Abstract
Pathogen selection is postulated to drive MHC allelic diversity at loci for antigen presentation. However, readily apparent MHC infectious disease associations are rare in most species. The strong link between MHC-B haplotype and the occurrence of virally induced tumors in the chicken provides a means for defining the relationship between pathogen selection and MHC polymorphism. Here, we verified a significant difference in resistance to gallid herpesvirus-2 (GaHV-2)-induced lymphomas (Marek's disease) conferred by two closely-related recombinant MHC-B haplotypes. We mapped the crossover breakpoints that distinguish these haplotypes to the highly polymorphic BG1 locus. BG1 encodes an Ig-superfamily type I transmembrane receptor-like protein that contains an immunoreceptor tyrosine-based inhibition motif (ITIM), which undergoes phosphorylation and is recognized by Src homology 2 domain-containing protein tyrosine phosphatase (SHP-2). The recombinant haplotypes are identical, except for differences within the BG1 3′-untranslated region (3′-UTR). The 3′-UTR of the BG1 allele associated with increased lymphoma contains a 225-bp insert of retroviral origin and showed greater inhibition of luciferase reporter gene translation compared to the other allele. These findings suggest that BG1 could affect the outcome of GaHV-2 infection through modulation of the lymphoid cell responsiveness to infection, a condition that is critical for GaHV-2 replication and in which the MHC-B haplotype has been previously implicated. This work provides a mechanism by which MHC-B region genetics contributes to the incidence of GaHV-2-induced malignant lymphoma in the chicken and invites consideration of the possibility that similar mechanisms might affect the incidence of lymphomas associated with other oncogenic viral infections.
Keywords: disease resistance gene, GaHV-2 induced lymphoma, Gallus gallus, Marek's disease
There is an astounding breadth of genetic diversity at the MHC, a region found within the genomes of all jawed vertebrates. Despite the seeming likelihood that pathogen selection has an important role in driving allelic diversity at MHC class I and class II loci, associations are rarely found between infectious diseases and particular MHC alleles or haplotypes. The presence of multiple polymorphic class I and class II gene family members within MHC haplotypes in many species could contribute to the difficulty in observing MHC disease associations. In the chicken, the MHC-B region has an exceptionally strong role in genetic resistance to Marek's disease (MD) caused by gallid herpesvirus-2 (GaHV-2) and to Rous sarcoma virus (RSV)-induced tumors (1, 2). Resistance to these diseases maps to the MHC-B subregion marked by a highly expressed classical MHC class I gene (3–6). It has been suggested that strong MHC-B disease associations are apparent as the result of alleles at this single locus for classical class I antigen presentation, BF2, contributing to immune responses either independently or in concert with closely-linked transporter associated with antigen-processing (TAP) genes (6, 7). Antigen presenting molecules encoded by different BF2 alleles bind quite different classes of peptides (7–10) and thereby likely selectively influence adaptive immune responses to antigen, but the gene or genes that provide MHC-linked resistance to virally induced tumors in chickens are not known. Recently, the crossover breakpoint in the recombinant haplotype originally used to map MD resistance to the MHC-B subregion marked by BF2 was localized, which revealed that 27 genes lie within the region between the crossover breakpoint and the BF2 locus (4, 11). Thus, it is now evident that many genes are candidates for providing the long noted MHC-B haplotype-linked resistance to MD. Overall, the MHC region in chicken stands out in contrast to the MHC in mammals. The chicken MHC is compact and segmented into the MHC-B and MHC-Y regions in which the MHC class I and class II loci reside, as well as genes that changed or moved later in evolutionary time in other species (6, 12–15). Meiotic recombination within MHC-B is rare (16), but several recombinant haplotypes are available that are suitable for further investigation of MHC-B-linked disease resistance.
BR2 and BR4 are two recombinant haplotypes within pedigreed matings designed to identify duplicate recombinant haplotypes originating from independent crossover events between the same parent haplotypes (Fig. 1A). Such recombinant haplotypes were sought as a means for defining which genes within the MHC-B region confer resistance to MD (16). When tested in a small challenge trial with GaHV-2 virus at the fourth backcross generation in the development of congenic lines, birds bearing BR2 and BR4 differed significantly in the incidence of MD (17). BR2 and BR4 also apparently contribute differently to the influence of MHC-B on the regression of RSV-induced tumors (18). These observations suggest that, although indistinguishable by the serologically-defined erythroid BG and class I BF antigens used to isolate them, BR2 and BR4 are likely different as the result of meiotic recombination breakpoints having occurred at different locations within the region separating the genes encoding these serological markers. The breakpoints apparently surround a gene that influences disease resistance.
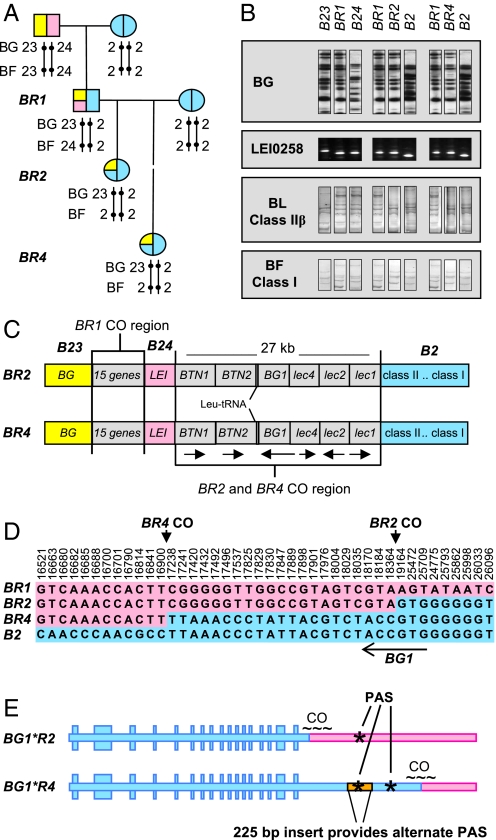
Fig. 1.
Derivation of the BR2 and BR4 MHC-B recombinant haplotypes and the genetic differences between them. (A) BR2 and BR4 haplotypes were recovered in crosses made between animals bearing BR1 (a recombinant haplotype derived from B23 and B24) and B2 haplotypes when atypical segregation of antigens encoded by the BG gene cluster and BF (MHC class I) genes were detected in serological typing of progeny. (B) The crossover breakpoint regions in BR2 and BR4 were narrowed to a region between LEI0258 and the BLB loci. The BG gene cluster, about 100 kb upstream from LEI0258, was typed by restriction fragment patterns; the LEI0258 by PCR product length; and the BF and BL loci by single strand conformation polymorphism. Images for each locus were obtained from single gels and are duplicated and rearranged as needed here to aid comparisons. (C) Diagram (not to scale) of the MHC-B, illustrating the genes located within the 27-kb crossover breakpoint region (arrows indicate direction of transcription). (D) Map of SNPs within the 27-kb region that define the crossover (CO) breakpoints in BR2 and BR4 as distinct, with the BR2 crossover occurring within the 3′-end of BG1, which is encoded on the complementary strand, as indicated by the arrow. Positions are numbered based on GenBank AB268588. E) The BG1 gene sequence is presented 5′ to 3′, illustrating the consequences of crossover breakpoints on the structure of the BG1*R2 and BG1*R4 alleles and the presence of a 225-bp insert in BG1*R4 that provides an alternate polyadenylation signal sequence (PAS, indicated by *).
Results
A Larger Challenge Trial Confirmed the Difference Between BR2 and BR4 in Conferring Resistance to Marek's Lymphoma.
We conducted a GaHV-2 challenge trial using the 003.R2 and 003.R4 lines, now more inbred than previously as the result of 10 backcross generations of breeding, to verify the previous observation suggesting that BR2 and BR4 differ in their influence in MD (17). Pedigree-hatched chicks were inoculated with the highly virulent RB1B strain of GaHV-2 and observed for the formation of tumors over 12 weeks. Genotypes were obtained for each bird and combined with disease data at the conclusion of the trial. A difference in MD mortality between the lines was evident during the trial and was consistent with the difference seen in MD incidence upon gross examination of all birds at completion of the trial. MD incidence was significantly lower in line 003.R2 birds compared to line 003.R4 birds (Table 1). Inclusion of the available histopathological findings increased the incidence of MD overall, but the two lines remained significantly different (39% in 003.R2 versus 61% in 003.R4, P < 0.0001). Tumors in the 003.R2 and 003.R4 lines were present predominantly in heart, kidney, liver, spleen, and gonad. The difference in tumor incidence between lines 003.R2 and 003.R4 in this trial, after 10 generations of backcrossing, is essentially identical to that reported for the fourth backcross generation (17). Thus, it appears that although BR2 and BR4 are identical for MHC-B serological markers for BG and BF, they differ significantly in the capacity to confer resistance to MD. This observation supports our hypothesis that the BR2 and BR4 haplotypes have different crossover breakpoints bounding a gene conferring disease resistance.
Table 1.
Congenic lines 003.R2 and 003.R4 birds differ in the incidence of Marek's disease tumors following infection with GaHV-2
| Line | MD mortality during trial | K2 | Total MD Incidence | K2 |
|---|---|---|---|---|
| 003.R2 | 10% (14/140) | b, c | 19% (27/140) | b, c |
| 003.R4 | 39% (51/130) | b | 47% (61/130) | b |
| Specific pathogen free | 66% (86/131) | 95% (124/131) |
The incidence of Marek's disease that occurred during the trial, as indicated by early mortality, and the total incidence detected upon gross examination at the conclusion of the trial differed significantly between the 003.R2 and 003.R4 lines. Specific pathogen free (SPF) birds, infected secondarily by inhalation of shed virus, served as controls to gauge the success of the initial infection. b = P < 0.0001 vs SPF; c = P < 0.0001 vs 003.R4
The Difference Between MHC-BR2 and MHC-BR4 Maps to BG1.
To define the crossover breakpoints, we initially assayed BR2, BR4, and the parental haplotypes B2 and BR1 (a recombinant haplotype derived earlier from B23 and B24, Fig. 1A) for BG restriction fragment patterns, LEI0258 microsatellite PCR-product lengths, and MHC class I and class II single strand conformation polymorphism patterns. Alleles in BR2 and BR4 were assigned based on matches with alleles in the parental haplotypes (Fig. 1B). Consistent with earlier serological typing, BR2 and BR4 were found to share MHC class I and class II alleles with the B2 parent haplotype (Fig. 1B). The restriction fragment patterns of the BG genes for BR2 and BR4 resembled those of the B23 portion of the BR1 parent haplotype, confirming earlier serological identification that the BG gene cluster originated from the BR1 parent haplotype. BR2 and BR4 were identical at the microsatellite marker LEI0258, and matched the BR1 parent haplotype. Thus, the crossover breakpoints producing BR2 and BR4 occurred downstream of LEI0258, narrowing further investigation to seven genes—B-BTN1, B-BTN2, tRNA-leu, BG1 [a structurally distinct member of the BG gene family, located about 100-kb distant from the cluster of BG genes encoding BG antigens (11, 19)], and Blec4, Blec2, and Blec1 (Fig. 1C).
We resolved the BR2 and BR4 crossover breakpoints through sequencing. Single nucleotide polymorphism (SNP) differences revealed closely positioned, but distinct, crossover breakpoints in the vicinity of the BG1 gene (encoded on the complementary strand) (Fig. 1D). In the formation of the BR4 haplotype, crossover between the BR1 and B2 parent haplotypes occurred somewhere within a 338-bp interval defined by SNPs 16900 and 17238. These are, respectively, the last SNP matching BR1 and first SNP matching B2 in the BR4 haplotype. The BR2 breakpoint occurred a short distance away, within an 800-bp region between SNPs 18364 and 19164 (Fig. 1D). The BR4 breakpoint is outside the BG1 coding region; therefore, the BG1*R4 allele is entirely identical to the BG1 allele in the B2 parent haplotype. In contrast, three SNPs (see SNPs 18170, 18184, and 18364 in Fig. 1D) mapped the breakpoint for BR2 to within the 3′-untranslated region (3′-UTR) of the BG1 gene. Thus, although identical to the BG1*B4 allele in its coding region, BG1*R2 possesses a 3′-UTR derived from the BR1 parent haplotype.
Sequencing revealed an additional difference between BG1*R2 and BG1*R4. The 3′-UTR of the BG1*R4 and the BG1 allele in the parent B2 haplotype contain a 225-bp insert that is absent from BG1*R2. This insert provides an alternate polyadenylation signal (PAS) (Fig. 1E, sequences in Fig. S1). BLAST searches showed that the 225-bp insert is essentially identical to the 3′-UTR within a retroviral Pro-Pol-dUTPase polyprotein-encoding sequence that is found at multiple sites throughout the Gallus gallus genome (Fig. S2). The PAS within the insert is the dominant signal used in the production of BG1*R4 transcripts, as shown by 3′-RACE analysis of cDNA (Fig. 2A). Less common, longer BG1*R4 transcripts arise from the use of the endogenous BG PAS that is displaced downstream as a result of the viral insertion. The 3′-RACE products from BG1*BR2 transcripts were of a single size, consistent with the single PAS in this allele. We found BR2 and BR4 are completely identical across a 61-kb interval of MHC-B (see GenBank FJ770459 and FJ770460), aside from the crossover breakpoint region. This includes identity at the MHC-B loci involved in class I and class II antigen processing and presentation, as well as Blec2, which encodes an inhibitory receptor expressed on natural killer cells, and BLec1, which encodes a putative activating receptor. In addition, typing of eight individual birds from each line with 2702 SNPs evenly-distributed across the genome showed that although some heterozygosity remains in the 003.R2 and 003.R4 lines, there is no evidence for any other region of the genome becoming fixed by chance during the production of the 003.R2 and 003.R4 congenic lines (Table S1).
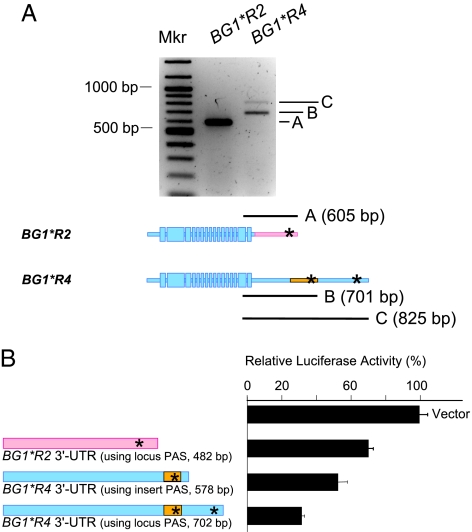
Fig. 2.
The 3′-untranslated regions of BG1*R2 and BG1*R4 differ in length and in influence on the translation of mRNA. (A) 3′-RACE reactions revealed length differences between the BG1*R2 and BG1*R4 cDNAs as a result of differences in the length of the 3′-UTR. The BG1*R2 allele provides 3′-RACE products of a single length, consistent with the anticipated size of 605 bp. The products formed in the BG1*R4 3′-RACE reactions are consistent with BG1*R4 transcripts produced primarily through use of the polyadenylation signal (PAS; indicated by *) introduced within the inserted sequence (701 bp) and secondarily from the PAS that is inherent in the locus but shifted by the inserted sequence (825 bp). (B) 3′-UTRs derived from BG1*R2 and BG1*R4 cDNA were cloned downstream of the firefly luciferase gene and tested in dual luciferase expression assays in chicken LMH cells stimulated with phorbol myristate acetate. All three 3′-UTRs inhibited luciferase activity compared to the vector-only control. The BG1*R4 3′-UTRs were consistently significantly more inhibitory than the 3′-UTR from the BG1*R2 allele.
The 3′-UTR Difference Between BG1*R2 and BG1*R4 Had Little Influence on Transcription but Produced Consistent Differences in Assays for Translation.
To determine whether the 3′-UTR differences found in BG1*R2 and BG1*R4 affect gene expression, we tested the relative transcription efficiency of the BG1*R2 and BG1*R4 alleles in quantitative PCR assays. Although BG1 was more highly expressed in some tissues than others, we found no significant differences in BG1 ΔCT values between paired tissue samples across the 003.R2 and 003.R4 lines (Table S2). These data indicate that the 3′-UTR difference has little effect on BG1 transcription. We then tested the 3′-UTRs for their effect on translation in dual luciferase reporter assays. Translation of firefly luciferase in LMH cells in conjunction with the shorter and longer BG1*R4 3′-UTRs was consistently repressed as compared to expression with the BG1*R2 3′-UTR (Fig. 2B), suggesting that differences in BG1 translation in vivo could be the basis for the differences in MD between the 003.R2 and 003.R4 lines.
The BG1 Transmembrane Protein Is Phosphorylated on Tyrosine in Pervanadate-Treated Cells and Co-Precipitates SHP-2.
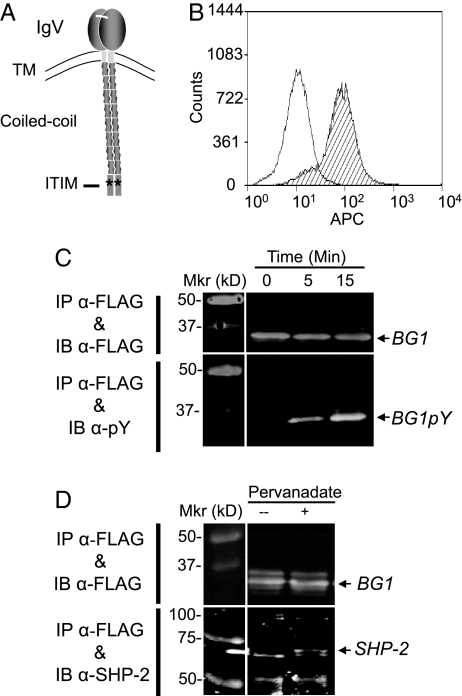
BG1 encodes a dimerizing 32-kDa type I transmembrane protein bearing an IgV-like ectodomain and a cytoplasmic tail made up of a coiled-coil region and a C-terminal domain containing a single immunoreceptor tyrosine-based inhibition motif (ITIM) (Fig. 3A). Therefore, BG1 is similar to receptors that bear single IgV-like ectodomains known to attenuate lymphoid cell activation via cytoplasmic ITIM signaling (20). To determine whether BG1 is capable of signaling, we expressed a full-length FLAG epitope-tagged BG1 cDNA clone in LMH cells. At least a portion of the BG1 expressed reached the cell surface (Fig. 3B). As suggested by its structure, FLAG-BG1 underwent tyrosine phosphorylation in the presence of pervanadate, as illustrated in BG1 immunoblots developed with 4G10, a monoclonal antibody specific for phosphotyrosine (Fig. 3C). Further, the protein tyrosine phosphatase SHP-2 (Src homology 2 domain phosphatase-1) co-precipitated with phosphorylated BG1 (Fig. 3D). Although the BG1 protein remains to be fully analyzed, the data presented here indicate that BG1 reaches the cell surface, can be phosphorylated on tyrosine, and that, in turn, it associates with the signaling phosphatase SHP-2. These are all features consistent with BG1 functioning as a surface receptor that limits cell activation.
Fig. 3.
BG1 has the structure of a tyrosine-based inhibitory receptor. (A) Depiction of the predicted structure of BG1 based on the sequence in Fig. S3. (B) Flow cytometry of LMH cells stably expressing FLAG-BG1 shows that BG1 reaches the cell surface. Cells were stained with the anti-FLAG monoclonal antibody M2 and anti-mouse IgG–allophycocyanin (APC) (striped histogram) and with anti-mouse IgG-APC alone (open histogram) and analyzed by flow cytometry gating on the DAPI-negative cell population. (C) BG1 is phosphorylated on tyrosine. LMH cells stably expressing FLAG-BG1 were lysed following 0-, 5-, and 15-min treatments with pervanadate, a tyrosine phosphatase inhibitor. BG1 was immunoprecipitated (IP) from total cell lysates using the anti-FLAG antibody M2 and then immunoblotted (IB) from a 12% polyacrylamide gel and probed with either M2 or the monoclonal anti-phosphotyrosine antibody 4G10. (D) The tyrosine phosphatase SHP-2 co-precipitates with BG1. FLAG-BG1 was immunoprecipitated (IP) from LMH cells stably expressing FLAG-BG1 with M2 using cultures without (–) or with treatment with pervanadate for 15 min (+), electrophoresed, transferred from a either a 9% or 12% gel, and then immunoblotted either with monoclonal M2 (12% gel) or polyclonal anti-SHP-2 C-18 (9% gel) antibodies. IRDye 800CW secondary antibodies were used to develop the blots.
Discussion
The selection of rare recombinants originating from the same parent haplotypes has provided two MHC-B haplotypes, BR2 and BR4, which differ in their contribution to genetic resistance to lymphomas induced by infection with GaHV-2. When tested at the fourth backcross generation in the production of congenic lines, the difference in tumor incidence between BR2 and BR4 line birds was 22% (17). As reported here, when tested in a substantially larger trial conducted after six additional generations of backcrossing, the difference in GaHV-2 tumor incidence between the 003.R2 and 003.R4 lines remained the same. Thus, the results indicate that genetic variability within the lines in regions outside MHC-B, which was no doubt greater at the fourth backcross generation than at the tenth, had little influence on the difference in the incidence of lymphomas between the lines in the two challenge trials. The findings in this study map the basis for the difference observed between 003.R2 and 003.R4 chickens in GaHV-2 lymphomas to BG1.
As noted in a recent comprehensive review by Calnek (21), the pathology that ensues following infection with GaHV-2 is complex, with a variety of cells responding in different ways at different times to the presence of the virus. In natural infections, GaHV-2 is transmitted by the inhalation of poultry dander laden with enveloped GaHV-2. The virus is transported from the respiratory tract to lymphoid organs. Infection of lymphocytes, viral proliferation, and subsequent necrosis in the spleen, bursa and thymus trigger the migration of additional lymphocytes, granulocytes, and macrophages to these organs and acute inflammatory responses ensue. Cytolytic infection occurs mostly in B cells, but also appears in some T cells and in some epithelial tissues as infected lymphoid cells disseminate. There is a fairly abrupt shift from cytolytic to latent infection, with latency developing mostly in T cells. The period of latent infection is short in genetically susceptible birds, and the virus soon emerges to begin a second cytolytic phase of infection. Transformation of activated CD4+ T cells soon occurs. Tumors and death typically follow within weeks. There are a number of points in the course of infection at which an inhibitory molecule, the role suggested for BG1, could influence the outcome of GaHV-2 infection.
Numerous investigations into the difference between MHC-B-resistant (B21-resistant lines N or N2a) and -susceptible (B19-susceptible lines P or P2a) lines provide intriguing data to consider in light of identification of BG1 as a candidate gene affecting MD. There is little evidence that MHC-B genotype-associated differences exist during the initial phase of infection, aside from the slightly delayed appearance of splenomegaly in MHC-B-resistant birds and occasional subtle differences in cytokine profiles [reviewed by (21); see also (22–24)]. However, by the end of the first 7–10 days post inoculation (dpi), substantial differences in MD pathogenesis become clearly evident. There is an abrupt decrease in the viral load in the white cells in resistant birds (23) that is accompanied by lower levels of infected T cells (25), relatively higher levels of virus-neutralizing antibodies (26), fewer AV37+ cells (22), and the absence of induction of the proinflammatory cytokines IL-6 and IL-18, which in contrast occurs in susceptible birds during the same interval (23). In the absence of complicating stress or infection, the genetically resistant birds remain healthy, with the only sign of active infection being the shedding of virus from the feather follicle.
Part of the change in MD pathogenesis in genetically resistant birds that occurs at 7–10 dpi could be due to differential development of specific cytotoxic T lymphocytes (CTLs) directed against viral antigens. Specific CTL responses in MD-resistant B21 homozygous birds to the GaHV-2 transcription regulatory protein ICP4 may be a factor in genetic resistance, particularly as immunity develops following vaccination (27). At the same time, there are clearly other genetic factors contributing in responses to MD. The generally low immune responsiveness of MHC-B-resistant birds lead Calnek and colleagues to suggest that progression of MD tumors is, contrary to logic, associated with greater immune activation or reactivity (21, 28). This proposal is especially interesting in light of the apparent role of BG1 in inhibitory signaling. Many studies provide data showing greater immune responsiveness in MHC-B genetically susceptible birds. For example, lymphocytes from MD-susceptible Line P (MHC-B19) are more responsive to mitogens (ConA and PHA) than birds from MHC-B-resistant Line N birds (MHC-B21) (29, 30). When infected in vitro lymphocytes from MHC-B-susceptible lines produce significantly more infected T cells compared to lymphocytes from MHC-B-resistant lines (31). Elevated tumor formation in the presence of allogeneic immune responses suggests, in another way, that immune activation promotes tumor formation (32). The striking difference in transcription of the proinflammatory IL-6 and IL-18 cytokine genes between MHC-B genetically resistant and susceptible lines suggests these cytokines drive the enhanced immune responses contributing to increased tumors (23). Although it remains to be tested, allelic differences at BG1 might contribute to these differences in activation noted between MHC-B-resistant (B21/B21) and susceptible (B19/B19) genotypes. BG1 isoforms encoded in resistant haplotypes might be more effective at signaling and contribute to the dampened immune responses associated with MD resistance. It will be interesting to determine the network within which BG1 molecules function and to explore the possibility that similar mechanisms affect the incidence of human lymphomas associated with oncogenic viral infection.
BG1*R2 and BG1*R4 represent only two of many BG1 alleles. BG1*R2, BG1*R4, the BG1 alleles in the parent haplotypes B2 and BR1, and the grandparent haplotype B24 constitute a subgroup of structurally distinct BG1 alleles. The coding region in this group is truncated by the presence of an early stop codon that eliminates 20 amino acids from the C terminus of the final domain of BG1 found in all other isoforms for which sequences are published. The early stop codon concomitantly increases the length of the 3′-UTR (Fig. S1). Truncation of the coding region could, in itself, alter the function of the BG1 protein compared to other isoforms (33). We have found that the presence of the 225-bp insert in the 3′-UTR of BG1*R4 allele affects gene function such that the BG1*R4 allele might best be considered a mutant allele due to the deleterious effect of the insert. So far, the 225-bp insert is unique to BR4 and the B2 parent haplotype among haplotypes for which sequence data are available. It represents only one of many structural differences found among BG1 isoforms.
Other BG1 alleles differ in a number of ways (19). In addition to a few synonymous and non-synonymous nucleotide substitutions, prominent variations in coding sequences appear as result of duplications of a set of four exons and adjacent introns. These variations result in different subgroups of BG1 alleles that possess one, two or four copies of this exon/intron “quartet” encoding a portion of the BG1 cytoplasmic tail. Another group of BG1 alleles lacks the penultimate BG1 exon that contains the ITIM-coding sequence (19). While it has not yet been determined whether the latter structural variation contributes to disease incidence, it is notable that two haplotypes (B5 and B15) associated with MD susceptibility and with RSV tumor progression lack this ITIM encoding exon (34). Additional study of BG1 structural variants is needed to determine whether they contribute to the disease associations observed among the different MHC-B haplotypes in which they are found and whether they reflect the results of pathogen selection.
Even though the BG1*R2 and BG1*R4 alleles have different 3′-UTRs, we found little evidence that these alleles were differentially transcribed. In contrast, the dual luciferase assays indicate that the 3′-UTR differences affect posttranscriptional regulation (Fig. 2B). Investigations are underway to identify the mechanism by which the 225-bp insert in the 3′-UTR of BG1*R4 affects translation. The presence of a 3′-UTR derived from a retroviral sequence could put BG1*R4 under the control of regulators of retroviral expression that might occur through binding of sequence-specific small RNAs or proteins. Such regulators, particularly if they are microRNAs, could result in BG1*R4 expression being more greatly affected in some tissues than others. Hence, expression in vivo could vary between tissues, with the differences being either more modest or greater than those observed in the LMH cell luciferase assays illustrated in Fig. 2B. Studies are ongoing to define the mechanism influencing translation of the BG1*R4 allele, including investigation of several miRNA target sequences harbored within the 225-bp insert.
Other polymorphic loci within MHC-B, in addition to BG1, may also contribute to the differences in the incidence of GaHV-2 lymphoma observed between MHC-B haplotypes. Different MHC class I and class II alleles, some better expressed than others, likely contribute to the effectiveness of adaptive immune responses to GaHV-2. An important series of experiments with MHC-B congenic lines has shown that MHC-B haplotypes differentially influence the effectiveness of vaccinal (adaptive) immunity (35–37). The MHC-B haplotypes providing the greatest protective immunity in GaHV-2 challenge trials following vaccination differ between vaccine serotypes, suggesting that allelic differences in antigen presentation have a critical role in the development of long-lasting immunity. Importantly, from the perspective of understanding the contribution of BG1, haplotypes providing the best protection following vaccination in this series of experiments were not always the same as those providing protection in the absence of immunization. Perhaps allelic differences at BG1 affect activation early in immune responses, while class I and class II allelic differences contribute primarily to the effectiveness of memory-based adaptive immunity.
Materials and Methods
Origin of BR2 and BR4 Haplotypes and the Production of Congenic Lines.
The BR2 and BR4 recombinant haplotypes were derived from a lineage of birds at Northern Illinois University originating from a mating between a male of B23/B24 genotype (derived from a strain of New Hampshire chickens) and females of the Avian Disease and Oncology Laboratory White Leghorn inbred line 72 (B2/B2 genotype) (16, 38, 39). The first recombinant haplotype that was recovered was designated BR1 and contained elements of B23 and B24 (as revealed by serological typing reagents) (Fig. 1A). The BR2 and BR4 recombinant haplotypes were recovered in subsequent matings between BR1/B2 males and line 72 (B2/B2) females.
Congenic line birds carrying BR2 and BR4 were produced at the University of New Hampshire by introducing the BR2 and BR4 haplotypes into the genetic background of the highly inbred line UCD003 and selecting progeny bearing BR2 and BR4 by serological typing over ten backcross generations. To ensure that the lines would not be lost, several individuals were selected at each generation for the next backcross. Hence, alleles at some loci across the genome continue to segregate at random in lines 003.R2 and 003.R4. The congenic lines were established by matings among the recombinant progeny within each line and homozygous birds selected to carry the line forward. The lines are maintained in two to three small breeding colonies, each containing a single homozygous sire and 10–15 homozygous dams per sire at each generation.
Viral Challenge Trials of Birds in BR2 and BR4 Congenic Lines.
BR2 and BR4 congenic line chicks were pedigree-hatched at University of California, Davis, double wing-banded, and challenged intra-abdominally with 500 plaque-forming units (pfu) of the very virulent RB1B strain of GaHV-2 (a serotype 1 virus also called MDV1) (40) at two days-of-age. MD virus is classified into three serotypes. Only Serotype I is oncogenic. Serotype I strains are divided into four pathotypes based on their capacity to cause MD in vaccinated birds. The “very virulent” category is the next to the most virulent category in this scale (41). Intra-abdominal inoculation broadly mimics natural infection by inhalation (42), and is the preferred inoculation route to achieve greater uniformity in GaHV-2 viral challenge trials. The RB1B inocula were propagated through not more than two passages on chick kidney culture cells. Five replicate trials were conducted at UC Davis under the supervision of a board-certified avian pathologist. In all trials, specific pathogen free (SPF) chicks (Charles River Laboratories) of a similar age were housed with the inoculated birds to serve as controls for GaHV-2 infectivity. The chicks were checked at least once per day for clinical signs of disease (paralysis, depression or anorexia) and euthanized if unable to stand and feed. Carcasses of any animals that died in advance of trial termination were collected and frozen for necropsy at the end of the trial at 12 weeks when all remaining birds were euthanized. At necropsy, all 270 birds were examined for signs of tumors. Skin, oral cavity, eyes, thymus, sciatic nerve, vagus nerve, brachial plexus, and all visceral organs were examined for gross evidence of tumors. Tissues were collected from nearly all birds found negative upon gross examination for microscopic examination; however, 37 carcasses (23 from 00.3R2 and 14 from 003.R4) were unavailable for histopathological examination. Histological sections were examined for evidence of pleomorphic nodules containing lymphocytes with increased nuclear to cytoplasmic ratios and for the appearance of infiltration or invasion of adjacent tissues. All animal experiments were carried out under institutionally approved protocols. All BR2 and BR4 birds in the challenge trial (and their parents) were MHC-B haplotyped by PCR (See SI Materials and Methods). Disease incidence (number of birds positive versus number of birds negative) for 003.R2, 003.R4 and SPF controls was compared using the χ2 test.
Mapping BR2 and BR4 Crossover Breakpoints and Sequence Differences.
A combination of methods was used narrow the region in BR2 and BR4 containing the crossover breakpoints (Table S3). Birds were typed for the BG gene family using Southern hybridizations, as previously described (43). BF and BL types were determined using single strand conformation polymorphism (44). Microsatellite LEI0258 types were revealed by PCR, as previously published (45). Sequencing of BR1, B2, BR2, and BR4 DNA was carried out to identify the crossover breakpoints. Once the crossover breakpoints were localized, the MHC-B regions in BR2 and BR4 were fully sequenced across a 61-kb region (See SI Materials and Methods).
Assays.
Standard methods were used for 3′-RACE, reverse-transcriptase real-time quantitative PCR, and firefly/Renilla dual luciferase reporter assays. These and assays for the expression of FLAG-BG1, tyrosine phosphorylation and phosphatase association are described in detail in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Lei Zhang for help with flow cytometry, Garrett Larson for helpful discussions of SNP typing, Roshni Patel for assistance with the genome-wide SNP assay, Kathy Reisinger for excellent support with figures, and Adam Bailis and Keely Walker for critical reading of this manuscript. This work was supported in part by National Cancer Institute Grant R21 CA105426 and U.S. Department of Agriculture National Research Initiative Competitive Grants Program Grants 2004-35205-14203 and 2006-35205-16678.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The nucleotide sequences reported in this paper have been deposited in the GenBank database (Accession Nos. FJ770457 - FJ770460).
This article contains supporting information online at www.pnas.org/cgi/content/full/0906776106/DCSupplemental.
References
- 1.Briles WE, Stone HA, Cole RK. Marek's disease: Effects of B histocompatibility alloalleles in resistant and susceptible chicken lines. Science. 1977;195:193–195. doi: 10.1126/science.831269. [DOI] [PubMed] [Google Scholar]
- 2.Taylor RL., Jr Major histocompatibility (B) complex control of responses against Rous sarcomas. Poult Sci. 2004;83:638–649. doi: 10.1093/ps/83.4.638. [DOI] [PubMed] [Google Scholar]
- 3.Plachy J, Benda V. Location of the gene responsible for Rous sarcoma regression in the B-F region of the B complex (MHC) of the chicken. Folia Biol (Praha) 1981;27:363–368. [PubMed] [Google Scholar]
- 4.Briles WE, Briles RW, Taffs RE, Stone HA. Resistance to a malignant lymphoma in chickens is mapped to subregion of major histocompatibility (B) complex. Science. 1983;219:977–979. doi: 10.1126/science.6823560. [DOI] [PubMed] [Google Scholar]
- 5.Hepkema BG, et al. Mapping of susceptibility to Marek's disease within the major histocompatibility (B) complex by refined typing of White Leghorn chickens. Anim Genet. 1993;24:283–287. doi: 10.1111/j.1365-2052.1993.tb00312.x. [DOI] [PubMed] [Google Scholar]
- 6.Kaufman J, et al. The chicken B locus is a minimal essential major histocompatibility complex. Nature. 1999;401:923–925. doi: 10.1038/44856. [DOI] [PubMed] [Google Scholar]
- 7.Kaufman J, Volk H, Wallny HJ. A “minimal essential Mhc” and an “unrecognized Mhc”: Two extremes in selection for polymorphism. Immunol Rev. 1995;143:63–88. doi: 10.1111/j.1600-065x.1995.tb00670.x. [DOI] [PubMed] [Google Scholar]
- 8.Wallny HJ, et al. Peptide motifs of the single dominantly expressed class I molecule explain the striking MHC-determined response to Rous sarcoma virus in chickens. Proc Natl Acad Sci USA. 2006;103:1434–1439. doi: 10.1073/pnas.0507386103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koch M, et al. Structures of an MHC class I molecule from B21 chickens illustrate promiscuous peptide binding. Immunity. 2007;27:885–899. doi: 10.1016/j.immuni.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 10.Sherman MA, et al. Mass spectral data for 64 eluted peptides and structural modeling define peptide binding preferences for class I alleles in two chicken MHC-B haplotypes associated with opposite responses to Marek's disease. Immunogenetics. 2008;60:527–541. doi: 10.1007/s00251-008-0302-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shiina T, et al. Extended gene map reveals tripartite motif, C-type lectin, and Ig superfamily type genes within a subregion of the chicken MHC-B affecting infectious disease. J Immunol. 2007;178:7162–7172. doi: 10.4049/jimmunol.178.11.7162. [DOI] [PubMed] [Google Scholar]
- 12.Miller MM, et al. Assignment of Rfp-Y to the chicken major histocompatibility complex/NOR microchromosome and evidence for high-frequency recombination associated with the nucleolar organizer region. Proc Natl Acad Sci USA. 1996;93:3958–3962. doi: 10.1073/pnas.93.9.3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salomonsen J, et al. Two CD1 genes map to the chicken MHC, indicating that CD1 genes are ancient and likely to have been present in the primordial MHC. Proc Natl Acad Sci USA. 2005;102:8668–8673. doi: 10.1073/pnas.0409213102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller MM, et al. Characterization of two avian MHC-like genes reveals an ancient origin of the CD1 family. Proc Natl Acad Sci USA. 2005;102:8674–8679. doi: 10.1073/pnas.0500105102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delany ME, Robinson CM, Goto RM, Miller MM. Architecture and organization of chicken microchromosome 16: Order of the NOR, MHC-Y and MHC-B subregions. J Heredity. 2009 doi: 10.1093/jhered/esp044. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 16.Briles WE, Briles RW. A search for “duplicate” recombinants between B–F and B–G regions in the chicken B complex. Anim. Blood Groups Biochem Genet. 1980;11:38–39. [Google Scholar]
- 17.Schat KA, Taylor RL, Jr, Briles WE. Resistance to Marek's disease in chickens with recombinant haplotypes to the major histocompatibility (B) complex. Poult Sci. 1994;73:502–508. doi: 10.3382/ps.0730502. [DOI] [PubMed] [Google Scholar]
- 18.White EC, Briles WE, Briles RW, Taylor RL., Jr Response of six major histocompatibility (B) complex recombinant haplotypes to Rous sarcomas. Poult Sci. 1994;73:836–842. doi: 10.3382/ps.0730836. [DOI] [PubMed] [Google Scholar]
- 19.Hosomichi K, et al. Contribution of mutation, recombination, and gene conversion to chicken MHC-B haplotype diversity. J Immunol. 2008;181:3393–3399. doi: 10.4049/jimmunol.181.5.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murphy KM, Nelson CA, Sedy JR. Balancing co-stimulation and inhibition with BTLA and HVEM. Nat Rev Immunol. 2006;6:671–681. doi: 10.1038/nri1917. [DOI] [PubMed] [Google Scholar]
- 21.Calnek BW. Pathogenesis of Marek's disease virus infection. Curr Top Microbiol Immunol. 2001;255:25–55. doi: 10.1007/978-3-642-56863-3_2. [DOI] [PubMed] [Google Scholar]
- 22.Burgess SC, Basaran BH, Davison TF. Resistance to Marek's disease herpesvirus-induced lymphoma is multiphasic and dependent on host genotype. Vet Pathol. 2001;38:129–142. doi: 10.1354/vp.38-2-129. [DOI] [PubMed] [Google Scholar]
- 23.Kaiser P, Underwood G, Davison F. Differential cytokine responses following Marek's disease virus infection of chickens differing in resistance to Marek's disease. J Virol. 2003;77:762–768. doi: 10.1128/JVI.77.1.762-768.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jarosinski KW, Njaa BL, O'Connell PH, Schat KA. Pro-inflammatory responses in chicken spleen and brain tissues after infection with very virulent plus Marek's disease virus. Viral Immunol. 2005;18:148–161. doi: 10.1089/vim.2005.18.148. [DOI] [PubMed] [Google Scholar]
- 25.Fabricant J, Ianconescu M, Calnek BW. Comparative effects of host and viral factors on early pathogenesis of Marek's disease. Infect Immun. 1977;16:136–144. doi: 10.1128/iai.16.1.136-144.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Calnek BW. Influence of age at exposure on the pathogenesis of Marek's disease. J Natl Cancer Inst. 1973;51:929–939. doi: 10.1093/jnci/51.3.929. [DOI] [PubMed] [Google Scholar]
- 27.Omar AR, Schat KA. Syngeneic Marek's disease virus (MDV)-specific cell-mediated immune responses against immediate early, late, and unique MDV proteins. Virology. 1996;222:87–99. doi: 10.1006/viro.1996.0400. [DOI] [PubMed] [Google Scholar]
- 28.Schat KA, Chen CL, Shek WR, Calnek BW. Surface antigens on Marek's disease lymphoblastoid tumor cell lines. J Natl Cancer Inst. 1982;69:715–720. [PubMed] [Google Scholar]
- 29.Calnek BW, Adene DF, Schat KA, Abplanalp H. Immune response versus susceptibility to Marek's disease. Poult Sci. 1989;68:17–26. doi: 10.3382/ps.0680017. [DOI] [PubMed] [Google Scholar]
- 30.Lee LF, Bacon LD. Ontogeny and line differences in the mitogenic response of chicken lymphocytes. Poult Sci. 1983;62:579–584. doi: 10.3382/ps.0620579. [DOI] [PubMed] [Google Scholar]
- 31.Calnek BW, Schat KA, Ross LJ, Chen CL. Further characterization of Marek's disease virus-infected lymphocytes. II. In vitro infection. Int J Cancer. 1984;33:399–406. doi: 10.1002/ijc.2910330319. [DOI] [PubMed] [Google Scholar]
- 32.Calnek BW, Lucio B, Schat KA, Lillehoj HS. Pathogenesis of Marek's disease virus-induced local lesions. 1. Lesion characterization and cell line establishment. Avian Dis. 1989;33:291–302. [PubMed] [Google Scholar]
- 33.Hosomichi K, et al. Contribution of mutation, recombination, and gene conversion to chicken Mhc-B haplotype diversity. J Immunol. 2008;181:3393–3399. doi: 10.4049/jimmunol.181.5.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bacon LD. Influence of the major histocompatibility complex on disease resistance and productivity. Poult Sci. 1987;66:802–811. doi: 10.3382/ps.0660802. [DOI] [PubMed] [Google Scholar]
- 35.Bacon LD, Witter RL. Influence of turkey herpesvirus vaccination on the B-haplotype effect on Marek's disease resistance in 15. B-congenic chickens. Avian Dis. 1992;36:378–385. [PubMed] [Google Scholar]
- 36.Bacon LD, Witter RL. Influence of B-haplotype on the relative efficacy of Marek's disease vaccines of different serotypes. Avian Dis. 1993;37:53–59. [PubMed] [Google Scholar]
- 37.Bacon LD, Witter RL. Serotype specificity of B-haplotype influence on the relative efficacy of Marek's disease vaccines. Avian Dis. 1994;38:65–71. [PubMed] [Google Scholar]
- 38.Briles WE, Briles RW. Genetic recombination within the B blood group system of chickens. Conf Intl Soc Animal Blood Group Res. 1976:8. [Google Scholar]
- 39.Briles WE, Briles RW. Some recent recombinants at the B locus. In: Benedict AA, editor. Avian Immunology. Advances in Experimental Medicine and Biology. Vol 88. New York: Plenum Press; 1977. pp. 299–307. [DOI] [PubMed] [Google Scholar]
- 40.Schat KA, Calnek BW, Fabricant J, Abplanalp H. Influence of oncogenicity of Marek' disease virus on evaluation of genetic resistance. Poult Sci. 1981;60:2559–2566. doi: 10.3382/ps.0602559. [DOI] [PubMed] [Google Scholar]
- 41.Witter RL. Increased virulence of Marek's disease virus field isolates. Avian Dis. 1997;41:149–163. [PubMed] [Google Scholar]
- 42.Islam AF, Walkden-Brown SW, Burgess SK, Groves PJ. Marek's disease in broiler chickens: Effect of route of infection and herpesvirus of turkey-vaccination status on detection of virus from blood or spleen by polymerase chain reaction, and on weights of birds, bursa and spleen. Avian Pathol. 2001;30:621–628. doi: 10.1080/03079450120092116. [DOI] [PubMed] [Google Scholar]
- 43.Miller MM, Abplanalp H, Goto R. Genotyping chickens for the B–G subregion of the major histocompatibility complex using restriction fragment length polymorphisms. Immunogenetics. 1988;28:374–379. doi: 10.1007/BF00364237. [DOI] [PubMed] [Google Scholar]
- 44.Goto RM, et al. Single-strand conformation polymorphism (SSCP) assays for major histocompatibility complex B genotyping in chickens. Poult Sci. 2002;81:1832–1841. doi: 10.1093/ps/81.12.1832. [DOI] [PubMed] [Google Scholar]
- 45.Fulton JE, et al. Molecular genotype identification of the Gallus gallus major histocompatibility complex. Immunogenetics. 2006;58:407–421. doi: 10.1007/s00251-006-0119-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.