Abstract
To elucidate the genetic architecture of familial schizophrenia we combine linkage analysis with studies of fine-level chromosomal variation in families recruited from the Afrikaner population in South Africa. We demonstrate that individually rare inherited copy number variants (CNVs) are more frequent in cases with familial schizophrenia as compared to unaffected controls and affect almost exclusively genic regions. Interestingly, we find that while the prevalence of rare structural variants is similar in familial and sporadic cases, the type of variants is markedly different. In addition, using a high-density linkage scan with a panel of nearly 2,000 markers, we identify a region on chromosome 13q34 that shows genome-wide significant linkage to schizophrenia and show that in the families not linked to this locus, there is evidence for linkage to chromosome 1p36. No causative CNVs were identified in either locus. Overall, our results from approaches designed to detect risk variants with relatively low frequency and high penetrance in a well-defined and relatively homogeneous population, provide strong empirical evidence supporting the notion that multiple genetic variants, including individually rare ones, that affect many different genes contribute to the genetic risk of familial schizophrenia. They also highlight differences in the genetic architecture of the familial and sporadic forms of the disease.
Keywords: rare mutations, chromosome 13q34, chromosome 1p36, RAPGEF gene family
Schizophrenia (SCZ) is a chronic, psychiatric disorder that has an estimated worldwide prevalence of ≈1%. The genetic architecture of the disease remains largely unknown. The strongest predictor of SCZ is having an affected first-degree relative (1). In addition to the familial forms, nonfamilial (sporadic) forms of the disease also exist (1, 2). The exact proportion of each form is largely unknown, but it is thought that at least 60% of cases are sporadic (3, 4). Genome-wide linkage scans have been conducted to identify loci harboring rare mutations/variants that increase susceptibility to familial SCZ. Loci have been identified on almost every chromosome, but only a few regions have been replicated across studies. One such region is near the telomere of chromosome 13q (5–12). Across studies the region of peak linkage is broad, from 13q12 to 13q34. This region has also been linked to bipolar disorder (13). In addition to linkage studies, a number of earlier (14) as well as more recent studies (15–18) have provided strong evidence supporting the importance of rare structural mutations/variants in SCZ vulnerability. Rare inherited structural lesions, in particular, are expected to be prominent in familial SCZ, but their full contribution to transmitted liability in familial SCZ cases has not been examined so far in a systematic manner (19).
To understand the genetic architecture of familial SCZ and the pattern of transmission of rare risk alleles in affected families, we combine high-resolution linkage analysis with studies of fine-level chromosomal variation in families recruited from the Afrikaner population in South Africa, a genetically and environmentally homogeneous population who have descended from mostly Dutch immigrants who settled in South Africa beginning in 1652 (20). In addition to the genetic homogeneity, the Afrikaners are valuable for genetic studies because they present a close-knit family structure and offer the potential to perform detailed genealogical analysis, which affords reliable discrimination of familial and nonfamilial forms of the disease and facilitates family-based genetic studies. Here, using approaches designed to detect risk variants with relatively low-frequency and high penetrance, we provide strong empirical evidence supporting the notion that multiple genetic variants, including individually rare ones that affect many different genes, contribute to the genetic risk of familial SCZ.
Results
Patient Cohorts.
We performed a genome-wide survey of rare inherited copy number variants (CNVs) in a total of 182 individuals, consisting of 48 probands with familial SCZ [positive disease history in a first-degree (n = 33) or second-degree (n = 15) relative; Fig. S1] and both of their biological parents, as well as all additional affected relatives that were available for genotyping. Of the 48 probands, 40 are diagnosed as affected in the narrow category and eight in the broad category (see Methods). The familial cases cohort was compared to a control cohort (n = 159 triad families) as well as to a cohort enriched in sporadic cases (n = 152 triad families), defined as cases with negative family history of SCZ in a first- or second-degree relatives, also recruited from the Afrikaner community as previously described (15). In that respect, it should be noted that there were no significant differences in the average number of first- or second-degree relatives among families with and without family history. Specifically, in the 48 families with positive family history of SCZ in first- or second-degree relatives reported here, the average proband sibship was comprised of 3.4, the average maternal sibship of 4.3, and the average paternal sibship of 4.2 individuals. In the cohort enriched in sporadic cases (15), these numbers are 3.3, 4.3, and 4.6, respectively. Negative or positive family history or availability of additional affecteds was not a screening criterion (see Methods).
For our linkage studies, we genotyped 479 subjects from 130 families. Sixty-nine families are informative. In these, 112 individuals are diagnosed as affected in the narrow category, and 128 individuals are classified as broadly affected. In the 54 informative families with at least two affected members, there are 60 and 79 affected relative pairs for the narrow and broad affection categories, respectively (Table S1); this is 43% more affected relative pairs than in our previous, 9-cM, linkage study (12). A subset of the families (67%) used in the CNV studies (n = 32) was also included in the linkage scan. The appropriate Institutional Review Boards and Ethics Committees at University of Pretoria and Columbia University have approved all procedures for this study.
Genome-Wide Survey of Rare Inherited CNVs.
We surveyed single nucleotide polymorphisms (SNPs) and CNVs using the Affymetrix Genome-Wide Human SNP 5.0 arrays and used intensity and genotype data from both SNP and CN probes to identify autosomal deletions and duplications as described previously (15). The estimated rare inherited mutation rate was compared to the collective rate of inherited CNVs among sporadic cases and unaffected individuals from the same population (15). Rare inherited CNVs detected in familial cases and their parents were considered only if they involved at least 10 consecutive probe sets (average resolution of ≈30 kb) and did not show ≥50% overlap with a CNV detected in any parental chromosome (other than those of the biological parents) in the familial, sporadic, or control cohorts (n = 1,432 chromosomes). Using these criteria, we identified 24 rare inherited CNVs in 19 familial cases affecting 52 genes (Tables S2 and S3). The frequency of carriers of rare inherited structural lesions is ≈40% (19 out of 48) in our cohort of familial cases as compared to the ≈20% (32 out of 159) collective rate of inherited CNVs among unaffected individuals from the same population (15) (relative enrichment 1.97, Fisher's Exact Test P = 0.01) (Table 1). Cases and controls carry on average 0.5 (24 CNVs in 48 cases) and 0.2 (32 in 159 controls) rare CNVs per person, respectively, a ≈2-fold difference in rare CNV burden. It should be noted that our population-specific filtering process is preferable to the one based on the diverse set of CNVs present in the database of genomic variants (DGV) (16) because DGV includes samples that have not been screened for psychiatric phenotypes and likely includes several pathogenic variants, and in addition, CNV frequency and disease-penetrance may vary across human populations (21). Nevertheless, essentially identical enrichment (relative enrichment 1.85, 27% vs. 14.5%, Fisher's Exact Test P = 0.05) was obtained upon further filtering that removed rare inherited variants overlapping with DGV (hg18 version 4). Familial clustering offers an important validation measure for all identified CNV regions (22). Nevertheless, we also confirmed, in four out of four tested families, the observed patterns of inheritance using an independent approach, the multiplex ligation-dependent probe amplification (MLPA) assay (23) (see SI Methods and Fig. S2).
Table 1.
Increased frequency of rare inherited genic CNVs in familial SCZ
| Cohort | No. of families | Total rare inherited CN mutation carriers |
Genic inherited CN mutation carriers |
Non-genic inherited CN mutation carriers |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | % |
P |
n | % |
P |
n | % |
P |
|||||
| vs. controls | vs. sporadic | vs. controls | vs. sporadic | vs. controls | vs. sporadic | ||||||||
| Familial | 48 | 19 | 39.6 | 0.012 | NS | 17 | 35.4 | 0.001 | 0.007 | 2 | 4.2 | NS | NS |
| Sporadic | 152 | 46 | 30.3 | 24 | 15.8 | 22 | 14.5 | ||||||
| Controls | 159 | 32 | 20.1 | 21 | 13.2 | 11 | 6.9 | ||||||
Statistical significance was determined using Fisher's exact probability test; NS = non-significant.
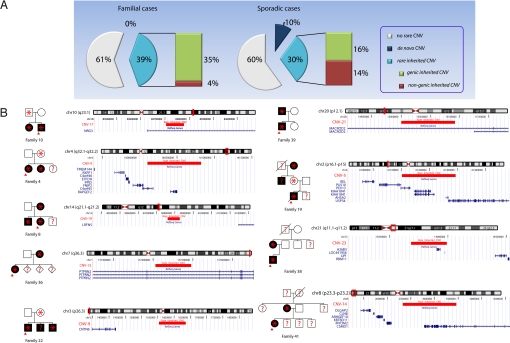
Two lines of evidence suggest that the observed ≈2-fold enrichment of rare inherited CNVs in familial SCZ has a bone fide pathogenic basis. First, the observed enrichment in inherited structural mutations is highly specific, since we did not find any enrichment of de novo structural lesions in the same familial cases cohort (15). Second, as would be expected for pathogenic lesions, the enrichment in inherited CNVs among familial cases is confined exclusively to CNVs overlapping at least one gene, either partly or in its entirety (herein referred to as genic CNVs). Specifically, when we analyzed CNVs separately according to their gene composition, we found that individuals with familial SCZ were ≈2.7 times as likely as controls to harbor rare genic CNVs (relative enrichment 2.7, Fisher's Exact Test P = 10−3, Table 1). Essentially identical enrichment in genic CNVs (relative enrichment 2.6, 25% vs. 9.5%, Fisher's Exact Test P = 0.012) was obtained upon further filtering that removed rare inherited variants overlapping with DGV (hg18 version 4). In contrast, there was no significant difference in the proportion of cases versus controls carrying rare nongenic CNVs (Table 1). Moreover, no such enrichment in inherited genic CNVs was observed in the cohort enriched for sporadic cases (relative enrichment 1.2, P = 0.52), where the contribution of inherited CNVs appears to be relatively minor. In that respect, it is noteworthy that while the overall frequency of carriers of all rare structural variants is the same between the familial and sporadic cases (15) (≈40%), the type of variants is markedly different (Fig. 1A). Sporadic SCZ is characterized by a marked enrichment of rare de novo mutations and only a modest increase in the rate of rare inherited CNVs, which do not appear to preferentially affect genes. By contrast, familial SCZ is characterized by enrichment in rare inherited genic CNVs (predicted to have higher penetrance), while de novo mutations are less prominent.
Fig. 1.
Inherited CNVs in families with SCZ. (A) Frequency distribution of rare CNVs identified in familial and sporadic cases of SCZ at the resolution afforded by the Affymetrix Genome-Wide Human SNP 5.0 arrays. There is a ≈20% basal rate of inherited CNVs in unaffected controls, while the overall frequency of carriers of all rare CNVs is the same between familial and sporadic cases (≈40%). (B) Rare inherited CNVs showing co-segregation with the clinical diagnosis in the respective affected families. For each CNV, the structure of the affected family, as well as the genomic position of the CNV, are indicated. Affected individuals are marked in black. Probands are indicated by red arrows. Individuals who carry rare CNVs are indicated by an asterisk. Individuals where no genotype information is available are indicated by question marks.
None of the rare CNVs found in familial cases are present in control chromosomes. To identify which of these CNVs are most likely to be pathogenic, we investigated the relationship between rare inherited CNVs and disease status within each family. For 12 of the 19 CNV carriers, DNA samples and genotypic information were available for at least one more affected relative. In nine of these 12 families, the CNV segregated to all genotyped affected members (only two of the 23 affected members of these nine families were not available for genotyping). Thus, CNVs in these nine families showed clear co-segregation with the clinical diagnosis, in a manner consistent with incomplete penetrance models (i.e., present also in some unaffected family members) (Fig. 1B). The remaining three CNVs segregated only to one of the genotyped affected members in each family. Given the ≈20% basal rate of inherited CNVs in unaffected controls (15), these are likely to be neutral variants. Alternatively, these cases may be indicative of more complex modes of inheritance, such as bilineal transmission of risk alleles. To exclude that the observed pattern of co-inheritance of CNVs and diagnosis in the informative families is due to chance alone, we conducted a simulation study where we disrupted the relationship between CNVs and diagnosis by permuting the diagnosis, while keeping constant the pedigree structure, the inherited pattern of CNVs and the number of affected individuals in each family. Co-inheritance of CNVs and diagnosis in nine out of 12 families was observed only once in 10,000 permutation runs (empirical P value = 0.0001) and, therefore, it is unlikely to be due to chance.
The nine CNVs segregating to all genotyped affected members within the respective families, alter 12 known genes: PEX13, KIAA1841, AHSA2, USP34, C4orf45, RAPGEF2, PTPRN2, CSMD1, NRG3, MACROD2, A26B3, and LOC441956, all of which are candidates for follow-up studies. Convergence with previous studies highlights at least two of these genes as particularly worthy of follow-up. Specifically, the neuregulin-3 (NRG3) gene is implicated by a 73.6-kb-long duplication in the first intron, which may cause regulatory deficits. An overlapping CNV has been reported previously in one case with SCZ (24) (Table S2). Three SNPs in the same intron were recently associated with SCZ-related quantitative traits (24). The homologous gene, NRG1, is also a well-known candidate gene (25). In addition, the RAPGEF2 gene, encoding for a GTP exchange factor, is implicated by a 716.4-kb duplication that encompasses this gene. Mutation of the RAPGEF2 ortholog in mice affects the formation of the cerebral cortex, reduces the threshold for the induction of epileptic seizures and results in commissural fiber defects (26). Previously, we have identified in a patient with SCZ a de novo exonic microdeletion affecting another member of the RAPGEF family (RAPGEF6) located within a SCZ susceptibility locus at chromosome 5q (15). Mutations in another member of this family (RAPGEF4) have been described in autism (27).
We examined the familial cases for differences in clinical and phenotypic variables (history of developmental delays or learning disabilities, mental retardation, age at onset, and disease severity) that may discriminate inherited CN mutation carriers. We focused our analysis on probands from families where CNVs show apparent co-segregation with the illness and are therefore more likely to be pathogenic. Among them, the male to female ratio is identical to that of the entire familial cases cohort, and there is no statistically significant evidence for parental origin effects. None of these probands had a history of developmental delays or learning disabilities, or presence of mental retardation. In addition, there was no difference in the age at disease onset between these probands and noncarriers of rare CNVs, but we found some suggestive differences in indices of disease severity, including co-morbid substance abuse (67% versus 23%, P = 0.03, uncorrected), duration of illness (141.6 months versus 92.2 months, P = 0.19), and number of hospitalizations (4.9 compared to 2.9, P = 0.13), indicating a more debilitating or treatment-resistant form of illness among CNV carriers.
High-Density Linkage Analysis.
Previously, a low-density (9-cM) linkage scan in our Afrikaner sample identified suggestive evidence for linkage at 13q34 and 1p36 (maximum nonparametric LOD scores 2.99 and 2.23, respectively) (12). Here we genotyped 2005 di-, tri-, and tetra-nucleotide repeat microsatellite markers, in one of a few linkage scans of psychiatric disorders to attain this level of genome-wide coverage. The average inter-marker distance was 1.9 cM (±1.4), and the average heterozygosity for the autosomal markers was 0.71 (±0.12) resulting in an information content of 0.84 (±0.046) (Figs. S3, S4). We conducted both parametric and nonparametric analyses. For our parametric analyses, we used the algorithm implemented in LAMP to estimate the disease allele frequency and genotype penetrances by maximum likelihood. We used the optimized parameters to calculate model maximized LOD (MOD) scores, which are more powerful than LOD scores when the disease parameters are unknown (28).
Singlepoint Parametric Results.
We found three markers with a MOD score of at least 3.0 in either affection category: D13S285 on 13q34, D9S50 on 9p13, and D21S270 on 21q22. The highest MOD scores for both affection statuses are for D13S285, located at 127 cM. For the narrow affection status the MOD score is 3.30 and for the broad classification is 3.67. D9S50 at 60 cM has a MOD score of 3.56 for the broad classification and 2.51 for the narrow. For D21S270 at 46 cM, the MOD score for the narrow classification is 3.0 and for the broad is 1.88 (Table S4).
Multipoint Parametric Results.
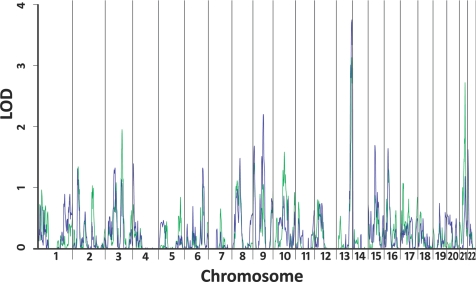
For our multipoint parametric analysis (Fig. 2), the maximum MOD scores for both affection classifications are also on 13q34. The maximum MOD score for the narrow affection status is 3.13 at 126 cM. When we repeated the linkage analyses on 1,000 simulated data sets, we calculated an empirical P value of 0.093. For the broad affection status, the highest MOD score is 3.76 at 131 cM, near D13S293, and the 1-MOD region spans from 115 cM to the q terminus. The empirical P value for the broad affection status alone is 0.025. When we include both the narrow and broad classifications in our simulations, 42 of the data sets resulted in a MOD score greater than or equal to 3.76. This empirical genome-wide P value of 0.042 meets the criteria for a significant linkage result (29). For the broad phenotype, the maximum likelihood estimate for the disease penetrance for an individual with two copies of the disease allele is 1.0, for an individual with one copy is 0.073, and for an individual with no copies is 0.005. Although the disease allele frequency is estimated to be fairly rare (fd = 0.030), the relative risk is very high (RR = 13.77). In addition to the 13q locus, we identified linkage peaks on chromosomes 21q22 at 46 cM, near D21S1900, with a 1-MOD interval from 41 to 48 cM and on 9q21 at 85 cM, near D9S1877, with a 1-MOD interval from 80 to 96 cM (Table 2).
Fig. 2.
MOD score analysis. Green line shows MOD scores for the narrow classification. Blue line shows MOD scores for the broad classification.
Table 2.
Parametric multipoint MOD scores > 1.5 for either SCZ status (multiplicative model)
| Chr | Position (1-MOD) | Nearest Marker | Aff | MOD | Freq of A | RR | Pen of AA | Pen of Aa | Pen of aa |
|---|---|---|---|---|---|---|---|---|---|
| 3q21 | 137 cM (128–147) | D3S1589 | N | 1.95 | 0.39 | 99.84 | 0.065 | 0.00065 | 6.5 × 10−6 |
| B | 1.13 | 0.53 | 99.97 | 0.035 | 0.00035 | 3.5 × 10−6 | |||
| 9p24 | 12 cM (3–19) | D9S1686 | N | 1.40 | 0.01 | 9.86 | 0.77 | 0.078 | 0.0079 |
| B | 1.68 | 0.01 | 11.03 | >.99 | 0.091 | 0.0082 | |||
| 9q21 | 85 cM (80–96) | D9S1877 | N | 1.05 | <.01 | 10.44 | >.99 | 0.096 | 0.0092 |
| B | 2.20 | <.01 | 10.72 | >.99 | 0.093 | 0.0087 | |||
| 10q22 | 92 cM (77–112) | D10S537 | N | 1.58 | 0.01 | 11.32 | >.99 | 0.088 | 0.0078 |
| B | 0.71 | 0.17 | 4.04 | 0.072 | 0.018 | 0.0044 | |||
| 13q34 | 131 cM (115-qter) | D13S293 | N | 3.13 | 0.03 | 14.76 | >.99 | 0.068 | 0.0046 |
| B | 3.76 | 0.03 | 13.77 | >.99 | 0.073 | 0.0053 | |||
| 15q21 | 56 cM (54–65) | D15S1022 | N | 0.87 | <.01 | 10.35 | >.99 | 0.097 | 0.0094 |
| B | 1.69 | <.01 | 10.39 | >.99 | 0.096 | 0.0093 | |||
| 16p13 | 31 cM (27–45) | D16S3047 | N | 1.31 | 0.17 | 5.60 | 0.10 | 0.018 | 0.0032 |
| B | 1.64 | 0.10 | 5.64 | 0.14 | 0.026 | 0.0046 | |||
| 21q22 | 46 cM (41–48) | D21S1900 | N | 2.72 | 0.36 | 99.95 | 0.073 | 0.00073 | 7.4 × 10−6 |
| B | 1.18 | 0.52 | 99.86 | 0.037 | 0.00037 | 3.7 × 10−6 | |||
| 22q11 | 3 cM (2–9) | D22S420 | N | 1.47 | 0.44 | 99.99 | 0.049 | 0.00050 | 5.0 × 10−6 |
| B | 1.62 | 0.45 | 99.99 | 0.048 | 0.00048 | 4.8 × 10−6 |
‘A’ is the disease allele, ‘a’ is the non-disease allele. 1-MOD, region in which the MOD score is within 1 MOD score of the highest MOD score; Aff, affection classification; N, narrowly affected; B, broadly affected; Freq, allele frequency; RR, relative risk based on a 1% prevalence of SCZ; Pen, penetrance of SCZ for the given genotype. Therefore, Pen of AA is the probability of having SCZ, given two copies of the disease allele.
Singlepoint Nonparametric Results.
The highest LOD score is for marker D1S2885 at 46 cM on 1p36. The LOD score for the narrow classification is 2.30, and is the genome-wide maximum at 2.87 for the broad classification. D13S285, at 127 cM on 13q34, has elevated LOD scores for both the narrow and broad classifications; the LOD scores are 2.80 and 2.70, respectively. Also noteworthy are three adjacent markers on 21q22 with LOD scores greater than 2.0 using the narrow category: D21S1900 (LOD = 2.30), D21S1919 (LOD = 2.55), and D21S270 (LOD = 2.31). For the broad category these three markers have LOD scores ranging from 1.20 to 1.53 (Table S5).
Multipoint Nonparametric Results.
The maximum multipoint LOD score is located about 2 cM away from D13S285 at 125 cM on 13q34. At this location, the LOD score for the narrow classification is 2.65; for the broad classification the LOD score is slightly higher at 2.66. The 1-LOD interval around this peak extends from 119 cM to the q terminus. However, the empirical significance based on 1,000 simulations is not significant (P = 0.25). The nonparametric multipoint analysis also provides evidence for linkage at 21q22. The LOD scores are 2.16 for the narrow and 0.83 for the broad classification (Table S6). This is the region where three markers showed evidence for linkage in the singlepoint analysis.
Targeted Analyses in Families Nonlinked to Chromosome 13.
Twenty-five of our families in our sample exhibited a MOD score of <0.0 at the location of our strongest linkage peak, at 131 cM on chromosome 13q34 near marker D13S293. To examine evidence for additional susceptibility loci, we carried out a series of analysis targeted at these 25 families (Table 3). Notably, the maximum MOD score in these 25 families occurs on chromosome 1p36 at 35 cM (≈1 cM from D1S2644), near the peak that was identified in our previous 9-cM scan. The MOD score for the narrow classification at that position is 3.21 (empirical P value = 0.15); the MOD score for the broad classification is 1.74. The nonparametric LOD scores in the region (both at ≈29 cM, near D1S2697) were only 1.32 and 0.54 for the narrow and broad classifications, respectively. The MOD score analysis results suggest a dominant mode of inheritance, with the estimated disease allele frequency of 0.003 and penetrances/genotype relative risks of 1.0, 0.93, and 0.004.
Table 3.
Parametric linkage to chromosome 1p36 (dominant model)
| Family group | Number of families | Position | Nearest Marker | Narrow MOD | Broad MOD |
|---|---|---|---|---|---|
| All* | 69 | 35 cM | D1S2826 | 1.28 | 0.20 |
| Unlinked to 13q | 25 | 35 cM | D1S2826 | 3.21 | 1.74 |
| Linked to 13q | 43 | 35 cM | D1S2826 | 0.12 | 0.00 |
*One family had a MOD score of 0.000 and was not used in the subset analysis.
Exploration of the Relation Between Linkage Signals and CNVs.
We first assessed the contribution of copy number mutations to the two primary and consistent among studies linkage signals at 13q34 and 1p36. Whole genome CNV annotation, conducted using dCHIP program, identified 788 putative CNVs in the 241 cases included in our scan. In the 13q34 region, there was only one CNV identified, which did not overlap with any gene (Table S7). This CNV was found in two out of 224 cases (0.89%) and three out of 361 parents of unaffected controls (0.83%). In the 1p36 region, there were seven CNVs including six genomic gains and one loss (Table S7). There was no statistical difference in the frequencies of these CNVs between cases and controls. None of the identified CNVs was present in families linked to the respective linkage loci. Thus, at the level of resolution of our scan, we could not identify any CNV that accounts for the linkage signals identified.
Because only a subset of the families used in the linkage studies (n = 32) were also included in the CNV scan and additional, yet unidentified rare CNV carrying families are likely to be part of the linkage cohort, it is not possible at this point to conclusively evaluate the effect that removal of families, which carry at least one rare CNV, has on linkage. Nevertheless, preliminary analysis shows that, despite the decrease in the sample size, after removing CNV carrying families (n = 14, eight of them with CNVs co-segregating with the clinical diagnosis), the evidence for linkage in 13q34 remained unchanged (MOD = 3.77). To estimate the expected change, under the assumption that the families removed from the analysis are contributing to the 13q34 signal, we randomly removed 14 families from among the ones showing linkage to 13q34. In 100 trials, the MOD scores varied from 0.53 to 2.76 for the narrow definition, with an average of 1.50, and from 0.63 to 2.99 for the broad definition, with an average of 1.74. This notable reduction in the MOD score suggests that at least compared to the families linked to 13q34, the rare CNV containing families may represent a largely distinct subset of genetic liability. Interestingly, our exploratory analysis shows that in a number of loci, weak linkage signals are amplified following removal of rare CNV families despite decrease in power (for example, at 3q21 MOD increases from 1.95 to 2.3 for the narrow classification, at 9p24 MOD increases from 1.68 to 2.13 for the broad classification, and at 16p13 MOD increases from 1.64 to 2.3 for the broad classification). These may reflect true linkage signals, which are masked by the heterogeneity introduced by the rare CNV families.
Discussion
Our results offer a comprehensive picture of the genetic architecture of familial SCZ in a relatively homogeneous population. Our chromosomal variation analysis provides evidence for a role of inherited structural lesions in familial SCZ and highlights some important differences in the genetic architecture of familial and nonfamilial forms of the disease. The majority of the identified inherited CNVs, co-segregate with disease in a manner consistent with necessary but not always sufficient genetic “hits,” which lead to a disease state only in combination with additional, inherited structural or sequence variation or environmental factors. Patient stratification suggests that inherited CNVs do not correlate with history of developmental delays, learning disabilities, or presence of mental retardation, but appear to be enriched in more debilitating or treatment-resistant form of illness. Although our study is statistically underpowered to prove the involvement of any specific CNV, analysis of co-segregation with the clinical diagnosis, as well as convergence with previous studies, highlights a number of genes, gene families, and related pathways (such as the contactin family, the CSMD1, ADARB1, RXFP2, LRFN5, and NRG3 genes) as particularly worthy of follow-up (see SI Text). In particular, we provide additional evidence strongly supporting a previously unknown role of a family of Rap1 guanine nucleotide exchange factors (RAPGEF family) and Rap1-mediated processes (30) in psychiatric disorders.
Our linkage analyses indicated one or more genes that increases susceptibility to SCZ on chromosome 13q34 in our broadly affected individuals. It is noteworthy that three of the four other SCZ linkage scans that identified a LOD score greater than 2.0 at 13q34 included schizoaffective disorder, bipolar type in the affection category (7–10). That our broad classification also includes this diagnosis supports the hypothesis that one or more genes on 13q34 increases susceptibility to SCZ and bipolar spectrum disorders. Based on the results of this study and previous ones, there is considerable evidence for linkage to 13q. When we analyzed families that do not show evidence for linkage to 13q34, we identified another linkage peak on 1p36. This result was much stronger when we used the narrow definition of SCZ that did not include schizoaffective disorder, bipolar subtype. Compared to the 13q34 locus, this could indicate different causal alleles and mechanisms in subjects with and without symptoms of bipolar disorder. Notably, at the level of resolution of our scan, our analysis indicates that CNVs within both linkage signal regions are likely to be neutral and unlikely to account for the linkage signals identified. Finally, although the sample sizes are relatively small, our findings suggest that compared to the families linked to 13q34, the rare CNV carrying families represent a largely distinct subset of genetic liability. CNV carrying families may be a source of heterogeneity and future studies will focus on identifying all rare CNVs as putative risk loci and as a tool to stratify the samples to reduce genetic heterogeneity for linkage analyses and improve detection of weak signals.
Irrespective of the pathogenic potential and the precise mode of action of each risk locus, our results highlight the difference in the genetic architecture of the familial and sporadic forms of the disease and support the notion that multiple genetic variants, including individually rare ones (often unique to a single patient) that affect many different genes contribute to the genetic risk of familial SCZ. This heterogeneity (present to some degree even in founder populations) is consistent with the hypothesis that there are many genes that contribute to SCZ and may account for past and present difficulties in finding bone fide genetic variants. Because there are significant clinical similarities of SCZ cases diagnosed in Afrikaners and those diagnosed in more heterogeneous populations (such as the U.S.) (20), our results are likely to have general implications regarding the genetic architecture of SCZ.
Samples and Methods
Cohorts.
Both affected and control families were recruited and diagnosed as part of our ongoing, large-scale genetic study of SCZ in the Afrikaner population in South Africa, as previously described (12, 15, 20) (see also SI Methods). For our linkage study, affected subjects were classified as either narrowly or broadly affected. The narrow diagnosis includes subjects with SCZ or schizoaffective disorder-depressive type, as previously described (12). The broad diagnosis includes all individuals classified as affected under the narrow definition as well as individuals with schizoaffective disorder-bipolar type. Compared to our previous classification (12), it is more encompassing than LCI, but not as broad as LCII. For our CNV studies, the criteria for inclusion in the affected cohort are: (i) Afrikaner heritage; (ii) proband meeting full diagnostic criteria for SCZ or schizoaffective disorder; and (iii) both biological parents alive and willing to participate. It should be noted that presence of negative or positive family history or availability of additional affected relatives is not a screening criterion. Nevertheless, for all recruited subjects, detailed information about family history of any psychiatric or medical illness was solicited from at least three sources (proband and each participating parent) by two independent raters [the nursing sister, who completes the Medical and Personal History form with each study participant and also draws a detailed pedigree for at least three to four generations for each family, as well as the psychiatrist who administers the Diagnostic Instrument for Genetic Studies (DIGS) to the proband]. In addition, since we also trace the ancestry of all recruited families, we routinely use several informants in each family to inquire about all relatives' names, date and place of birth, and death and psychiatric status. Because of the close-knit family structure of the Afrikaner families and the availability of detailed psychiatric records over several generations due to the large catchment area and long-term care provided by the local recruiting hospital, we are typically able to obtain information about psychiatric status for at least three to four generations removed from the proband. For any relative identified as possibly having symptoms of SCZ or schizoaffective disorder, or a history of treatment or hospitalization for a psychiatric condition, every effort was made to include that relative in the study, if alive and willing to participate, by obtaining a blood sample and administering an in-person diagnostic interview. In a few instances where the exact nature of a reported psychiatric diagnosis in a first- or second-degree relative could not be substantiated (i.e., because the person was not alive or access to records was not possible), the family history status was left unknown. Such families are not considered in the present study or in the Xu et al. study (15). Finally, in addition to being matched by ancestry, a subset of the control families (three informants per family inquiring for up to three generations) completed a detailed self-report questionnaire that inquired about several psychiatric conditions, including psychosis, phobias, anxiety, and depression (see also SI Methods).
Genotyping procedures, linkage analysis and CNV identification, and verification are outlined in detail in the SI Methods.
Supplementary Material
Acknowledgments.
We thank Alexandra Abrams-Downey and Yan Sun for expert technical assistance. This work was supported in part by National Institute of Mental Health Grant MH061399 (to M.K.) and MH077235 (to J.A.G.) and the Lieber Center for Schizophrenia (SCZ) Research at Columbia University Medical Center (CUMC).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908584106/DCSupplemental.
References
- 1.Gottesman II. Schizophrenia Genesis. New York, NY: W.H. Freeman and Company; 1991. [Google Scholar]
- 2.Griffiths TD, et al. Minor physical anomalies in familial and sporadic schizophrenia: The Maudsley family study. J Neurol Neurosurg Psychiatry. 1998;64:56–60. doi: 10.1136/jnnp.64.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kendler KS, Diehl SR. The genetics of schizophrenia: A current, genetic-epidemiologic perspective. Schizophr Bull. 1993;19:261–285. doi: 10.1093/schbul/19.2.261. [DOI] [PubMed] [Google Scholar]
- 4.Gottesman II, Erlenmeyer-Kimling L. Family and twin strategies as a head start in defining prodromes and endophenotypes for hypothetical early-interventions in schizophrenia. Schizophr Res. 2001;51:93–102. doi: 10.1016/s0920-9964(01)00245-6. [DOI] [PubMed] [Google Scholar]
- 5.Lin MW, et al. Suggestive evidence for linkage of schizophrenia to markers on chromosome 13q14.1-q32. Psychiatr Genet. 1995;5:117–126. doi: 10.1097/00041444-199505030-00004. [DOI] [PubMed] [Google Scholar]
- 6.Lin MW, et al. Suggestive evidence for linkage of schizophrenia to markers on chromosome 13 in Caucasian but not Oriental populations. Hum Genet. 1997;99:417–420. doi: 10.1007/s004390050382. [DOI] [PubMed] [Google Scholar]
- 7.Blouin JL, et al. Schizophrenia susceptibility loci on chromosomes 13q32 and 8p21. Nat Genet. 1998;20:70–73. doi: 10.1038/1734. [DOI] [PubMed] [Google Scholar]
- 8.Shaw SH, et al. A genome-wide search for schizophrenia susceptibility genes. Am J Med Genet. 1998;81:364–376. doi: 10.1002/(sici)1096-8628(19980907)81:5<364::aid-ajmg4>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 9.Brzustowicz LM, et al. Linkage of familial schizophrenia to chromosome 13q32. Am J Hum Genet. 1999;65:1096–1103. doi: 10.1086/302579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Camp NJ, et al. Genomewide multipoint linkage analysis of seven extended Palauan pedigrees with schizophrenia, by a Markov-chain Monte Carlo method. Am J Hum Genet. 2001;69:1278–1289. doi: 10.1086/324590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faraone SV, et al. Linkage of chromosome 13q32 to schizophrenia in a large veterans affairs cooperative study sample. Am J Med Genet. 2002;114:598–604. doi: 10.1002/ajmg.10601. [DOI] [PubMed] [Google Scholar]
- 12.Abecasis GR, et al. Genomewide scan in families with schizophrenia from the founder population of Afrikaners reveals evidence for linkage and uniparental disomy on chromosome 1. Am J Hum Genet. 2004;74:403–417. doi: 10.1086/381713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Badner JA, Gershon ES. Meta-analysis of whole-genome linkage scans of bipolar disorder and schizophrenia. Mol Psychiatry. 2002;7:405–411. doi: 10.1038/sj.mp.4001012. [DOI] [PubMed] [Google Scholar]
- 14.Karayiorgou M, et al. Schizophrenia susceptibility associated with interstitial deletions of chromosome 22q11. Proc Natl Acad Sci USA. 1995;92:7612–7616. doi: 10.1073/pnas.92.17.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu B, et al. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat Genet. 2008;40:880–885. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- 16.Walsh T, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 17.Stefansson H, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maher BS, Riley BP, Kendler KS. Psychiatric genetics gets a boost. Nat Genet. 2008;40:1042–1044. doi: 10.1038/ng0908-1042. [DOI] [PubMed] [Google Scholar]
- 20.Karayiorgou M, et al. Phenotypic characterization and genealogical tracing in an Afrikaner schizophrenia database. Am J Med Genet B. 2004;124:20–28. doi: 10.1002/ajmg.b.20090. [DOI] [PubMed] [Google Scholar]
- 21.Jakobsson M, et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- 22.Ionita-Laza I, Laird NM, Raby BA, Weiss ST, Lange C. On the frequency of copy number variants. Bioinformatics. 2008;24:2350–2355. doi: 10.1093/bioinformatics/btn421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vorstman JA, et al. MLPA: A rapid, reliable, and sensitive method for detection and analysis of abnormalities of 22q. Hum Mutat. 2006;27:814–821. doi: 10.1002/humu.20330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen PL, et al. Fine mapping on chromosome 10q22–q23 implicates Neuregulin 3 in schizophrenia. Am J Hum Genet. 2009;84:21–34. doi: 10.1016/j.ajhg.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stefansson H, et al. Neuregulin 1 and susceptibility to schizophrenia. Am J Hum Genet. 2002;71:877–892. doi: 10.1086/342734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bilasy SE, et al. Dorsal telencephalon-specific RA-GEF-1 knockout mice develop heterotopic cortical mass and commissural fiber defect. Eur J Neurosci. 2009;29:1994–2008. doi: 10.1111/j.1460-9568.2009.06754.x. [DOI] [PubMed] [Google Scholar]
- 27.Bacchelli E, et al. Screening of nine candidate genes for autism on chromosome 2q reveals rare nonsynonymous variants in the cAMP-GEFII gene. Mol Psychiatry. 2003;8:916–924. doi: 10.1038/sj.mp.4001340. [DOI] [PubMed] [Google Scholar]
- 28.Greenberg DA, Abreu P, Hodge SE. The power to detect linkage in complex disease by means of simple LOD-score analyses. Am J Hum Genet. 1998;63:870–879. doi: 10.1086/301997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lander E, Kruglyak L. Genetic dissection of complex traits: Guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 30.Kawasaki H, et al. A family of cAMP-binding proteins that directly activate Rap1. Science. 1998;282:2275–2279. doi: 10.1126/science.282.5397.2275. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.