Abstract
Polysaccharides comprise an extremely important class of biopolymers that play critical roles in a wide range of biological processes, but the synthesis of these compounds is challenging because of their complex structures. We have developed a chemoenzymatic method for regioselective deprotection of monosaccharide substrates using engineered Bacillus megaterium cytochrome P450 (P450BM3) demethylases that provides a highly efficient means to access valuable intermediates, which can be converted to a wide range of substituted monosaccharides and polysaccharides. Demethylases displaying high levels of regioselectivity toward a number of protected monosaccharides were identified using a combination of protein and substrate engineering, suggesting that this approach ultimately could be used in the synthesis of a wide range of substituted mono- and polysaccharides for studies in chemistry, biology, and medicine.
Keywords: biocatalysis, demethylation, evolution, P450, polysaccharide
Complex carbohydrates comprise an extremely important class of biopolymers that play critical roles in a number of biological processes, including inflammation, cancer metastasis, and bacterial and viral infection (1). Unlike nucleic acids and proteins, oligosaccharides can be highly branched, and their biosynthesis proceeds in a non–template-directed fashion to produce complex mixtures of closely related structures that are difficult, if not impossible, to separate (2). Because subtle changes in the structures of these compounds can alter their properties dramatically (3), detailed studies of oligosaccharide-mediated events require the use of homogeneous synthetic material (4). Oligosaccharide synthesis traditionally has been accomplished through the installation and removal of various orthogonally reactive protecting groups to differentiate each position of a monosaccharide substrate, and a similar procedure is used to synthesize glycosides and substituted monosaccharides in which 1 hydroxyl group is used as a handle for the introduction of small molecules or other functional groups (5). Despite numerous advances in the synthesis of these materials during the past decade, their preparation remains an arduous task (6).
Researchers have sought to simplify the preparation of suitably protected monosaccharides with 1-pot synthetic procedures, but these approaches require considerable synthetic expertise and are limited in terms of substrate scope (7). Elimination of protecting groups altogether also has been demonstrated using enzymes to affect the glycosylation of various substrates (8). The utility of this approach could be expanded greatly by identifying or engineering additional enzymes (9), but the preparation of substituted monosaccharides lies beyond its scope. Chemoenzymatic regioselective deprotection of a globally protected monosaccharide is a potentially practical alternative to these approaches. This approach has been demonstrated using lipases to deprotect peracylated monosaccharides, but the range of products reported has been limited thus far to those accessible with known enzymes, and no attempt has been made to improve or alter the biocatalyst activity or regioselectivity (10). Although careful control of reaction conditions permits regioselective acyl migration to produce other products (11), non-selective migration remains problematic, and the generality of the lipase method seems to be limited.
The generality and utility of a chemoenzymatic method for the synthesis of monosaccharide derivatives would be expanded greatly if each site of a given substrate could be deprotected using a unique catalyst. This approach would require not only a protecting group resistant to migration under the deprotection conditions but also a catalyst capable of removing this group with high levels of chemo- and regioselectivity. We hypothesized that cytochrome P450-catalyzed heteroatom demethylation might provide a solution to both these problems. Although chemical removal of methyl groups requires harsh and unselective conditions, this enzyme-catalyzed reaction proceeds via insertion of oxygen into a methyl C-H bond to generate a hemiacetal intermediate that spontaneously decomposes to release formaldehyde and an alcohol under ambient conditions (Fig. 1A) (12). One of our groups (F.H.A.) has used variants of Bacillus megaterium cytochrome P450BM3 (BM3) to hydroxylate a wide range of non-natural substrates with high levels of regioselectivity (13). If this enzyme could be engineered to accept permethylated monosaccharides, the challenging task of regioselectively demethylating these substrates could be possible. Because BM3 is amenable to protein engineering, the activity, regioselectivity, and substrate scope of these reactions could be optimized using directed evolution (14) to generate BM3 demethylases for the production of any desired monosaccharide derivative.
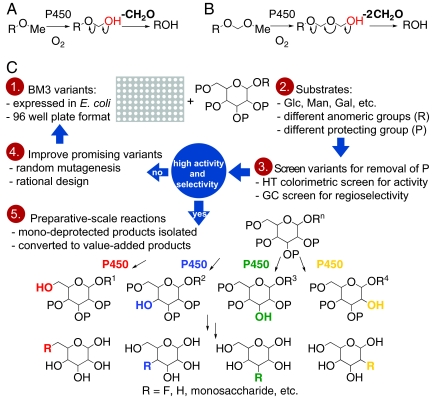
Fig. 1.
(A) P450-catalyzed removal of methyl (Me) and (B) methoxymethyl (MOM) groups. (C) Method used to screen and optimize activity of BM3 demethylases. Libraries of BM3 variants were screened for activity on Me- or MOM-protected substrates. The activity of BM3 variants in cell lysate was assayed by colorimetric detection of formaldehyde (CH2O) released during the deprotection, and regioselectivity was determined by GC analysis of the reaction mixtures. Promising enzymes were optimized by directed evolution to provide enzymes suitable for preparative scale. Mono-demethylated products were converted to a variety of value-added products.
Results
We therefore screened libraries of BM3 variants on hand in our laboratory to determine the scope of their reactivity toward a number of common pentamethyl hexoses (Fig. 1C). A compilation library comprised of 96 BM3 variants previously engineered for a variety of applications was used to survey functional enzymes rapidly (15). Because many of these variants were related along an evolutionary lineage by mutation of 1–3 residues, we also used a targeted library of 1,024 BM3 variants having up to 10 mutations in the active site of the parent variant, 9–10A. Demethylase activity first was determined using a high-throughput (HT) colorimetric assay for formaldehyde released during the reactions (Fig. 1A; see supporting information (SI) Appendix). The most active enzymes then were re-expressed, purified, and used to catalyze the same reaction under controlled conditions to ensure the accuracy and precision of catalyst loadings. Finally, the reaction mixtures were analyzed by gas chromatography (GC) to establish the regioselectivity of demethylation. Promising variants were optimized by incorporation of random mutations and screening to provide total turnover numbers (TTN, ca. 1,000) and regioselectivities (> 80%) sufficient for practical gram-scale preparation of regioselectively deprotected monosaccharides. This approach also was used to identify variants capable of removing the methoxymethyl (MOM) group from MOM-protected monosaccharides to enable additional synthetic applications (Fig. 1B).
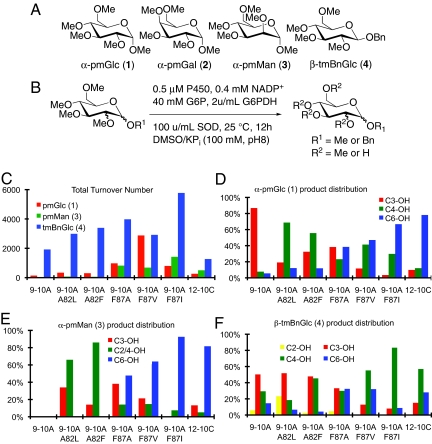
Initially, 4 substrates, 1,2,3,4,6-pentamethyl-α-glucopyranoside (1), 1,2,3,4,6-pentamethyl-α-galactopyranoside (2), 1,2,3,4,6-pentamethyl-α-mannopyranoside (3), and 1-benzyl-2,3,4,6-tetramethyl-β-glucopyranoside (4) (Fig. 2A) were examined. The last of these compounds, 4, was included to determine if a substrate-engineering approach (16) could lead to alternative regio selectivities, and authentic samples of each of the possible mono-demethylation products for 1–4 were synthesized to serve as GC standards for product identification. We first screened the compilation library for activity on these substrates (Fig. 2B). Pleasingly, we found that a number of enzymes in this small library were active on substrates 1, 3, and 4 in the HT assay, and GC analysis of these same reactions conducted with purified enzymes under optimized conditions (17) revealed that, except for C-1, nearly every position of the monosaccharide substrates investigated was accessible (Fig. 2 C–F). This broad range of regioselectivities contrasts with the narrow scope of existing chemoenzymatic methods and suggested that BM3-catalyzed demethylation could provide a general approach to monosaccharide elaboration if the yields and regioselectivities observed for these enzymes could be improved using directed evolution. Furthermore, the significant differences in both activity and regioselectivity between the methyl- and benzyl-protected glucose substrates 3 and 4 indicated that varying the anomeric substituent could be used to identify suitable enzyme/substrate combinations without the need for protein engineering. Indeed, variant 9–10A F87I catalyzed the demethylation of both 3 and 4 with suitably high yield (> 1,000 TTN) and regioselectivity (> 80%) to warrant investigation of these reactions on a preparative scale, as discussed later.
Fig. 2.
(A) Structures of substrates used in initial screens. (B) Optimized reaction conditions for demethylation reactions. (C) TTN for enzymes (see SI Appendix for sequences) identified in HT screen on substrates 1, 3, and 4 and (D–F) product distributions (% of total product) for demethylation of substrates 1, 3, and 4 by BM3 variants determined by GC analysis of crude product mixtures.
Although these results were very promising, we were concerned about the lack of reactivity observed with galactose substrate 2, especially given the frequency with which this monosaccharide is found in natural systems (1). To address this issue, we examined the activities of the compilation library enzymes on 4 additional tetramethyl galactose derivatives with a variety of anomeric substituents in the β configuration, including methyl (Me, 5), benzyl (Bn, 6), benzoyl (Bz, 7), and acetyl (Ac, 8). Once again, 9–10A F87I displayed high demethylation activity, this time on the 1-β-benzyl galactose substrate 6, and nearly quantitative conversion and high selectivity for the 4-position was confirmed by NMR spectroscopy. This result further established the utility of substrate engineering to achieve highly selective demethylation reactions and confirmed that derivatives of 3 of the most common hexoses were compatible with our enzymatic demethylation method.
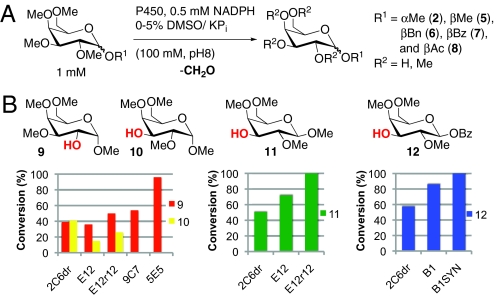
To broaden the generality of the method further, we explored the activities of the active-site library enzymes on α- and β-pentamethyl galactose substrates 2 and 5 because galactose derivatives had proven most resistant to demethylation by the small set of enzymes surveyed thus far (Fig. 3A). A number of hits were obtained in the HT assay, and 8 of the most active variants were purified and re-screened against substrates 2, 5, 6, 7, and 8. Colorimetric analysis of formaldehyde produced in these reactions confirmed the activity observed on 2 and 5 but also indicated that 7 is a compatible substrate (see SI Appendix). Six variants catalyzed the demethylation of these substrates with moderate efficiency (ca. 30% conversion based on formaldehyde concentration), indicating that active-site engineering could extend further the scope of the reaction.
Fig. 3.
(A) Reaction conditions used for colorimetric analysis. (B) Yields for conversion of 2 to 9 + 10, 5 to 11, and 7 to 12 determined by GC analysis of crude reaction mixtures using indicated BM3 variants. The 1 mM (0.1 mol%) BM3 variant was used except for the conversion of 7 to 12 where 0.5 mM (0.05 mol%) B1SYN were used.
GC analysis of the crude reaction mixtures obtained with 2 revealed that variant 2C6dr provided the highest conversion of substrate, producing a 1:1 mixture of O-2 (39%) and O-3 (41%) demethylation (compounds 9 and 10). Although the activity of 2C6dr on 5 and 7 was slightly lower under these reaction conditions, we observed nearly exclusive O-3 demethylation with these compounds to give 11 (50%) and 12 (57%), as confirmed by NMR spectroscopy and x-ray analysis, respectively. These results indicated that the remaining 2 secondary hydroxyl positions of galactose could be accessed, but both the conversion and regioselectivity of these transformations required improvement for practical synthetic applications.
A combination of random and site-directed mutagenesis of the gene for 2C6dr led to the identification of variant E12r12, which had improved efficiency on substrates 2 (50% increase in regioselectivity) and 5 (100% conversion) because of active-site mutation A180V (see SI Appendix). Variant 5E5 was obtained from an additional round of random mutagenesis of the gene for E12r12 and recombination of improved variants, and this enzyme provided high regioselectivity in the conversion of 2 to 9 (96%, single isomer) because of active-site mutation V87G. A similar engineering effort led to the generation of B1SYN, which had improved activity on 7 dramatically and enabled quantitative demethylation of this substrate even at 0.05 mol% enzyme loading. Fig. 3B shows the improvement in the conversion of 2 to a mixture of 9 and 10, of 5 to 11, and of 7 to 12 using the evolved BM3 variants. In each case, the activity or regioselectivity of 2C6dr was improved sufficiently by mutations identified from 1 or 2 rounds of error-prone PCR and screening to enable efficient demethylation of the 2- and 3-positions of suitable β-galactose derivatives. Given the range of activities and regioselectivities observed for BM3 variants identified from screening both the compilation and active-site libraries (Fig. 2 C–F), directed evolution should provide a rapid means for optimizing demethylases well beyond the examples shown here.
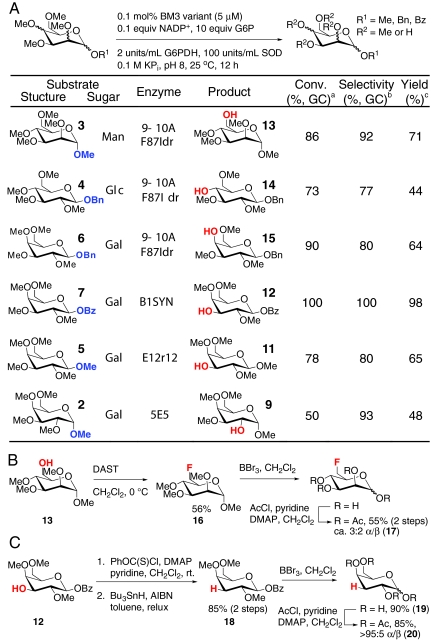
Next, the synthetic utility of the engineered demethylases was demonstrated by their application in preparative-scale reactions (Fig. 4A). The high conversions and regioselectivities observed previously in small-scale reactions translated well into the 100-mg scale investigated, and the desired products, 9–15, were isolated in high yield using 0.05–0.1 mol% enzyme loading. These mono-demethylated compounds could be converted to a variety of biologically relevant products using established chemical transformations. Fluorination frequently is used to modify the pharmacokinetic properties of molecules for biological studies or for PET imaging (18). The Arnold group previously reported the tandem demethylation/fluorination of a variety of small molecules (15), and this procedure proved applicable for the fluorination of 13 to provide 16. The latter compound could be demethylated in quantitative yield and per-acetylated to provide 17 in 55% yield over 2 steps. Deoxy sugars also are important components of a variety of small-molecule therapeutics (19), and deoxygenation of 12 using Barton conditions proceeded smoothly to provide 18 in good yield (20). This product was demethylated in excellent yield (19) and was acetylated (20) to facilitate handling and characterization.
Fig. 4.
(A) Regioselective deprotection of substrates 11–15. a, Conversion of starting material determined by GC analysis of crude reaction mixture. b, Percent of desired product relative to additional products determined by GC analysis of crude reaction extracts. c, Isolated yield of pure product. d, 2 mM substrate used; 0.05 mol% B1SYN used. (B) Fluorination and global deprotection of 13. (C) Deoxygenation and global deprotection of 12.
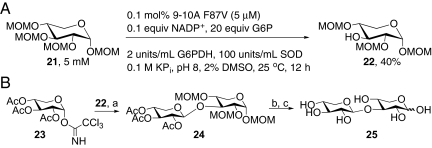
To augment the utility of our method further, we explored the regioselective deprotection of MOM-protected monosaccharides in the context of a chemoenzymatic disaccharide synthesis (Fig. 1B). The ability to remove the MOM group is highly valuable for the synthesis of complex molecules, because the conditions required for its removal are much milder than those required for removal of methyl groups and therefore are compatible with sensitive functionality (21). A number of substrates with differing sugar scaffolds, anomeric substituents, and anomeric configurations were synthesized, and the compilation library was screened for activity on these substrates. High activity was observed on a number of MOM-protected pentoses without the need for any protein engineering, and GC analysis of reactions catalyzed by the most active variants revealed that 9–10A F87V catalyzed the removal of a single MOM group from α-xylose substrate 21. Importantly, the product, 22, could be isolated in moderate yield on a preparative scale, and removal of the 3-MOM group was confirmed by NMR spectroscopy (Fig. 5A). Compound 22 could be glycosylated with trichloroacetimidate donor 23 to give disaccharide 24 in high yield (22). Global deprotection of 24 under mild conditions provided disaccharide 25, a component of the promising immune adjuvant, QS-21 (Fig. 5B) (23).
Fig. 5.
(A) Regioselective removal of 3-MOM group from substrate 21. (B) Synthesis of QS21-A disaccharide 25. a, TMSOTf, 4Å MS, 88%; b, NaOMe; c, 2 M HCl, 55% (2 steps)
Discussion
Regioselective deprotection of monosaccharide substrates using engineered P450BM3 demethylases provides a highly efficient means to access valuable intermediates that can be converted to a wide range of substituted monosaccharides and polysaccharides. Demethylases with activity on a number of monosaccharides and different positions on a given monosaccharide were identified using a combination of protein and substrate engineering, suggesting that this procedure ultimately could be used to functionalize regioselectively any monosaccharide at any desired position. The ability of these enzymes (i.e., 9–10A F87Idr) to catalyze efficient demethylation of various substrates with high regioselectivity should greatly simplify the expansion of the method to additional substrates.
These results also highlight the efficiency with which BM3 variants can insert oxygen into the C-H bonds of methyl ethers to catalyze heteroatom demethylation under conditions far more mild than those associated with chemical methods. Furthermore, the ability of the demethylases to situate substrates in a particular orientation to favor hydroxylation of a particular methyl group over 3 or 4 others located within only a few angstroms indicates the exquisite control imparted by supramolecular interactions. Unlike synthetic supramolecular catalysts (24), the activity and selectivity of BM3 variants can be tuned readily to suit a particular application. Such control enables efficient hydroxylation of C-H bonds lacking any significant stereoelectronic differentiation and augments the means by which synthetic chemists can target otherwise indistinguishable C-H bonds (25).
All these factors result in a procedure that greatly simplifies the synthesis of selectively deprotected monosaccharide derivatives compared with conventional procedures that require extensive synthetic manipulation. The extension of this technology to additional synthetically useful protecting groups, such as the MOM ether, renders the method compatible with sensitive glycosidic linkages and greatly expands the scope of the reaction. We anticipate that this chemoenzymatic approach could greatly simplify the synthesis of a wide range of polysaccharides for studies in chemistry, biology, and medicine.
Materials and Methods
Experimental Procedures and Characterization.
Complete experimental procedures and characterization can be found in the online SI Appendix.
BM3 Expression and Purification.
For preparative bioconversions, P450 BM3 demethylases were used in purified form. Enzyme batches were prepared as follows. We inoculated 2 L TBamp with an overnight culture (100 mL, LBamp) of recombinant E. coli DH5α cells harboring a pCWori plasmid encoding for the P450 variant under the control of Plac promoter. At an OD600 of 1.8 (ca. 3–4 h), the incubation temperature was reduced to 25 °C (30 min), and the cultures were induced by adding isopropyl-beta-D-thiogalactopyranoside to a final concentration of 0.1 mM. The cultures were allowed to continue at this temperature for another 24 h. After the cells were harvested by centrifugation (4 °C, 15 min, 3000 × g), the cell pellet was resuspended in 25 mM Tris-HCl buffer (pH 8.0), and cells were disrupted by sonication (4 × 1 min, 50% duty cycle). Cell debris was removed by centrifugation for 20 min at 4 °C and 20,000 × g; the resulting cell lysate was loaded onto a Q resin, and the column was washed with 3 column volumes of 25 mM Tris-HCl (pH 8.0), 150 mM NaCl. Bound protein was eluted with 25 mM Tris-HCl (pH 8.0), 340 mM NaCl and was concentrated using Millipore Centricon tubes. After buffer exchange with 100 mM KPi (pH 8.0), protein samples were frozen and stored at −80 °C. Protein concentration was determined in duplicate from CO-difference spectra as previously described. Yields typically ranged between 100 and 500 mg protein/L, depending on the variant.
Preparative-Scale Bioconversions.
Potassium phosphate buffer (100 mM, pH 8) and a solution of the desired substrate in phosphate buffer or DMSO (1 equiv, 5 mM final concentration) was added to a 100 × 50 mm crystallizing dish (i.e., ChemGlass #CG-8276–100). A solution of NADP+ (0.1 equiv, 0.5 mM final concentration), glucose-6-phosphate (10 equiv, 50 mM final concentration), and glucose-6-phosphate dehydrogenase (2 units/mL final reaction volume) was added. A solution of the appropriate BM3 variant (0.001 equiv, 5 mM final concentration) was added, the dish was covered loosely with aluminum foil, and the contents were stirred using a magnetic stir bar. The moderate stir rate was used to allow efficient O2 transfer to the reaction mixture while avoiding foaming. The reaction progress could be monitored by analyzing aliquots of the reaction mixture as described for the medium-scale bioconversions. After 12 h, the pH of the reaction mixture was adjusted to 3–4, and the solution was saturated with NaCl to precipitate most of the protein from solution. The resulting suspension then was filtered through Celite (Aldrich), which was rinsed with methylene chloride to provide a biphasic mixture. The mixture was transferred to a separatory funnel and extracted with 4 × 75 mL CH2Cl2. The combined organic extracts were dried with sodium sulfate, filtered, and concentrated. The resulting residue was purified by SiO2 chromatography eluting with ethyl acetate/hexanes.
Supplementary Material
Acknowledgments.
This work was supported by the Jacobs Institute for Molecular Medicine (F.H.A.). J.C.L. is supported by U.S. National Institutes of Health Fellowship 1F32GM079932–01. S.B. was supported by the Deutsche Forschungsgemeinschaft (DFG) Grant no. BA3486/1–1. C.H.W, C.S.B., Y.F., and W.A.G. were supported by the U.S. National Institutes of Health Grants GM044154 and AI072155. C.S.B. also was supported by the U.S. National Institutes of Health Fellowship 1F32GM073500–02S01.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates have been deposited in the Cambridge Structural Database, Cambridge Crystallographic Data Centre, Cambridge CB2 1EZ, United Kingdom (CSD reference no. 673244).
This article contains supporting information online at www.pnas.org/cgi/content/full/0908954106/DCSupplemental.
References
- 1.Varki A, et al., editors. Essentials of Glycobiology. New York: Cold Spring Harbor Laboratory Press; 1999. [PubMed] [Google Scholar]
- 2.Laine RA. Information capacity of the carbohydrate code. Pure Appl Chem. 1997;69:1867–1873. [Google Scholar]
- 3.Jenkins N, Parekh RB, James DC. Getting the glycosylation right: Implications for the biotechnology industry. Nat Biotechnol. 1996;14:975–981. doi: 10.1038/nbt0896-975. [DOI] [PubMed] [Google Scholar]
- 4.Bennett CS, Wong CH. Chemoenzymatic approaches to glycoprotein synthesis. Chemical Society Reviews. 2007;36:1227–1238. doi: 10.1039/b617709c. [DOI] [PubMed] [Google Scholar]
- 5.Tsuchiya T. Reactions at oxygen atoms in carbohydrates. Glycoscience. 2001;1:117–194. [Google Scholar]
- 6.Zhu X, Schmidt RR. New principles for glycoside-bond formation review. Angewandte Chemie (International Edition in English) 2009;48:1900–1934. doi: 10.1002/anie.200802036. [DOI] [PubMed] [Google Scholar]
- 7.Wang C-C, et al. Regioselective one-pot protection of carbohydrates. Nature. 2007;446:869–899. doi: 10.1038/nature05730. [DOI] [PubMed] [Google Scholar]
- 8.Thibodeaux CJ, Melancon CE, Liu HW. Natural product sugar biosynthesis and enzymatic glycodiversification. Angewandte Chemie (International Edition in English) 2008;47:9814–9859. doi: 10.1002/anie.200801204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams GJ, Zhang C, Thorson JS. Expanding the promiscuity of a natural-product glycosyltransferase by directed evolution. Nature Chemical Biology. 2007;3:657–662. doi: 10.1038/nchembio.2007.28. [DOI] [PubMed] [Google Scholar]
- 10.Horrobin T, Tran CH, Crout D. Esterase-catalysed regioselective 6-deacylation of hexopyranose per-acetates, acid-catalysed rearrangement to the 4-deprotected products and conversions of these into hexose 4- and 6-sulfates. J Chem Soc Perkin Trans. 1998;1:1069–1080. [Google Scholar]
- 11.Filice M, et al. Preparation of linear oligosaccharides by a simple monoprotective chemoenzymatic approach. Tetrahedron. 2008;64:9286–9292. [Google Scholar]
- 12.Guengerich FP. Common and uncommon cytochrome P450 reactions related to metabolism and chemical toxicity. Chem Res Toxicol. 2001;14:612–640. doi: 10.1021/tx0002583. [DOI] [PubMed] [Google Scholar]
- 13.Lewis JC, Arnold FH. Catalysts on demand: Selective oxidations by laboratory-evolved cytochrome P450 BM3. Chimia. 2009;63:309–312. [Google Scholar]
- 14.Arnold FH. Design by directed evolution. Acc Chem Res. 1998;31:125–131. [Google Scholar]
- 15.Rentmeister A, Arnold FH, Fasan R. Chemo-enzymatic fluorination of unactivated organic compounds. Nature Chemical Biology. 2008;5:26–28. doi: 10.1038/nchembio.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lairson LL, Watts AG, Wakarchuk WW, Withers SG. Using substrate engineering to harness enzymatic promiscuity and expand biological catalysis. Nature Chemical Biology. 2006;2:724–728. doi: 10.1038/nchembio828. [DOI] [PubMed] [Google Scholar]
- 17.Wichmann R, Vasic-Racki D. Cofactor regeneration at the lab scale. Advances in Biochemical Engineering/Biotechnology. 2005;92:225–260. doi: 10.1007/b98911. [DOI] [PubMed] [Google Scholar]
- 18.Muller K, Faeh C, Diederich F. Fluorine in pharmaceuticals: Looking beyond intuition. Science. 2007;317:1881–1886. doi: 10.1126/science.1131943. [DOI] [PubMed] [Google Scholar]
- 19.Kirschning A, Bechthold AFW, Rohr J. Chemical and biochemical aspects of deoxysugars and deoxysugar oligosaccharides. Top Curr Chem. 1997;188:1–84. [Google Scholar]
- 20.McDonough MJ, Stick RV, Tilbrook DMG, Watts AG. An investigation into the synthesis of some molecules related to methyl acarviosin. Aust J Chem. 2004;57:233–241. [Google Scholar]
- 21.Benneche T. α-Monohalo ethers in organic synthesis. Synthesis. 1995:1–27. [Google Scholar]
- 22.Mori M, Ito Y, Ogawa T. Synthetic studies on cell-surface glycans. 62. Total synthesis of the mollu-series glycosyl ceramides α-D-man-p-(1→3)-β-D-man-p-(1→4)-β-D-glc-p-(1→1)-cer and α-D-man-p-(1→3)-[β-D-xyl-p-(1→2)]-β-D-man-p-(1→4)-β-D-glc-p-(1→1)-cer. Carbohydr Res. 1990;195:199–224. doi: 10.1016/0008-6215(90)84167-s. [DOI] [PubMed] [Google Scholar]
- 23.Kensil CR. Saponins as vaccine adjuvants. Critical Reviews in Therapeutic Drug Carrier Systems. 1996;13:1–55. [PubMed] [Google Scholar]
- 24.Fiedler D, Leung DH, Bergman RG, Raymond KN. Selective molecular recognition, C-H bond activation, and catalysis in nanoscale reaction vessels. Acc Chem Res. 2005;38:349–358. doi: 10.1021/ar040152p. [DOI] [PubMed] [Google Scholar]
- 25.Chen MS, White MC. A predictably selective aliphatic C-H oxidation reaction for complex molecule synthesis. Science. 2007;318:783–787. doi: 10.1126/science.1148597. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.