antibody (Cell Signaling Technology), anti-GAPDH monoclonal antibody (Abcam), anti-JNK polyclonal antibody (Cell Signaling Technology), anti-phospho-JNK (Thr-183/Tyr-185) polyclonal antibody (Cell Signaling Technology), anti-IRE1α monoclonal antibody (Cell Signaling Technology), anti-phospho-IRE1α (Ser-724) polyclonal antibody (Affinity Bioreagents), anti-PERK polyclonal antibody (Lifespan), anti-phospho-PERK (Thr-980) monoclonal antibody (Cell Signaling Technology), and rabbit anti-ATF6α polyclonal antibody that we generated by using the recombinant mouse ATF6 (1–294 aa) fragment as an antigen.
ELISA.
HIF-1α protein and VEGF-A protein were quantified by using commercial ELISA kits (R&D Systems and BioVendor, respectively) in accordance with the manufacturer's instructions. Total protein level was used as an internal control in the analysis. The results were expressed as mean ± SEM from triplicate experiments using the extract from three independent tissues and cell cultures.
Supplementary Material
Acknowledgments.
We thank Robert Farese, Jr. (Gladstone Institute, CA), for providing the RF8 ES cells used to generate gene-targeting mice, and Mio Tokuda, Michiko Saito, Misato Chiba, Hisayo Masuda, Junko Iida, Miki Matsumura, Kazuaki Takahashi, and Tomoko Ichisaka for technical assistance. This work was supported by grants from RIKEN (to T.I.), JST (to T.I.), the Mochida Memorial Foundation (to T.I.), the Suzuken Memorial Foundation (to T.I.), the Takeda Science Foundation (to T.I.), the Naito Memorial Foundation (to T.I.), and MEXT (No. 14037240 to K.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.B. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0903775106/DCSupplemental.
References
- 1.Cox JS, Shamu CE, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- 2.Mori K, Ma W, Gething MJ, Sambrook J. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 1993;74:743–756. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- 3.Cox JS, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- 4.Shamu ES, Walter P. Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J. 1996;15:3028–3039. [PMC free article] [PubMed] [Google Scholar]
- 5.Calfon M, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 6.Kimata Y, et al. Two regulatory steps of ER-stress sensor Ire1 involving its cluster formation and interaction with unfolded proteins. J Cell Biol. 2007;179:75–86. doi: 10.1083/jcb.200704166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tirasophon W, Welihinda AA, Kaufman RJ. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 1998;12:1812–1824. doi: 10.1101/gad.12.12.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang XZ, et al. Cloning of mammalian Ire1 reveals diversity in the ER stress responses. EMBO J. 1998;17:5708–5717. doi: 10.1093/emboj/17.19.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iwawaki T, et al. Translational control by the ER transmembrane kinase/ribonuclease IRE1 under ER stress. Nat Cell Biol. 2001;3:158–164. doi: 10.1038/35055065. [DOI] [PubMed] [Google Scholar]
- 10.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 11.Urano F, et al. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- 12.Yoshida H, Haze K, Yanagi H, Yura T, Mori K. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. J Biol Chem. 1998;273:33741–33749. doi: 10.1074/jbc.273.50.33741. [DOI] [PubMed] [Google Scholar]
- 13.Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 14.Harding HP, et al. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 15.Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell. 1999;10:3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ye J, et al. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell. 2000;6:1355–1364. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- 17.Lee K, et al. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 2002;16:452–466. doi: 10.1101/gad.964702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu J, et al. ATF6alpha optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell. 2007;13:351–364. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 19.Yamamoto K, et al. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6α and XBP1. Dev Cell. 2007;13:365–376. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 20.Harding HP, et al. Diabetes mellitus and exocrine pancreatic dysfunction in Perk-/- mice reveals a role for translational control in secretory cell survival. Mol Cell. 2001;7:1153–1163. doi: 10.1016/s1097-2765(01)00264-7. [DOI] [PubMed] [Google Scholar]
- 21.Zhang K, et al. The unfolded protein response sensor IRE1alpha is required at 2 distinct steps in B cell lymphopoiesis. J Clin Invest. 2005;115:268–281. doi: 10.1172/JCI21848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen X, et al. Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell. 2001;107:893–903. doi: 10.1016/s0092-8674(01)00612-2. [DOI] [PubMed] [Google Scholar]
- 23.Iwawaki T, Akai R, Kohno K, Miura M. A transgenic mouse model for monitoring endoplasmic reticulum stress. Nat Med. 2004;10:98–102. doi: 10.1038/nm970. [DOI] [PubMed] [Google Scholar]
- 24.Lescisin KR, Varmuza S, Rossant J. Isolation and characterization of a novel trophoblast-specific cDNA in the mouse. Genes Dev. 1988;2:1639–1646. doi: 10.1101/gad.2.12a.1639. [DOI] [PubMed] [Google Scholar]
- 25.Steingrimsson E, Tessarollo L, Reid SW, Jenkins NA, Copeland NG. The bHLH-Zip transcription factor Tfeb is essential for placental vascularization. Development. 1998;125:4607–4616. doi: 10.1242/dev.125.23.4607. [DOI] [PubMed] [Google Scholar]
- 26.Metcalfe LD, Schmitz AA, Pelka JR. Rapid preparation of fatty acid esters from lipids for gas chromatographic analysis. Anal Chem. 1966;38:514–515. [Google Scholar]
- 27.Wu L, et al. Extra-embryonic function of Rb is essential for embryonic development and viability. Nature. 2003;421:942–947. doi: 10.1038/nature01417. [DOI] [PubMed] [Google Scholar]
- 28.Ferrara N, et al. Heterozygous embryonic lethality induced by targeted inactivation of the VEGF gene. Nature. 1996;380:439–442. doi: 10.1038/380439a0. [DOI] [PubMed] [Google Scholar]
- 29.Abcouwer SF, Marjon PL, Loper RK, Vander Jagt DL. Response of VEGF expression to amino acid deprivation and inducers of endoplasmic reticulum stress. Invest Ophthalmol Vis Sci. 2002;43:2791–2798. [PubMed] [Google Scholar]
- 30.Liu Y, Cox SR, Morita T, Kourembanas S. Hypoxia regulates vascular endothelial growth factor gene expression in endothelial cells. Identification of a 5′ enhancer. Circ Res. 1995;77:638–643. doi: 10.1161/01.res.77.3.638. [DOI] [PubMed] [Google Scholar]
- 31.Forsythe JA, et al. Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol Cell Biol. 1996;16:4604–4613. doi: 10.1128/mcb.16.9.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kozak KR, Abbott B, Hankinson O. ARNT-deficient mice and placental differentiation. Dev Biol. 1997;191:297–305. doi: 10.1006/dbio.1997.8758. [DOI] [PubMed] [Google Scholar]
- 33.Adelman DM, Gertsenstein M, Nagy A, Simon MC, Maltepe E. Placental cell fates are regulated in vivo by HIF-mediated hypoxia responses. Genes Dev. 2000;14:3191–3203. doi: 10.1101/gad.853700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tallquist MD, Soriano P. Epiblast-restricted Cre expression in MORE mice: a tool to distinguish embryonic vs. extra-embryonic gene function. Genesis. 2000;26:113–115. doi: 10.1002/(sici)1526-968x(200002)26:2<113::aid-gene3>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 35.Galabova-Kovacs G, et al. Essential role of B-Raf in ERK activation during extraembryonic development. Proc Natl Acad Sci USA. 2006;103:1325–1330. doi: 10.1073/pnas.0507399103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takeda K, et al. Placental but not heart defects are associated with elevated hypoxia-inducible factor alpha levels in mice lacking prolyl hydroxylase domain protein 2. Mol Cell Biol. 2006;26:8336–8346. doi: 10.1128/MCB.00425-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soares MJ, et al. Differentiation of trophoblast endocrine cells. Placenta. 1996;17:277–289. doi: 10.1016/s0143-4004(96)90051-x. [DOI] [PubMed] [Google Scholar]
- 38.Achen MG, Gad JM, Stacker SA, Wilks AF. Placenta growth factor and vascular endothelial growth factor are co-expressed during early embryonic development. Growth Factors. 1997;15:69–80. doi: 10.3109/08977199709002113. [DOI] [PubMed] [Google Scholar]
- 39.Groskopf JC, Syu LJ, Saltiel AR, Linzer DI. Proliferin induces endothelial cell chemotaxis through a G protein-coupled, mitogen-activated protein kinase-dependent pathway. Endocrinology. 1997;138:2835–2840. doi: 10.1210/endo.138.7.5276. [DOI] [PubMed] [Google Scholar]
- 40.Vuorela P, et al. Expression of vascular endothelial growth factor and placenta growth factor in human placenta. Biol Reprod. 1997;56:489–494. doi: 10.1095/biolreprod56.2.489. [DOI] [PubMed] [Google Scholar]
- 41.Reimold AM, et al. An essential role in liver development for transcription factor XBP-1. Genes Dev. 2000;14:152–157. [PMC free article] [PubMed] [Google Scholar]
- 42.Lee AH, Chu GC, Iwakoshi NN, Glimcher LH. XBP-1 is required for biogenesis of cellular secretory machinery of exocrine glands. EMBO J. 2005;24:4368–4380. doi: 10.1038/sj.emboj.7600903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iyer NV, et al. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev. 1998;12:149–162. doi: 10.1101/gad.12.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kotch LE, Iyer NV, Laughner E, Semenza GL. Defective vascularization of HIF-1alpha-null embryos is not associated with VEGF deficiency but with mesenchymal cell death. Dev Biol. 1999;209:254–267. doi: 10.1006/dbio.1999.9253. [DOI] [PubMed] [Google Scholar]
- 45.Yun H, Lee M, Kim SS, Ha J. Glucose deprivation increases mRNA stability of vascular endothelial growth factor through activation of AMP-activated protein kinase in DU145 prostate carcinoma. J Biol Chem. 2006;280:9963–9972. doi: 10.1074/jbc.M412994200. [DOI] [PubMed] [Google Scholar]
- 46.Lipson KL, et al. Regulation of insulin biosynthesis in pancreatic beta cells by an endoplasmic reticulum-resident protein kinase IRE1. Cell Metab. 2006;4:245–254. doi: 10.1016/j.cmet.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 47.Özcan U, et al. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science. 2004;306:457–461. doi: 10.1126/science.1103160. [DOI] [PubMed] [Google Scholar]
- 48.Katayama T, et al. Presenilin-1 mutations downregulate the signalling pathway of the unfolded-protein response. Nat Cell Biol. 1999;1:479–485. doi: 10.1038/70265. [DOI] [PubMed] [Google Scholar]
- 49.Nishitoh H, et al. ASK1 is essential for endoplasmic reticulum stress-induced neuronal cell death triggered by expanded polyglutamine repeats. Genes Dev. 2002;16:1345–1355. doi: 10.1101/gad.992302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Drogat B, et al. IRE1 signaling is essential for ischemia-induced vascular endothelial growth factor-A expression and contributes to angiogenesis and tumor growth in vivo. Cancer Res. 2007;67:6700–6707. doi: 10.1158/0008-5472.CAN-06-3235. [DOI] [PubMed] [Google Scholar]
- 51.Iwawaki T, Akai R. Analysis of the XBP1 splicing mechanism using endoplasmic reticulum stress-indicators. Biochem Biophys Res Commun. 2006;350:709–715. doi: 10.1016/j.bbrc.2006.09.100. [DOI] [PubMed] [Google Scholar]
- 52.Farley FW, Soriano P, Steffen LS, Dymecki SM. Widespread recombinase expression using FLPeR (flipper) mice. Genesis. 2000;28:106–110. [PubMed] [Google Scholar]
- 53.Ramirez-Solis R, Davis AC, Bradley A. Gene targeting in embryonic stem cells. Methods Enzymol. 1993;225:855–878. doi: 10.1016/0076-6879(93)25054-6. [DOI] [PubMed] [Google Scholar]
- 54.Meiner VL, et al. Disruption of the acyl-CoA:cholesterol acyltransferase gene in mice: Evidence suggesting multiple cholesterol esterification enzymes in mammals. Proc Natl Acad Sci USA. 1996;93:14041–14046. doi: 10.1073/pnas.93.24.14041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.