Abstract
Expression of enzymatically inactive caspase-8, or deletion of caspase-8 from basal epidermal keratinocytes, triggers chronic skin inflammation in mice. Unlike similar inflammation resulting from arrest of nuclear factor κB activation in the epidermal cells, the effect induced by caspase-8 deficiency did not depend on TNF, IL-1, dermal macrophage function, or expression of the toll-like receptor adapter proteins MyD88 or TRIF. Both interferon regulatory factor (IRF) 3 and TANK-binding kinase were constitutively phosphorylated in the caspase-8–deficient epidermis, and knockdown of IRF3 in the epidermis-derived cells from these mice abolished the expression of up-regulated genes. Temporal and spatial analyses of the alterations in gene expression that result from caspase-8 deficiency reveal that the changes are initiated before birth, around the time that cornification develops, and occur mainly in the suprabasal layer. Finally, we found that caspase-8–deficient keratinocytes display an enhanced response to gene activation by transfected DNA. Our findings suggest that an enhanced response to endogenous activators of IRF3 in the epidermis, presumably generated in association with keratinocyte differentiation, contributes to the skin inflammatory process triggered by caspase-8 deficiency.
Members of the caspase cysteine-protease family can be activated by a variety of death-inducing agents to cleave death-related cellular proteins, thereby initiating programmed cell death. The caspases also serve other nonapoptotic cellular functions, of whose physiological significance and mechanisms little is known (Lamkanfi et al., 2007).
The epidermal keratinocytes of the skin are programmed to undergo spontaneous cell death while progressing along their pathway from the basal layer of the epidermis, a process which results in formation of the skin's cornified layer (Candi et al., 2005). These cells express all the known caspases and can be induced by ligands of the TNF family to die in a caspase-dependent manner. Activation of this caspase-mediated cell death process apparently contributes to pathological damage of the epidermis in conditions such as toxic epidermal necrolysis (Viard et al., 1998), sunburn (Zhuang et al., 2000), eczematous dermatitis (Trautmann et al., 2000), and skin ulceration caused by parasitic infection (Eidsmo et al., 2005) or Pemphigus foliaceus (Li et al., 2009). Whether the apoptotic caspases also mediate the nonpathological cell death process that causes cornification has been a matter of controversy. Early studies suggested that caspase-3 is proteolytically processed and activated in the cornifying epidermis and that this activation is required for the programmed death of cells associated with the cornification (Weil et al., 1999; Allombert-Blaise et al., 2003). In subsequent studies, however, such activation could not be confirmed, nor could processing of caspase-6 or -7 be discerned in the differentiating epidermal cells (Lippens et al., 2000; Rendl et al., 2002). More recent evidence points to a role for caspase-3 in controlling the rate of differentiation of the epidermal keratinocytes during embryonic development (Okuyama et al., 2004). This function, however, appears to be independent of the role of this enzyme in death induction and does not seem to involve its proteolytic processing (Fischer et al., 2005). The only caspase shown conclusively to participate in the cornification process is caspase-14, which is quite selectively expressed and activated in the differentiating and cornifying layers of the epidermis and the hair follicles. Its participation, however, does not occur through activation of an apoptotic process and is not needed for the cornification itself but rather for associated processes such as hydration of the skin via proteolytic processing of filaggrin into free hygroscopic amino acids (Denecker et al., 2008).
Caspase-8 is known mainly for its role as the proximal caspase in the cell death pathway activated by receptors of the TNF/NGF family. In this paper, we show that this enzyme is not required for the differentiation-associated death of the epidermal keratinocytes. However, its deficiency in these cells disrupts epidermal homeostasis and triggers a severe cutaneous inflammation associated, as in other inflammatory skin diseases, with aberrant epidermal growth and differentiation.
Generation of inflammatory mediators in the epidermis is shown in this study to be initiated before birth and independently of the adapter proteins for the toll-like receptors (TLRs), ruling out the possibility that the triggering involves either pathogen-associated molecular patterns (PAMPs) or new antigens to which the mice become exposed upon birth. Participation of a trigger that is endogenous to the keratinocytes is further indicated by our data showing that the inflammation is largely independent of dermal macrophages. We also show that the inflammation is independent of the major inflammatory mediators TNF, IL-1α, and IL-1β, thus differing from a similar disease caused by arrest of the canonical NF-κB activation pathway in the keratinocytes that crucially depends on TNF and on macrophage function (Pasparakis et al., 2002; Gugasyan et al., 2004; van Hogerlinden et al., 2004; Omori et al., 2006; Sayama et al., 2006).
We present evidence that an IFN regulatory factor (IRF) 3 activation pathway is constitutively activated in the caspase-8–deficient epidermis and that, consistently, the genes that are overexpressed in this enzyme-deficient epidermis include many that are known to be IRF3 dependent. Our spatial analysis of the expression of inflammatory genes in the caspase-8–deficient epidermis indicated that its location is largely suprabasal, suggesting that the trigger for its activation is related to the differentiation/cornification process. We found, moreover, that transfection of caspase-8–deficient keratinocytes with DNA triggers activation of these genes more effectively than in caspase-8–expressing cells. These findings lead us to suggest that certain endogenous activators of the response to foreign nucleic acids are generated in the epidermis, possibly in association with the cornification process, and that amplification of such signaling by the absence of caspase-8 contributes to the chronic skin inflammation caused by caspase-8 deficiency in the epidermis.
RESULTS
Mice expressing enzymatically inactive caspase-8 develop inflammation in various organs, including skin
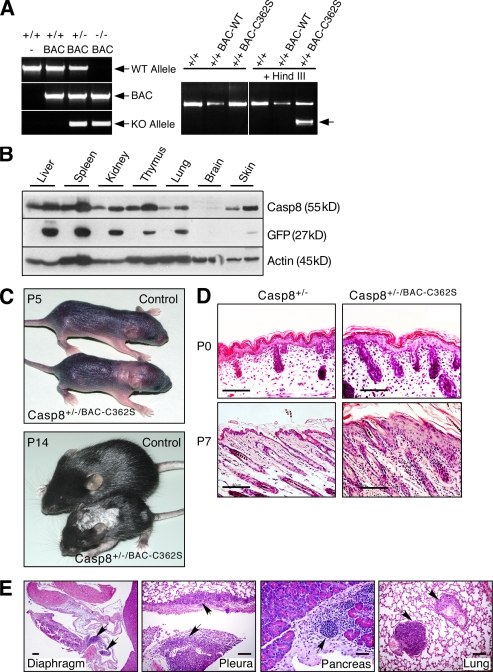
To investigate structure–function relationships for the various effects of caspase-8 in vivo, we used bacterial artificial chromosome (BAC)–mediated transgenesis to produce mice that express, besides their endogenous caspase-8 alleles, an additional copy of the caspase-8 gene or various mutants thereof (Fig. 1 A and not depicted; Kang et al., 2008). These transgenes were expressed ubiquitously and manifested the same tissue-specific expression pattern as that observed for the endogenous gene (Fig. 1 B).
Figure 1.
Mice expressing an enzymatically inactive caspase-8 transgene develop a skin disorder. (A) PCR genotyping of the caspase-8 BAC transgenes. Left, genotyping of WT mice and of mice expressing the WT caspase-8 transgene and either WT or KO endogenous caspase-8 alleles. Right, restriction analysis distinguishing the enzymatically inactive caspase-8 transgene (BAC-C362S) from WT (BAC-WT) transgene and endogenous allele. The arrow points to the HindIII fragment specifically generated in the BAC-C362S transgene. (B) Western blot analysis of caspase-8 in Casp-8+/+/BAC-WT transgenic mouse tissues, relative to the WT (right and left lanes, respectively, for each pair), demonstrating its increased expression in all transgenic mouse tissues (top). Internal ribosome entry site (IRES)–aided expression of GFP cDNA (inserted into the BAC under control of the caspase-8 promoter) is proportional to that of endogenous caspase-8 (middle). Immunoblot analysis of β-actin confirmed equal loading of tissue extracts from the two mice (bottom). (C) General appearance of control and transgenic mice. At P5, pallor and thickening in the interscapular area and dorsally at the caudal head indicate early lesions. At P14, the mutant is markedly smaller, with scaling, loss of hair, and a small lesion on the left thigh. Lesion distribution is multifocal and random. (D) Microscopy of the skin. At P0, skin samples from WT and mutant mice are similar. At P7, the latter demonstrate epidermal hyperplasia and multifocal dermal inflammatory cellular infiltration. Hematoxylin and eosin (H&E); Bars, 100 µm. (E) Transgenic mouse specimens demonstrate multifocal cellular inflammatory infiltration in diaphragmatic parietal pleura, lung visceral pleura, and interstitium of pancreas and lung (arrows). Bars, 100 µm. The data presented in B, C, D, and E are representative of analyses of four, five, three, and three mice, respectively.
Mice that expressed, in addition to the endogenous caspase-8 alleles, an active site mutant of the protein devoid of enzymatic activity (Casp-8+/+/BAC-C362S) appeared normal. However, deletion of one of the two endogenous caspase-8 alleles from these mice (Casp-8+/−/BAC-C362S) resulted in their development of a chronic inflammatory skin disorder. Cutaneous lesions characterized by focal thickening and scaling appeared 3–5 d after birth and gradually spread, covering large areas (Fig. 1 C). Histological analysis of the lesions revealed extensive epidermal hyperplasia and marked inflammatory cellular infiltration of the dermis (Fig. 1 D). Further examination revealed that leukocytes had also infiltrated various internal organs including lung, pancreas, and pleura (Fig. 1 E, arrows).
Specific deletion of caspase-8 from epidermal keratinocytes results in a similar skin disorder
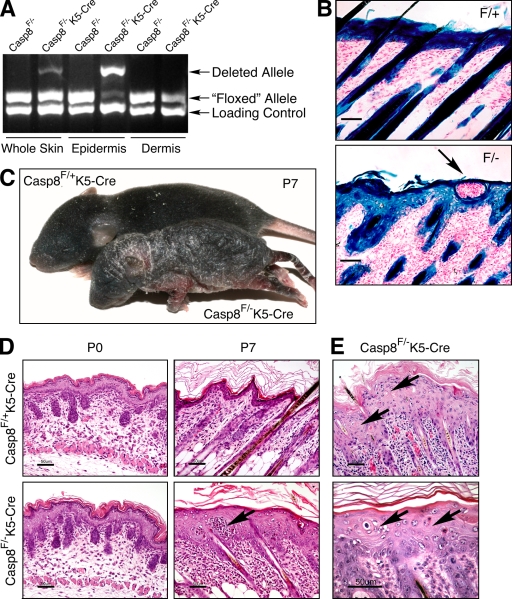
To examine whether the cutaneous abnormality caused by the expression of enzymatically inactive caspase-8 reflects a functional role of this enzyme in the epidermal cells themselves, we crossed mice carrying a conditional caspase-8 allele with mice expressing Cre under control of the keratin 5 (K5) promoter. In this way, we obtained mice in which caspase-8 expression was specifically eliminated in the keratinocytes (Fig. 2, A and B). The progeny (Casp-8F/−K5-Cre) appeared normal at birth but, 3 d later (i.e., at postnatal day 3 [P3]), began to develop a skin disease similar to that of the Casp-8+/−/BAC-C362S mice (Fig. 2 C). They failed to thrive and died by P7. Consistently with the known activity of the K5 promoter in the squamous mucosa of the stomach, some of the mice also exhibited severe gastric inflammation (Fig. S1). However, death of the mice did not correlate with the occurrence of gastritis or with decreased food uptake and thus was apparently the result of another cause.
Figure 2.
Epidermis-specific KO of caspase-8. (A) Quantitative assessment of WT and deleted caspase-8 alleles in Casp-8F/−K5-Cre mice by genomic PCR. (B) X-gal staining for β-galactosidase activity in the skins of ROSA26-reporter Casp-8F/+K5-Cre (F/+) and ROSA26-reporter Casp-8F/−K5-Cre (F/−) mice, demonstrating specific Cre-induced recombination in the superficial and follicular epidermis and hair follicles. Bars, 50 µm. (C) General appearance of Casp-8F/+K5-Cre and Casp-8F/−K5-Cre mice at P7. The mutant is much smaller than the WT. The skin is unevenly thickened with marked alopecia and hyperkeratosis. (D) Microscopy of the skin. At P0, the skin of the Casp-8F/−K5-Cre mice is indistinguishable in appearance from the skin of the Casp-8F/+K5-Cre mice. At P7 there is marked epidermal hyperplasia and widespread dermal inflammatory cellular infiltration in the Casp-8F/−K5-Cre mouse. Arrows in B and D point to intraepidermal eosinophilic pustules. H&E; Bars, 50 µm. (E) The affected epidermis contains numerous dyskeratotic foci, consisting of maloriented keratinocytes often forming a whorl around a central keratinocyte which has become prematurely keratinized (arrows). H&E; Bars, 50 µm. The data presented in B, C, D, and E are representative of analyses of 4, 350, 30, and 30 mice, respectively.
The skin in affected sites of the Casp-8F/−K5-Cre mice displayed pronounced epidermal hyperplasia, as well as orthokeratotic and milder parakeratotic hyperkeratosis with loss of the stratum granulosum. Intraepidermal pustules were not uncommon and numerous cells were dyskeratotic (Fig. 2, D and E).
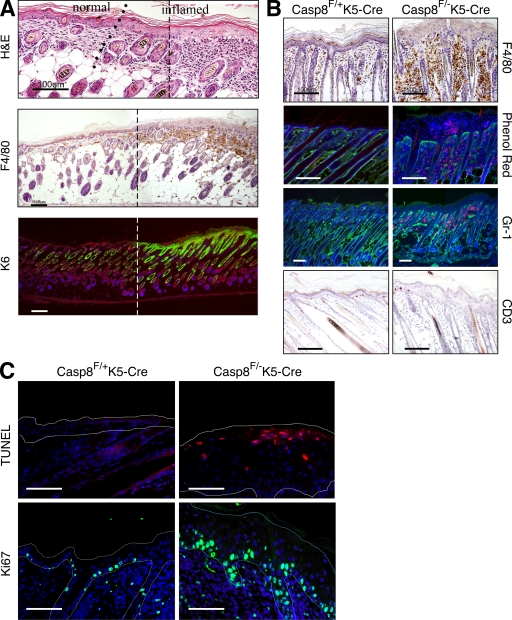
Epidermal hyperplasia was restricted to sites of inflammation (Fig. 3 A). In the dermis these sites exhibited severe diffuse infiltration of inflammatory cells, including eosinophils, macrophages, and granulocytes, with no significant increase in B lymphocytes (indicated by staining with anti-CD45R antibody; unpublished data), and a marked decrease in epidermal CD3+ T lymphocytes (Fig. 3 B). Eosinophils also accumulated within intraepidermal pustules (Fig. 2, B and D; and Fig. 3 B). The superficial and follicular epidermis expressed K6 specifically in affected areas (Fig. 3 A).
Figure 3.
Assessment of inflammation in the skin of Casp-8F/−K5-Cre mice. (A) Multifocal cutaneous inflammation at P5. To allow continuous viewing of the transition area between inflamed and unaffected skin, two overlapping images of adjacent regions in a skin section covering such an area were obtained, and a panoramic image was then constructed using a conventional Photoshop (Adobe) Photomerge routine. The broken line marks the site at which the two pictures were merged. The line of dots in the top indicates the approximate location of the transition point. The micrographs demonstrate epidermal hyperplasia, as well as increased dermal cellularity in the inflamed skin (top, H&E) as a result of infiltration of eosinophils and F4/80-positive cells (mononuclear phagocytes, middle). Interfollicular expression of K6, which is indicative of epidermal hyperproliferation (bottom), is restricted to the thickened epidermal region. Bars, 100 µm. (B) Staining for macrophages (anti-F4/80 antibody), eosinophils (phenol red uptake), granulocytes (anti–Gr-1 antibody), and T lymphocytes (anti-CD3 antibody) in skin sections from Casp-8F/+K5-Cre and Casp-8F/−K5-Cre mice at P7. Note that eosinophils also accumulate within pustules in the epidermis of the Casp-8F/−K5-Cre mice (also marked by arrows in Fig. 2, B and D). Bars, 100 µM. (C) TUNEL staining (top) for assessing apoptosis and Ki67 staining (bottom) for determining cell proliferation rates in the Casp-8F/+K5-Cre and Casp-8F/−K5-Cre epidermis at P7. Fine broken lines delineate approximate contours of the epidermis. Scale bar, 50 µm. The data presented in A, B, and C are representative of analyses of 10, 20, and 4 mice, respectively.
To investigate the cellular function of caspase-8 in the cutaneous epidermal keratinocytes, we first examined whether its deficiency affects cell death, the most thoroughly documented function of caspase-8. Cornification seemed unharmed. Moreover, outside-in dye penetration tests consistently failed to reveal any defect in epidermal barrier function, even as late as P7 (unpublished data). No conspicuous aberration was observed in the expression or localization of E-cadherin (Fig. S2), the major mediator of intracellular adhesion in epithelial tissues and an essential contributor to epidermal barrier function in vivo (Tunggal et al., 2005). Terminal transferase-mediated dUTP nick-end labeling (TUNEL) revealed no decrease and, at a late stage of the pathology, a substantial increase of death in the caspase-8–deficient epidermis, mainly in its suprabasal layers (Fig. 3 C, top). Assessment of the cell proliferation rate by staining for the Ki67 antigen (Fig. 3 C, bottom) or by bromodeoxyuridine incorporation (not depicted) disclosed considerably enhanced proliferation of keratinocytes in the basal layer. Ki67-positive cells were also found in abundance suprabasally.
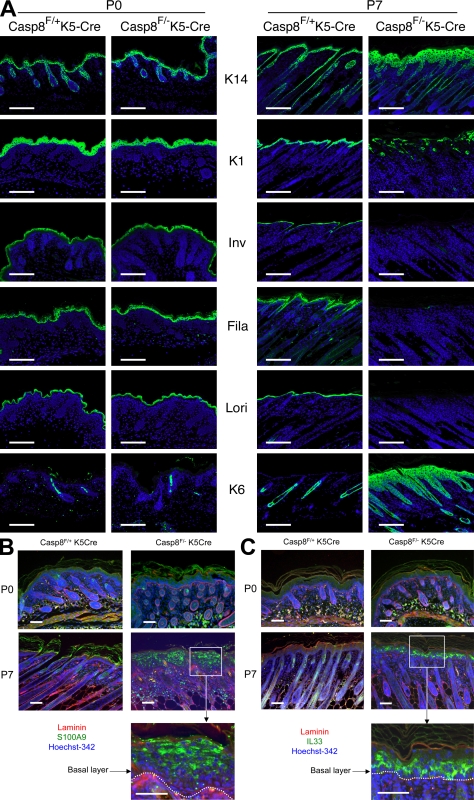
Immunohistological analysis of the caspase-8–deficient skin revealed aberrations of the expression patterns of proteins that characterize distinct steps in keratinocyte differentiation, a common observation in acute cutaneous inflammation. Whereas normally the expression of K14 is restricted to the basal layer, in our mutant mice it was also expressed suprabasally (Fig. 4 A). Moreover, the expression of proteins that mark the intermediate (K1) and late differentiation stages (loricrin and filaggrin) was considerably reduced. The reduction was progressive to the extent that in the lesions of P7 mice those proteins were barely discernible (Fig. 4 A). The deficiency of filaggrin in the epidermis of these mice seemed consistent with the scaly skin and loss of the granular layer, two characteristic features of human skin pathologies resulting from loss-of-function mutations of the filaggrin gene (McGrath, 2008).
Figure 4.
Spatial analysis of the effects of caspase-8 deficiency on protein and gene expression in the epidermis. (A) Immunohistochemical characterization of the caspase-8–deficient epidermis. At P7 (right), immunostaining shows expression of the basal layer marker K14 throughout the Casp-8F/−K5-Cre epidermis. Staining for the intermediate (K1 and involucrin [Inv]) and late (filaggrin [Fila] and loricrin [Lori]) differentiation markers shows either their disorganized localization or their almost complete absence in the Casp-8F/−K5-Cre lesion areas. Also shown is the ubiquitous expression of K6, a general marker for epidermal hyperproliferation. None of these changes was discernible at P0 (left) or P1 (not depicted). Bars, 100 µm. (B and C) In situ hybridization for spatial analysis of up-regulated inflammatory genes in the caspase-8–deficient epidermis. Skin samples of Caspase-8F/− K5-Cre and Caspase-8F/+K5-Cre littermates at P0 and P7 were hybridized with DIG-labeled antisense RNA probe for mRNA of S100a9 (B) or IL33 (C) and then subjected to fluorescence detection (Cy2–anti-DIG, green). Nuclei were counterstained with Hoechst 342 (blue). Rabbit anti-laminin was used to visualize the basal membrane (red), highlighted in the magnified presentation of the insets by dotted lines and detected with Cy3-anti–rabbit Ig. Bars, 50 µm. The data presented in A, B, and C are representative of analyses of 10, 5, and 5 mice, respectively.
Gene expression analysis reveals that visible pathological changes in the skin are preceded by prenatally initiated generation of inflammatory mediators in the epidermis
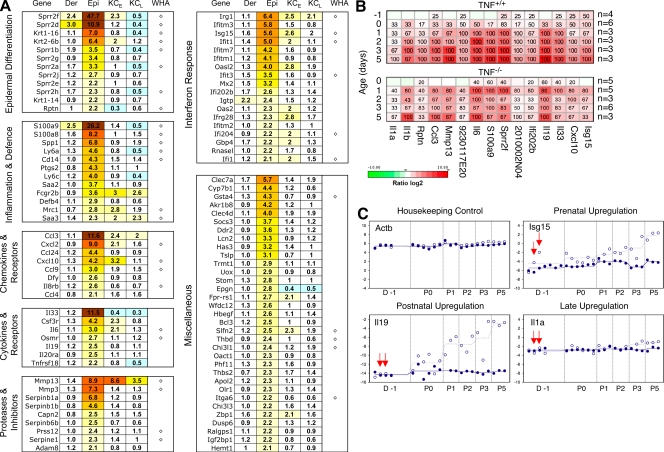
In seeking clues to the molecular changes underlying the inflammatory process in the skin and its trigger, we compared patterns of gene expression in the skin of Casp-8F/−K5-Cre and WT mice at different times before and after birth. Numerous genes were found to be up-regulated in the Casp-8F/−K5-Cre skin, most of them in the epidermis. The up-regulated genes included several groups encoding proteins that play a role in inflammation (Fig. 5 A), and nearly half were genes encoding proteins shown to be induced during epidermal wound healing in neonatal mice (WHA; Cooper et al., 2004). A rather large subgroup of the up-regulated genes were encoders of proteins known to participate in the IFN response (Fig. 5 A and Fig. S3) and to depend on activation of the transcription factor IRF3 (Grandvaux et al., 2002; Andersen et al., 2008).
Figure 5.
Expression-profiling analysis of the Casp-8F/−K5-Cre skin. (A) Functional groups of genes overexpressed in caspase-8 KO skin. Total RNA was extracted from the dermis (Der) or epidermis (Epi) or from cultured keratinocytes at the first and third passages (KCE and KCL, respectively) derived from Casp-8F/−K5-Cre and Casp-8F/+K5-Cre mice at P3. Epidermal or dermal total RNA from two or three mice of each genotype was pooled for genome-wide expression analysis using Agilent microarrays (see Materials and methods). Ratios of mRNA expression in Casp-8F/−K5-Cre and Casp-8F/+K5-Cre samples are presented as heat maps with the corresponding values recorded in the cells. Depicted ratios from keratinocyte samples (KCE and KCL) represent geometric mean values from two microarray experiments. The 100 most strongly up-regulated genes in the caspase-8–deficient mouse epidermis at P3 were depicted and categorized into functional groups according to published references. In addition to the flagged spots, defined as described in Materials and methods, we also excluded Y-chromosomal and poorly annotated transcripts from the list. WHA, genes previously found to be induced during epidermal wound healing in neonatal mice (Cooper et al., 2004). (B and C) Kinetic analysis of gene-expression changes in the caspase-8–null epidermis and assessment of their dependence on TNF. (B) RNA isolated from the epidermal tissues of 22 pairs of Casp-8F/+K5-Cre and Casp-8F/−K5-Cre mice (from eight litters) and from the epidermal tissues of 22 pairs of TNF−/−Casp-8F/+K5-Cre/TNF−/−Casp-8F/−K5-Cre mice (from seven litters) was used for real-time RT-PCR determination of the expression of 15 selected genes representative of most of the functional groups identified in A. For heat map visualization, the log2-transformed expression ratios were ordered according to increasing consistency of changes in mRNA expression in the TNF+/+ mice (horizontal axis). Percentages of Casp-8F/−K5-Cre mice at a given age with a significantly up-regulated transcript relative to its mean expression in all Casp-8F/+K5-Cre age-matched mice are recorded in the heat map boxes. n, number of experimental/control mice pairs analyzed for the given age (also see Materials and methods). (C) RNA was isolated from the epidermis of Casp-8F/−K5-Cre mice (empty circles, values for individual mice born on different dates; broken lines, mean values for all individuals analyzed at a particular age) and of Casp-8F+K5-Cre mice (filled circles and solid lines) 1 d before birth (D-1) and at different times postnatally. For each mouse, mRNA expression of the indicated four genes was analyzed by real-time PCR. Samples from pairs of Casp-8F/−K5-Cre and Casp-8F+K5-Cre mice were tested simultaneously; these pairs are depicted at identical horizontal positions. (In the case of Isg15, four additional D-1 WT samples were analyzed.) Shown are the −Δ cycle-threshold (Ct) values obtained by subtraction of the Ct value of each PCR product from the Ct value of the housekeeping gene Hprt1. Red arrows point to values for two Casp-8F/−K5-Cre mice that showed significant up-regulation of Isg15 expression before birth.
Kinetic analysis disclosed that although initial signs of the pathological change in the skin could be discerned only at about P3, expression of many of the induced genes was enhanced in the Casp-8F/−K5-Cre epidermis before that time. Some were found to be elevated prenatally, 1 d (Fig. 5, B and C [D-1]) and even 2 d (not depicted) before birth.
Cultured keratinocytes derived from the Casp-8F/−K5-Cre mouse epidermis generate inflammatory mediators, but the generation gradually abates
To examine the mechanism underlying the up-regulation of inflammatory genes in the Casp-8F/−K5-Cre epidermis, we tried to establish whether this up-regulation occurs autonomously in the caspase-8–deficient keratinocytes or depends on extracellular stimuli. By applying gene array analysis, we found that some of the inflammatory genes that were up-regulated in the Casp-8F/−K5-Cre epidermis were still activated in keratinocytes derived from that epidermis shortly after their culture was initiated. However, in the course of passaging of the cells this increased expression gradually abated (Fig. 5 A, KCE versus KCL). These findings suggested that in addition to the caspase-8 deficiency in the keratinocytes, some other molecular determinants that occur in the skin and dissipate under our cell culturing conditions contribute to activation of inflammatory genes in these cells.
The disease caused by caspase-8 deficiency in keratinocytes is triggered independently of TNF, IL-1, the TLR adapter proteins, or macrophages
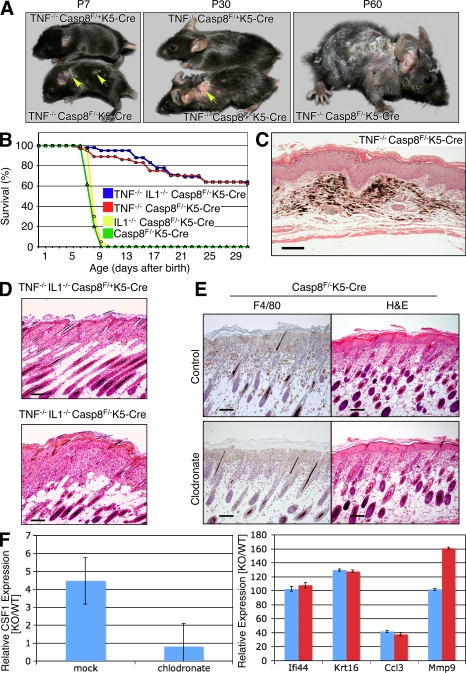
Cells of the immune system and mediators generated by them play crucial roles in various inflammatory skin disorders of humans and in their experimental models (Kupper and Fuhlbrigge, 2004). This is true for an inflammatory skin disorder that was described in mice with mutations preventing the expression or activation of NF-κB transcription factors in keratinocytes (Pasparakis et al., 2002; Gugasyan et al., 2004; van Hogerlinden et al., 2004; Omori et al., 2006; Sayama et al., 2006) and that closely resembles the one seen here in Casp-8F/−K5-Cre mice. The former disorder was found to be fully dependent on accumulation of macrophages in the dermis, as well as on TNF whose expression in the macrophages is high (Pasparakis et al., 2002; Omori et al., 2006). To determine whether TNF or some other macrophage-produced mediator is also responsible for triggering the skin disease in the Casp-8F/−K5-Cre mice, we first assessed the effect of deletion of the TNF or the TNFR1 gene from these mice by crossing them with the corresponding KO mouse strains. Such deletion dramatically increased survival of the offspring (Fig. 6 B and not depicted). Their skin disease was not prevented, however, but only delayed for a few days. In fact, because these mice survived and their skin condition kept worsening, disease severity eventually exceeded that seen in the TNF-proficient Casp-8F/−K5-Cre mice (Fig. 6 A and not depicted). Histologically, late-stage disease in TNF−/−Casp-8F/−K5-Cre mice was characterized by widespread and severe dermal fibrosis, adnexal loss, and persistence of melanin pigment in the dermis, either in melanocytes or in melanophages, after adnexal destruction (pigmentary incontinence; Fig. 6 C). Cutaneous scarring was of such a magnitude that it led to severe contraction and rigidity of the skin, causing the mice to be in permanent kyphosis (Fig. S4). Deletion of the TNFR2 gene in addition to TNFR1 caused no further delay in onset of the skin disorder (unpublished data).
Figure 6.
Limited effect of deficiency of TNF, IL-1, or macrophage depletion on cutaneous disease development in the caspase-8–null skin. (A) Gross appearance of TNF−/−Casp-8F/+K5-Cre and TNF−/−Casp-8F/−K5-Cre mice at different ages. (Arrows point to skin lesions at P7 and P30.) There is progressive hyperkeratosis, scaliness, and hair loss, with ulceration in some cases (middle). (B) Survival curves for colonies of Casp-8F/−K5-Cre (green; 62 mice), IL1α−/−IL1β−/−Casp-8F/−K5-Cre (yellow; 55 mice), TNF−/−Casp-8F/−K5-Cre (red; 37 mice), and IL1α−/−IL1β−/−TNF−/−Casp-8F/−K5-Cre mice (blue; 42 mice). (C) Microscopic features of late-stage disease in a TNF−/−Casp-8F/−K5-Cre mouse at P60. There is diffuse epidermal hyperplasia, dermal fibrosis, adnexal loss, and accumulation of melanocytes and melanophages (pigmentary incontinence). H&E; Bar, 100 µm. (D) Microscopic appearance of the skin of IL1α−/−IL1β−/−TNF−/−Casp-8F/+K5-Cre and IL1α−/−IL1β−/−TNF−/−Casp-8F/−K5-Cre mice at P7. H&E; Bars, 100 µm. (E) Macrophage content (anti-F4/80 staining, left) and microscopic appearance (H&E, right) of the skin of Casp-8F/−K5-Cre mice injected with control liposomes (top) and clodronate-containing liposomes (bottom) at P5. Bars, 100 µM. (F) Effect of macrophage depletion on gene expression changes in the Casp-8F/−K5-Cre epidermis. To deplete dermal macrophages, mice were injected subcutaneously every other day, starting at P0, with clodronate-loaded liposomes or PBS-loaded liposomes (control). RNA was isolated from the epidermis at P5 and analyzed for expression of CSF1, a macrophage marker (left), and of four inflammation markers found to be increased in Casp-8F/−K5-Cre mice (right). Mean values ± SD for three experimental/control mice pairs are shown. The data presented in A, C, D, E, and F are representative of analyses of 10, 4, 7, 15, and 5 mice, respectively. The data in E and F are the results of three independent experiments in both cases.
Like the limited effect of TNF deficiency on development of the skin disease, the absence of TNF had little effect on gene expression in the epidermis. It resulted only in moderation of gene induction, with no evident change in the pattern of the induced genes (Fig. 5 B, compare bottom and top).
An increase in IL-1, which, like TNF, is known to play a major role in the induction of inflammation, was reported to accompany the cutaneous inflammation that results from the arrest of NF-κB activation in the epidermis (Pasparakis et al., 2002). We found that the messenger RNAs (mRNAs) of IL-1α and IL-1β were also increased in the epidermis of our Casp-8F/−K5-Cre mice (Fig. 5 B and not depicted). However, simultaneous deletion of the genes for IL-1α and IL-1β had no effect on the skin pathology in these mice (not depicted), nor did it affect their survival (Fig. 6 B). Deletion of these genes from Casp-8F/−K5-Cre mice that were also deficient in TNF led to a slight further increase in their survival (Fig. 6 B) but did not alleviate the cutaneous inflammation (Fig. 6 D and not depicted).
To determine whether signaling by TLRs contributes to the initiation of the skin disease in the Casp-8F/−K5-Cre mice, we crossed these mice with KO mice for MyD88 and for TRIF, which encode two adapter proteins that mediate TLR-dependent signaling. Neither the rate nor the extent of development of cutaneous inflammation in the Casp-8F/−K5-Cre mice nor the onset of their death was affected by deficiency of either of these proteins (unpublished data).
In mice with an inflammatory skin disease resulting from IKK2 deficiency in the keratinocytes, depletion of macrophages from the dermis by subcutaneous injection of clodronate-containing liposomes abolished the symptoms (Stratis et al., 2006). However, no such effect on the kinetics or extent of development of cutaneous inflammation was detectable in our Casp-8F/−K5-Cre mice (Fig. 6 E and not depicted). Moreover, such treatment did not significantly affect either the pattern or the extent of changes in gene expression in the epidermis (Fig. 6 F and not depicted).
Thus, unlike in the case of the inflammatory skin disease that results from deficiency of the NF-κB activation pathway in keratinocytes, the findings described in this section indicated that triggering of the cutaneous inflammation that results from caspase-8 deficiency in the Casp-8F/−K5-Cre mice occurs independently of TNF or of any other mediator produced by the dermal macrophages. It also does not depend on IL-1–induced or TLR-induced signaling.
The skin disease caused by caspase-8 deficiency in keratinocytes is associated with constitutive signaling for IRF3 activation
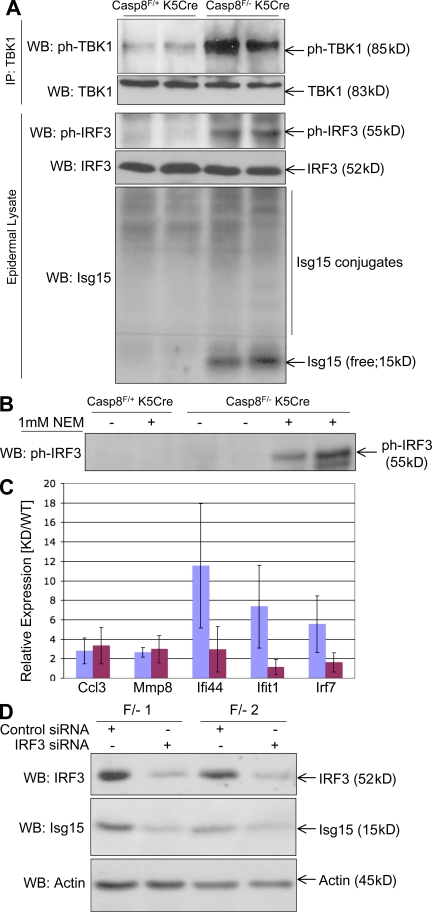
Western analysis of IRF3 in the Casp-8F/−K5-Cre mouse epidermis revealed that this transcription factor is phosphorylated as early as P1 (Fig. 7, A and B). Moreover, the Casp-8F/−K5-Cre epidermis was found to contain the phosphorylated form of TANK-binding kinase (TBK; TBK1), which is known to activate IRF3 (Fig. 7 A), and to have heightened expression of Isg15 (Fig. 7 A), an IRF3-dependent ubiquitin-related polypeptide. This was consistent with our finding that the Isg15 gene was activated in the Casp-8F/−K5-Cre mice epidermis even before birth (Fig. 5, A–C). The analysis also revealed enhanced conjugation of this polypeptide to cellular proteins, a hallmark of the antiviral response (Sadler and Williams, 2008). Although expression of the IRF7 transcript was also enhanced in the Casp-8F/−K5-Cre epidermis, Western analysis of these epidermal cells failed to show any expression of the IRF7 protein in them (unpublished data).
Figure 7.
IRF3 is constitutively activated in the Casp-8F/−K5-Cre epidermis. (A) Western blot analysis of lysates of the epidermis of two Casp-8F/+K5-Cre and two Casp-8F/−K5-Cre littermates at P3 for phosphorylation of TBK1 (ph-TBK1, enriched by immunoprecipitation with anti-TBK antibody) and of IRF3 (ph-IRF3), as well as for expression of IRF3 and of the IRF3-induced protein Isg15, and conjugation of Isg15 to the cellular proteins. (B) Western analysis of ph-IRF3, as in A, for mouse littermates at P1. As illustrated, the small amount of ph-IRF3 in the lysates of the Casp-8F/−K5-Cre epidermis at that date could be discerned only when the cells were lysed in the presence of the sulphhydryl blocking agent N-ethylmaleimide. (C and D) Activation of inflammatory genes in early passage Casp-8F/−K5-Cre keratinocytes is dependent on the presence of IRF3. (C) Relative expression (means ± SD of the indicated gene transcripts in Casp-8F/−K5-Cre mRNA to the same transcripts in Casp-8F/+K5-Cre mRNA) assayed in keratinocytes from three pairs of P1 littermates that were transfected, 1 d after plating, with either control siRNA (blue) or IRF3 siRNA (maroon). (D) Western analysis of the effect of IRF3 siRNA transfection on IRF3 expression and on expression of Isg15 in two of the Casp-8F/−K5-Cre keratinocyte samples used in C. Analysis of the expression of β-actin served as a loading control. The data presented in panels A, B, C, and D are representative of findings obtained in three, two, four, and four independent experiments, respectively.
Knockdown of IRF3 in cultured Casp-8F/−K5-Cre keratinocytes abolished the up-regulation of Isg15, as well as the activation of three out of five other tested genes chosen from those which were still up-regulated in these cells after they were plated for culturing (Fig. 7, C and D). These findings indicated that the constitutive activation of IRF3 in the Casp-8F/−K5-Cre epidermis contributes to the up-regulation of inflammatory mediators in the epidermal cells.
Generation of inflammatory mediators in the epidermis occurs mainly in its suprabasal layer
In seeking clues to the nature of the endogenous stimulus that triggers gene up-regulation in the epidermis, we applied in-situ hybridization to determine the epidermal site at which two of the most strongly up-regulated genes, those encoding the alarmin S100a9 and the cytokine IL33, are activated. As shown in Fig. 4 (B and C), although caspase-8 deletion is induced in the basal cell layer of the epidermis, neither of these genes was expressed at that site but rather was expressed in the higher layers. Expression of S100a9 could also be discerned in some distinct cells in the dermis. These findings suggested that activation of inflammatory genes in the epidermis of the Casp-8F/−K5-Cre mice is triggered by molecular changes that occur suprabasally.
Gene activation by transfected DNA is enhanced in caspase-8–deficient keratinocytes
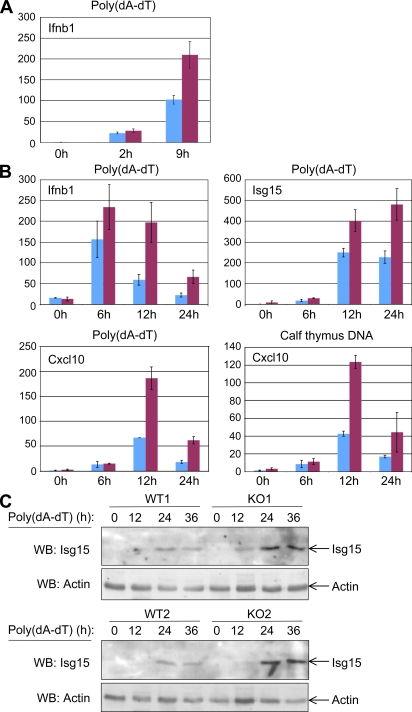
To examine the possibility that damage-associated molecular patterns (DAMPs) generated during cornification contribute to the induction of skin inflammation, we attempted to reproduce the effect of DNA that might be released to the cytoplasm when the nuclei of the cornifying cells break up. We did this by transfecting caspase-8–deficient keratinocytes with DNA. As shown in Fig. 8 (A and B), both knockdown of caspase-8 in vitro and KO of the caspase-8 gene in vivo significantly enhanced the induction of inflammatory cytokines in the cultured DNA-transfected keratinocytes. Expression of the Isg15 protein was also greatly enhance in these cultures (Fig. 8 C). These findings indicate that caspase-8 deficiency facilitates the cellular response to foreign nucleic acid.
Figure 8.
Caspase-8–deficient keratinocytes display an enhanced response to transfected DNA. (A and B) Effect on gene activation. (A) Effect of caspase-8 knockdown. Relative expression of the IFNβ gene (ratio of the IFN-β transcript in caspase-8 knockdown keratinocytes to the same transcripts in mock-transfected cells) assayed in keratinocytes from one pair of P1 littermates that were transfected in duplicate, 1 d after plating, with either control siRNA (blue) or caspase-8 siRNA (maroon) and, 72 h later, transfected with poly(dA-dT) for the indicated times. Shown are mean expression ratios ± SD for the respective duplicate transfections. (B) Effect of caspase-8 KO. Expression of the indicated gene transcripts at different times after DNA transfection of keratinocytes from Casp-8F/+K5-Cre mouse and Casp-8F/−K5-Cre mouse epidermis from two pairs of P1 littermates, 1 d after plating. Cells were transfected either with poly(dA-dT) or with calf thymus DNA, as described in Materials and methods. (C) Effect on Isg15 expression. Western analysis of the expression of Isg15 protein at different times after poly(dA-dT) transfection of keratinocytes from Casp-8F/+K5-Cre and Casp-8F/−K5-Cre epidermis from two pairs of P1 littermates, 1 d after plating, as in A. Analysis of the expression of β-actin served as a loading control. The data presented in each of the panels are representative of findings obtained in two independent experiments.
DISCUSSION
Caspase-8 is known mainly for its role in the induction of cell death. Our investigation of the effect of caspase-8 deficiency on the epidermis disclosed no decrease in the effectiveness of the cornification. This finding is consistent with the results of several earlier studies that cast doubt on the supposition that caspases participate in the programmed cell death process that causes cornification (Lippens et al., 2000; Rendl et al., 2002; Fischer et al., 2005). We found, however, that epidermal deficiency of caspase-8 did have a pronounced effect on skin homeostasis in that it gave rise to a chronic inflammatory disease associated with a hyperproliferative state. In a previous study we found that caspase-8 deficiency in hepatocytes promotes chronic inflammation in the liver after partial hepatectomy (Ben Moshe et al., 2007, 2008). The present study shows that widespread inflammation also develops in mice that express an enzymatically inactive caspase-8 allele. Collectively, these findings suggest that apart from its roles in mediating death and, in certain tissues, also growth and survival, caspase-8 also acts to restrict inflammation.
As yet, little is known of mechanisms that underlie nonapoptotic functions of caspase-8. Nevertheless, there are some clues to the mechanism responsible for the nonapoptotic function of caspase-8 described in the present study. We previously reported that death induction by caspase-8 is compromised in BAC transgenic mice in which the aspartate residue upstream of the initial self-processing site in caspase-8 is mutated, yet these mice show no overt pathology in the skin or other tissues (Kang et al., 2008). Thus, unlike the apoptotic function of caspase-8, and similarly to various nonapoptotic functions of this enzyme, processing of caspase-8 is not required for its antiinflammatory activity. However, our present finding that the mere expression of an enzymatically inactive caspase-8 allele causes inflammation in the skin and in other tissues, apparently because of its interference with the function of coexpressed enzymatically active caspase-8 molecules, suggests that the restriction of inflammation by this enzyme, even though it is independent of its self-processing, does depends on its enzymatic activity. Moreover, the dominant-negative effect of the inactive molecules on the function of the enzymatically active coexpressed molecules suggests that the mechanism by which caspase-8 restricts inflammation depends, as in the case of caspase-8 induction of cell death, on the association of two or more caspase-8 molecules with one another.
Although caspase-3 reportedly also contributes to development of the epidermis through a function that is not apoptotic, this contribution, unlike that of caspase-8, takes place only prenatally (Okuyama et al., 2004). The fact that the skin of capase-3–deficient mice is normal in appearance (Leonard et al., 2002) further suggests that the functional role of caspase-3 in the epidermis is distinct from that of caspase-8. Yet another caspase, caspase-14, serves an important role in controlling epidermal differentiation (Denecker et al., 2007). However, we observed no deficiency in expression or processing of caspase-14 in the caspase-8–deficient epidermis (Fig. S5), suggesting that the function of caspase-8 in the epidermis is also distinct from that of caspase-14.
To obtain further clues to the mechanisms underlying the inflammation caused by caspase-8 deficiency, as well as to suggest potential treatments for skin diseases that might be related to that deficiency, it is important to identify the agents that trigger this inflammation. TNF is known to play a role in the pathogenesis of psoriasis, a skin disease that in some respects resembles the disease seen in our mice (Schottelius et al., 2004); in addition, this cytokine is critically involved in a skin disease of mice with epidermal-specific deletion of genes encoding proteins that participate in NF-κB activation (Pasparakis et al., 2002; Gugasyan et al., 2004; van Hogerlinden et al., 2004; Omori et al., 2006; Sayama et al., 2006). We therefore investigated the extent to which TNF contributes to the skin inflammation caused by caspase-8 deficiency. Our results showed that although TNF expression facilitates development of the disease, it is not crucial for it. We further established that IL-1α and IL-1β, both of which are prominent players in a variety of inflammatory diseases, do not contribute to the mouse cutaneous inflammation. Notably, this last finding refutes the recent suggestion that the induced generation of IL-1α mediates the inflammatory skin disease inflicted by caspase-8 deficiency in the epidermis (Lee et al., 2009). In further disagreement with mechanisms suggested in that study, we also found that this disease was not affected when the mice were injected with a pharmacological inhibitor of the p38 MAP kinase (SB203580; unpublished data), which was suggested by those authors to trigger the generation of IL-1α. In addition, pharmacological depletion of macrophages accumulating in the dermis in both diseases, which protects against the disease caused by interference with NF-κB activation (Stratis et al., 2006), had no effect on our mice. It thus appears that whereas the disease caused by interference with NF-κB activation develops through a dialogue between the keratinocytes and cells of the immune system, the disease caused by caspase-8 deficiency in the keratinocytes reflects a cell-autonomous function of these epidermal cells.
We found that IRF3 and TBK1 (a protein kinase that participates in IRF3 activation) are constitutively phosphorylated in the epidermis of the Casp-8F/−K5-Cre mice and that genes known to be induced by IFN and IRF3 are activated in the epidermis. Phosphorylated IRF3 could be discerned in the epidermis as early as P1, significantly before any pathological change became apparent (the limited sensitivity of western analysis precluded earlier assessment), strongly suggesting that the phosphorylation is a cause rather than a consequence of the disease.
IRF3 is known to be activated by extracellular molecular patterns associated with pathogens (PAMPs). In addition, cells possess mechanisms that also allow this transcriptional factor to be activated in response to intracellular molecular patterns associated with pathogens, or with the molecular changes resulting from cell damage (DAMPs). Some endogenous cellular components can activate these mechanisms even in the absence of pathogens (Chi and Flavell, 2008; Okabe et al., 2008; Stetson et al., 2008). Because the disease in our Casp-8F/−K5-Cre mice became apparent only a few days after birth (even though the gene deletion dictated by the expression of Cre under the K5 promoter occurs prenatally), it would have been reasonable to suspect that it was triggered by antigens and PAMPs, to which the mice were exposed for the first time after birth. This possibility, however, was ruled out by our findings that the generation of inflammatory mediators in the epidermis begins before birth and that the disease was not affected by deletion of MyD88 or TRIF, genes encoding the major adapter proteins for the TLRs which signal for the effects of extracellular PAMPs.
We showed in this paper that caspase-8–deficient keratinocytes exhibit enhanced gene activation by transfected DNA. Recently, we observed that cells deficient in caspase-8 also respond more effectively to viral RNA and that caspase-8 binds to components of the RIG-I signaling complex that mediates cellular responses to intracellular foreign ribonucleic acid, suppressing the complex-induced IRF3 activation but having little effect on the ability of the complex to activate NF-κB (unpublished data). These findings suggest that caspase-8 acts to restrain excessive activation of pathways of response to intracellular pathogenic components and to DAMPs and that its failure to so in the Casp-8F/−K5-Cre keratinocytes contributes to the triggering of the cutaneous disease in Casp-8F/−K5-Cre mice.
The nature of the endogenous cellular components that trigger the disease remains to be further defined. This study suggested that they are generated suprabasally in the epidermis and that their generation is initiated before birth (the time at which cornification starts; Mack et al., 2005) and is greatly enhanced after birth (when cornification reaches its maximal extent). Our findings raise the possibility that generation of these endogenous activators is associated with the differentiation process that leads to the cornification.
MATERIALS AND METHODS
Mice
All experiments in this study were performed with mice on the C57BL/6 background. Mouse strains carrying a knocked-out caspase-8 allele (Casp8−/+ [Varfolomeev et al., 1998]) or a conditional caspase-8 allele (Casp8F/+ [Kang et al., 2004]) and their use in obtaining tissue-specific deletion of the caspase-8 gene (Kang et al., 2004) have been described previously. Mice deficient in TNF (strain B6; 129S6-Tnftm1Gkl/J), TNF-R1 and TNF-R2 (strain B6; 129S-Tnfrsf1atm1Imx Tnfrsf1btm1Imx/J), or TRIF (C57BL/6J-Ticam1Lps2/J [Hoebe et al., 2003]) and the ROSA26 reporter strain (B6.129S4-Gt(ROSA)26Sortm1Sor/J) were obtained from The Jackson Laboratory. Mice expressing Cre under control of the K5 promoter (Ramirez et al., 2004) were obtained from J. Jorcano (Centro de Investigaciones Energéticas, Medioambientales y Tecnológicas, Madrid, Spain). Mice deficient in IL-1α and IL-1β (Yamada et al., 2000) were obtained from Y. Iwakura (Research Institute of Tuberculosis, Kiyose, Tokyo), and mice deficient in MyD88 were provided by S. Akira (Osaka University, Osaka, Japan). All animal protocols were approved by the Ethical Care Committee of the Weizmann Institute of Science.
Antibodies
Rabbit anti-cytokeratin 1, 6, and 14, as well as involucrin, filaggrin, and loricrin antibodies, were purchased from Covance. Rat anti-F4/80 and anti-CD3 were obtained from AbD Serotec, rat anti-Ki67 from Dako, rat anti–mouse Ly6G (Gr1) from eBioscience, rabbit anti-IRF3, rabbit anti–mouse Isg15, and anti–caspase-14 from Santa Cruz Biotechnology, Inc., rabbit anti-TBK1 and rabbit phospho (ph)-IRF3 from Cell Signaling Technology, mouse monoclonal anti–ph-TBK1 from BD, goat anti-TNF from R&D Systems, anti–mouse caspase-8 from Enzo Biochem, Inc., anti–β-actin and anti-laminin from Sigma Aldrich, and Cy2–anti-DIG and Cy3-anti–rabbit Ig from Jackson ImmunoResearch Laboratories. Immunoperoxidase polymer anti–rat was obtained from Nichirei.
Depletion of macrophages from the skin
To deplete skin macrophages, we injected the mice subcutaneously, at 10 sites over the back, with clodronate-loaded liposomes or, as a control, with PBS-loaded liposomes (100 µl/mouse; gift of N. van Rooijen, Vrije Universiteit, Amsterdam, Netherlands) every second day, starting from the day of birth (P0).
Histology and immunostaining
Skin and other organs were fixed in 10% phosphate-buffered formalin, pH 7.4, embedded in paraffin, cut into 4-µm sections, and stained with H&E. Adjacent sections were used for immunostaining with the indicated antibodies according to the manufacturers' instructions. For fluorescent detection we used Cy-2– or Cy-3–coupled secondary antibodies (Jackson ImmunoResearch Laboratories). To visualize nuclei, we further counterstained sections with Hoechst 33342 (Sigma-Aldrich). For chromogenic detection, sections were stained with immunoperoxidase polymer anti–rat antibodies and developed with 3,3′-diaminobenzidine (Sigma-Aldrich) for 10 min. A TUNEL staining kit (Roche Diagnostics) was used to detect apoptotic cells. Frozen sections (10-µm thick) were stained with X-Gal (5-bromo-4-chloro-3-indolyl-d-galactoside) or with phenol red (Ain et al., 2002). All histological and immunohistological analyses shown are representative of at least five sections examined.
In situ hybridization
Probe synthesis and in situ RNA hybridization were performed essentially as described (Darby and Hewitson, 2006). In brief, 6-µm skin sections were prepared from paraffin-embedded tissue blocks. After deparaffinization, slides were treated with 10 µg/ml proteinase-K for 15 min, fixed in 4% paraformaldehyde, and acetylated. The slides were then hybridized with sense or antisense DIG-labeled RNA probe overnight at 65°C. The next day, the slides were washed in saline-sodium citrate buffer and incubated with Cy2-labeled anti-DIG antibody for 1 h at room temperature. The data shown are representative of analysis of at least five different sections.
Western analysis of epidermal proteins
The epidermis and dermis were pulled apart after incubation for 2–3 s at 65°C. Epidermis samples were lysed in RIPA buffer (20 mM Tris-HCl, pH 7.5, 50 mM NaCl, 0.5% NP-40, 4 mM EDTA, 0.1% SDS, and 0.5% sodium deoxycholate) supplemented with 1 mM PMSF, 50 mM NaF, 1 mM sodium vanadate, 80 mM β-glycerophosphate, and a cocktail of protease inhibitors (Roche). For detection of ph-IRF3 in the epidermis at P1, we further supplemented the lysis buffer with 1 mM N-ethylmaleimide (Sigma-Aldrich). This greatly facilitated detection, probably by blocking the function of some potent protein phosphatases which had remained active in spite of the presence of phosphatase inhibitors in the lysis buffer.
Keratinocyte culture
Skin was removed from the mice and incubated overnight at 4°C in 2.4 U/ml Dispase (Roche) to separate the epidermis from the dermis. The keratinocytes were then dissociated by incubation of the epidermis for 10 min in 0.1% trypsin, recovered by centrifugation, and plated on tissue culture dishes in keratinocyte serum-free medium containing 50 µg/ml bovine pituitary extract and 5 ng/ml of epidermal growth factor (Invitrogen) supplemented with antibiotics. For passaging, cells were detached by incubation in PBS containing 0.1% trypsin, 0.5 mM EDTA, and 0.1% glucose and replated in keratinocyte serum-free medium.
Genotyping of mice
Deletion of caspase-8 from the skins of Casp-8F/−K5-Cre and Casp-8F/+K5-Cre mice was quantitatively assessed by PCR, as previously described (Kang et al., 2004).
RNA interference
Keratinocytes were plated in 12-well plates at 50,000 cells/well and transfected 1 d later with 500 nmol of small interfering RNA (siRNA) duplex per well (Accell; Thermo Fisher Scientific). After 72 h the cells were processed for analysis.
DNA transfection
Keratinocytes were transfected either with 1 µg/ml poly(deoxyadenylic-thymidylic) (poly(dA-dT); Sigma-Aldrich) or calf thymus DNA (Sigma-Aldrich) using Lipofectamine 2000 (Invitrogen) as specified by the manufacturer.
Real-time PCR
We isolated RNA from epidermis and dermis samples (peeled apart after brief heating as described in Western analysis of epidermal proteins) using the RNeasy Fibrous Tissue mini kit, and from cultured keratinocytes using the RNeasy kit (QIAGEN). 1 µg of total RNA was reverse transcribed into complementary DNA (cDNA) using 50 U BioScript RNase H Low (Bioline). Aliquots (20 ng, 1:50 dilution) of this reaction mixture were used to amplify cDNAs with Assays-on-Demand (Applied Biosystems) on an ABI7500 real-time PCR system. The Ct value for each individual PCR product was calculated from duplicate measurements by means of the instrument's software, and the Ct values obtained were normalized by subtracting the Ct values obtained for Hprt1 as a housekeeping gene. The resulting mean −ΔCt values from WT or KO mice were then used to calculate relative changes of mRNA expression as the ratio (R) of mRNA expression, according to the following equation: R(log2) = (mean − ΔCt(caspase-8−/−)) − (mean − ΔCt(WT)). The increase in gene expression for individual KO mice was defined as significant if the −ΔCt value obtained for a given gene and a given KO mouse was greater than the mean −ΔCt + 2 SD of all WT mice of corresponding ages. Heat maps were visualized by importing log2-transformed ratio data into the Mayday program (http://www.zbit.uni-tuebingen.de/pas/mayday/mayday.html).
Microarray experiments
The Mouse Oligo Microarray V2 (G4121B, design ID 13326; Agilent Technologies) used in this study contained oligonucleotide probes directed against 19,307 endogenous mRNAs. RNA was used to prepare Cy3- or Cy5-labeled cRNA by oligo-(dT)-T7–primed double-stranded cDNA synthesis (cDNA Synthesis System; Roche), followed by in vitro transcription and labeling (Amino Allyl MessageAmp II kit; Applied Biosystems) as directed by the manufacturer. For the cRNA mixture, the fragment length, yield, and labeling efficiency were analyzed by a 2100 Bioanalyzer (Agilent Technologies) and a NanoDrop ND1000 spectrophotometer. cRNA populations (differentially labeled with 3 µg each of Cy3 and Cy5) were combined for cohybridization onto a single microarray. cRNA fragmentation, hybridization, and washing steps were performed according to Two-Color Microarray-Based Gene Expression Analysis Protocol V5.0.1 (Agilent Technologies; for details see http://www.agilent.com).
Hybridized and washed arrays were scanned on a 428 Array Scanner (Affymetrix). Data were extracted using of Imagene V5.0 software (BioDiscovery). Fluorescence intensity values were normalized according to LOWESS-like calculations. Data from qualitatively suboptimal spots were flagged and excluded from further analysis if CVsignal mean > 0.20 or if SMglobal background corrected/SMlocal background corrected > 1.3, where CVsignal mean is the coefficient of variance deduced from signal values for all pixels of a spot and SMglobal background corrected/SMlocal background corrected is the ratio of a global background-corrected signal mean value of a spot divided by its local background-corrected signal mean value.
mRNA expression analysis of cultured keratinocytes was performed using Mouse Whole Genome 4 × 44k Microarrays (G4122F, design ID 014868; Agilent Technologies). Sample processing, hybridization, data extraction, and normalization were performed exactly as recommended by the manufacturer. The microarray-based data discussed in this publication have been deposited at NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession no. GSE17365.
Modification of a BAC encompassing caspase-8 and generation of BAC-transgenic mice
Modification of a BAC encompassing the caspase-8 gene.
RP24-238B22, a BAC clone encompassing the caspase-8 gene (RPCI-24 mouse [C57BL/6] BAC library) was obtained from CHORI (BACPAC Resources). DH10B bacteria harboring the RP24-238B22 BAC clone were grown in LB medium containing 12.5 µg/ml chloramphenicol. The presence of caspase-8 in the bacterial colonies was verified by PCR using oligonucleotides exon8F-8R, exon1F-1R, and 5′UTRF-R (the oligonucleotides used are listed in Table S1). The BAC clone was modified essentially as previously described (Gong et al., 2002; Sparwasser et al., 2004) using the pDelsac shuttle vector both for deletion of the SacB gene from the RP24-238B22 clone and for modification of the caspase-8 gene.
Four modifications (A–D) were introduced (Fig. S6, scheme). (A) To monitor the expression of transgenic BAC in vivo, we placed an IRES sequence and the GFP ORF flanked by the two sequences corresponding to the FRT recombination site downstream of the stop codon of caspase-8. To introduce this sequence into the shuttle vector, we inserted the homology arms upstream and downstream of the stop codon (BoxA and BoxB; Fig. S5, scheme), cloned from the RP24-238B22 DNA by PCR, into the SalI–EcoRI and SpeI–NotI sites of the pBC FRT-IRES/GFP-FRT vector. The AF-AR and BF-BR oligonucleotides were used for generation of the Caspase-8 FRT-IRES-GFP construct, and the AF-AR2 and BR-BR oligonucleotides were used for generation of the Caspase-8/T7 tag/FRT-IRES-GFP construct. The composite insert was then transferred to the shuttle vector. (B) To aid the monitoring of transgenic BAC expression, we fused the T7 epitope tag (MASMTGGQQMG) to the C terminus of caspase-8 by introducing the T7 coding sequence to the EcoRI site in pBC FRT-IRES/GFP-FRT containing BoxA (derived from the AF-AR2 PCR product) and BoxB of (A). (C) To facilitate genotyping of the transgenic mice, we exchanged a 20-nt sequence within the first caspase-8 intron for a unique sequence (with equal nucleotide proportions). BoxC and BoxD were amplified by the use of primers CF-CR and DF-DR, respectively. We then fused these two PCR products by using them as templates in a second PCR, in which the CF and DR primers were used.
The data presented in this study were obtained with mice that express the BAC with all of these three (A–C) modifications. Several distinct mouse strains harboring the WT BAC and two strains harboring the mutant BAC (see Generation and analysis of BAC-transgenic mouse) were generated. Identical findings were obtained with mice expressing BAC clones in which only the first modification (IRES/GFP) was introduced. (D) A catalytically inactive caspase-8 BAC transgenic mouse was generated by replacement of the active-site cysteine at position 362 with serine (C362S). BoxI and BoxJ, which border this sequence, were amplified and fused by PCR using primers Box1S-IR and JF-Box2AS. All of the modification cassettes (A–D) were cloned into the AscI–NotI sites of the pDelsac shuttle vector.
500 ng of the shuttle vector was transformed into 50 µl of electrocompetent BAC-containing bacteria using a MicroPulser Electroporator (Bio-Rad Laboratories) at 1.8 kV. After electroporation, the cells were incubated for 1 h at 37°C in 1 ml of SOC medium and then selected in LB medium containing 12.5 µg/ml chloramphenicol and 30 µg/ml ampicillin overnight. The cells were diluted and grown on solid LB medium containing 12.5 µg/ml chloramphenicol and 50 µg/ml ampicillin. Co-integrates were identified by direct PCR on individual colonies.
Two or three positive bacterial colonies of each kind were then cultured in 5% sucrose to select for resolved BACs, and this was followed by further selection in which we picked ampicillin-sensitive colonies. Positive colonies were validated by direct PCR (TB3F-TB3R and TB4F-TB4R for AB or ATBCD and 1130F-1370R and MutCF-1370R for CD) and for IJ by HindIII restriction analysis after PCR performed with the IF2-JR1 primer pair.
Isolation of BAC DNA for microinjection.
BAC DNA was isolated by double acetate precipitation and cesium chloride gradient ultracentrifugation. After being washed with ethanol, the BAC DNA was dissolved in TE buffer, linearized by digestion with PI–SceI endonuclease (NEB), and drop dialyzed for 6 h against microinjection buffer (10 mM Tris, pH 7.5, 0.1 mM EDTA, pH 8.0, and 100 mM NaCl) by floating on a 0.025 µm membrane filter disc (Millipore). The quality and quantity of BAC DNA were assessed by pulse-field gel electrophoresis (PFEG; 5 V/cm, 120° angle, linear ramping time 5−120 s for 24−30 h) in 1% agarose using the CHEF-DR III PFEG system (Bio-Rad Laboratories). The DNA was diluted to 2 ng/µl in microinjection buffer and mixed with an equal volume of 2× polyamine (60 mM spermine and 140 mM spermidine in injection buffer) for the injection.
Generation and analysis of BAC-transgenic mouse.
1 ng/µl DNA was injected into the pronuclei of fertilized oocytes derived from CBF1 or C57BL/6 mice. Transgenic mice were identified both by PCR analysis (using primers Mut CF and IN1-1730R) of genomic DNA prepared from tail biopsies and by FACS analysis for GFP expression in peripheral blood leukocytes. Catalytically inactive caspase-8 transgenic mice were distinguished from mice harboring active caspase-8 by the use of primers IF2-JR1 for PCR followed by digestion with HindIII.
For Western blot analysis of extracts of various tissues of the BAC transgenic mice, tissue lysates were prepared by homogenization in lysis buffer (1% SDS, 1 mM sodium orthovanadate, and 10 mM Tris, pH 7.4) followed by boiling for 5 min. Protein concentration was determined with the BCA protein assay kit (Thermo Fisher Scientific). Aliquots of 25 µg of protein were then analyzed by SDS–PAGE and immunoblotted with anti–mouse caspase-8 (3B10; Enzo Biochem, Inc.), anti-GFP, and anti–β-actin monoclonal antibodies (Sigma-Aldrich).
Online supplemental material
Fig. S1 shows the gastric inflammation in Casp-8F/−K5-Cre mice. Fig. S2 shows the expression pattern of E-cadherin in the Casp-8F/−K5-Cre epidermis. Fig. S3 presents an assessment of the expression of several IRF3 target genes in the Casp-8F/−K5-Cre epidermis. Fig. S4 compares radiographs of TNF−/−Casp-8F/+K5-Cre and TNF−/−Casp-8F/−K5-Cre mice. Fig. S5 shows western analysis of the expression of caspase-14 in the Casp-8F/−K5-Cre epidermis. Fig. S6 is a diagrammatic presentation of the strategy for BAC modification in this study. Table S1 lists the oligonucleotides used for BAC transgenesis. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20090616/DC1.
Acknowledgments
We thank Drs. Nathaniel Heintz, Shiaoching Gong, Gérard Eberl, Nathalie Uyttersprot, and Yael Pewzner-Jung for their advice and help with BAC transgenesis, Karen Laviv for assistance in BAC DNA modification, Golda Damari for BAC DNA microinjection, Dr. Jose Jorcano for the K5-Cre transgenic mice, Dr. Shizuo Akira for the MyD88-deficient mice, Dr. Marina Gartsbein for advice on culturing of keratinocytes, Shoshana Grossfeld for maintenance of the mice, Jadi Natan, Tamara Berkutzki, and Calanit Raanan for assistance with histology, Inna Kolesnik, Tatiana Shalevich, and Dvir Mintz for genotyping the mice, Heike Schneider for performance of microarray and real-time PCR experiments, Kelly Jackson (University Health Network Microarray Centre, Toronto, Canada) for microarray hybridization and data analysis, and Dr. Nico van Rooijen for the generous gift of clodronate-loaded liposomes. Our thanks are also due to Drs. Cord Brakebusch, Uri Gat, Tamar Tennenbaum, and Dina Ron for helpful advice and discussions and to Shirley Smith for scientific editing. Special thanks to Professor Israel Vlodavsky for assistance. Microarray data analyses were performed using BRB ArrayTools developed by Dr. Richard Simon and Amy Peng Lam.
This work was supported in part by grants from Ares Trading SA (Switzerland), a Center of Excellence Grant from the Flight Attendant Medical Research Institute, and the Kekst Family Center for Medical Genetics and the Shapell Family Center for Genetic Disorders Research at The Weizmann Institute of Science.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- BAC
- bacterial artificial chromosome
- cDNA
- complementary DNA
- Ct
- cycle-threshold
- DAMP
- damage-associated molecular pattern
- IRES
- internal ribosome entry site
- IRF
- IFN regulatory factor
- K5
- keratin 5
- mRNA
- messenger RNA
- PAMP
- pathogen-associated molecular pattern
- siRNA
- small interfering RNA
- TBK
- TANK-binding kinase
- TLR
- toll-like receptor
- TUNEL
- terminal transferase-mediated dUTP nick-end labeling
References
- Ain R., Tash J.S., Soares M.J. 2002. A simple method for the in situ detection of eosinophils.J. Immunol. Methods. 260:273–278 doi:10.1016/S0022-1759(01)00526-9 [DOI] [PubMed] [Google Scholar]
- Allombert-Blaise C., Tamiji S., Mortier L., Fauvel H., Tual M., Delaporte E., Piette F., DeLassale E.M., Formstecher P., Marchetti P., Polakowska R. 2003. Terminal differentiation of human epidermal keratinocytes involves mitochondria- and caspase-dependent cell death pathway.Cell Death Differ. 10:850–852 doi:10.1038/sj.cdd.4401245 [DOI] [PubMed] [Google Scholar]
- Andersen J., VanScoy S., Cheng T.F., Gomez D., Reich N.C. 2008. IRF-3-dependent and augmented target genes during viral infection.Genes Immun. 9:168–175 doi:10.1038/sj.gene.6364449 [DOI] [PubMed] [Google Scholar]
- Ben Moshe T., Barash H., Kang T.B., Kim J.C., Kovalenko A., Gross E., Schuchmann M., Abramovitch R., Galun E., Wallach D. 2007. Role of caspase-8 in hepatocyte response to infection and injury in mice.Hepatology. 45:1014–1024 doi:10.1002/hep.21495 [DOI] [PubMed] [Google Scholar]
- Ben Moshe T., Kang T.-B., Kovalenko A., Barash H., Abramovitch R., Galun E., Wallach D. 2008. Cell-autonomous and non-cell-autonomous functions of caspase-8.Cytokine Growth Factor Rev. 19:209–217 doi:10.1016/j.cytogfr.2008.04.012 [DOI] [PubMed] [Google Scholar]
- Candi E., Schmidt R., Melino G. 2005. The cornified envelope: a model of cell death in the skin.Nat. Rev. Mol. Cell Biol. 6:328–340 doi:10.1038/nrm1619 [DOI] [PubMed] [Google Scholar]
- Chi H., Flavell R.A. 2008. Innate recognition of non-self nucleic acids.Genome Biol. 9:211 doi:10.1186/gb-2008-9-3-211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper L., Johnson C., Burslem F., Martin P. 2004. Wound healing and inflammation genes revealed by array analysis of ‘macrophageless’ PU.1 null mice.Genome Biol. 6:R5 doi:10.1186/gb-2004-6-1-r5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darby I.A., Hewitson T.D. 2006. In Situ Hybridization Protocols. Humana Press Inc., New Jersey: 269 pp [Google Scholar]
- Denecker G., Hoste E., Gilbert B., Hochepied T., Ovaere P., Lippens S., Van den Broecke C., Van Damme P., D'Herde K., Hachem J.P., et al. 2007. Caspase-14 protects against epidermal UVB photodamage and water loss.Nat. Cell Biol. 9:666–674 doi:10.1038/ncb1597 [DOI] [PubMed] [Google Scholar]
- Denecker G., Ovaere P., Vandenabeele P., Declercq W. 2008. Caspase-14 reveals its secrets.J. Cell Biol. 180:451–458 doi:10.1083/jcb.200709098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eidsmo L., Nylen S., Khamesipour A., Hedblad M.A., Chiodi F., Akuffo H. 2005. The contribution of the Fas/FasL apoptotic pathway in ulcer formation during Leishmania major-induced cutaneous Leishmaniasis.Am. J. Pathol. 166:1099–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer H., Rossiter H., Ghannadan M., Jaeger K., Barresi C., Declercq W., Tschachler E., Eckhart L. 2005. Caspase-14 but not caspase-3 is processed during the development of fetal mouse epidermis.Differentiation. 73:406–413 [DOI] [PubMed] [Google Scholar]
- Gong S., Yang X.W., Li C., Heintz N. 2002. Highly efficient modification of bacterial artificial chromosomes (BACs) using novel shuttle vectors containing the R6Kgamma origin of replication.Genome Res. 12:1992–1998 doi:10.1101/gr.476202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandvaux N., Servant M.J., tenOever B., Sen G.C., Balachandran S., Barber G.N., Lin R., Hiscott J. 2002. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes.J. Virol. 76:5532–5539 doi:10.1128/JVI.76.11.5532-5539.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gugasyan R., Voss A., Varigos G., Thomas T., Grumont R.J., Kaur P., Grigoriadis G., Gerondakis S. 2004. The transcription factors c-rel and RelA control epidermal development and homeostasis in embryonic and adult skin via distinct mechanisms.Mol. Cell. Biol. 24:5733–5745 doi:10.1128/MCB.24.13.5733-5745.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoebe K., Du X., Georgel P., Janssen E., Tabeta K., Kim S.O., Goode J., Lin P., Mann N., Mudd S., et al. 2003. Identification of Lps2 as a key transducer of MyD88-independent TIR signalling.Nature. 424:743–748 doi:10.1038/nature01889 [DOI] [PubMed] [Google Scholar]
- Kang T.B., Ben-Moshe T., Varfolomeev E.E., Pewzner-Jung Y., Yogev N., Jurewicz A., Waisman A., Brenner O., Haffner R., Gustafsson E., et al. 2004. Caspase-8 serves both apoptotic and nonapoptotic roles.J. Immunol. 173:2976–2984 [DOI] [PubMed] [Google Scholar]
- Kang T.B., Oh G.S., Scandella E., Bolinger B., Ludewig B., Kovalenko A., Wallach D. 2008. Mutation of a self-processing site in caspase-8 compromises its apoptotic but not its nonapoptotic functions in bacterial artificial chromosome-transgenic mice.J. Immunol. 181:2522–2532 [DOI] [PubMed] [Google Scholar]
- Kupper T.S., Fuhlbrigge R.C. 2004. Immune surveillance in the skin: mechanisms and clinical consequences.Nat. Rev. Immunol. 4:211–222 doi:10.1038/nri1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamkanfi M., Festjens N., Declercq W., Vanden Berghe T., Vandenabeele P. 2007. Caspases in cell survival, proliferation and differentiation.Cell Death Differ. 14:44–55 doi:10.1038/sj.cdd.4402047 [DOI] [PubMed] [Google Scholar]
- Lee P., Lee D.J., Chan C., Chen S.W., Ch'en I., Jamora C. 2009. Dynamic expression of epidermal caspase 8 simulates a wound healing response.Nature. 458:519–523 doi:10.1038/nature07687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard J.R., Klocke B.J., D'Sa C., Flavell R.A., Roth K.A. 2002. Strain-dependent neurodevelopmental abnormalities in caspase-3-deficient mice.J. Neuropathol. Exp. Neurol. 61:673–677 [DOI] [PubMed] [Google Scholar]
- Li N., Zhao M., Wang J., Liu Z., Diaz L.A. 2009. Involvement of the apoptotic mechanism in pemphigus foliaceus autoimmune injury of the skin.J. Immunol. 182:711–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippens S., Kockx M., Knaapen M., Mortier L., Polakowska R., Verheyen A., Garmyn M., Zwijsen A., Formstecher P., Huylebroeck D., et al. 2000. Epidermal differentiation does not involve the pro-apoptotic executioner caspases, but is associated with caspase-14 induction and processing.Cell Death Differ. 7:1218–1224 doi:10.1038/sj.cdd.4400785 [DOI] [PubMed] [Google Scholar]
- Mack J.A., Anand S., Maytin E.V. 2005. Proliferation and cornification during development of the mammalian epidermis.Birth Defects Res. C Embryo Today. 75:314–329 doi:10.1002/bdrc.20055 [DOI] [PubMed] [Google Scholar]
- McGrath J.A. 2008. Filaggrin and the great epidermal barrier grief.Australas. J. Dermatol. 49:67–73 doi:10.1111/j.1440-0960.2008.00443.x [DOI] [PubMed] [Google Scholar]
- Okabe Y., Kawane K., Nagata S. 2008. IFN regulatory factor (IRF) 3/7-dependent and -independent gene induction by mammalian DNA that escapes degradation.Eur. J. Immunol. 38:3150–3158 doi:10.1002/eji.200838559 [DOI] [PubMed] [Google Scholar]
- Okuyama R., Nguyen B.C., Talora C., Ogawa E., Tommasi di Vignano A., Lioumi M., Chiorino G., Tagami H., Woo M., Dotto G.P. 2004. High commitment of embryonic keratinocytes to terminal differentiation through a Notch1-caspase 3 regulatory mechanism.Dev. Cell. 6:551–562 doi:10.1016/S1534-5807(04)00098-X [DOI] [PubMed] [Google Scholar]
- Omori E., Matsumoto K., Sanjo H., Sato S., Akira S., Smart R.C., Ninomiya-Tsuji J. 2006. TAK1 is a master regulator of epidermal homeostasis involving skin inflammation and apoptosis.J. Biol. Chem. 281:19610–19617 doi:10.1074/jbc.M603384200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasparakis M., Courtois G., Hafner M., Schmidt-Supprian M., Nenci A., Toksoy A., Krampert M., Goebeler M., Gillitzer R., Israel A., et al. 2002. TNF-mediated inflammatory skin disease in mice with epidermis-specific deletion of IKK2.Nature. 417:861–866 doi:10.1038/nature00820 [DOI] [PubMed] [Google Scholar]
- Ramirez A., Page A., Gandarillas A., Zanet J., Pibre S., Vidal M., Tusell L., Genesca A., Whitaker D.A., Melton D.W., Jorcano J.L. 2004. A keratin K5Cre transgenic line appropriate for tissue-specific or generalized Cre-mediated recombination.Genesis. 39:52–57 doi:10.1002/gene.20025 [DOI] [PubMed] [Google Scholar]
- Rendl M., Ban J., Mrass P., Mayer C., Lengauer B., Eckhart L., Declerq W., Tschachler E. 2002. Caspase-14 expression by epidermal keratinocytes is regulated by retinoids in a differentiation-associated manner.J. Invest. Dermatol. 119:1150–1155 doi:10.1046/j.1523-1747.2002.19532.x [DOI] [PubMed] [Google Scholar]
- Sadler A.J., Williams B.R. 2008. Interferon-inducible antiviral effectors.Nat. Rev. Immunol. 8:559–568 doi:10.1038/nri2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayama K., Hanakawa Y., Nagai H., Shirakata Y., Dai X., Hirakawa S., Tokumaru S., Tohyama M., Yang L., Sato S., et al. 2006. Transforming growth factor-beta-activated kinase 1 is essential for differentiation and the prevention of apoptosis in epidermis.J. Biol. Chem. 281:22013–22020 doi:10.1074/jbc.M601065200 [DOI] [PubMed] [Google Scholar]
- Schottelius A.J., Moldawer L.L., Dinarello C.A., Asadullah K., Sterry W., Edwards C.K., III 2004. Biology of tumor necrosis factor-alpha- implications for psoriasis.Exp. Dermatol. 13:193–222 doi:10.1111/j.0906-6705.2004.00205.x [DOI] [PubMed] [Google Scholar]
- Sparwasser T., Gong S., Li J.Y., Eberl G. 2004. General method for the modification of different BAC types and the rapid generation of BAC transgenic mice.Genesis. 38:39–50 doi:10.1002/gene.10249 [DOI] [PubMed] [Google Scholar]
- Stetson D.B., Ko J.S., Heidmann T., Medzhitov R. 2008. Trex1 prevents cell-intrinsic initiation of autoimmunity.Cell. 134:587–598 doi:10.1016/j.cell.2008.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratis A., Pasparakis M., Rupec R.A., Markur D., Hartmann K., Scharffetter-Kochanek K., Peters T., van Rooijen N., Krieg T., Haase I. 2006. Pathogenic role for skin macrophages in a mouse model of keratinocyte-induced psoriasis-like skin inflammation.J. Clin. Invest. 116:2094–2104 doi:10.1172/JCI27179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautmann A., Akdis M., Kleemann D., Altznauer F., Simon H.U., Graeve T., Noll M., Bröcker E.B., Blaser K., Akdis C.A. 2000. T cell-mediated Fas-induced keratinocyte apoptosis plays a key pathogenetic role in eczematous dermatitis.J. Clin. Invest. 106:25–35 doi:10.1172/JCI9199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunggal J.A., Helfrich I., Schmitz A., Schwarz H., Günzel D., Fromm M., Kemler R., Krieg T., Niessen C.M. 2005. E-cadherin is essential for in vivo epidermal barrier function by regulating tight junctions.EMBO J. 24:1146–1156 doi:10.1038/sj.emboj.7600605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hogerlinden M., Rozell B.L., Toftgård R., Sundberg J.P. 2004. Characterization of the progressive skin disease and inflammatory cell infiltrate in mice with inhibited NF-kappaB signaling.J. Invest. Dermatol. 123:101–108 doi:10.1111/j.0022-202X.2004.22706.x [DOI] [PubMed] [Google Scholar]
- Varfolomeev E.E., Schuchmann M., Luria V., Chiannilkulchai N., Beckmann J.S., Mett I.L., Rebrikov D., Brodianski V.M., Kemper O.C., Kollet O., et al. 1998. Targeted disruption of the mouse Caspase 8 gene ablates cell death induction by the TNF receptors, Fas/Apo1, and DR3 and is lethal prenatally.Immunity. 9:267–276 doi:10.1016/S1074-7613(00)80609-3 [DOI] [PubMed] [Google Scholar]
- Viard I., Wehrli P., Bullani R., Schneider P., Holler N., Salomon D., Hunziker T., Saurat J.H., Tschopp J., French L.E. 1998. Inhibition of toxic epidermal necrolysis by blockade of CD95 with human intravenous immunoglobulin.Science. 282:490–493 doi:10.1126/science.282.5388.490 [DOI] [PubMed] [Google Scholar]
- Weil M., Raff M.C., Braga V.M. 1999. Caspase activation in the terminal differentiation of human epidermal keratinocytes.Curr. Biol. 9:361–364 doi:10.1016/S0960-9822(99)80162-6 [DOI] [PubMed] [Google Scholar]
- Yamada H., Mizumo S., Horai R., Iwakura Y., Sugawara I. 2000. Protective role of interleukin-1 in mycobacterial infection in IL-1 alpha/beta double-knockout mice.Lab. Invest. 80:759–767 [DOI] [PubMed] [Google Scholar]
- Zhuang L., Wang B., Sauder D.N. 2000. Molecular mechanism of ultraviolet-induced keratinocyte apoptosis.J. Interferon Cytokine Res. 20:445–454 doi:10.1089/10799900050023852 [DOI] [PubMed] [Google Scholar]