Abstract
The unfolded protein response (UPR) is a stress response pathway that is driven by the increased load of unfolded proteins in the endoplasmic reticulum of highly secretory cells such as plasma cells (PCs). X box binding protein 1 (XBP1) is a transcription factor that mediates one branch of the UPR and is crucial for the development of antibody-secreting PCs. PCs represent only one class of terminally differentiated B cells, however, and little is known about the role for XBP1 in the other class: memory B cells. We have developed an XBP1fl/fl CD19+/cre conditional knockout (XBP1CD19) mouse to build upon our current understanding of the function of XBP1 in PC differentiation as well as to explore the role of XBP1 in memory cell development. Using this model, we show that XBP1CD19 mice are protected from disease in an autoantibody-mediated mouse lupus model. We also identify a novel developmental stage at which B cells express the traditional PC marker CD138 (syndecan-1) but have yet to undergo XBP1-dependent functional and morphological differentiation into antibody-secreting cells. Finally, we show that memory B cells develop normally in XBP1CD19 mice, demonstrating that XBP1-mediated functions occur independently of any memory cell lineage commitment.
Proper protein folding is essential for normal immunoglobulin synthesis and secretion in antibody-secreting plasma cells (PCs). The mammalian unfolded protein response (UPR) is a signaling pathway that responds to ER stress that is induced by the accumulation of unfolded proteins within the ER lumen (Todd et al., 2008). Inositol-requiring transmembrane kinase/endonuclease 1 (IRE1) is an ER-localized transmembrane protein that senses unfolded proteins and serves to activate X-box binding protein 1 (XBP1), a member of the CREB/ATF basic leucine zipper family of transcription factors and a crucial mediator of one branch of the UPR (Yoshida et al., 2001; Calfon et al., 2002; Lee et al., 2002). IRE1 possesses endoribonuclease activity that excises a 26-nt sequence from XBP1 messenger RNA (mRNA). This event, termed XBP1 splicing, shifts the reading frame to excise a premature stop codon, resulting in a full-length functional XBP1 protein product (Yoshida et al., 2001; Calfon et al., 2002; Lee et al., 2002). XBP1 then translocates to the nucleus where it induces target genes involved in protein synthesis and secretion (Shaffer et al., 2004; Lee et al., 2005).
XBP1 activation is crucial to the normal function and survival of highly secretory cells such as exocrine gland acinar cells and Paneth cells (Kaser et al., 2008). Using XBP1/RAG2−/− lymphoid chimeric mice, we have shown previously that XBP1 expression is required for normal PC development and function as well (Reimold et al., 2001). Chimeric mice demonstrated markedly reduced serum Ig levels and impaired Ig response to immunization. This defect was the result of a failure of cells to up-regulate UPR-related XBP1 target genes during terminal B cell differentiation (Iwakoshi et al., 2003; Shaffer et al., 2004). The requirement of XBP1 for PC differentiation raises the possibility that disruption of XBP1 activity may represent a potential therapeutic target for the treatment of autoimmune diseases, such as systemic lupus erythematosus (SLE), in which autoantibodies may be directly pathogenic.
Little is known about the role of XBP1 in the development and function of another important B cell population: memory B cells. These B cells have been exposed to antigen, undergo a germinal center (GC) reaction, and function to differentiate rapidly into antibody-secreting B cells after reexposure to cognate antigen (for review see McHeyzer-Williams and McHeyzer-Williams, 2005). They do not express CD138 (syndecan-1), a cell surface glycoprotein which has traditionally been used a marker for Ig-secreting PCs (Sanderson and Børset, 2002). Memory B cells also function to replete the pool of long-lived antibody-secreting PCs (for review see McHeyzer-Williams and McHeyzer-Williams, 2005), which may represent a persistent source of autoantibodies (Neubert et al., 2008).
The recent development of a XBP1flox conditional KO (cKO) mouse (Hetz et al., 2008) has prompted us to explore further the function of XBP1 in terminal B cell development. CD19 expression is limited to B cells and occurs in pro–B cells and throughout the remaining stages of B cell development (Zhou et al., 1991; Hardy and Hayakawa, 2001). We therefore bred the XBP1flox cKO mouse to a mouse expressing cre recombinase under the control of CD19 promoter to delete XBP1 selectively from B cells (Rickert et al., 1997). In the current series of experiments, we use XBP1flox/flox CD19cre/+ (XBP1CD19) cKO mice first to confirm the functional PC defects in XBP1/RAG2−/− lymphoid chimeric mice (Reimold et al., 2001). In addition, we now report that XBP1CD19 mice were protected from mouse lupus. We also show, unexpectedly, that the number of cells with the traditional B220int CD138+ PC phenotype is normal in XBP1CD19 mice as compared with XBP1flox/flox CD19+/+ (XBP1+/+) controls. Finally, we report that XBP1CD19 mice have normal populations of memory B cells, demonstrating that this alternative differentiation pathway for B cells is not dependent on the XBP1 branch of the UPR.
RESULTS AND DISCUSSION
Initial characterization of XBP1CD19 cKO mice
To determine the efficiency of XBP1 deletion in the B cells in XBP1CD19 mice, splenocytes from these mice were isolated and separated into B220+ and B220− populations using magnetic beads coupled to an anti-B220 mAb. Southern blotting demonstrated efficient and specific deletion of XBP1 in the B220+ (B cell) fraction (Fig. S1 A). There were no differences in cell surface expression of IgM and IgD between XBP1CD19 and XBP1+/+ splenocytes (Fig. S1 B). In accord with the known function of XBP1 in PC development, basal Ig levels were markedly reduced in XBP1CD19 mice (Fig. S1 C). Furthermore, these mice exhibited impaired Ig responses to the T cell–dependent antigen nitrophenol (NP)-KLH upon primary or secondary stimulation (Fig. S2, A and B, respectively). A similar defect was observed in mice immunized with the T cell–independent antigen NP-ficoll (Fig. S2 C).
XBP1 up-regulates the expression of UPR-related target genes that function collectively in protein synthesis and secretion. XBP1 targets include the ER-associated degradation protein EDEM, ER-localized chaperones like ER-localized Dnajb9 (ERdj4), the ER translocon component Sec61α, and proteins, such as protein disulfide isomerase (PDI), that aid disulfide bond formation (Todd et al., 2008). Using B220+ splenocytes isolated from XBP1CD19 and XBP1+/+ mice, we next measured the expression of XBP1 target genes in these B cells after culture for 0–3 d in the presence or absence of 10 µg/ml LPS (Fig. S3 A). As expected, quantitative PCR analysis demonstrated that, by day 3 of culture, there were significant increases in the expression of the aforementioned XBP1 target genes in XBP1+/+ B cells. These same target genes were up-regulated to a lesser degree or not at all in XBP1CD19 B cells. In addition, XBP1CD19 B cells did not demonstrate an appropriate up-regulation of μS, which normally allows for newly synthesized IgM to be secreted. Importantly, B cells from XBP1CD19 and XBP1+/+ mice showed no difference in their modest increase in expression of Ig heavy chain binding protein (BiP), another UPR related gene which is not directly downstream of XBP1.
XBP1 deletion did not affect B cell proliferation or viability in response to LPS treatment. Both were comparable between XBP1CD19 and XBP1+/+ B cells as measured by exclusion of 7-amino-actinomycin D (7-AAD) and annexin V (Fig. S3 B) or by exclusion of trypan blue (Fig. S3 C). In contrast, XBP1 deletion led to markedly reduced secretion of IgM by LPS-stimulated XBP1CD19 B cells (Fig. S3 D). Collectively, these data demonstrate that XBP1 deletion leads to defects in the UPR that impair normal synthesis and secretion of immunoglobulin without affecting B cell viability or proliferative properties. Importantly, these initial experiments in XBP1CD19 mice recapitulated the known roles of XBP1 in PC development and function, as has been shown previously using XBP1/RAG2−/− lymphoid chimeric mice (Reimold et al., 2001).
We next investigated the impact of XBP1 deficiency on the broader network of transcriptional regulators that direct PC differentiation (for review see Calame et al., 2003). The transcription factors Bcl-6 (B cell lymphoma 6) and paired box gene 5 (Pax5) repress B lymphocyte–induced maturation protein 1 (Blimp1) and IFN regulatory factor 4 (IRF4), respectively, and promote B cell proliferation and the GC reaction. On the contrary, Blimp1 and IRF4 normally arrest cell cycle progression and the GC reaction to promote and sustain PC development (Calame et al., 2003). Furthermore, Blimp1 and IRF4 permit the expression of XBP1 (Shaffer et al., 2004; Klein et al., 2006). For Blimp1, this regulatory event is accomplished by Blimp1-mediated repression of Pax5, which derepresses XBP1 to ensure an appropriate UPR in anticipation of Ig production and secretion (Shaffer et al., 2004). Despite the failure to differentiate into Ig-secreting PCs, LPS-stimulated XBP1CD19 B cells displayed a typical PC signature of these transcription factors: down-regulation of Pax5 and up-regulation of Blimp1 and IRF4 mRNAs. Interestingly, Blimp1 and IRF4 were substantially overexpressed in LPS-stimulated XBP1CD19 B cells compared with XBP1+/+ controls (Fig. S3 A). These data suggest that XBP1 deletion did not alter the genetic program of PC development. Furthermore, failure to up-regulate Ig secretion in the absence of XBP1 appears to promote the PC developmental program, supporting the hypothesis that a mechanism of feedback inhibition exists whereby XBP1 down-regulates Blimp1 and IRF4 expression in LPS-stimulated B cells (Hu et al., 2009).
Mice with XBP1-deficient B cells are protected from mouse lupus
BALB/c mice develop anti–double-stranded DNA (dsDNA) autoantibodies and immune complex deposits in glomeruli when immunized with a DNA mimotope, the decapeptide DWEYSVWLSN on a polylysine backbone (MAP-DWEYS; Putterman and Diamond, 1998). Furthermore, when treated with LPS to disrupt the blood brain barrier, MAP-DWEYS–immunized mice develop lesions in the hippocampus of the brain, a process which is attributable to excitatory cell death induced by stimulatory autoantibodies bound to neuronal N-methyl-d-aspartic acid (NMDA) receptors (Kowal et al., 2004). Anti-NMDA receptor autoantibodies have also been identified in the cerebrospinal fluid of humans with SLE, and this mouse system has been used as a model of human central nervous system lupus (DeGiorgio et al., 2001; Kowal et al., 2006).
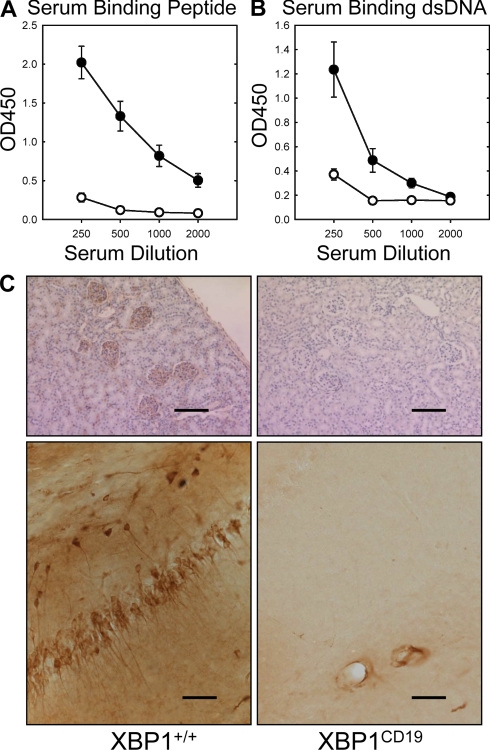
We next sought to use this mouse model of SLE to test whether XBP1CD19 mice were capable of developing autoantibodies and associated pathological findings of mouse lupus. XBP1CD19 and litter-matched XBP1+/+ mice were immunized with MAP-DWEYS three times over a 1-mo period, as described in Materials and methods. Serum anti-peptide and anti-dsDNA antibodies were readily detected in XBP1+/+ control mice but were only weakly present in XBP1CD19 mice (Fig. 1, A and B). These same mice were then treated with 3 mg/kg LPS and killed 4–7 d later for immunohistochemistry (IHC) analysis of kidney and brain (Fig. 1 C). IHC revealed abundant IgG in glomeruli of five out of five control mice and in the hippocampus of two out of five controls, but none was present in any of the XBP1CD19 mouse tissues. Thus, mice with B cells deficient in XBP1 were protected from autoantibody production and disease expression in this mouse model of SLE.
Figure 1.
Female XBP1+/+ and XBP1CD19 mice were immunized i.p. with 100 µg MAP-DWEYS in CFA followed by booster immunizations with 100 µg MAP-DWEYS in IFA on days 14 and 28. (A and B) On day 35, serum was tested for anti-DWEYS (peptide; A) and anti-dsDNA (B) antibodies by ELISA of four serum dilutions. In both panels, each circle represents the mean ± SEM value for XBP1+/+ (closed circles) or XBP1CD19 (open circles) mice. (C) XBP1+/+ and XBP1CD19 mice from A and B were treated i.p. with 3 mg/kg LPS on days 52 and +54. Shown are representative images of IgG-specific IHC of kidney (top) and hippocampus (bottom) of mice 4–7 d after LPS treatment. XBP1+/+ mice demonstrated anti-IgG binding to neurons in the CA1 region of the hippocampus. There was no anti-IgG binding in any of the XBP1CD19 animals. Bars: (kidney) 200 µm; (hippocampus) 100 µm. All panels represent results from a single experiment with five mice per group. A second independent experiment reproduced these findings (not depicted).
This observation is reminiscent of a recent compelling study in which two lupus-prone mouse strains (NZB/W F1 and MRL/lpr) were treated with bortezomib, a selective inhibitor of the 26S proteosome which is used therapeutically to treat human myeloma, a PC malignancy (Neubert et al., 2008). When treated with bortezomib, these lupus-prone mice lacked autoantibodies, were devoid of mature PCs, and were protected from mouse lupus. Proteasome inhibitors suppress ER-associated degradation pathways such that misfolded proteins accumulate in cells, cause ER stress, and induce the UPR (Obeng et al., 2006). Chronic ER stress, however, causes cell cycle arrest and cellular apoptosis (Zinszner et al., 1998; Brewer and Diehl, 2000). Proteasome inhibitors like bortezomib are toxic to myeloma cells, in part through targeting the activity of IRE1 (Lee et al., 2003; Obeng et al., 2006). These findings raise the intriguing notion that inhibition of XBP1 or the UPR in general may represent a potential therapeutic target in the treatment of autoantibody-mediated diseases like SLE.
The developmental defect in XBP1-deficient PCs occurs after CD138 expression
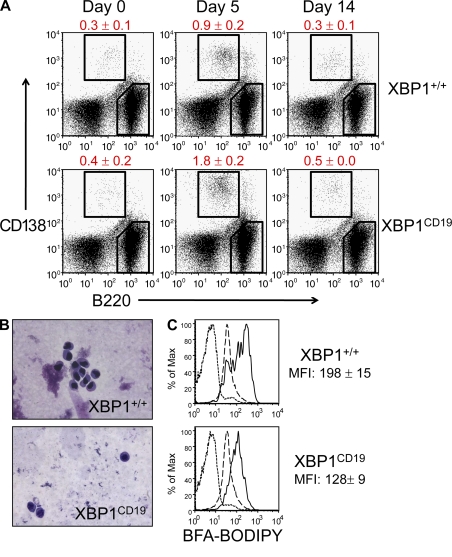
XBP1CD19 mice allow us to explore further the developmental stage at which PC differentiation arrests. Complicating this analysis, however, is the observation that PCs down-regulate many B cell surface markers such as B220, CD19, and CD20 (Hardy and Hayakawa, 2001). Traditionally, cell surface expression of CD138 by B cells has been viewed as a marker for antibody-secreting PCs (B220int CD138+ cells; Sanderson and Børset, 2002). Accordingly, we immunized XBP1CD19 and XBP1+/+ mice with NP-KLH to measure the distribution of B220 and CD138 staining over time. Surprisingly, even though XBP1CD19 mice had markedly impaired Ig responses to immunization (Fig. S2), both XBP1CD19 and XBP1+/+ mice had comparable numbers of B220int CD138+ splenocytes (Fig. 2 A). In XBP1CD19 mice, these cells lacked the expanded ER and perinuclear cuff of Golgi found in normal PCs (Fig. 2 B). This difference was quantifiable; B220int CD138+ cells from XBP1CD19 mice demonstrated less intensive staining by brefeldin A boron-dipyrromethane (BODIPY), which selectively binds to secretory ER and Golgi content (Fig. 2 C) (Hetz et al., 2008).
Figure 2.
XBP1+/+ and XBP1CD19 mice were immunized i.p. with a single 100-µg dose of NP-KLH in alum. (A) Representative flow cytometry of cell surface CD138 (y axis) and B220 (x axis) expression in splenocytes isolated from XBP1+/+ and XBP1CD19 mice at days 0, 5, and 14 after immunization. Mean percentage ± SEM of B220int CD138+ cells is indicated. (B) B220int CD138+ cells were isolated by FACS from a subset of splenocytes at day 5 after immunization. Cytospin preparations of cells were stained with modified Wright-Giemsa stain. (C) Splenocytes were isolated from mice on day 5 after immunization and incubated with Brefeldin A BODIPY, which selectively stains ER and Golgi organelles. Shown are representative flow cytometry histograms for B220int CD138+ cells (solid line), B220+ CD138− cells (long dashed line), and unstained cells (short dashed line). Brefeldin A-BODIPY mean fluorescence intensity (MFI) ± SEM is indicated for B220int CD138+ cells (no difference in B220+ CD138− cells). All experiments were performed with individual mice in at least three independent experiments.
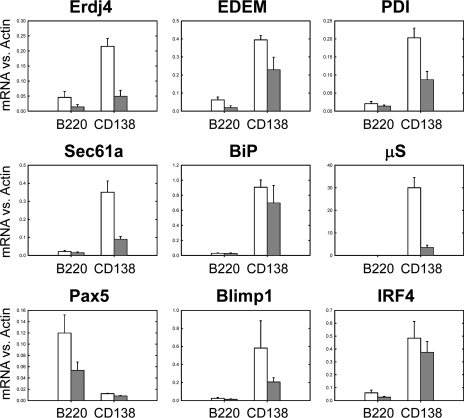
5 d after NP-KLH immunization, splenocytes from XBP1CD19 and XBP1+/+ mice were isolated and FACS sorted into B220int CD138+ and B220+ CD138− cell populations, from which RNA was purified for quantitative PCR analysis. In accord with what is known about PCs, B220int CD138+ cells from XBP1+/+ mice expressed much greater levels of the XBP1 target genes ERdj4, EDEM, PDI, and Sec61 as compared with B220+ CD138− cells from the same mice (Fig. 3). There were also increased levels of BiP, μS, Blimp1, and IRF4 and reduced levels of Pax5. These differences were also observed, but to a much lesser degree, in the two sorted XBP1CD19 B cell populations. When comparing B220int CD138+ cells from XBP1CD19 and XBP1+/+ mice, XBP1 target genes and μS were reduced in expression in cKO cells, and levels of BiP and Pax5 were similar. Unlike what was observed in LPS-stimulated B cells, however, levels of Blimp1 and IRF4 were not elevated in cells from XBP1CD19 mice. This supports the hypothesis that there are multiple pathways of B cell differentiation depending on the specific exogenous stimulus. Indeed, in vitro LPS-stimulated B cells behave differently than in vivo immunogen-stimulated B cells, and this is likely caused, in part, by the absence of T helper cell activity as well as by the different activation pathways involved (for review see McHeyzer-Williams and McHeyzer-Williams, 2005).
Figure 3.
XBP1+/+ and XBP1CD19 mice were immunized i.p. with a single 100-µg dose of NP-KLH in alum. At day 5 after immunization, splenocytes were isolated and sorted by FACS for B220+ CD138− (B220) and B220int CD138+ (CD138) populations. Panels show relative expression of indicated mRNA versus actin as measured by RT-PCR of cDNA prepared from indicated cell populations. In all panels, each bar represents the mean ± SEM value of individual XBP1+/+ (open bar) and XBP1CD19 (solid bar) mice in three independent experiments.
Collectively, our data show that XBP1CD19 B cells can survive and differentiate to the developmental stage at which they express CD138, at which point they developmentally arrest. This occurs before achieving normal PC morphology and functional activity because of a failure to up-regulate XBP1-dependent UPR target genes. CD138 is a member of the syndecan family of heparin sulfate proteoglycans and is comprised of a transmembrane core protein covalently bonded to heparin sulfate moieties (Sanderson and Yang, 2008). CD138 also exists as part of the bone marrow stroma and in a soluble form that may be shed from cell membranes. Cell surface–bound CD138 impairs invasion of myeloma cells through collagen gels (Liebersbach and Sanderson, 1994), facilitates cell–cell aggregation (Stanley et al., 1995), and promotes myeloma adhesion to collagen in the extracellular matrix (Ridley et al., 1993). These functions have been hypothesized to explain the organ selectivity of myeloma cells, and CD138 is being investigated as a potential therapeutic target in myeloma (Sanderson and Yang, 2008).
The role of CD138 in normal PCs, however, is less well defined, although it may have similar bone marrow homing functions in PCs as in myeloma cells. CD138 is expressed on the cell surface of PCs and bone marrow–resident B cell precursors. It is not present on the surface of mature naive B cells in the bone marrow, circulation, or secondary lymphoid organs (Sanderson et al., 1989). CD138 expression has been closely correlated with Ig secretion (Lalor et al., 1992), and CD138+ plasmablasts have been previously described (Angelin-Duclos et al., 2000), but the existence of a non–Ig-secreting CD138+ cell population has not been formally demonstrated until now. The converse has been seen, however, in that Blimp1-deficient B cells do not express CD138 but can secrete small amounts of Ig (Shapiro-Shelef et al., 2003; Kallies et al., 2007). However, this represents an earlier (initial) developmental stage of PC differentiation in that these cells are unable to attain the large secretory capabilities or phenotype of mature PCs (Kallies et al., 2007). IRF4-deficient B cells also fail to express CD138 (Klein et al., 2006).
Our findings therefore indicate that in developing PCs, Blimp1 and IRF4 expression precedes CD138 expression, which is then followed by XBP1-directed UPR transcriptional events and large-scale immunoglobulin secretion. This differs from our previously published results in which CD138 was not detected on the surface of XBP1-deficient B cells 9 d after immunization with 2,4-dinitrophenyl (DNP)-albumin in XBP1/RAG2−/− lymphoid chimeric mice (Reimold et al., 2001). One limitation of the XBP1/RAG2−/− lymphoid chimeric model, however, is that it cannot ascribe this difference solely to an intrinsic deficiency of XBP1 in B cells. With the recent discovery that XBP1 is important for the normal development and function of dendritic cells (Iwakoshi et al., 2007), one could conjecture that XBP1 function in non–B cell populations might in part explain the reduced numbers of CD138+ cells in the chimeric mice. Thus, the XBP1CD19 cKO system provides some refinement in our understanding of the relationship between XBP1 and CD138 expression.
Post-GC memory cells and preplasma memory cells are intact in mice with B cells that lack XBP1
Memory B cells make up the second major population of terminally differentiated B cells (for review see McHeyzer-Williams and McHeyzer-Williams, 2005). They are long-lived antigen-specific cells that reside mainly in peripheral tissues, including in the lung, gut, and peripheral lymphoid organs. When reexposed to cognate antigen, memory B cells proliferate, and a subset differentiates into antibody-secreting PCs. Memory B cell subpopulations can be distinguished phenotypically as post-GC B cells (B220+ CD138−) or preplasma memory B cells (B220− CD79b+ CD138−; for review see McHeyzer-Williams and McHeyzer-Williams, 2005).
Little is known about the transcriptional programs that regulate B cell differentiation into memory cells, including whether there is any role for XBP1 or the UPR in this process. Neither Blimp1 nor IRF4 are required for the development of post-GC memory B cells (Shapiro-Shelef et al., 2003; Klein et al., 2006). Blimp1-deficient B cells, however, fail to differentiate into preplasma memory B cells (Shapiro-Shelef et al., 2003). Furthermore, IRF4-deficient post-GC B cells can expand upon reencounter with cognate antigen (a memory cell–driven response), but they fail to differentiate into PCs (Klein et al., 2006). Thus, neither of these transcription factors appears to be crucial for development of the memory B cell population de novo.
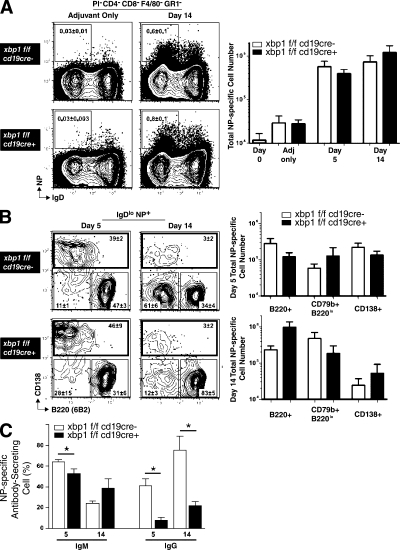
We have already demonstrated that secondary Ig responses are markedly blunted in XBP1CD19 mice (Fig. S2). Using a well established protocol to study memory B cells (Shapiro-Shelef et al., 2003), we next sought to answer whether the defect in secondary memory responses in XBP1CD19 mice is solely attributable to impaired antibody secretion by PCs or whether there may also be independent deficiencies in memory B cell populations themselves. Mice were immunized with 400 µg NP-KLH in Ribi adjuvant and splenocytes were isolated at day 5 or 14. Cells were stained with antibodies specific for CD4, CD8, F4/80 (a macrophage marker), GR1 (a granulocyte and monocyte marker), B220 (CD45R), CD138, IgD, GL7, CD38, and Ig λ light chain. In addition, cells were stained with propidium iodide (PI) to determine viability and allophycocyanin (APC)-conjugated NP to identify antigen-specific B cells. Viable post-GC NP-specific B cells were identified as those cells that excluded PI, were [CD4, CD8, F4/80, GR1]neg, and demonstrated a NP+IgDlo phenotype. The expansion of these cells after immunization was comparable between XBP1CD19 and XBP1+/+ mice (Fig. 4 A).
Figure 4.
Memory B cells are present in normal numbers in XBP1CD19 mice. (A, left) Representative expression of IgD and NP on [PI, CD4, CD8, F4/80, GR1]neg splenocytes of day-14 mice immunized with Ribi (adjuvant) only (left) or NP-KLH in Ribi (right). Shown are XBP1fl/fl CD19+/+ (XBP1+/+) mice (top) and XBP1fl/fl CD19cre/+ (XBP1CD19) littermates (bottom). The inset represents the percentage of cells within profile (mean ± SEM; n = 3). (A, right) Total number of IgDlo NP-specific B cells from day 0 (naive BALB/c), adjuvant only (day 14 Ribi only), day 5, and day 14 NP-KLH in Ribi immunized XBP1+/+ mice (white columns) and XBP1CD19 (black columns) mice (mean ± SEM; n = 3). (B, left) Representative expression of B220 and CD138 at the surface of IgDlo NP-specific B cells at days 5 (left) and 14 (right) from XBP1+/+ mice (top) and XBP1CD19 littermates (bottom). The inset represents the percentage of cells within profile (mean ± SEM; n = 3). (B, right) Total number of NP-specific B cells compartment B220+, CD79b+ B220lo, or CD138+ at day 5 (top) or day 14 (bottom) immunized XBP1+/+ (white columns) and XBP1CD19 (black columns) mice (mean ± SEM; n = 3; unpaired Student's t test; P > 0.05 for all data points). (C) [PI, CD4, CD8, F4/80, GR1]neg IgDlo NP+ CD138+ splenocytes of day-5 and day-14 immunized mice were sorted directly ex vivo into NP-specific Ig (IgM or IgG) revealing ELISPOT assays. Positives were scored manually under a dissection microscope. Each assay was done in triplicate from three separate animals (mean ± SEM; n = 3; unpaired Student's t test; *, P ≤ 0.05).
These NP-specific B cells were further assayed for expression of B220 and CD138 to identify B220+ CD138− post-GC memory cells, B220lo CD79b+ CD138− preplasma memory cells, and B220int CD138+ cells with a PC phenotype (Fig. 4 B). At day 5 after immunization, there were no differences in the relative or absolute numbers of the subpopulations between XBP1CD19 and XBP1+/+ mice. At day 14, there were some differences in the relative numbers of these cells, but the absolute numbers were not significantly different because the number of splenocytes and absolute number of NP-specific cells were up to twofold greater in XBP1CD19 mice. There were no differences in the total number of antigen-specific GC versus post-GC events at day 14 as measured by CD138neg GL7hi CD38lo GC B cells versus CD138neg GL7lo CD38hi post-GC B cells (Fig. S4 A). There were also no differences between XBP1CD19 and XBP1+/+ mice when assayed for the repertoire of λ light chain expression by antigen-specific B cells (Fig. S4 B). ELISPOT was used to measure the ability of FACS-purified B220int CD138+ cells to secrete IgM and IgG. At days 5 and 14, the number of NP-specific IgG-secreting B cells was significantly reduced in XBP1CD19 mice as compared with controls (Fig. 4 C). Similar results were obtained for NP-specific IgM-secreting cells at day 5 but not at day 14. These assays reinforce the notion that XBP1CD19 B cells are able to differentiate into B220int CD138+ cells but lack the normal capacity for Ig secretion.
Thus, XBP1CD19 B cells are capable of differentiating normally into post-GC or preplasma memory B cell populations. These observations again place XBP1-mediated events downstream of Blimp1, which is crucial for the development of preplasma memory B cell (Shapiro-Shelef et al., 2003). These data also show for the first time that the XBP1-driven branch of the UPR is unnecessary for the development of memory B cells themselves. Thus, the defects in Ig production during secondary antigen challenge in XBP1CD19 mice are solely the result of a failure of formation of antibody-secreting PCs and not of B cell memory itself.
Summary
Collectively, this analysis of B cells from XBP1CD19 cKO mice expands our understanding of the crucial role of XBP1 in terminal B cell development as it pertains to PC development, autoantibody-mediated autoimmune disease, and memory cell development. Deletion of XBP1 leads to failed up-regulation of the UPR in developing PCs with subsequent deficiencies in antibody responses to antigen. Interestingly, the experiments presented here identify and characterize a previously unrecognized population of antibody hyposecreting B220int CD138+ B cells in XBP1CD19 mice, demonstrating that XBP1-mediated UPR events occur after the developmental stage at which CD138 is expressed. Importantly, we establish that memory B cell populations are unaffected by XBP1 deficiency such that XBP1-related activity can be situated independent of the memory B cell lineage commitment. Finally, XBP1CD19 mice are protected from autoantibody-mediated mouse lupus. This has therapeutic implications from a clinical perspective in that inhibition of XBP1 or the UPR in general potentially represents a therapeutic target in the treatment of SLE.
MATERIALS AND METHODS
Mice and immunization.
The creation of XBP1flox mice harboring loxP sites in the first and second intron of the XBP1 gene has been described previously (Hetz et al., 2008; Kaser et al., 2008; Lee et al., 2008). These mice were backcrossed 10 generations with BALB/c mice, which was the background strain for all experiments described in these studies. XBP1flox/flox mice were bred to CD19cre/+ BALB/c mice that express cre recombinase under the control of the CD19 promoter (The Jackson Laboratory; Rickert et al., 1997). Two generations of breeding yielded XBP1CD19 cKO mice and XBP1+/+ control mice. All mice were born in Mendelian ratios and developed normally. Wild-type BALB/c mice were purchased from Taconic. Experimental protocols were approved by the Standing Committee on Animals at the Harvard Medical School and were designed with institutional and National Institutes of Health guidelines for the humane use of animals.
To measure serum antibody responses to immunization, 6–8-wk-old XBP1CD19 and XBP1+/+ mice were injected i.p. with 100 µg NP-KLH (Biosearch Technologies) emulsified in alum (Thermo Fisher Scientific) or 25 µg NP-ficoll (Biosearch Technologies). Blood was collected from mice via retroorbital bleed at the time of immunization and weekly for 2 wk. To measure secondary serum antibody responses, some mice that had received NP-KLH were reimmunized with 100 µg NP-KLH in alum 8 wk after primary immunization, and additional blood samples were obtained at the time of reimmunization and weekly for 2 wk. For NP-specific B cell studies, 8–14-wk-old mice were immunized subcutaneously with 400 µg NP-KLH in Ribi adjuvant (laboratory formulation based on Baldridge and Crane [1999]).
B cell isolation and culture.
Single cell suspensions of splenocytes were prepared from 6–8-wk-old mice, and erythrocytes were removed by osmotic lysis with NH4Cl buffer (Sigma-Aldrich) as previously described (Iwakoshi et al., 2003). Splenic B cells were purified using B220+ magnetic bead selection (Miltenyi Biotech) and confirmed to have >90% purity by flow cytometry for cell surface CD19 expression. For some B cell preparations, Southern blotting was used to determine efficacy of XBP1 deletion in B220+ lymphocytes. The remaining B220+ splenocytes from XBP1CD19 and XBP1+/+ mice were cultured at 106 cells/ml for 0–3 d in the presence or absence of 10 µg/ml LPS (Sigma-Aldrich) in complete media containing RPMI 1640 supplemented with 10% FBS (Hyclone Laboratories), 2 mM glutamine, 50 U/ml penicillin, 50 µg/ml streptomycin, 100 mM Hepes, nonessential amino acids, 1 mM sodium pyruvate, and 50 µM β-mercaptoethanol. Cells were harvested on days 0–3 of culture for RNA extraction and quantitative PCR analysis. In addition, on day 3 of culture, supernatants were tested by enzyme-linked immunosorption assay ELISA to quantify IgM production, cells were harvested for viability assay by annexin V and 7-AAD staining, and cell counts by exclusion of trypan blue.
Flow cytometry and cell sorting.
Splenocytes were prepared as described in the previous section. Non–NP-specific flow cytometry and FACS were performed using antibodies specific for mouse IgM, IgD, B220, CD19, or CD138 (BD) as previously described (Iwakoshi et al., 2007). To measure cell viability, cells were stained with PE-conjugated annexin V and 7-AAD according to manufacturer instructions (BD). Viable cells exclude both stains. To measure the cellular content of ER and Golgi organelles, cells were stained with Brefeldin A–BODIPY (Invitrogen) at 0.2 µg/ml at 37°C for 40 min before flow cytometric analysis as previously described (Hetz et al., 2008). Cytospin samples of FACS-sorted B220int CD138+ cells were prepared from XBP1CD19 and XBP1+/+ mice and stained with modified Wright-Giemsa stain according to the manufacturer's instructions (Sigma-Aldrich).
For NP-specific B cell studies, spleens were harvested at day 5 or 14 after immunization and single cell suspensions were prepared as previously described (McHeyzer-Williams et al., 2000). Cell suspensions were labeled for flow cytometry at 2 × 108 cells/ml on ice for 45 min with the following fluorophore-labeled mAbs or Ag: FITC-11.26 (anti-IgD; McHeyzer-Williams laboratory), FITC-GL7 (BD), PE-281.2 (anti-CD138/syndecan-1; BD), PE-90 (anti-CD38; BD), Cy5PE-H129.19 (anti-CD4; BD), Cy5PE-53-6.7 (anti-CD8; BioLegend), Cy5PE-F4/80 (eBioscience), Cy5PE-RB6-8C5 (anti-Ly6G/GR1), Cy7PE-6B2 (antiCD45R/B220), APC-NP (4-hydroxy-5-iodo-3-nitrophenyl; McHeyzer-Williams laboratory), and biotin-HM79b (anti-CD79b/Igβ; McHeyzer-Williams laboratory). Streptavidin-Cy7APC (BD) was used as a second-step revealing reagent. Cells were washed and resuspended in 2 µg/ml PI for dead cell exclusion in PBS with 5% FCS for analysis. Samples were analyzed using a FACS Vantage SE (BD) and data analyzed using Flow Jo software (Tree Star, Inc.). Profiles are presented as 5% probability contours with outliers.
ELISA and ELISPOT assays.
ELISA was used to quantify IgM levels in cell culture supernatants and levels of IgM, IgG1, IgG2a, IgG2b, IgG3, and IgA in the serum of XBP1CD19 and XBP1+/+ mice as previously described (Reimold et al., 2001). To measure the relative amounts of NP-specific serum antibodies in mice immunized with NP-KLH or NP-ficoll, plates were coated with 25 µg/ml NP(25)-BSA (Biosearch Technology) overnight at 4°C. To standardize and quantify relative amounts of NP-specific IgG responses, all experimental samples were compared with a standardized dilution of pooled serum obtained from XBP1+/+ mice immunized with NP-KLH or NP-ficoll and bled on day 14. NP-specific ELISA was otherwise performed as previously described (Reimold et al., 2001). Serum anti-peptide and anti-dsDNA antibodies were tested by ELISA as previously described (Kowal et al., 2004).
ELISPOT for detection of NP-specific Ig was performed as described previously (McHeyzer-Williams et al., 1993; McHeyzer-Williams et al., 2000). 96-well multiscreen membrane filtration plates (Millipore) were coated with 25 µg/ml NP-BSA overnight at 4°C. The wells were washed and incubated with RPMI 1640 and 5% FCS, and 25–50 [PI, CD4, CD8, F4/80, GR1]neg IgDlo NP+ CD138+ cells were sorted directly into each well and incubated for >18 h at 37°C in 5% CO2. The wells were washed before the addition of horseradish peroxidase goat anti–mouse IgM or IgG (SouthernBiotech) for 4 h and developed using 3-amino-9-ethylcarbazole (Sigma-Aldrich). Wells were manually scored by a person blinded to the test conditions using a dissection microscope. Each assay was done in triplicate wells from three separate animals.
Quantitative PCR.
Total RNA was isolated from cells using TRIzol reagent (Invitrogen), and complementary DNA (cDNA) was synthesized from RNA using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems). Quantitative real-time PCR reactions using SYBR green fluorescent reagent were run in an Mx3005P system (Agilent Technologies) and analyzed as previously described (Iwakoshi et al., 2007). Primer sequences are listed in Table S1.
Mouse lupus.
Using an established model of mouse lupus (Kowal et al., 2004), 6–8-wk-old XBP1CD19 and XBP1+/+ mice were immunized i.p. with 100 µg MAP-DWEYS emulsified in CFA (Sigma-Aldrich). Booster immunizations with 100 µg MAP-DWEYS in IFA (Sigma-Aldrich) were given on days 14 and 28. Serum was obtained from these mice on day 35 and tested for anti-peptide and anti-dsDNA antibodies as previously described (Kowal et al., 2004). There were five mice per group. To disrupt the blood brain barrier in the hippocampus, mice were given i.p. injections of 3 mg/kg LPS (Escherichia coli, 055:B5; Sigma-Aldrich) on days 52 and 54. Mice were then killed 4–7 d after LPS treatment to analyze tissues for histological evidence of mouse SLE. After cardiac perfusion, brains and kidneys were isolated and tissues were immunostained for IgG as previously described (Putterman and Diamond, 1998; Kowal et al., 2004).
Online supplemental material.
Table S1 lists primer sequences used for quantitative real-time PCR. Fig. S1 shows XBP1 deletion and basal immunoglobulin production in XBP1CD19 mice. Fig. S2 shows immunoglobulin production in immunized XBP1CD19 mice. Fig. S3 shows in vitro analysis of XBP1-deficient B cells. Fig. S4 shows additional characterization of memory cells in XBP1CD19 mice. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20090738/DC1.
Acknowledgments
We would like to acknowledge Dorothy Zhang and RoseAnn Berlin for their expertise in processing the histological samples, Landy Kangaloo for laboratory assistance, and Marisol Avellaneda for assistance with manuscript submission.
This study was supported by National Institutes of Health grant AI32412 (L.H. Glimcher), The Ellison Foundation (L.H. Glimcher), National Institutes of Health grant T32 AR007530 (D.J. Todd), American College of Rheumatology Fellowship Training Award (D.J. Todd), the American Heart Association Investigator award (A.-H. Lee) and National Institutes of Health grant AI047231 (M.G. McHeyzer-Williams).
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- 7-AAD
- 7-amino-actinomycin D
- APC
- allophycocyanin
- BiP
- Ig heavy chain binding protein
- Blimp1
- B lymphocyte-induced maturation protein 1
- BODIPY
- boron-dipyrromethane
- cDNA
- complementary DNA
- cKO
- conditional KO
- dsDNA
- double-stranded DNA
- ERdj4
- ER-localized Dnajb9
- GC
- germinal center
- IHC
- immunohistochemistry
- IRE1
- inositol-requiring transmembrane kinase/endonuclease 1
- IRF4
- IFN regulatory factor 4
- mRNA
- messenger RNA
- NP
- nitrophenol
- Pax5
- paired box gene 5
- PC
- plasma cell
- PDI
- protein disulfide isomerase
- PI
- propidium iodide
- SLE
- systemic lupus erythematosus
- UPR
- unfolded protein response
- XBP1
- X box binding protein 1
References
- Angelin-Duclos C., Cattoretti G., Lin K.I., Calame K. 2000. Commitment of B lymphocytes to a plasma cell fate is associated with Blimp-1 expression in vivo.J. Immunol. 165:5462–5471 [DOI] [PubMed] [Google Scholar]
- Baldridge J.R., Crane R.T. 1999. Monophosphoryl lipid A (MPL) formulations for the next generation of vaccines.Methods. 19:103–107 doi:10.1006/meth.1999.0834 [DOI] [PubMed] [Google Scholar]
- Brewer J.W., Diehl J.A. 2000. PERK mediates cell-cycle exit during the mammalian unfolded protein response.Proc. Natl. Acad. Sci. USA. 97:12625–12630 doi:10.1073/pnas.220247197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calame K.L., Lin K.-I., Tunyaplin C. 2003. Regulatory mechanisms that determine the development and function of plasma cells.Annu. Rev. Immunol. 21:205–230 doi:10.1146/annurev.immunol.21.120601.141138 [DOI] [PubMed] [Google Scholar]
- Calfon M., Zeng H., Urano F., Till J.H., Hubbard S.R., Harding H.P., Clark S.G., Ron D. 2002. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA.Nature. 415:92–96 doi:10.1038/415092a [DOI] [PubMed] [Google Scholar]
- DeGiorgio L.A., Konstantinov K.N., Lee S.C., Hardin J.A., Volpe B.T., Diamond B. 2001. A subset of lupus anti-DNA antibodies cross-reacts with the NR2 glutamate receptor in systemic lupus erythematosus.Nat. Med. 7:1189–1193 doi:10.1038/nm1101-1189 [DOI] [PubMed] [Google Scholar]
- Hardy R.R., Hayakawa K. 2001. B cell development pathways.Annu. Rev. Immunol. 19:595–621 doi:10.1146/annurev.immunol.19.1.595 [DOI] [PubMed] [Google Scholar]
- Hetz C., Lee A.H., Gonzalez-Romero D., Thielen P., Castilla J., Soto C., Glimcher L.H. 2008. Unfolded protein response transcription factor XBP-1 does not influence prion replication or pathogenesis.Proc. Natl. Acad. Sci. USA. 105:757–762 doi:10.1073/pnas.0711094105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C.C., Dougan S.K., McGehee A.M., Love J.C., Ploegh H.L. 2009. XBP-1 regulates signal transduction, transcription factors and bone marrow colonization in B cells.EMBO J. 28:1624–1636 doi:10.1038/emboj.2009.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakoshi N.N., Lee A.H., Vallabhajosyula P., Otipoby K.L., Rajewsky K., Glimcher L.H. 2003. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1.Nat. Immunol. 4:321–329 doi:10.1038/ni907 [DOI] [PubMed] [Google Scholar]
- Iwakoshi N.N., Pypaert M., Glimcher L.H. 2007. The transcription factor XBP-1 is essential for the development and survival of dendritic cells.J. Exp. Med. 204:2267–2275 doi:10.1084/jem.20070525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallies A., Hasbold J., Fairfax K., Pridans C., Emslie D., McKenzie B.S., Lew A.M., Corcoran L.M., Hodgkin P.D., Tarlinton D.M., Nutt S.L. 2007. Initiation of plasma-cell differentiation is independent of the transcription factor Blimp-1.Immunity. 26:555–566 doi:10.1016/j.immuni.2007.04.007 [DOI] [PubMed] [Google Scholar]
- Kaser A., Lee A.H., Franke A., Glickman J.N., Zeissig S., Tilg H., Nieuwenhuis E.E., Higgins D.E., Schreiber S., Glimcher L.H., Blumberg R.S. 2008. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease.Cell. 134:743–756 doi:10.1016/j.cell.2008.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein U., Casola S., Cattoretti G., Shen Q., Lia M., Mo T., Ludwig T., Rajewsky K., Dalla-Favera R. 2006. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination.Nat. Immunol. 7:773–782 doi:10.1038/ni1357 [DOI] [PubMed] [Google Scholar]
- Kowal C., DeGiorgio L.A., Nakaoka T., Hetherington H., Huerta P.T., Diamond B., Volpe B.T. 2004. Cognition and immunity; antibody impairs memory.Immunity. 21:179–188 doi:10.1016/j.immuni.2004.07.011 [DOI] [PubMed] [Google Scholar]
- Kowal C., Degiorgio L.A., Lee J.Y., Edgar M.A., Huerta P.T., Volpe B.T., Diamond B. 2006. Human lupus autoantibodies against NMDA receptors mediate cognitive impairment.Proc. Natl. Acad. Sci. USA. 103:19854–19859 doi:10.1073/pnas.0608397104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalor P.A., Nossal G.J.V., Sanderson R.D., McHeyzer-Williams M.G. 1992. Functional and molecular characterization of single, (4-hydroxy-3-nitrophenyl)acetyl (NP)-specific, IgG1+ B cells from antibody-secreting and memory B cell pathways in the C57BL/6 immune response to NP.Eur. J. Immunol. 22:3001–3011 doi:10.1002/eji.1830221136 [DOI] [PubMed] [Google Scholar]
- Lee K., Tirasophon W., Shen X., Michalak M., Prywes R., Okada T., Yoshida H., Mori K., Kaufman R.J. 2002. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response.Genes Dev. 16:452–466 doi:10.1101/gad.964702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.H., Iwakoshi N.N., Anderson K.C., Glimcher L.H. 2003. Proteasome inhibitors disrupt the unfolded protein response in myeloma cells.Proc. Natl. Acad. Sci. USA. 100:9946–9951 doi:10.1073/pnas.1334037100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.H., Chu G.C., Iwakoshi N.N., Glimcher L.H. 2005. XBP-1 is required for biogenesis of cellular secretory machinery of exocrine glands.EMBO J. 24:4368–4380 doi:10.1038/sj.emboj.7600903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.-H., Scapa E.F., Cohen D.E., Glimcher L.H. 2008. Regulation of hepatic lipogenesis by the transcription factor XBP1.Science. 320:1492–1496 doi:10.1126/science.1158042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebersbach B.F., Sanderson R.D. 1994. Expression of syndecan-1 inhibits cell invasion into type I collagen.J. Biol. Chem. 269:20013–20019 [PubMed] [Google Scholar]
- McHeyzer-Williams L.J., McHeyzer-Williams M.G. 2005. Antigen-specific memory B cell development.Annu. Rev. Immunol. 23:487–513 doi:10.1146/annurev.immunol.23.021704.115732 [DOI] [PubMed] [Google Scholar]
- McHeyzer-Williams M.G., McLean M.J., Lalor P.A., Nossal G.J. 1993. Antigen-driven B cell differentiation in vivo.J. Exp. Med. 178:295–307 doi:10.1084/jem.178.1.295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHeyzer-Williams L.J., Cool M., McHeyzer-Williams M.G. 2000. Antigen-specific B cell memory: expression and replenishment of a novel b220(-) memory b cell compartment.J. Exp. Med. 191:1149–1166 doi:10.1084/jem.191.7.1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubert K., Meister S., Moser K., Weisel F., Maseda D., Amann K., Wiethe C., Winkler T.H., Kalden J.R., Manz R.A., Voll R.E. 2008. The proteasome inhibitor bortezomib depletes plasma cells and protects mice with lupus-like disease from nephritis.Nat. Med. 14:748–755 doi:10.1038/nm1763 [DOI] [PubMed] [Google Scholar]
- Obeng E.A., Carlson L.M., Gutman D.M., Harrington W.J., Jr., Lee K.P., Boise L.H. 2006. Proteasome inhibitors induce a terminal unfolded protein response in multiple myeloma cells.Blood. 107:4907–4916 doi:10.1182/blood-2005-08-3531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putterman C., Diamond B. 1998. Immunization with a peptide surrogate for double-stranded DNA (dsDNA) induces autoantibody production and renal immunoglobulin deposition.J. Exp. Med. 188:29–38 doi:10.1084/jem.188.1.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimold A.M., Iwakoshi N.N., Manis J., Vallabhajosyula P., Szomolanyi-Tsuda E., Gravallese E.M., Friend D., Grusby M.J., Alt F., Glimcher L.H. 2001. Plasma cell differentiation requires the transcription factor XBP-1.Nature. 412:300–307 doi:10.1038/35085509 [DOI] [PubMed] [Google Scholar]
- Rickert R.C., Roes J., Rajewsky K. 1997. B lymphocyte-specific, Cre-mediated mutagenesis in mice.Nucleic Acids Res. 25:1317–1318 doi:10.1093/nar/25.6.1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley R.C., Xiao H., Hata H., Woodliff J., Epstein J., Sanderson R.D. 1993. Expression of syndecan regulates human myeloma plasma cell adhesion to type I collagen.Blood. 81:767–774 [PubMed] [Google Scholar]
- Sanderson R.D., Børset M. 2002. Syndecan-1 in B lymphoid malignancies.Ann. Hematol. 81:125–135 doi:10.1007/s00277-002-0437-8 [DOI] [PubMed] [Google Scholar]
- Sanderson R.D., Yang Y. 2008. Syndecan-1: a dynamic regulator of the myeloma microenvironment.Clin. Exp. Metastasis. 25:149–159 doi:10.1007/s10585-007-9125-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson R.D., Lalor P., Bernfield M. 1989. B lymphocytes express and lose syndecan at specific stages of differentiation.Cell Regul. 1:27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer A.L., Shapiro-Shelef M., Iwakoshi N.N., Lee A.H., Qian S.B., Zhao H., Yu X., Yang L., Tan B.K., Rosenwald A., et al. 2004. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation.Immunity. 21:81–93 doi:10.1016/j.immuni.2004.06.010 [DOI] [PubMed] [Google Scholar]
- Shapiro-Shelef M., Lin K.I., McHeyzer-Williams L.J., Liao J., McHeyzer-Williams M.G., Calame K. 2003. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells.Immunity. 19:607–620 doi:10.1016/S1074-7613(03)00267-X [DOI] [PubMed] [Google Scholar]
- Stanley M.J., Liebersbach B.F., Liu W., Anhalt D.J., Sanderson R.D. 1995. Heparan sulfate-mediated cell aggregation. Syndecans-1 and -4 mediate intercellular adhesion following their transfection into human B lymphoid cells.J. Biol. Chem. 270:5077–5083 [DOI] [PubMed] [Google Scholar]
- Todd D.J., Lee A.H., Glimcher L.H. 2008. The endoplasmic reticulum stress response in immunity and autoimmunity.Nat. Rev. Immunol. 8:663–674 doi:10.1038/nri2359 [DOI] [PubMed] [Google Scholar]
- Yoshida H., Matsui T., Yamamoto A., Okada T., Mori K. 2001. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor.Cell. 107:881–891 doi:10.1016/S0092-8674(01)00611-0 [DOI] [PubMed] [Google Scholar]
- Zhou L.J., Ord D.C., Hughes A.L., Tedder T.F. 1991. Structure and domain organization of the CD19 antigen of human, mouse, and guinea pig B lymphocytes. Conservation of the extensive cytoplasmic domain.J. Immunol. 147:1424–1432 [PubMed] [Google Scholar]
- Zinszner H., Kuroda M., Wang X., Batchvarova N., Lightfoot R.T., Remotti H., Stevens J.L., Ron D. 1998. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum.Genes Dev. 12:982–995 doi:10.1101/gad.12.7.982 [DOI] [PMC free article] [PubMed] [Google Scholar]