Abstract
Septins constitute a group of GTP-binding proteins involved in cytokinesis and other essential cellular functions. They form heterooligomeric complexes that polymerize into nonpolar filaments and are dynamic during different stages of the cell cycle. Posttranslational modifications and interacting partners are widely accepted regulators of septin filament function, but the contribution of nucleotide is undefined due to a lack of detailed structural information. Previous low-resolution structures showed that the G domain assembles into a linear polymer with 2 different interfaces involving the N and C termini and the G binding sites. Here we report the crystal structure of SEPT2 bound to GppNHp at 2.9 Å resolution. GTP binding induces conformational changes in the switch regions at the G interfaces, which are transmitted to the N-terminal helix and also affect the NC interface. Biochemical studies and sequence alignment suggest that a threonine, which is conserved in certain subgroups of septins, is responsible for GTP hydrolysis. Although this threonine is not present in yeast CDC3 and CDC11, its mutation in CDC10 and CDC12 induces temperature sensitivity. Highly conserved contact residues identified in the G interface are shown to be necessary for Cdc3–10, but not Cdc11–12, heterodimer formation and cell growth in yeast. Based on our findings, we propose that GTP binding/hydrolysis and the nature of the nucleotide influence the stability of interfaces in heterooligomeric and polymeric septins and are required for proper septin filament assembly/disassembly. These data also offer a first rationale for subdividing human septins into different functional subgroups.
Keywords: filament, GTP-binding protein, X-ray crystallography
Septins are GTP-binding proteins that form oligomers and filaments in vitro and in vivo. They were identified first as cell division cycle mutants in yeast Saccharomyces cerevisiae (1) and later in all eukaryotes. Multiple septin genes have been identified in eukaryotic genomes, ranging from 5 in yeast to 14 in humans. These genes can be subdivided into different groups according to sequence conservation, but the functional significance of these subgroups is unclear (2, 3). Deletion and mutation studies in yeast septins have shown incomplete cell division, suggesting an important role for septins in cytokinesis (4, 5). Moreover, septins also play important roles in secretion, membrane remodeling, and cytoskeleton dynamics (6, 7). The hallmark of septin proteins is a conserved central G domain flanked by variable N and C termini, with the C-terminal ends predicted as coiled coils. Septins from endogenous sources are purified as heterooligomeric complexes, which can form filaments and ring-like structures under appropriate conditions (8, 9). Such complexes also can be isolated by recombinant coexpression in Escherichia coli, and they can assemble into homooligomers and heterooligomers and form filaments (4, 8–10).
We previously solved the crystal structure of the SEPT2 G domain in its GDP-bound state and the low-resolution structure of an oligomeric septin complex consisting of SEPT2/6/7. These structures revealed that filament formation involves conserved interactions between adjacent nucleotide-binding sites (G interface) and N- and C-terminal extensions (NC interface) of the protomers (11). These studies, complemented by electron microscopy studies of septin complexes from human, Caenorhabditis elegans, and S. cerevisae, demonstrated a universal principle for the assembly of septins (11–13). The asymmetric heterotetramers, heterohexamers, or heterooctamers formed by 2, 3, or 4 different septins associate to form nonpolar septin filaments. A linear hexamer also has been demonstrated for the SEPT3/5/7 complex isolated from rat brain (14).
But despite these structural breakthroughs in understanding the filamentous assembly of septins, the role of the guanine nucleotide in septin assembly and function remains elusive (15). Purified septin complexes are saturated with GDP and GTP in a defined and consistent ratio, ≈2:1 for the SEPT2/6/7 complex and 3:1 for the yeast Cdc3/10/11/12 complex. Native or recombinant heterooligomeric complexes demonstrate a very slow GTPase reaction (8–10). In vivo, nucleotide exchange experiments also have shown that guanine nucleotides bound to yeast septins do not turn over during a cell cycle (16). Compared with septin complexes, monomeric septins with little or no bound nucleotide have higher nucleotide exchange and hydrolysis rates (17, 18). Although the details differ, phylogenetic analyses of septin sequences indicate that they can be grouped into different subclasses (2, 3). Whether these differences can be related to different biochemical and structural properties, such as preferred binding of GDP versus GTP and/or preference for G or NC type interfaces, is unknown, however.
Fluorescence recovery after photobleaching (FRAP) and polarization microscopy experiments suggest that the septin filaments are highly dynamic during certain stages of the cell cycle (19, 20). Although the dynamics of septin filaments are believed to be regulated by phoshorylation, a contribution of GTP hydrolysis to this process cannot be ruled out. The juxtaposition of 2 G domains via their nucleotide-binding sites in the septin filaments is reminiscent of other G proteins activated by dimerization (GADs), such as MnmE and hGBP1 (21–23). On the other hand, mutational analysis of septins based on the Ras·GTP hydrolysis mechanism revealed no observable phenotype (24–26), leading to the conclusion that GTP hydrolysis of septins has no influence on septin assembly or dynamics (24). Based on the structural alignment of SEPT2-GDP with Toc34 protein, Weirich et al. (6) proposed the possible involvement of an invariant histidine in engaging the nucleotide across the G dimer. Because ≈1 mol of GTP is always present in a septin complex, it is unlikely that this invariant histidine is involved in GTP hydrolysis. Whether there are any structural consequences for the binding of GDP/GTP to septins is also unclear.
To address these questions, we set out to investigate structural differences between GDP- and GTP-bound septins and to identify the residues possibly responsible for GTP hydrolysis. The SEPT2/6/7 complex contains both GDP-bound and GTP-bound septins, but differences between them cannot be identified in the filament structure due to the low (4 Å) resolution. Here we solved the structure of SEPT2 G domain in its active state (GppNHp-bound form) and analyzed the contribution of active site residues by mutational analysis in vitro and in vivo. This allowed us to propose a structural classification of different septin subunits that might explain their nonrandom distribution in septin complexes. This study represents an important step forward in understanding the role of guanine nucleotides in septin filament assembly and dynamics.
Results
Structural Differences Between GDP-Bound and GTP-Bound Septins.
To gain insight into the GTP-bound structure of septin, mouse SEPT2 (1-315) (hereinafter SEPT2–315) was purified following a protocol described previously (11). The protein-bound nucleotide (mostly GDP) was exchanged to GppNHp, and crystallization trials were performed. Because we could not obtain diffraction-quality crystals, we turned our attention to an N-terminal–deleted construct, SEPT2 (33-306) (hereinafter ΔNSEPT2). The domain boundaries introduced to avoid oligomerization across the NC interface are based on the previous crystal structure of N-terminally–deleted SEPT2·GDP (PDB ID: 2QNR). Well-diffracting crystals were obtained and used to solve and refine the structure of ΔNSEPT2 to 2.9 Å resolution (see Materials and Methods and Table S1).
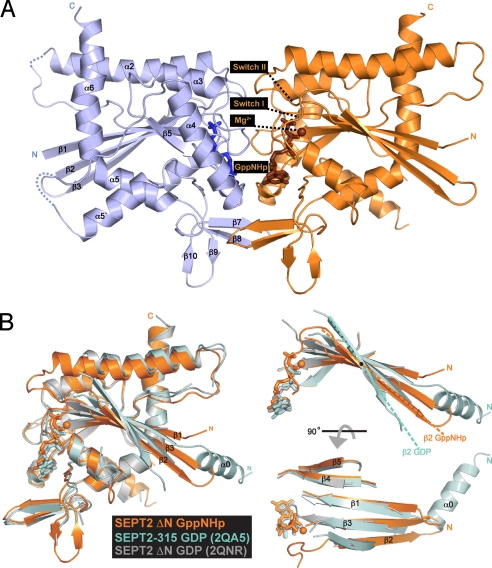
As expected and unlike the previous structure (2QA5), ΔNSEPT2 does not form filaments in the crystal. As in solution, in the crystal it exists as dimer with subunits facing one another across the nucleotide-binding site, forming what is known as the G interface (11). The structure revealed new elements not previously observed in the GDP-bound structure of SEPT2–315 (Fig. 1A). In contrast to the earlier structure, and analogous to structural studies on Ras proteins (27), the switch I and II regions are now ordered and visible, as is a magnesium ion coordinating the nucleotide. In a previously disordered region, we now see an additional antiparallel β-turn (β9 and β10) looped in between the antiparallel β7–β8 and α5. The G dimer interface of ΔNSEPT2·GppNHp has a larger buried surface than the GDP structure (2,485 Å2 vs. 1,852 Å2), suggesting that GTP induces tighter binding between subunits. The crystal structure also shows that the G dimer is slightly tilted at right angles ≈10° away from the filament axis [supporting information (SI) Fig. S1]. This tilting could be due to either the presence of GTP or the absence of filamentous assembly in the crystal.
Fig. 1.
Structure of SEPT2·GppNHp. (A) Ribbon model of the overall structure of the SEPT2 dimerized across the nucleotide-binding site. New elements observed are labeled and the GppNHp and magnesium are colored in brown. (B) Superimposition of the SEPT2·GppNHp structure (gold) with the previous structures of SEPT2·GDP [PDB ID 2QA5 (cyan) and 2QNR (gray)].
Although the overall structures of GTP-bound and GDP-bound septin look similar, superimposition of the structures reveals important differences (Fig. 1B). Whereas the α-helices remain unchanged, the central β-strands show an angular difference of ≈20°. The largest effect is on beta strands β2 and 3, which are tilted by 20° with respect to the strands of GDP structure (Fig. 1B). There is a partial movement of the N-terminal half of β1 to remain hydrogen-bound with β3. Because β2 and β3 are in close proximity to the switch regions, and because the GDP-bound structures of SEPT2 (1–315) (11) and N-terminally–deleted SEPT2 (33–306) (PDB ID: 2QNR) do not exhibit this angular difference, the γ-phosphate, not the deletion, appears to be the driving force for the β-strand torsion (Fig. 1B). From this structure, it is clear that the first strands of the β-sheet would clash with the position of α0, an important element in the NC interface observed in the previous structures. This suggests that the binding of triphosphate modulates both the G and NC interfaces simultaneously.
The SEPT2 G Interface.
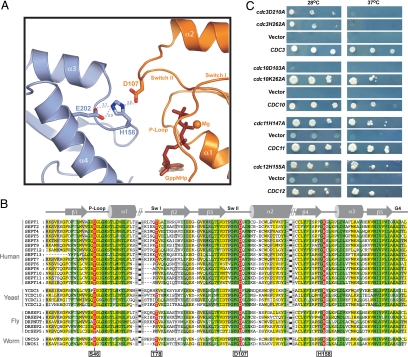
The formation of a G interface of ΔNSEPT2·GppNHp observed in the crystal is consistent with previous studies (11). In the triphosphate structure, the G interface is stabilized by an interaction between Asp-107 from switch II and His-158 of the neighboring subunit, which in turn is stabilized by Glu-202, suggesting that His-158 is protonated (Fig. 2A). Asp-107 and His-158 are highly conserved between yeast and man, whereas Glu-202 can be replaced by either glutamine or aspargine (Fig. 2B). Because Asp-107 is adjacent to switch II and this interaction is not seen in the GDP structure, we assume that this interaction is induced due to the presence of GTP.
Fig. 2.
Sequence alignment and interface analysis. (A) Part of the SEPT2·GppNHp G interface showing residues from the protomers required for tight interface formation, shown as ball and stick, with other structural elements as ribbons. (B) Sequence alignment of the complete set of septins from various organisms, highlighting the residues analyzed and discussed in the text. (C) Role of interface residues in vivo, using yeast complementation to introduce single copies of the corresponding WT and mutant septins described in Materials and Methods.
To study the importance of the GTP-induced interaction between Asp-107 and His-158, we checked the functionality of these residues in yeast. The His-158 equivalent residue was found in the 3 essential yeast septins CDC3, CDC11, and CDC12, except in CDC10, the very rare exception from total conservation. These residues were mutated to Ala, as were the residues in CDC3 and CDC10 equivalent to Asp-107. The yeast complementation assay (Fig. 2C) demonstrated that the mutation of the conserved Asp in Cdc10 and Cdc3 has a dramatic effect on temperature-sensitive growth. The Cdc10(D103A) mutant is more severely affected, because it does not grow even at the permissible temperature (Fig. 2C). The His-to-Ala mutations in Cdc11 and Cdc12 exhibited no observable growth phenotype, whereas Cdc3(H262A) was temperature-sensitive. Strikingly, the Cdc10(K155A) showed no observable phenotype, suggesting that the essential Asp-103 in Cdc10 creates a different type of interaction in the interface.
To further corroborate the importance of these interface residues in stabilizing the G interface, we performed in vitro coexpression experiments. Mutations were introduced in Cdc3 and Cdc12 and coexpressed with Cdc10 and Cdc11, respectively (see SI Text). The His-to-Ala mutation in the G interface was seen to have a dramatic effect on formation of Cdc3–10 heterodimer, but not of Cdc11–12 heterodimer (Fig. S2). In general, our findings indicate that Cdc3 and Cdc10, which constitute the central part of the yeast septin octamer (13), are more sensitive to mutations in the interface.
The Nucleotide-Binding Pocket.
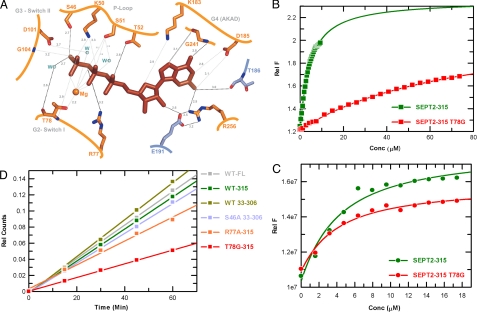
The guanine nucleotide is completely buried inside the dimer interface and is inaccessible to solvent. It is bound in a canonical fashion with 4 of the 5 conserved G domain signature motifs involved in binding (Fig. 3A). The β- and γ-phosphates are surrounded by the P-loop GxxxGKS/T motif, the conserved lysine (Lys-50) and serine (Ser-51) of which contact the β- and γ-phosphate and the Mg2+ ion, respectively. An additional theronine, Thr-52, which is totally invariant in septins, creates a hydrogen bond to the α-phosphate. This interaction has not been observed in any other G protein except Toc34, a close homologue of septin (6, 28). Here a serine rather than a threonine causes a similar interaction with the α-phosphate (29). Thr-78 in switch I, homologous to Thr-35 in Ras, coordinates the Mg2+ ion and forms a main hydrogen bond with the γ-phosphate. This threonine (Thr-78), together with the invariant glycine (Gly-104) from the switch II DxxG motif, mediate the universal switch mechanism. Their main chain amides form hydrogen bonds with the γ-phosphate, which are released after GTP hydrolysis and Pi release in what has been termed the “loaded spring” mechanism (27). Because Thr-78 is highly conserved and Gly-104 is totally conserved in all septins (Fig. 2B), this universal mechanism would be assumed to operate in most, if not all, septins. One of the G protein signature motifs is N/TKxD, which is replaced by AKAD in septins. Asp-185 from the AKAD motif of SEPT2 forms the canonical double-bifurcated hydrogen bond with the guanine base, which is the most important element of the guanine-versus-adenine specificity.
Fig. 3.
Biochemical analyses of active site residues in SEPT2. (A) Details of the nucleotide-binding site, showing GppNHp and surrounding residues, with the 2 protomers color-coded as in Fig. 1A. (B and C) Affinity for GppNHp (B) and GDP (C) measured by the increase in fluorescence obtained using 500 nM of the corresponding mant nucleotide and adding increasing amounts of WT and mutant SEPT2 as indicated. The data are fitted to a quadratic binding equation to produce the Kd values given in Table S2. (D) The GTPase reaction measured with 10 μM WT and mutant SEPT2 using 150 μM radioactive GTP mixture in a charcoal assay as described in Materials and Methods. The amount of Pi released was measured and plotted against time.
Along with the canonical G protein elements, several novel types of interactions are involved in GTP binding by the SEPT2 dimer. The guanine and ribose moieties of the nucleotide are sandwiched in the dimer interface and have contact with both protomers (Fig. S3). The N3, O6, and N7 atoms of the guanine base are hydrogen-bound to Arg-256 and the main chain amide and carbonyl group of Gly-241, respectively (Fig. 3A). The main chain carbonyl of Thr-186 and the side chain carboxyl of Glu-191 from the other protomer have additional contacts with the guanine base and ribose, respectively. Glu-191 from the other protomer also stabilizes Arg-256. The nucleotide-binding site in the interface of 2 G domains is reminiscent of other GADs (23), for which the high-affinity binding site and/or the enzymatic machinery for GTP hydrolysis requires the presence of both subunits. This also might explain the observations that isolated septins have a low affinity for nucleotides and that no nucleotide dissociation is detected in the septin oligomers (9, 16).
Concerning the GTPase reaction, the residues in the neighborhood of the γ-phosphate are candidate residues relevant for catalysis. Ser-46 from the P-loop (Gly-12 position in Ras) is involved in hydrogen bonding with the O3G of the γ-phosphate and is conserved as Ser or Thr in septins. Arg-77 from switch I is located near the phosphates. Although Arg-77 does not have direct interactions with the β- and γ-phosphates, it potentially could function as an “arginine finger.” Such an arginine is found only in SEPT2, however, and thus it can be excluded as a relevant residue (Fig. 2B). A water molecule is located 2.7 Å away from the γ-phosphate, in a position for an in-line attack on the γ-phosphate. It is hydrogen-bound with the main chain carbonyl of Thr-78 and thus could be responsible for polarizing the attacking water. Alternatively, if repositioned, the hydroxyl group of Ser-46, the group next closest to the water (≈4.3 Å), also could serve as a catalytic residue, as has been observed for Ser-73 in the interface of the hGBP1 dimer (Fig. S4) (21).
Biochemical Studies of SEPT2 Mutants.
To investigate whether the differences in nucleotide states in SEPT2/6/7 hexamer is due to the active site residues, we set out to replace the corresponding residues. Sequence alignment revealed that Ser-46 in the P loop is almost totally conserved in all septins, while threonine of switch I (Thr-35 in Ras), which is almost universally conserved in G proteins, is present in only about half of the human septins. It is not present in the SEPT6 group, and the residues around it are surprisingly poorly conserved (Fig. 2B). Comparing SEPT2 and SEPT6 revealed that the residues equivalent to Ser-46, Arg-77, and Thr-78 in SEPT2 are replaced by Thr, Pro, and Gly, respectively, in SEPT6 (Fig. 2B). Thus, we mutated Ser-46 to Thr and Ala, Thr-78 to Gly, and Arg-77 to Ala in SEPT2–315. The most drastic variation of Ser-46 was found in yeast Cdc3 with an aspartate at the equivalent position. To explore the results of that replacement, we made the S46D mutation in SEPT2–315.
We tested the purified soluble SEPT2 mutants for guanine nucleotide binding using equilibrium titration experiments with fluorescently labeled mant nucleotides, as described in Materials and Methods (Fig. 3 B and C). The dissociation constants are summarized in Table S2. Apart from the S46D mutant, the proteins tested show a similar affinity for GDP, and only the T78G mutant has a significantly reduced (≈21-fold) affinity for GTP (Fig. 3B). Deletion of the N terminus has no significant influence on nucleotide affinity. S46A in the ΔNSEPT2 construct has a 2- to 3-fold effect on the affinity for GDP or GTP (Table S2).
We performed multi-turnover GTP hydrolysis assays with WT and mutant proteins using radioactive GTP in the charcoal assay, as described in Materials and Methods. Testing the GTP hydrolysis rates of various SEPT2 constructs revealed that SEPT2-fl and the 2 deletion constructs have similar GTPase rates (Fig. 3D). Among the mutants, R77A is similar to WT, indicating that the (nonconserved) arginine plays no role in GTP hydrolysis (Fig. S4). Ser-46 is close to the active site but does not seem to play a similar role in the catalysis of SEPT2; neither the S46T nor, surprisingly, the more drastic S46D mutation have any notable effect (Fig. S5). The only significant effect on catalysis comes from the T78G mutation, which reduces the rate of hydrolysis by 2-fold. Thr-78 has various interactions in the active site (Fig. 3A), and the failure of any of these could lead to reduced GTP hydrolysis by T78G. Thr-78 is not present in a special subgroup of human septins (Fig. 2B), which might explain why SEPT6 in the SEPT2/6/7 complex retains bound GTP.
We also tested the influence of His-158 on GTP hydrolysis, because it is close to the active site and is responsible for the tight interaction in the G dimer interface. If anything, there is a slight increase in the GTP hydrolysis of the SEPT2 H158A mutant (Fig. S5). Therefore, the conserved histidine is unlikely to be involved in GTP hydrolysis, as suggested by Weirich et al. (6).
Analysis of Active Site Residues in Vivo.
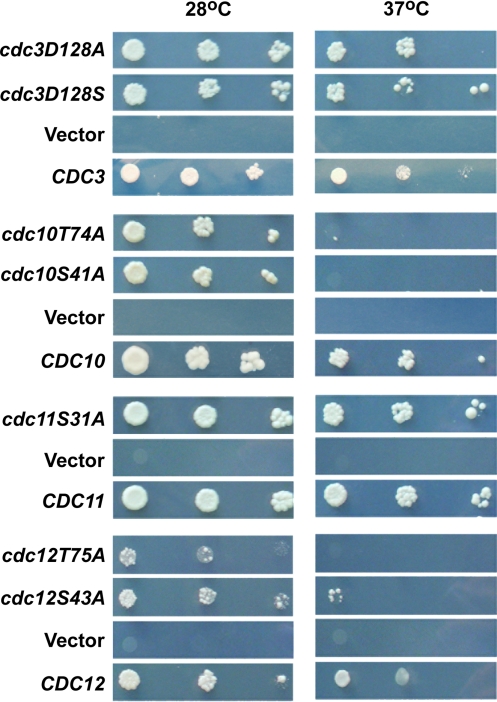
To assess the in vivo functional significance of the Thr-78 and Ser-46 residues, we investigated the effect of corresponding mutations in yeast. Similar to the SEPT6 group in human septins, CDC3 and CDC11 septins do not have a residue corresponding to Thr-78, whereas CDC10 and CDC12 do have such a residue (Fig. 2B). Thus, we constructed Cdc10(T74A) and Cdc12(T75A) in addition to Cdc10(S41A), Cdc11(S31A), and Cdc12(S43A) (Table S3). Finally, the P-loop Asp-128 of CDC3 was mutated to Ser and Ala, respectively.
Haploid deletion strains were complemented using plasmids with WT and mutant septin genes. The complementation assays revealed that different yeast septins are differentially sensitive to the mutations. For the septins carrying a residue equivalent to Ser-46, only Cdc10 is sensitive to the S41A mutation, while Cdc12 with S43A is only weakly sensitive and Cdc11 with S31A is not at all sensitive to the mutation (Fig. 4). We also checked the viability of Cdc12(S43V) and found no phenotype (not shown), in line with previous mutational studies (25). Similarly, the D128S and D128A mutations of Cdc3 show no obvious growth phenotype. This indicates that the Ser-46 equivalent residue does not have an essentially general role in septins, but is required for the function of Cdc10. The presence of a residue equivalent to Thr-78 is essential in both Cdc10 and Cdc12. The T74A mutation in CDC10 shows temperature sensitivity, and the Cdc12(T75A) mutant does not even grow at the permissive temperature (Fig. 4). Assuming that Thr-78 is required for GTP binding and/or hydrolysis, the yeast data suggest that GTP binding and/or hydrolysis is essential for Cdc10 and Cdc12, but not for Cdc3 and Cdc11, consistent with previous biochemical studies (25).
Fig. 4.
Yeast complementation assay. Role of active site residues in vivo, using yeast complementation to introduce single copies of the corresponding WT and mutant septins described in Materials and Methods.
Discussion
The previous structure of the SEPT2–315 in the GDP-bound form showed a linear polymer with alternating G and NC interfaces. Only by deleting N-terminal sequences and thus preventing filament formation were we able to crystallize SEPT2 in the triphosphate form. The structure of ΔNSEPT2·GppNHp shows elements of the protein that previously were not visible or unstructured, such as the switch regions that form the main chain canonical γ-phosphate contacts of G proteins (27). Here those contacts seem to stabilize the dimerization interface and the binding of nucleotide. Important for filament formation is the observation that the 2 outermost β-strands, β2 and β3, change their orientation and elongate somewhat toward the position occupied by helix α0 seen in the GDP structure (Fig. 1B). These β-strands clash with α0 in the GTP-bound state and might push the α0 to destabilize the NC interface (Fig. S6). This also explains why no well-diffracting crystals could be obtained for SEPT2–315-GppNHp. Thus, even though filament formation seems to occur independent of the septin nucleotide state (9), our structural analysis suggests that the presence of the γ-phosphate influences the structure and stability not only of the G, but also of the NC interface, at least for some septins.
Dimerization across the active site is a mechanism used by many GAD proteins (23). While the details differ, the unifying principle in GTP hydrolysis of GADs is the insertion and/or stabilization of residues in the active site of one protomer by the other (21, 22). Although a similar principle seems to be operating in the case of the SEPT2 dimer, and potential catalytic residues have been identified here, only the Thr-78 mutation has a significant effect on the GTPase reaction. While this might suggest that a GTPase reaction is not required for septin function, the in vivo yeast data show that the residues equivalent to Thr-78 are required for Cdc10 and Cdc12. Cdc3 and Cdc11 do not have a residue corresponding to Thr-78. Because replacements equivalent to Thr-78 are found in many septins, the mutations are unlikely to cause significant structural changes. We assume that GTP hydrolysis is important for the function of at least some septins. Because GTP hydrolysis of the oligomeric complexes is slow, it might imply that other factors in the cell are involved in regulating the GTPase reaction, as observed for other GADs (23). Indeed, a recent study of the GTPase activity of Drosophila septins shows that Orc6 enhances GTP hydrolysis (30). The possibility that the observed reduction of GTP affinity also affects septin function cannot be ruled out, however.
Considering the dynamic changes in septin assembly during the life cycle of yeast, as evidenced by FRAP (19) and fluorescence polarization experiments (20), GTPase likely is relevant in the context of the biological function. The structure of the SEPT2/6/7 filament indicates that SEPT6 in the SEPT6–SEPT2 G interface retains GTP, while the SEPT7–SEPT7 interface shows 2 molecules of GDP, confirming that the complex is consistently isolated with a GDP:GTP ratio of 2:1. The Cdc3/10/11/12 octameric complex is similarly purified with a defined GDP:GTP ratio of 3:1 (4, 9). Although a recent electron microscopy structural analysis of the octameric yeast complex could identify the alternating G and NC interfaces, it could not identify which of the yeast septins retains GTP (13). Mutational studies in yeast have shown that the GTP-induced contacts in Cdc3 and Cdc10 are essential. Because Cdc3 and Cdc10 form the core of the octameric complex and interact via the G interface (13), and because Cdc3 does not contain a Thr residue in the switch I, we suggest that the presence of GTP in Cdc3 might stabilize the Cdc3–Cdc10 heterodimer interface, thereby forming a rigid core in which different functional octamers are formed during mitosis and sporulation (31).
The available data suggest that some septins are able to hydrolyze GTP but others are not. This may be related to the phylogenetic analysis in which human septins were classified into 4 different subgroups according to Kinoshita (2), along with a more detailed analysis of all known animal and fungal isoforms (3). It also has been suggested that septins can form complexes in various combinations as long as septins from 3 different subgroups are combined (2). Some of the mammalian septin complexes purified so far, including SEPT2/6/7 (10, 11), SEPT7/9b/11 (32), and SEPT3/5/7 (14), seem to support this hypothesis of group-specific combinatorial complex formation. Although the precise nature of functional and structural differences cannot be deduced from our present knowledge of structure–function relationships, we can assume that the ability to hydrolyze GTP is a distinctive feature for classification. Thus, we can predict that the members of the human SEPT6 (2) or 1B group (3) consisting of septins 6, 8, 10, and 11, which do not contain Thr-78, are not GTPase competent. Each of the yeast septins has been classified into a different subgroup. Yeast complementation assays show that Cdc10 and Cdc12 require the switch I threonine residue for in vivo function (presumably for GTP hydrolysis), while Cdc3 and Cdc11 do not. C. elegans has only 2 septin genes, which belong to the 1B and 2B groups as classified by Pan et al. (3); the 5 Drosophila septins have been assigned to the 3 subgroups represented by human SEPT2, 6, and 7 (3).
SEPT2 in the GDP-bound form and in the absence of cognate partner SEPT6 has the ability to pair with itself using both the G and NC interfaces (11). But in the GTP-bound form, it is apparently not able to maintain the NC interface, as is evident from the β-strand torsion. Although a SEPT2 G dimer presumably is not formed in vivo, the structural differences observed between GDP-bound and GTP-bound SEPT2 have implications for a physiological septin complex such as SEPT2/6/7, where SEPT2 dimerizes via NC and forms a G interface with SEPT6·GTP. It thus appears that a mixed interface between GDP-bound and GTP-bound septins is not only tolerated, but is even favorable, because the structural analysis suggests that this interface is tighter. The SEPT2/6/7 structure also reveals that SEPT6 specifically pairs with SEPT7 due to the extended N terminus of SEPT6, which wraps around the latter. Furthermore, the interaction between certain pairs of septins such as SEPT6 and SEPT7, is favored by the close juxtaposition of the C-terminal coiled coils in the NC interfaces. Along the same lines, this may indicate that the GDP–GDP interface of SEPT7 in the septin filament is the least stable interaction and might be the reason why the filament disassembly to the hexameric SEPT 7/6/2/2/6/7 unit is favored. The human SEPT7 is unique in terms of constituting the edge of the oligomeric unit. It remains to be tested whether SEPT13 belonging to the same 2B subgroup (3) can replace SEPT7 in the hexameric unit.
Our structural analysis leads us to conclude that the inability of certain septins to hydrolyze GTP could have a major role in dictating the rules for septin pairing and assembly. While the presence of the γ-phosphate further stabilizes the G interface via additional contacts from the switch II residue Asp-107, the NC interface appears to be destabilized due to the β-strand torsion induced by the switch I theronine (Thr-78). This counteracting function of switches would be released by GTP hydrolysis in SEPT2 and other septins with an intact switch I theronine. In the case of SEPT6 and other SEPT6-IB subgroup septins, where GTP is required for stabilization of the G interface, the switch I is considerably shortened and threonine is absent. In this case, GTP can stabilize the G interface with SEPT2 without compromising the NC interface with SEPT7. We thus conclude that the purpose of having different nucleotide states in septins is to allow structural transitions and recycling of septin filaments (7, 19, 20, 31) during particular stages of the cell cycle.
Materials and Methods
Detailed description of purification, crystalization, structure determination, biochemical, and in vivo experiments are described in SI Text published online.
Supplementary Material
Acknowledgments.
Data collection was done at the Swiss Light Source, beamline X10SA, Paul Scherrer Institute, Villigen, Switzerland; we thank the beam line staff for assistance. We also thank Anton Meinhart, Bernhard Loll, Ilme Schlichting, and Ingrid Vetter for data collection and crystallographic advice, and Dorothee Kuehlmann for technical assistance. M.F. is supported by Vedecká Grantová Agentúra, Ministerstva školstva Slovenskej republiky Grant 2/0038/08.
Footnotes
The authors declare no conflicts of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3FTQ).
This article contains supporting information online at www.pnas.org/cgi/content/full/0902858106/DCSupplemental.
References
- 1.Hartwell LH. Genetic control of the cell division cycle in yeast, IV: Genes controlling bud emergence and cytokinesis. Exp Cell Res. 1971;69:265–276. doi: 10.1016/0014-4827(71)90223-0. [DOI] [PubMed] [Google Scholar]
- 2.Kinoshita M. Assembly of mammalian septins. J Biochem. 2003;134:491–496. doi: 10.1093/jb/mvg182. [DOI] [PubMed] [Google Scholar]
- 3.Pan F, Malmberg RL, Momany M. Analysis of septins across kingdoms reveals orthology and new motifs. BMC Evol Biol. 2007;7:103. doi: 10.1186/1471-2148-7-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frazier JA, et al. Polymerization of purified yeast septins: Evidence that organized filament arrays may not be required for septin function. J Cell Biol. 1998;143:737–749. doi: 10.1083/jcb.143.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kinoshita M, et al. Nedd5, a mammalian septin, is a novel cytoskeletal component interacting with actin-based structures. Genes Dev. 1997;11:1535–1547. doi: 10.1101/gad.11.12.1535. [DOI] [PubMed] [Google Scholar]
- 6.Weirich CS, Erzberger JP, Barral Y. The septin family of GTPases: Architecture and dynamics. Nat Rev Mol Cell Biol. 2008;9:478–489. doi: 10.1038/nrm2407. [DOI] [PubMed] [Google Scholar]
- 7.Kinoshita M. Diversity of septin scaffolds. Curr Opin Cell Biol. 2006;18:54–60. doi: 10.1016/j.ceb.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 8.Field CM, et al. A purified Drosophila septin complex forms filaments and exhibits GTPase activity. J Cell Biol. 1996;133:605–616. doi: 10.1083/jcb.133.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farkasovsky M, Herter P, Voss B, Wittinghofer A. Nucleotide binding and filament assembly of recombinant yeast septin complexes. Biol Chem. 2005;386:643–656. doi: 10.1515/BC.2005.075. [DOI] [PubMed] [Google Scholar]
- 10.Sheffield PJ, et al. Borg/septin interactions and the assembly of mammalian septin heterodimers, trimers, and filaments. J Biol Chem. 2003;278:3483–3488. doi: 10.1074/jbc.M209701200. [DOI] [PubMed] [Google Scholar]
- 11.Sirajuddin M, et al. Structural insight into filament formation by mammalian septins. Nature. 2007;449:311–315. doi: 10.1038/nature06052. [DOI] [PubMed] [Google Scholar]
- 12.John CM, et al. The Caenorhabditis elegans septin complex is nonpolar. EMBO J. 2007;26:3296–3307. doi: 10.1038/sj.emboj.7601775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bertin A, et al. Saccharomyces cerevisiae septins: Supramolecular organization of heterooligomers and the mechanism of filament assembly. Proc Natl Acad Sci USA. 2008;105:8274–8279. doi: 10.1073/pnas.0803330105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lukoyanova N, Baldwin SA, Trinick J. 3D reconstruction of mammalian septin filaments. J Mol Biol. 2008;376:1–7. doi: 10.1016/j.jmb.2007.11.029. [DOI] [PubMed] [Google Scholar]
- 15.Mitchison TJ, Field CM. Cytoskeleton: What does GTP do for septins? Curr Biol. 2002;12:R788–R790. doi: 10.1016/s0960-9822(02)01295-2. [DOI] [PubMed] [Google Scholar]
- 16.Vrabioiu AM, Gerber SA, Gygi SP, Field CM, Mitchison TJ. The majority of the Saccharomyces cerevisiae septin complexes do not exchange guanine nucleotides. J Biol Chem. 2004;279:3111–3118. doi: 10.1074/jbc.M310941200. [DOI] [PubMed] [Google Scholar]
- 17.Mendoza M, Hyman AA, Glotzer M. GTP binding induces filament assembly of a recombinant septin. Curr Biol. 2002;12:1858–1863. doi: 10.1016/s0960-9822(02)01258-7. [DOI] [PubMed] [Google Scholar]
- 18.Huang Y, Surka MC, Reynaud D, Pace-Asciak C, Trimble WS. GTP binding and hydrolysis kinetics of human septin 2. FEBS J. 2006;273:3248–3260. doi: 10.1111/j.1742-4658.2006.05333.x. [DOI] [PubMed] [Google Scholar]
- 19.Dobbelaere J, Gentry MS, Hallberg RL, Barral Y. Phosphorylation-dependent regulation of septin dynamics during the cell cycle. Dev Cell. 2003;4:345–357. doi: 10.1016/s1534-5807(03)00061-3. [DOI] [PubMed] [Google Scholar]
- 20.Vrabioiu AM, Mitchison TJ. Structural insights into yeast septin organization from polarized fluorescence microscopy. Nature. 2006;443:466–469. doi: 10.1038/nature05109. [DOI] [PubMed] [Google Scholar]
- 21.Ghosh A, Praefcke GJK, Renault L, Wittinghofer A, Herrmann C. How guanylate-binding proteins achieve assembly-stimulated processive cleavage of GTP to GMP. Nature. 2006;440:101–104. doi: 10.1038/nature04510. [DOI] [PubMed] [Google Scholar]
- 22.Scrima A, Wittinghofer A. Dimerisation-dependent GTPase reaction of MnmE: how potassium acts as GTPase-activating element. EMBO J. 2006;25:2940–2951. doi: 10.1038/sj.emboj.7601171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gasper R, Meyer S, Gotthardt K, Sirajuddin M, Wittinghofer A. It takes two to tango: Regulation of G proteins by dimerization. Nat Rev Mol Cell Biol. 2009;10:423–429. doi: 10.1038/nrm2689. [DOI] [PubMed] [Google Scholar]
- 24.Versele M, Thorner J. Some assembly required: Yeast septins provide the instruction manual. Trends Cell Biol. 2005;15:414–424. doi: 10.1016/j.tcb.2005.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Versele M, Thorner J. Septin collar formation in budding yeast requires GTP binding and direct phosphorylation by the PAK, Cla4. J Cell Biol. 2004;164:701–715. doi: 10.1083/jcb.200312070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagaraj S, Rajendran A, Jackson CE, Longtine MS. Role of nucleotide binding in septin–septin interactions and septin localization in Saccharomyces cerevisiae. Mol Cell Biol. 2008;28:5120–5137. doi: 10.1128/MCB.00786-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 28.Leipe DD, Wolf YI, Koonin EV, Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol. 2002;317:41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- 29.Koenig P, et al. The GTPase cycle of the chloroplast import receptors Toc33/Toc34: implications from monomeric and dimeric structures. Structure. 2008;16:585–596. doi: 10.1016/j.str.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 30.Huijbregts RP, Svitin A, Stinnett MW, Renfrow MB, Chesnokov I. Drosophila Orc6 facilitates GTPase activity and filament formation of the septin complex. Mol Biol Cell. 2009;20:270–281. doi: 10.1091/mbc.E08-07-0754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McMurray MA, Thorner J. Reuse, replace, recycle: Specificity in subunit inheritance and assembly of higher-order septin structures during mitotic and meiotic division in budding yeast. Cell Cycle. 2009;8:195–203. doi: 10.4161/cc.8.2.7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagata K, Asano T, Nozawa Y, Inagaki M. Biochemical and cell biological analyses of a mammalian septin complex, Sept7/9b/11. J Biol Chem. 2004;279:55895–55904. doi: 10.1074/jbc.M406153200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.