Abstract
The E2A gene products, E12 and E47, are critical regulators of B cell development. However, it remains elusive whether E12 and E47 have overlapping and/or distinct functions during B lymphopoiesis. We have generated mice deficient for either E12 or E47 and examined their roles in B cell maturation. We show that E47 is essential for developmental progression at the prepro–B cell stage, whereas E12 is dispensable for early B cell development, commitment, and maintenance. In contrast, both E12 and E47 play critical roles in pre–B and immature B cells to promote immunoglobulin λ (Igλ) germline transcription as well as Igλ VJ gene rearrangement. Furthermore, we show that E12 as well as E47 is required to promote receptor editing upon exposure to self-antigen. We demonstrate that increasing levels of E12 and E47 act to induce Igλ germline transcription, promote trimethylated lysine 4 on histone 3 (H3) as well as H3 acetylation across the Jλ region, and activate Igλ VJ gene rearrangement. We propose that in the pre–B and immature B cell compartments, gradients of E12 and E47 activities are established to mechanistically regulate the sequential rearrangement of the Ig light chain genes.
B cells develop in a multistep process from hematopoietic stem cells (HSCs) and lymphoid-primed multipotent progenitors (LMPPs) into committed B cells that express a B cell receptor (BCR; Kee and Paige, 1995; Hardy et al., 2007). B cell development has been well characterized mainly based on the developmental stage-specific rearrangement of the Ig heavy chain (IgH) and light chain loci (Alt et al., 1984; Cedar and Bergman, 1999). In pro–B cells, IgH DJ rearrangement precedes that of VDJ joining. Upon expression of a productive VDJ gene rearrangement, a pre-BCR is formed that acts, in turn, to suppress the expression of the RAG1 and RAG2 proteins, and to promote the survival and proliferation of developing large pre–B cells. The proliferation phase is followed by cell-cycle arrest, during which RAG gene expression is reinduced to permit Ig light chain gene rearrangement. In the presence of self-reactivity, continued Ig light chain DNA recombination will replace primary Igκ VJ joints, ultimately generating BCRs with novel and innocuous specificities (Radic et al., 1993; Tiegs et al., 1993). Once a BCR has formed that lacks self-reactivity, tonic signaling mediated by the BCR will permanently inhibit RAG1 and RAG2 gene expression and promote positive selection. Positively selected B cells migrate to the peripheral lymphoid organs where they, upon interacting with pathogenic determinant, will undergo class switch recombination and somatic hypermutation, and differentiate into plasma or memory B cells (Gellert, 2002; Nemazee, 2006).
The specification and commitment of hematopoietic progenitors to the B cell lineage and their maturation into mature B-lineage cells requires the activities of multiple transcription factors, including E2A, early B cell factor (EBF), and Pax5 (Nutt and Kee, 2007). The E2A proteins E12 and E47, which arise through alternative splicing of the Tcfe2a gene, belong to a subset of helix-loop-helix (HLH) proteins named E proteins. E proteins are transcriptional regulators that contain an HLH dimerization domain, as well as a basic DNA binding region that is located immediately N terminal to the HLH domain. E proteins form homodimers or heterodimers with other E proteins or other members of the HLH family (Murre, 2005). These protein complexes have the ability to act either as transcriptional activators or repressors. In vertebrates, four E proteins have been identified. These include the E2A proteins, E12 and E47, which only differ in their basic DNA binding region, as well as HEB and E2-2, both of which give rise to distinct isoforms generated through alternative initiation of transcription (Corneliussen et al., 1991; Wang et al., 2006). E proteins also have the capacity to interact with antagonistic HLH proteins, named Id proteins. Id proteins contain an HLH domain but lack a basic region, and upon interacting with E proteins, inactivate their DNA binding activity (Benezra et al., 1990). E and Id proteins are widely expressed throughout the entire hematopoietic system. They play critical roles at virtually every step of hematopoiesis to promote developmental progression, expansion, and survival of developing lymphocytes (Lazorchak et al., 2005; Murre, 2005).
The E2A proteins are active at the HSC cell stage, where they are required for the maintenance of the stem cell pool (Yang et al., 2008; Semerad et al., 2009). They remain active during the development of HSCs into LMPPs, common lymphoid progenitors (CLPs), and prepro–B cells (Zhuang et al., 2004; Borghesi et al., 2005; Dias et al., 2008; Yang et al., 2008; Semerad et al., 2009). In the absence of E2A, the LMPP and CLP compartments are severely reduced and the pro–B cell compartment is completely absent (Dias et al., 2008; Semerad et al., 2009). In both the LMPP and CLP compartment, the E2A proteins have been postulated to act in concert with other transcriptional regulators, such as PU.1 and Ikaros, to promote further developmental progression (Singh et al., 2007; Nutt and Kee, 2007).
B cell development in E2A-ablated mice is arrested at the prepro–B cell stage before the onset of IgH DJ gene rearrangement (Bain et al., 1994; Zhuang et al., 1994). A similar block in B cell development is also observed in EBF-deficient mice, and recent data have demonstrated that the E2A proteins act upstream of EBF to promote B-lineage maturation (Lin and Grosschedl, 1995; Ikawa et al., 2004; Roessler et al., 2007). Specifically, forced EBF expression in E2A-deficient bone marrow cells relieves the block at the prepro–B cell stage (Seet et al., 2004). These data suggest that in the absence of E2A, B lymphopoiesis is blocked before the initiation of B cell development, owing to the inability to activate Ebf1 transcription in prepro–B cells. The E2A proteins are also required to promote a B-lineage program of gene expression in pro–B cells, because conditional deletion of Tcfe2a revealed that the E2A proteins act to maintain the expression of Ebf1 and Pax5 in pro–B cells. The block at the prepro–B cell stage in E2A-deficient bone marrow is also relieved by the enforced expression of Pax5, likely because of the ability of Pax5 to regulate Ebf1 expression (Lazorchak et al., 2006b; Kwon et al., 2008). In addition, multiple pro–B cell–specific genes contain binding sites for E2A, EBF, and Pax5 in putative promoter and enhancer regions, raising the possibility that the combined activities of E2A, EBF, and Pax5 initiate the B cell fate (Nutt and Kee, 2007).
Upon expression of a pre-BCR, E47 protein levels transiently decline, whereas Id3 levels are elevated (Schebesta et al., 2002; Quong et al., 2004). E12 protein levels have not been studied in detail. At the small pre–B cell stage, E47 levels increase again to induce Igκ VJ gene rearrangement (Quong et al., 2004). E47 expression remains high at the immature B cell stage when interaction of the BCR with self-antigen promotes receptor editing (Quong et al., 2004). During positive selection within the transitional B cell compartments, E47 levels decrease and then decline further at the IgM+IgD+ stage (Quong et al., 2004; Kwon et al., 2008). We have previously proposed a model in which high abundance of E47 enforces the immature B cell checkpoint (Quong et al., 2004). In the presence of self-reactivity, E47 levels remain high to promote continuous Igκ locus accessibility and receptor editing. The expression of an innocuous BCR would lead to a decline of E47 levels, suppressing further Ig light chain recombination and promoting developmental progression toward the mature B cell stage. The E2A proteins initiate Igκ VJ gene rearrangement by directly binding to the E-box sites located within the Igκ intronic (iEκ) and 3′ enhancers (Murre et al., 1989). Mutational analysis of the two E2A binding sites in iEκ affects Igκ rearrangement as severely as deletion of the entire enhancer, providing unambiguous evidence for a critical function of E-box sites in Igκ enhancer function (Greenbaum and Zhuang, 2002a; Inlay et al., 2004). Enforced expression of E47 or E12 readily promotes Igκ locus accessibility to the recombination machinery in nonlymphoid cells, and E2A-deficient pre–B cells are defective in their ability to undergo Igκ VJ gene rearrangement (Romanow et al., 2000; Goebel et al., 2001; Lazorchak et al., 2006a). Finally, secondary rearrangements and receptor editing are severely impaired in E2A-heterozygous mice (Quong et al., 2004).
Because the E2A proteins fulfill fundamental roles in so many aspects of the lymphoid branch of the hematopoietic system, it is important to determine in detail how they exert their unique effects. In particular, although the role of E2A proteins in lymphocyte development is now well established, it is less clear whether E12 and E47 have overlapping and redundant roles or whether they perform distinct functions. Previous studies have described the phenotypes in mice that lack E47 expression (Bain et al., 1997). B cell development in E47-ablated mice was blocked at a similar stage as in E2A-deficient mice, although low levels of Ebf1 and Pax5 transcripts were detectable (Bain et al., 1997). However, because of the applied gene targeting strategy, these mice showed a substantially lower abundance of E12, and it remained unclear whether the defects that were detected were caused by a decrease in E12 and E47 rather than E47 alone (Bain et al., 1997).
To determine the individual roles of E12 and E47 in B cell development, we have generated E12- and E47-deficient mice and analyzed them for abnormalities during B lymphopoiesis. We found that deletion of E47 leads to a block in early B cell development, whereas E12 is dispensable for B cell specification. During later stages of development, however, there is a critical dependence on E12 as well as E47 expression in the pre–B and immature B cell compartment. Specifically, using both in vitro and in vivo approaches, we show that E12 and E47 play a crucial role in promoting Igλ VJ gene rearrangement and receptor editing. Specifically, we demonstrate that the E2A proteins modulate trimethylated lysine 4 on histone 3 (H3K4me3) across the Igλ J regions. These data link E2A, epigenetic marking, and the recruitment of the RAG proteins in a common pathway. Finally, we propose a model in which a gradient of E12 and E47 acts to sequentially regulate Igk and Igλ VJ gene rearrangement.
RESULTS
Essential role for E47 but not E12 in early B cell development
To examine the individual roles of E12 and E47 in B cell development, mice were generated that were either deficient in E12 or E47 (Fig. 1 A). To generate E12−/− and E47−/− mice, exons encoding for either E12 or E47 were flanked with loxP sites by homologous recombination in embryonic stem (ES) cells (Fig. 1 A). Genomic DNA was isolated from targeted ES cells and analyzed by Southern blotting for correct targeting (Fig. 1 B). Mutant mice were generated from cells carrying the floxed E12 or E47 alleles and crossed with transgenic mice that express the Ella-cre transgene to delete the loxP-flanked DNA regions in the germline (Lakso et al., 1996). Mice carrying specific deletions in either E12 or E47 were identified using PCR on genomic DNA (Fig. 1 C).
Figure 1.
Generation of E12- and E47-deficient mice. (A) Structure of the targeted Tcfe2a locus. The alternatively spliced E12 and E47 exons, respectively, were flanked with loxP sites (indicated by arrows). In addition, a tetracysteine tag (black bar) was inserted into exon 20, 3′ of the coding region, to produce tagged E47 and E12 proteins. Furthermore, a neomycin (neo) resistance gene was introduced to screen for targeted ES cells. The EcoRI (E) fragments as well as the probe used for screening of the ES cells are indicated. After crossing of Tcfe2afl_E12- or Tcfe2afl_E47-bearing mice with a germline Cre-deleter strain (Ella-Cre), the corresponding Tcfe2aΔE12 or Tcfe2aΔE47 strains were obtained. (B) Southern blot analysis of EcoRI-digested DNA from correctly targeted ES cells of the indicated genotype. The position of the probe is shown in A. (C) PCR analysis of genomic DNA from mice of the indicated genotype. Primer positions are depicted in A.
To determine whether E12 and/or E47 is required to promote early B cell development, bone marrow derived from E12−/− and E47−/− mice was examined for cellularity as well as for the presence of B cell markers. In bone marrow and spleen derived from E47−/− mice, B cells were virtually absent (Fig. 2, A and B). In contrast, the number of B cells in E12−/− mice was not significantly altered as compared with wild-type mice (Fig. 2, A and B).
Figure 2.
Essential role for E47 but not E12 in early B cell development. (A and B) Representative FACS analysis of bone marrow (A) and spleens (B) from wild-type, E47−/−, and E12−/− mice. Numbers indicate percentages of B cells within the live cell fraction. Mean percentages of B cells (B220+CD19+) in the live cell gate as well as absolute B cell numbers in bone marrow (A) and spleens (B) are depicted. At least four independent experiments using one to three mice per group were performed. Horizontal bars represent the mean. ***, P < 0.001. (C) Flow cytometry of bone marrow B cells from wild-type, E47−/−, and E12−/− mice. Numbers indicate percentages of B cells within the live cell fraction (left) or the B220+CD43+ pro–B cell fraction (right). At least four independent experiments using one to three mice per group were performed. (D) Mean percentages of fraction A (HSA−BP-1−), fraction B (HSA+BP-1−), and fraction C (HSA+BP-1+) cells within the pro–B cell (B220+CD43+) compartment of wild-type, E47−/−, and E12−/− mice. At least four independent experiments using one to three mice per group were performed. ns, not significant.
Previous studies using E2A−/− mice have revealed that B cell development in these mice is blocked at the prepro–B cell stage, because the majority of the cells are arrested in Hardy fraction A (Bain et al., 1997). To more precisely determine the arrest in B cell development in E47−/− mice, bone marrow was isolated and analyzed by flow cytometry for the expression of B220, CD43, heat stable antigen (HSA), and BP-1 (Fig. 2 C; Hardy and Hayakawa, 2001). B cell development in E47−/− mice was partially blocked in fraction A and a substantial fraction of cells could be detected in fraction B, whereas virtually no B cells could be detected in fraction C (Fig. 2, C and D). As expected, the B220+c-kit+ compartment was also severely reduced in E47−/− bone marrow (Fig. S1). Interestingly, E12−/− mice showed a slightly higher percentage of fraction C cells among the B220+CD43+ population than wild-type controls (Fig. 2, C and D).
To determine whether the excision of the E47 exon affects E12 mRNA abundance, LMPPs and CLPs were isolated from wild-type and E47−/− bone marrow, and mRNA was isolated and examined for the expression of E12 by quantitative PCR (Fig. S2). Preliminary data show that in both the LMPP and CLP compartment, E12 levels in the E47-deficient cells were similar to that of their wild-type counterparts (Fig. S2). These data indicate that the block in early B cell development in E47−/− mice is specifically caused by the depletion of E47 expression. Collectively, these data show that E12 does not play a critical role in early B-lineage specification. In contrast, E47 is indispensable at the earliest stages of B cell commitment.
E12 is required to promote the development of B cells expressing Igλ
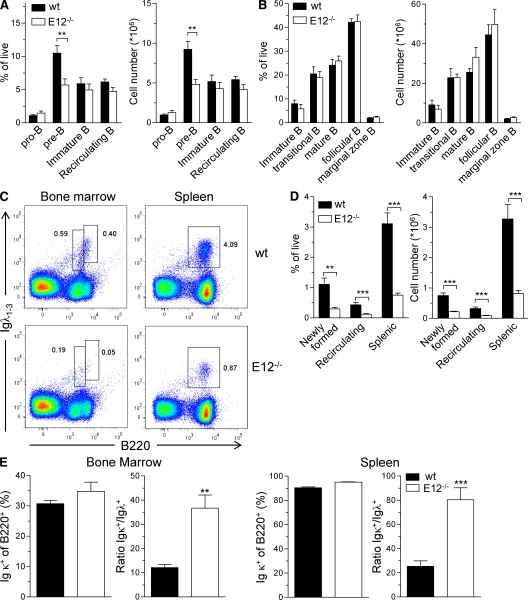
As described in the previous section, B cell development in E12−/− mice seems to be slightly blocked at the transition from Hardy fraction C to the small pre–B cell stage, raising the possibility that E12 is required during the small pre–B cell stage. To examine this possibility, bone marrow derived from E12−/− mice was analyzed for the abundance of pro–B cells (B220+IgM−c-kit+), pre–B cells (B220+IgM−CD25+), immature B cells (B220intermediateIgM+), and recirculating B cells (B220hiIgM+) using flow cytometry (Fig. 3 A). As predicted, the size of the pre–B cell compartment was slightly reduced in E12−/− bone marrow (Fig. 3 A). On the other hand, the pro–B, immature B, and mature B cell compartments were not altered in E12−/− bone marrow as compared with wild-type controls (Fig. 3 A). Similarly, the cellularity of immature B cells (B220+IgM+IgDlo), transitional B cells (B220+IgM+IgD+), mature B cells (B220+IgMloIgD+), follicular B cells (B220+CD23+CD21+), and marginal zone B cells (B220+CD23−CD21hi) in the spleen was similar in E12−/− as compared with wild-type mice (Fig. 3 B).
Figure 3.
E12 is required to promote the development of B cells expressing Igλ. Flow cytometric analysis of B cell populations in wild-type and E12−/− mice. (A) Mean percentages of depicted cell types within the live cell gate and total numbers of pro–B cells (B220+IgM−c-kit+), pre–B cells (B220+IgM−CD25+), immature B cells (B220intermediateIgM+), and recirculating B cells (B220hiIgM+) in the bone marrow. Six independent experiments using one mouse per group were performed. (B) Mean percentages of depicted cell types within the live cell gate and total numbers of immature B cells (B220+IgM+IgDlo), transitional B cells (B220+IgM+IgD+), mature B cells (B220+IgMloIgD+), follicular B cells (B220+CD23+CD21+), and marginal zone B cells (B220+CD23−CD21hi) in the spleen. Six independent experiments using one mouse per group were performed. (C) Representative FACS analysis of Igλ-expressing B cells in the bone marrow and spleen. Numbers indicate percentages of the depicted gates within the live cell fraction. Nine independent experiments using one mouse per group were performed. (D) Mean percentages and total numbers of newly formed Igλ+ B cells (B220intermediateIgλ1-3+) and recirculating Igλ+ B cells (B220hiIgλ1-3+) in the bone marrow as well as Igλ+ B cells (B220+Igλ1-3+) in the spleen as gated in C. Nine independent experiments using one mouse per group were performed. (E) Percentages of Igκ+ cells within the B220+ fraction as well as the ratio of Igκ+ to Igλ+ B cells in the bone marrow (left) and spleen (right). Six independent experiments using one mouse per group were performed. All bar graphs show means ± SEM. **, P ≤ 0.01; ***, P < 0.001 (values are not significantly different from wild-type controls if there is no p-value depicted).
Previous studies demonstrated a critical role for the E2A proteins in receptor editing (Quong et al., 2004). Furthermore, a substantial reduction in the fraction of Igλ-expressing cells was observed in E2A+/− mice. To determine whether E12 plays a critical role in the development and/or survival of Igλ-expressing cells, E12−/− bone marrow was analyzed for the presence of Igλ1-3+ cells by flow cytometry. Strikingly, the fraction of Igλ-expressing B cells was substantially reduced in E12−/− mice as compared with those of wild-type littermates (Fig. 3, C and D). Similarly, total numbers of Igλ+ cells in the bone marrow of E12−/− mice were significantly reduced as compared with those of wild-type mice (Fig. 3 D). Comparable reductions were observed among newly formed B220intermediate and recirculating B220hi bone marrow B cells as well as in splenic B cells (Fig. 3, C and D). In contrast, the fraction of Igκ-expressing cells was slightly elevated in the bone marrow as well as in the spleen of E12−/− mice, resulting in a much higher Igκ/Igλ ratio in E12−/− mice (Fig. 3 E). In summary, these data show that E12 plays a distinct and essential role in the developmental progression and/or survival of B cells that express Igλ.
Defective receptor editing in E12−/− mice
The aforementioned data show that the Igκ/Igλ ratio was severely perturbed in E12−/− B cells, raising the possibility that E12 plays a critical role in promoting Igλ VJ gene rearrangement upon exposure to self-antigen. To determine whether E12 plays a role in promoting receptor editing, bone marrow cells were isolated from wild-type and E12−/− mice and transplanted into recipients that constitutively express a κ-macroself transgene (Ait-Azzouzene et al., 2005). 6 wk after transplantation, bone marrow as well as spleen cells were harvested and examined for the expression of Igλ, Igκ, and B220 (Fig. S3). As shown previously, the bone marrow of recipients expressing the κ-macroself transgene that received wild-type cells showed an increased frequency of Igλ+ B cells as compared with recipients that lacked κ-macroself expression (Fig. S3 B). Similarly our preliminary results show that mice that received E12−/− bone marrow cells also have increased percentages of Igλ+ cells if recipients expressed the κ-macroself transgene as compared with nontransgenic control recipients (Fig. S3 B). This increase in Igλ-expressing cells was similar or even greater in the absence as compared with the presence of E12. However, the fraction of Igλ-expressing cells in κ-macroself recipients reconstituted with E12−/− cells was significantly lower as compared with those reconstituted with bone marrow cells from wild-type littermate controls (Fig. S3, A and B).
As expected, tolerance was maintained because no Igκ-expressing cells were detectable in the spleen of recipients expressing the κ-macroself antigen transplanted with either wild-type or E12−/− bone marrow (Fig. S3 C, bottom). The fraction of Igλ-expressing cells in the spleens of recipients that express the κ-macroself transgene was significantly lower in E12−/− bone marrow as compared with wild-type chimeras (Fig. S3, C and D, top). Furthermore, the overall decrease in the total number of splenic B cells was much more substantial in E12−/− chimeras as compared with controls (Fig. S3 D, bottom). Collectively, these data indicate that upon exposure to self-antigen, the induction of receptor editing does not fully require E12 activity; however, E12 critically modulates the magnitude of the Igλ response.
E47 and E12 abundance is limiting during the development of Igλ-expressing B cells
The observations described in the previous section demonstrate that E12 plays a critical role in the developmental progression of Igλ-expressing cells upon exposure to self-antigen. Because B cell development in E47−/− mice was blocked at the Hardy fraction A/B cell stage, we were not able to assess how the complete absence of E47 affects the developmental progression of B cells beyond the pro–B cell stage. To address this issue and to compare the individual roles of E12 and E47 at the pre–B and immature B cell stage, we examined E12+/− and E47+/− mice for the abundance of Igλ-expressing cells. Bone marrow cells derived from wild-type, E12+/−, E47+/−, and E12−/− mice were isolated and examined for the expression of Igλ. Both E12+/− and E47+/− mice showed a substantial reduction in the fraction of Igλ1-3+ B cells (Fig. 4, A and B). Similarly, the fraction of Igλ-expressing cells in the spleen derived from E12+/− and E47+/− mice was significantly reduced as compared to wild-type mice (Fig. 4 C).
Figure 4.
E47 and E12 abundance is limiting during the development of Igλ-expressing B cells. (A–C) Mean percentages and total numbers of newly formed Igλ+ B cells (B220intermediateIgλ1-3+; A) and recirculating Igλ+ B cells (B220hiIgλ1-3+; B) in the bone marrow as well as Igλ+ B cells (B220+Igλ1-3+; C) in the spleen as gated in Fig. 3 C from mice of the indicated genotypes. At least three independent experiments using one mouse per group were performed. (D) Quantitative PCR analysis of E12 and E47 transcripts in FACS-sorted pre–B cells (B220+IgM−CD25+) from the bone marrow of wild-type, E12+/−, E47+/−, and E12−/− mice. Levels were normalized to hypoxanthine guanine phosphoribosyl transferase 1 (Hprt1) transcripts and relative levels to wild-type controls are shown (2−ΔΔCt). Bar graphs show means ± SEM of three independent experiments. *, P ≤ 0.05; **, P ≤ 0.01; ***, P < 0.001 (values are not significantly different from wild-type controls if there is no p-value depicted). nd, not detectable.
Although the E12 and E47 exons were specifically excised in the null mutant mice, it remained possible that E12 and E47 cross-regulate each other. To explore this possibility, we isolated mRNA from sorted pre–B cells derived from wild-type, E12+/−, E47+/−, or E12−/− mice and determined the mRNA levels of E12 and E47 by quantitative PCR (Fig. 4 D). Pre–B cells derived from E47+/− showed slightly elevated levels of E12 mRNA (Fig. 4 D, left). In contrast, E47 levels in E12+/− pre–B cells were not significantly altered (Fig. 4 D, right). E12−/− pre–B cells, on the other hand, showed a modest reduction in E47 levels (Fig. 4 D, right). These data indicate that E47 levels are modestly but significantly reduced in E12−/− pre–B cells. On the other hand, E47 abundance is not significantly affected in E12+/− pre–B cells. Furthermore, these data indicate that depletion of E47 slightly but significantly elevates the expression of E12. Thus the defect in Igλ-expressing cells in E47+/− and E12+/− mice cannot be explained by abnormalities in the abundance of either E12 or E47. Collectively, these data demonstrate that both E12 and E47 play a critical role in the developmental progression of Igλ-expressing B cells.
E12 and E47 facilitate Igλ locus accessibility
Given that Igλ-expressing cells were less abundant in E12+/−, E47+/−, and E12−/− mice, we considered the possibility that E12 proteins act to regulate Igλ VJ gene rearrangement. To address this issue, DNA was isolated from sorted pre–B cells derived from wild-type, E12+/−, E47+/−, and E12−/− mice and analyzed by semiquantitative PCR for the presence of IgH DJ as well as Igκ and Igλ VJ gene rearrangements. IgH DJ and Igκ VJ joining is not perturbed in pre–B cells isolated from E47+/−, E12+/−, and E12−/− mice (Fig. S4). Recombining sequence (RS) rearrangements were only modestly affected in E12+/−, E47+/−, and E12−/− mice (Fig. 5 B, left and middle). In contrast, Igλ V1-J1 rearrangement was significantly reduced in pre–B cells isolated from both E12+/− and E47+/− mice (Fig. 5 B, right). As predicted, E12−/− pre–B cells showed a significant decrease in the levels of Igλ VJ gene rearrangements as compared with wild-type mice and, importantly, also with E47+/− pre–B cells (Fig. 5 B, right). These data indicate that both E12 and E47 act to promote Igλ VJ gene rearrangement.
Figure 5.
E12 and E47 facilitate Igλ locus accessibility. (A) Schematic diagram of the mouse IgH and light chain (Igκ and Igλ) loci. V, D, J, and C segments as well as the RS in Igκ are depicted as gray boxes and the enhancers are depicted as white boxes. Analyzed gene rearrangements are shown by arrows on top of the loci, whereas germline transcripts are shown by arrows under the gene locus. (B) Quantification of Southern blots from PCR analysis of Ig gene rearrangements in FACS-sorted pre–B cells (B220+IgM−CD25+) from mice of the indicated genotypes. PCR products from reactions of fourfold serial dilutions were electrophoresed on agarose gels and abundance was quantitated by Southern blotting using specific probes. Shown are means ± SEM relative to wild-type signal. *, P ≤ 0.05; ***, P < 0.001 (values are not significantly different from wild-type controls if there is no p-value depicted). Three independent experiments using one mouse per group were performed. (C) Quantitative PCR analysis of Igλ1 germline transcripts (Igλ1°) in FACS-sorted pre–B cells (B220+IgM−CD25+) from the bone marrow of wild-type, E12+/−, E47+/−, and E12−/− mice. Levels were normalized to Hprt1 transcripts and relative levels to wild-type controls are shown (2−ΔΔCt). Shown are means ± SEM. *, P ≤ 0.05; **, P ≤ 0.01; ***, P < 0.001 (values are not significantly different from wild-type controls if there is no p-value depicted). Three independent experiments using one mouse per group were performed. (D) ChIP assays using anti-E2A antibodies on MACS-purified total B220+ bone marrow cells from wild-type and E12−/− mice. Relative enrichment of the bound DNA over input was determined by quantitative PCR using specific primers for the Igλ enhancers Eλ1-3 and Eλ2-4. Shown are means ± SEM. Two independent experiments using one mouse per group were performed. IRS1, intronic recombination sequence 1.
To determine whether the decrease in Igλ VJ gene rearrangements in E12+/− and E47+/− bone marrow cells was caused by lower levels of RAG1 and/or RAG2 expression, mRNA was isolated from sorted pre–B cells and examined by quantitative PCR for the expression of RAG1/2. Both RAG1 and RAG2 mRNA levels were not significantly altered in E12+/− and E47+/− as compared with wild-type mice (Fig. S5). To determine whether a reduction in E12 and/or E47 affected Igλ germline transcription, mRNA from sorted pre–B cells was analyzed for the expression of Igλ1 germline transcripts. Consistent with the decrease in Igλ VJ gene rearrangements in E12+/− and E47+/− pre–B cells, Igλ1 germline transcript levels were significantly reduced as compared with wild-type (Fig. 5 C). As expected, a further reduction in Igλ germline transcription was observed in E12−/− pre–B cells (Fig. 5 C). Collectively, these data suggest that E12 and E47 activate Igλ VJ gene rearrangement by modulating Igλ locus accessibility to the recombination machinery.
Consistent with a role for E12 and E47 in modulating Igλ locus accessibility are previous observations indicating the presence of E12 and E47 binding sites in the Igλ enhancer regions (Rudin and Storb, 1992). To determine whether E12 and E47 directly interact with E-box sites present in the Igλ enhancer, B cells were isolated from wild-type and E12−/− bone marrow, cross-linked with formaldehyde, and immunoprecipitated with an anti-E2A antibody. Subsequently, DNA was isolated from the immunoprecipitates and examined for the presence of regions containing Eλ1-3 and Eλ2-4 using the appropriate primers. Both E12 and E47 interacted with the Igλ enhancer regions, Eλ1-3 and Eλ2-4, because the enrichment observed in wild-type bone marrow cells was severely reduced in E12−/− cells but not abolished (Fig. 5 D). Thus, we conclude that the residual binding to the Igλ enhancer regions is caused by E47 (Fig. 5 D). Collectively, these data demonstrate that both E12 and E47 interact with E-box sites present in the Igλ enhancers, activate Igλ germline transcription, modulate Igλ locus accessibility, and promote Igλ VJ gene rearrangement.
Attenuation of IL-7 signaling elevates E12 and E47 abundance
Previous observations have indicated that E47 abundance is low in cycling large pre–B cells but is elevated in small resting pre–B cells (Quong et al., 2004). Because Ig light chain gene rearrangement is initiated by pre-BCR signaling as well as attenuation of IL-7 signaling, we examined the temporal expression of E12 and E47 in bone marrow cells that express an innocuous BCR upon withdrawal of IL-7 signaling. In brief, B cells expressing the hen egg lysozyme transgene (HEL-Tg) were isolated from the bone marrow and grown for a period of 4 d in the presence of IL-7. Upon withdrawal of IL-7 from the B cell cultures, the abundance of Igλ germline transcripts as well as E12 and E47 transcript levels were determined at various time points using quantitative PCR. Upon attenuation of IL-7–mediated signaling, E12 and E47 transcript levels increased gradually (Fig. 6 A). Interestingly, the relative increase in E12 expression was modestly but consistently larger than that of E47 expression, with the ratio of E12 to E47 expression increasing over time (Fig. 6 A). Concomitant with the increase in E12 and E47 mRNA levels, we observed an increase in Igλ germline transcription consistent with a model in which E12 directly regulates Igλ germline transcription (Fig. 6 B). To determine whether E12 is required for the induction of Igλ germline transcription, E12+/+, HEL-Tg and E12+/−, HEL-Tg bone marrow cells were cultured in the presence of IL-7 for 4 d, followed by withdrawal of IL-7. RNA was isolated at the indicated time points and examined for the presence of Igλ germline transcription. Consistent with these observations, Igλ germline transcript levels were significantly reduced in E12+/− immature B cells (Fig. 6 B).
Figure 6.
Attenuation of IL-7 signaling elevates E12 and E47 abundance. (A) Quantitative PCR analysis of E47 and E12 transcripts at various time points after IL-7 withdrawal from cell cultures of HEL-Tg B cells. Levels were normalized to Hprt1 transcripts, and expression levels relative to before IL-7 withdrawal are determined (2−ΔΔCt). Shown are means ± SEM of three independent experiments. (B) Quantitative PCR analysis of Igλ1 germline transcripts at various time points after IL-7 withdrawal from B cell cultures generated from the bone marrow of HEL-Tg and E12+/−, HEL-Tg mice. Levels were normalized to Hprt1 transcripts and relative expression levels are determined (2−ΔCt). Shown are means ± SEM of three independent experiments.
E2A proteins modulate epigenetic marks across the Igλ J regions
Intriguing studies have recently indicated a correlation between epigenetic marking and antigen receptor assembly. Specifically, a noncanonical plant homeodomain located within the noncore region of RAG2 was shown to interact with H3K4me3 (Liu et al., 2007; Matthews et al., 2007; Ramón-Maiques et al., 2007). Furthermore, across the Jλ region, H3K4me3 and H3 acetylation (H3ac) were shown to be specifically enriched during the pre–B and immature B cell stages (Xu and Feeney, 2009). These observations raise the possibility that the E2A proteins regulate Igλ VJ gene rearrangement by promoting the recruitment of RAG2 to the recombination signal sequences by, directly or indirectly, modulating trimethylation of H3K4. To address this issue, B220+ bone marrow cells from wild-type and E12−/− mice were cultured in the presence of IL-7 and stem cell factor for 8 d to obtain a homogeneous culture of B220+CD19+CD43+ pro–B cells. After IL-7 withdrawal for 18 h, we examined the Jλ region for H3K4 trimethylation as well as H3ac by chromatin immunoprecipitation (ChIP; Fig. 7, A and B). Interestingly, both H3K4me3 and H3ac across the Jλ region were significantly reduced in the absence of E12 (Fig. 7, A and B).
Figure 7.
E2A modulates the abundance of H3K4me3 across the Igλ J regions. (A and B) ChIP assays using anti-H3K4me3 (A) or anti-H3ac (B) antibodies on pro–B cell cultures from wild-type and E12−/− mice that were deprived of IL-7 for 18 h. Relative enrichment of the bound DNA over input was determined by quantitative PCR using specific primers for the Igλ J1 region. Shown are means ± SEM of two independent experiments. *, P ≤ 0.05; **, P ≤ 0.01.
Collectively, these data indicate that E12 expression is elevated in immature B cells to initiate Igλ germline transcription and to promote Igλ locus accessibility. Based on these, as well as previous observations, we propose that in immature B cells initially, E12 and E47 abundance is low, preventing accessibility of the Igλ locus to the recombination machinery. However, increasing levels of E12 and E47 would eventually lead to relative high occupancy of E-box elements present in the Igλ enhancer, inducing Igλ germline transcription and permitting the activation of Igλ VJ gene rearrangement (Fig. 8). Furthermore, we suggest that the E2A proteins act to promote epigenetic marking of histone residues located at the Jλ regions to permit the recruitment of RAG2 to the recombination machinery.
Figure 8.
A gradient of E12 and E47 activity leads to the sequential activation of the Ig light chains. Attenuation of IL-7 signaling leads to an increase in E12 and E47 expression to initiate Ig light chain gene rearrangement. Signaling through a functional BCR suppresses E12/E47 expression to prevent further rearrangements and promote positive selection and maturation. However, in the absence of such a signal, the ratio of E12 to E47 reaches a certain threshold, which enables the cell to rearrange the Igλ loci. Failure to generate a functional BCR during a certain time window will lead to cell death, presumably caused by high E12 and E47 levels.
DISCUSSION
The Tcfe2a locus encodes for two highly related proteins that differ in their DNA binding region through alternative splicing of one exon (Murre et al., 1989; Sun and Baltimore, 1991). From previous studies, it is apparent that the transcription factor E2A is not only indispensable for commitment to the B cell lineage but fulfills critical functions throughout B lymphopoiesis that include maintenance, DNA recombination, receptor editing, marginal versus follicular zone development, and class switch recombination (Greenbaum and Zhuang, 2002b; Murre, 2005). We have previously generated E47-deficient mice that also showed substantially reduced abundance of E12 caused by the insertion of the neomycin gene. Thus, an important unresolved question is whether E12 and E47 fulfill distinct and/or overlapping roles during B lymphopoiesis. In this study, we have generated two mouse strains that selectively delete either the E12 or the E47 exon through the usage of the loxP system, enabling us to study their specific roles during B cell development.
Previous studies have shown that B cell development in E2A-deficient mice is completely blocked at the prepro–B cell stage (Bain et al., 1994; Zhuang et al., 1994). Overexpression of either E12 or E47 has the ability of triggering commitment to the B cell lineage, raising the possibility that both play a role in B cell specification and/or commitment (Bain et al., 1997). Our study shows that E47 is strictly required for progression beyond the prepro–B cell stage and that endogenous levels of E12 cannot compensate for its loss. In striking contrast, E12-deficient mice show almost normal numbers of mature B cells in the bone marrow and spleen. However, the pre–B cell compartment is slightly reduced in E12-deficient bone marrow. We note that the decrease in the pre–B cell compartment of E12-deficient bone marrow might be caused by the twofold decrease in E47 mRNA levels (Fig. 4 D). Thus, in the presence of endogenous levels of E47 protein, E12 is dispensable for the specification of the B cell fate. In turn, this suggests that E47 but not E12 activates a B lineage–specific program of gene expression.
In addition to regulating lineage decisions through the activation of gene expression of B cell–specific genes, the E2A proteins also play critical roles in regulating Ig gene rearrangements. Both E47 as well as E12 were originally identified as factors that interact with the κE2 site of the iEκ. Deletion of the two E-boxes in iEκ affects Igκ recombination to the same degree as deletion of the complete enhancer (Murre et al., 1989; Inlay et al., 2004). Forced E12 or E47 expression, in conjunction with RAG1 and RAG2, promotes Igκ VJ gene rearrangement in nonlymphoid cells (Romanow et al., 2000). Furthermore, Igκ VJ gene rearrangements in E2A-deficient pre–B cells are completely abolished (Lazorchak et al., 2006b; Kwon et al., 2008).
Collectively, these studies establish the E2A proteins as key regulators of Igκ VJ gene rearrangement. We show in this study, however, that Igκ VJ gene rearrangement is not affected in E12-deficient mice. Because B cell development in E47-deficient mice is completely blocked at the prepro–B cell stage and because we were not able to generate E47F/F mice that would permit the stage-specific deletion at the pre–B or immature B cell stage, we could not assess directly whether E47 is the critical component in promoting Igκ VJ DNA recombination. However, because E2A-deficient B cells are completely defective in Igκ VJ recombination and E12 activity is not required, we infer that E47 is the main factor required to promote Igκ VJ joining.
In contrast, we show in this study that both E47 and E12 are required for proper Igλ gene rearrangement. As described, E12-deficient B cells show a reduction in E47 mRNA levels that is similar to that observed in B cells from E47-heterozygous mice. Because E47-heterozygous B cells also have a significant defect in Igλ VJ gene rearrangement, E12-deficient B cells cannot be used to evaluate the role of E12 in Igλ VJ gene rearrangement. Therefore, E12-heterozygous B cells that show normal levels of E47 were used to unambiguously demonstrate a critical role for E12 in Igλ VJ gene rearrangement. We emphasize, however, that in addition to E12, E47 also plays a critical role in modulating Igλ VJ gene rearrangement.
There are several mechanisms that may underpin the decrease in Igλ-expressing cells in E12- or E47-deficient pre–B cells. First, it is conceivable that E12 and E47 act to promote the survival and/or life span of immature B cells, because previous studies have demonstrated a critical role for the E proteins in B cell survival (Kee et al., 2002; Lazorchak et al., 2006b). In this case, however, survival of in vitro–cultured E12+/− B cells expressing the HEL-Tg is not affected, yet they do not efficiently up-regulate Igλ germline transcripts. Thus, although we cannot exclude the possibility that the E2A proteins act, in part, to promote cell survival at the immature B cell stage in vivo, we consider it unlikely that this is their sole function. A second possibility is that RS deletion is regulated by E12 and E47. RS is a DNA element that carries a canonical recombination signal sequence. It is located ∼25 kbp downstream of the Igκ constant region and rearranges by DNA recombination to Vκ elements and to elements present in the Jκ-Cκ intronic region. The absence of RS deletion substantially interferes with the development of Igλ-producing cells (Vela et al., 2008). Thus, it is conceivable that Igλ production in E12-ablated bone marrow cells is perturbed because of a reduction in RS deletion. However, in this study we show that RS deletion is only modestly affected in E12-deficient pre–B and immature B cells carrying unmanipulated BCRs. A third possibility is that E12 and E47 are required to activate the expression of RAG1/2. However, E12- and E47-heterozygous pre–B cells did not show decreased levels of either RAG1 or RAG2 mRNA. A fourth possibility, and one that we favor, is that E12 and E47 binding to the Igλ enhancers directly allows the recombinase access to the recombination signal sequences.
It is now well established that the Ig light chain loci rearrange sequentially. Specifically, Igκ VJ rearrangements are initiated before the onset of Igλ VJ gene rearrangement (Nadel et al., 1990; Engel et al., 1999). It has remained unclear how the sequential rearrangement of the Ig light chain genes is achieved during the developmental progression of B-lineage cells. We suggest that E12 and E47 are critical players that regulate the sequential activation of Igκ versus Igλ loci. Previous observations have demonstrated that E47 abundance in large cycling pre–B cells is low but elevated during the transition toward the small pre–B cell stage (Quong et al., 2004). We also have previously proposed that at the pre–B cell stage, concurrent with the cessation of proliferation, E2A levels are elevated to initiate Igκ VJ gene rearrangement (Quong et al., 2004). In this study, we show that attenuation of IL-7 signaling elevates both E12 and E47 expression. We speculate that once pre–B cells migrate away from IL-7–expressing stroma, E47 as well as E12 levels increase to establish a gradient of E2A activity. This gradient of E2A activity promotes the sequential activation of Igκ versus Igλ VJ gene rearrangement. In such a model, relatively low levels of E47 would be sufficient to bind the E-box sites present in the Igκ enhancer and to promote Igκ locus accessibility. Initial levels of E47 are sufficient to achieve high occupancy binding to E-box sites present in the Igκ but not the Igλ locus. The differences in the degree of occupancy by E12 and E47 in the Igκ and Igλ enhancers might be caused by interactions with other regulators. IRF-4 is an attractive candidate because it has been shown to bind cooperatively with the E proteins to sites present in the Igκ enhancers (Johnson et al., 2008). Thus, initially, E47 levels might be limiting and act in concert with IRF-4 to drive Igκ VJ gene rearrangement but not to promote Igλ accessibility and DNA recombination. Later, during the pre–B to the immature B cell transition, the absence of an Igκ or Igλ chain or the expression of a self-reactive BCR would increase E12 and E47 abundance to levels that allow sufficient occupancy at sites present in the Igλ enhancers and, thus, drive Igλ VJ recombination. On the other hand, because high levels of E47 reduce cell size and ultimately promote cell death, we suggest that the increase in E2A levels restricts the time that is available for the B cells to generate a useful receptor (Fig. 8; Engel et al., 2001; Engel and Murre, 2004). If, however, an innocuous BCR is generated, E12 and E47 abundance declines, resulting in a decrease in Igκ and Igλ occupancy and the promotion of positive selection (Quong et al., 2004).
An intriguing question is how the E2A proteins modulate the assembly of such a diverse set of antigen receptor loci, and the data described in this study may help to illuminate the mechanism by which the E2A proteins contribute to antigen receptor recombination. Recently, it was shown that RAG2 associates with H3K4me3, establishing a direct link between epigenetic marking and antigen receptor assembly (Liu et al., 2007; Matthews et al., 2007; Ramón-Maiques et al., 2007). In this study, we demonstrate that E12 elevates the abundance of H3K4me3 across the Igλ J regions in B cells poised to undergo Igλ VJ gene rearrangement. Clearly, E12 is not the sole factor responsible for H3K4me3 because residual H3K4me3 remains across the Jλ region in E12-deficient B cells, and consequently, other factors must contribute as well. There are several possible mechanisms of how E12 might modulate the degree of H3K4me3. First, it is possible that E12 contributes directly to the recruitment of methyltransferase activity acting on H3K4. Alternatively, E12 may function indirectly by activating Igλ germline transcription, enabling methyltransferases and acetylases to travel with the RNA pol II complex, which then may lead to the trimethylation of H3K4. Collectively, these data have implications for the role of the E proteins in modulating the assembly of the large majority of antigen receptor loci (Murre, 2005). We suggest that the E proteins, directly or indirectly and in conjunction with other factors, act to promote epigenetic marking of antigen receptor loci across J regions to permit the recruitment of the RAG2 component of the recombinase.
In conclusion, this study demonstrates the multiple and unique functions displayed by the E2A isoforms E12 and E47. E47 is essential during B cell specification, commitment, and maintenance and it is necessary but not sufficient during the sequential rearrangement of the Igκ and Igλ loci. E12 and E47 are both required to promote Igλ VJ rearrangement by modulating, at least in part, the trimethylation of H3K4 across the Jλ region. Finally, we propose that in the pre–B and immature B cell compartments, a gradient of E12 and E47 activity is established to mechanistically regulate the sequential rearrangement of the Ig light chains.
MATERIALS AND METHODS
Generation of specific E12- and E47-deficient mice.
The linearized targeting vector, containing either the E12 or the E47 exon flanked by two loxP sites as well as a C-terminal tetracysteine tag (Martin et al., 2005), was electroporated into 129Sv/Ev ES cells, and correctly targeted clones were identified by Southern blot analyses using the enzymes and outside probe depicted in the figures. After injection into blastocysts and generation of chimeric mice the correctly germline-targeted mouse was backcrossed to Ella-cre mice to generate E12- and E47-deficient mice (Lakso et al., 1996). Mice were genotyped using primers in Table S1. All experiments using mice were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee of the University of California, San Diego (UCSD).
Flow cytometry and cell sorting.
FACS analyses were performed with 8–12-wk-old mice. Single cells were stained with various fluorochrome-conjugated antibodies purchased from eBioscience or BD. Data were collected with a flow cytometer (LSRII; BD) and were analyzed with FlowJo software (Tree Star, Inc.). Specific cell populations were either sorted on a FACSAria (BD) or isolated by magnetic cell sorting with anti-B220 or anti-biotin microbeads (Miltenyi Biotec) according to the manufacturer’s protocol. The following antibodies were used for flow cytometry: anti-B220 (RA3-6B2); CD19 (1D3); CD43 (R2/60); BP-1 (6C3); CD24/HSA (M1/69); CD117/c-kit (2B8); CD25 (PC61); IgM (II/41); IgD (11-26); CD21 (8D9); CD23 (B3B4); Igκ light chain (187.1); Igλ1, λ2, and λ3 light chain (R26-46); Flt3 (AF10.1); CD127/IL-7Rα (A7R34); Sca1 (D7); Ter119 (TER-119); Gr1 (RB6-8C5); CD11b/Mac1 (M1/70); and CD3ε (145-2C11) and CD11c (N418). For CLP/LMPP sorting, the lineage cocktail contained antibodies against B220, Ter119, Gr1, CD11b/Mac1, CD3ε, and CD11c. Unspecific antibody binding was prevented by preincubation with CD16/CD32 Fc blocking solution (eBioscience).
Quantitative RT-PCR.
Total RNA was isolated using the DNeasy kit (QIAGEN), and cDNA was prepared with oligo-dT primers using SuperScriptIII reverse transcription (Invitrogen). Quantitative PCR was performed using Brilliant SYBR Green Master Mix (Agilent Technologies) on the Mx3500P cycler (Agilent Technologies) using the gene-specific primers listed in Table S2 and a 60°C annealing temperature.
Bone marrow chimeras.
Recipients were either κ-macroself transgenic mice or littermate controls that carried the CD45.1 allele and have been previously described (Ait-Azzouzene et al., 2005). Recipients received 1,000 rads of γ irradiation from a Cs source the day before transfer. Bone marrow donors were all of the CD45.2 allotype. Bone marrow cell suspensions were depleted of T cells by treatment with biotinylated anti-Thy1.2 antibodies and anti-biotin microbeads (Miltenyi Biotec). 10 million cells were transferred i.v. per recipient. After 6 wk, recipients were killed and their lymphoid tissues were analyzed by flow cytometry. Only chimeras in which ≥98% of cells in the spleen were donor derived were included in the analysis.
PCR assays for Ig gene recombinations.
PCR reactions were performed in a final volume of 25 µl containing 4, 1, and 0.25 ng of genomic DNA using the primers listed in Table S3. Specific probes were also generated by PCR using the primers listed in Table S4, gel purified using the QIAquick Gel Extraction Kit (QIAGEN), and radioactively labeled using the Prime-It Kit (Agilent Technologies). Samples were amplified for 25 cycles of 94°C for 1 min, 60°C for 30 s, and 72°C for 1.5 min. PCR products were electrophoresed on 1.5% (0.8% for Igκ V-J) agarose gels, blotted on nylon membranes (Schleicher & Schuell), and hybridized with radioactive probes. Signals were detected with a Phosphorimager (GE Healthcare) and analyzed using ImageJ software (available at http://rsbweb.nih.gov/ij/).
ChIPs.
ChIP assays were performed as previously described (Agata et al., 2007). In brief, B cells were fixed with 1% formaldehyde for 10 min at room temperature, lysed, and sonicated. Samples were precleared for 1 h with protein G beads, and ChIP was performed overnight using anti-E2A (anti-E12 V18; Santa Cruz Biotechnology, Inc.), anti-H3K4me3 (Millipore), anti–acetyl H3K9/14 (Millipore), or control normal rabbit IgG (Santa Cruz Biotechnology, Inc.). After washing and elution of the beads, cross-linking was reversed and protein was digested by proteinase K treatment. ChIP DNA was isolated using QIAquick PCR Purification Kit (QIAGEN) and analyzed by quantitative PCR using Brilliant SYBR Green Master Mix on an Mx3500P and the gene-specific primers listed in Table S5.
In vitro HEL-Tg B cell cultures.
HEL-Tg mice, which bear rearranged IgH and Igκ light chain genes that encode an HEL-specific BCR, were obtained from the Jackson Laboratory. Whole bone marrow was isolated from 4–6-wk-old HEL-Tg mice, and red blood cells were lysed and cultured in RPMI 1640 medium with 10% (vol/vol) FCS, 100 U/ml penicillin-streptomycin, 2 mM l-glutamine, and 50 µM 2-mercaptoethanol in the presence of 1% conditioned supernatant of rIL-7–producing J558L cells for 4 d at 37°C in 5% CO2. Remaining myeloid cells as well as surface IgD+ B cells were depleted using magnetic cell sorting (Miltenyi Biotec), and purified B cells were cultured in the absence of IL-7. Cells were harvested at the time points indicated in the figures and used for quantitative PCR.
In vitro B cell cultures.
Total B cells from bone marrow were isolated using B220-coupled magnetic beads (Miltenyi Biotec) and cultured in OptiMEM medium with 10% (vol/vol) FCS, 100 U/ml penicillin-streptomycin, 2 mM l-glutamine, and 50 µM 2-mercaptoethanol in the presence of IL-7 and stem cell factor for 8 d at 37°C in 5% CO2. The remaining nonpro–B cells were depleted using magnetic cell sorting (Miltenyi Biotec), and purified B cells were cultured in the absence of IL-7 for 18 h before they were used for ChIP.
Statistical analysis.
Statistical significance was calculated using the two-tailed Student’s t test. P < 0.05 was considered significant.
Online supplemental material.
Fig. S1 shows c-kit staining in the bone marrow of E12- and E47-deficient mice. Fig. S2 shows E12 and E47 expression levels in LMPP and CLP cells. Fig. S3 is the analysis of bone marrow chimeras generated by transplanting wild-type and E12-deficient bone marrow cells into κ-macroself transgenic mice. Fig. S4 shows IgH and Igk gene rearrangement. Fig. S5 shows RAG1 and RAG2 expression levels in pre–B cells. Tables S1–S5 list primer sequences used for PCR. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20090756/DC1.
Acknowledgments
We thank Dr. S. Andre and E. Mercer for careful reading and editing of the manuscript, and members of the Murre laboratory for stimulating discussions; Dr. Y. Lin for technical advice with the ChIP experiments; E. Welinder and Dr. R. Månsson for assistance with the CLP and LMPP sorting; B. Duong for assistance with the receptor editing experiment; K. Ong for assistance with FACS sorting; and the UCSD Transgenic and Gene Targeting Core for producing E12- and E47-deficient mice.
This study was supported by the National Institutes of Health (C. Murre and D. Nemazee).
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- BCR
- B cell receptor
- ChIP
- chromatin immunoprecipitation
- CLP
- common lymphoid progenitor
- EBF
- early B cell factor
- ES
- embryonic stem
- H3
- histone 3
- H3ac
- H3 acetylation
- H3K4me3
- trimethylated lysine 4 on H3
- HEL-Tg
- hen egg lysozyme transgene
- HLH
- helix-loop-helix
- Hprt1
- hypoxanthine guanine phosphoribosyl transferase 1
- HSA
- heat stable antigen
- HSC
- hematopoietic stem cell
- iEκ
- Igκ intronic enhancer
- IgH
- Ig heavy chain
- LMPP
- lymphoid-primed multipotent progenitor
- RS
- recombining sequence
References
- Agata Y., Tamaki N., Sakamoto S., Ikawa T., Masuda K., Kawamoto H., Murre C. 2007. Regulation of T cell receptor beta gene rearrangements and allelic exclusion by the helix-loop-helix protein, E47.Immunity. 27:871–884 doi:10.1016/j.immuni.2007.11.015 [DOI] [PubMed] [Google Scholar]
- Ait-Azzouzene D., Verkoczy L., Peters J., Gavin A., Skog P., Vela J.L., Nemazee D. 2005. An immunoglobulin Cκ-reactive single chain antibody fusion protein induces tolerance through receptor editing in a normal polyclonal immune system.J. Exp. Med. 201:817–828 doi:10.1084/jem.20041854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alt F.W., Yancopoulos G.D., Blackwell T.K., Wood C., Thomas E., Boss M., Coffman R., Rosenberg N., Tonegawa S., Baltimore D. 1984. Ordered rearrangement of immunoglobulin heavy chain variable region segments.EMBO J. 3:1209–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain G., Maandag E.C., Izon D.J., Amsen D., Kruisbeek A.M., Weintraub B.C., Krop I., Schlissel M.S., Feeney A.J., van Roon M., et al. 1994. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements.Cell. 79:885–892 doi:10.1016/0092-8674(94)90077-9 [DOI] [PubMed] [Google Scholar]
- Bain G., Robanus Maandag E.C., te Riele H.P., Feeney A.J., Sheehy A., Schlissel M., Shinton S.A., Hardy R.R., Murre C. 1997. Both E12 and E47 allow commitment to the B cell lineage.Immunity. 6:145–154 doi:10.1016/S1074-7613(00)80421-5 [DOI] [PubMed] [Google Scholar]
- Benezra R., Davis R.L., Lassar A., Tapscott S., Thayer M., Lockshon D., Weintraub H. 1990. Id: a negative regulator of helix-loop-helix DNA binding proteins. Control of terminal myogenic differentiation.Ann. NY Acad. Sci. 599:1–11 doi:10.1111/j.1749-6632.1990.tb42359.x [DOI] [PubMed] [Google Scholar]
- Borghesi L., Aites J., Nelson S., Lefterov P., James P., Gerstein R. 2005. E47 is required for V(D)J recombinase activity in common lymphoid progenitors.J. Exp. Med. 202:1669–1677 doi:10.1084/jem.20051190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedar H., Bergman Y. 1999. Developmental regulation of immune system gene rearrangement.Curr. Opin. Immunol. 11:64–69 doi:10.1016/S0952-7915(99)80012-0 [DOI] [PubMed] [Google Scholar]
- Corneliussen B., Thornell A., Hallberg B., Grundström T. 1991. Helix-loop-helix transcriptional activators bind to a sequence in glucocorticoid response elements of retrovirus enhancers.J. Virol. 65:6084–6093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias S., Månsson R., Gurbuxani S., Sigvardsson M., Kee B.L. 2008. E2A proteins promote development of lymphoid-primed multipotent progenitors.Immunity. 29:217–227 doi:10.1016/j.immuni.2008.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel I., Murre C. 2004. E2A proteins enforce a proliferation checkpoint in developing thymocytes.EMBO J. 23:202–211 doi:10.1038/sj.emboj.7600017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel H., Rolink A., Weiss S. 1999. B cells are programmed to activate kappa and lambda for rearrangement at consecutive developmental stages.Eur. J. Immunol. 29:2167–2176 doi:10.1002/(SICI)1521-4141(199907)29:07<2167::AID-IMMU2167>3.0.CO;2-H [DOI] [PubMed] [Google Scholar]
- Engel I., Johns C., Bain G., Rivera R.R., Murre C. 2001. Early thymocyte development is regulated by modulation of E2A protein activity.J. Exp. Med. 194:733–745 doi:10.1084/jem.194.6.733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M. 2002. V(D)J recombination: RAG proteins, repair factors, and regulation.Annu. Rev. Biochem. 71:101–132 doi:10.1146/annurev.biochem.71.090501.150203 [DOI] [PubMed] [Google Scholar]
- Goebel P., Janney N., Valenzuela J.R., Romanow W.J., Murre C., Feeney A.J. 2001. Localized gene-specific induction of accessibility to V(D)J recombination induced by E2A and early B cell factor in nonlymphoid cells.J. Exp. Med. 194:645–656 doi:10.1084/jem.194.5.645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum S., Zhuang Y. 2002a. Identification of E2A target genes in B lymphocyte development by using a gene tagging-based chromatin immunoprecipitation system.Proc. Natl. Acad. Sci. USA. 99:15030–15035 doi:10.1073/pnas.232299999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum S., Zhuang Y. 2002b. Regulation of early lymphocyte development by E2A family proteins.Semin. Immunol. 14:405–414 doi:10.1016/S1044532302000751 [DOI] [PubMed] [Google Scholar]
- Hardy R.R., Hayakawa K. 2001. B cell development pathways.Annu. Rev. Immunol. 19:595–621 doi:10.1146/annurev.immunol.19.1.595 [DOI] [PubMed] [Google Scholar]
- Hardy R.R., Kincade P.W., Dorshkind K. 2007. The protean nature of cells in the B lymphocyte lineage.Immunity. 26:703–714 doi:10.1016/j.immuni.2007.05.013 [DOI] [PubMed] [Google Scholar]
- Ikawa T., Kawamoto H., Wright L.Y., Murre C. 2004. Long-term cultured E2A-deficient hematopoietic progenitor cells are pluripotent.Immunity. 20:349–360 doi:10.1016/S1074-7613(04)00049-4 [DOI] [PubMed] [Google Scholar]
- Inlay M.A., Tian H., Lin T., Xu Y. 2004. Important roles for E protein binding sites within the immunoglobulin κ chain intronic enhancer in activating Vκ Jκ rearrangement.J. Exp. Med. 200:1205–1211 doi:10.1084/jem.20041135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K., Hashimshony T., Sawai C.M., Pongubala J.M., Skok J.A., Aifantis I., Singh H. 2008. Regulation of immunoglobulin light-chain recombination by the transcription factor IRF-4 and the attenuation of interleukin-7 signaling.Immunity. 28:335–345 doi:10.1016/j.immuni.2007.12.019 [DOI] [PubMed] [Google Scholar]
- Kee B.L., Paige C.J. 1995. Murine B cell development: commitment and progression from multipotential progenitors to mature B lymphocytes.Int. Rev. Cytol. 157:129–179 doi:10.1016/S0074-7696(08)62158-0 [DOI] [PubMed] [Google Scholar]
- Kee B.L., Bain G., Murre C. 2002. IL-7Ralpha and E47: independent pathways required for development of multipotent lymphoid progenitors.EMBO J. 21:103–113 doi:10.1093/emboj/21.1.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon K., Hutter C., Sun Q., Bilic I., Cobaleda C., Malin S., Busslinger M. 2008. Instructive role of the transcription factor E2A in early B lymphopoiesis and germinal center B cell development.Immunity. 28:751–762 doi:10.1016/j.immuni.2008.04.014 [DOI] [PubMed] [Google Scholar]
- Lakso M., Pichel J.G., Gorman J.R., Sauer B., Okamoto Y., Lee E., Alt F.W., Westphal H. 1996. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage.Proc. Natl. Acad. Sci. USA. 93:5860–5865 doi:10.1073/pnas.93.12.5860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazorchak A., Jones M.E., Zhuang Y. 2005. New insights into E-protein function in lymphocyte development.Trends Immunol. 26:334–338 doi:10.1016/j.it.2005.03.011 [DOI] [PubMed] [Google Scholar]
- Lazorchak A.S., Schlissel M.S., Zhuang Y. 2006a. E2A and IRF-4/Pip promote chromatin modification and transcription of the immunoglobulin kappa locus in pre-B cells.Mol. Cell. Biol. 26:810–821 doi:10.1128/MCB.26.3.810-821.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazorchak A.S., Wojciechowski J., Dai M., Zhuang Y. 2006b. E2A promotes the survival of precursor and mature B lymphocytes.J. Immunol. 177:2495–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Grosschedl R. 1995. Failure of B-cell differentiation in mice lacking the transcription factor EBF.Nature. 376:263–267 doi:10.1038/376263a0 [DOI] [PubMed] [Google Scholar]
- Liu Y., Subrahmanyam R., Chakraborty T., Sen R., Desiderio S. 2007. A plant homeodomain in RAG-2 that binds hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement.Immunity. 27:561–571 doi:10.1016/j.immuni.2007.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin B.R., Giepmans B.N., Adams S.R., Tsien R.Y. 2005. Mammalian cell-based optimization of the biarsenical-binding tetracysteine motif for improved fluorescence and affinity.Nat. Biotechnol. 23:1308–1314 doi:10.1038/nbt1136 [DOI] [PubMed] [Google Scholar]
- Matthews A.G., Kuo A.J., Ramón-Maiques S., Han S., Champagne K.S., Ivanov D., Gallardo M., Carney D., Cheung P., Ciccone D.N., et al. 2007. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination.Nature. 450:1106–1110 doi:10.1038/nature06431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murre C. 2005. Helix-loop-helix proteins and lymphocyte development.Nat. Immunol. 6:1079–1086 doi:10.1038/ni1260 [DOI] [PubMed] [Google Scholar]
- Murre C., McCaw P.S., Baltimore D. 1989. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins.Cell. 56:777–783 doi:10.1016/0092-8674(89)90682-X [DOI] [PubMed] [Google Scholar]
- Nadel B., Cazenave P.A., Sanchez P. 1990. Murine lambda gene rearrangements: the stochastic model prevails over the ordered model.EMBO J. 9:435–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemazee D. 2006. Receptor editing in lymphocyte development and central tolerance.Nat. Rev. Immunol. 6:728–740 doi:10.1038/nri1939 [DOI] [PubMed] [Google Scholar]
- Nutt S.L., Kee B.L. 2007. The transcriptional regulation of B cell lineage commitment.Immunity. 26:715–725 doi:10.1016/j.immuni.2007.05.010 [DOI] [PubMed] [Google Scholar]
- Quong M.W., Martensson A., Langerak A.W., Rivera R.R., Nemazee D., Murre C. 2004. Receptor editing and marginal zone B cell development are regulated by the helix-loop-helix protein, E2A.J. Exp. Med. 199:1101–1112 doi:10.1084/jem.20031180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radic M.Z., Erikson J., Litwin S., Weigert M. 1993. B lymphocytes may escape tolerance by revising their antigen receptors.J. Exp. Med. 177:1165–1173 doi:10.1084/jem.177.4.1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramón-Maiques S., Kuo A.J., Carney D., Matthews A.G., Oettinger M.A., Gozani O., Yang W. 2007. The plant homeodomain finger of RAG2 recognizes histone H3 methylated at both lysine-4 and arginine-2.Proc. Natl. Acad. Sci. USA. 104:18993–18998 doi:10.1073/pnas.0709170104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler S., Györy I., Imhof S., Spivakov M., Williams R.R., Busslinger M., Fisher A.G., Grosschedl R. 2007. Distinct promoters mediate the regulation of Ebf1 gene expression by interleukin-7 and Pax5.Mol. Cell. Biol. 27:579–594 doi:10.1128/MCB.01192-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanow W.J., Langerak A.W., Goebel P., Wolvers-Tettero I.L., van Dongen J.J., Feeney A.J., Murre C. 2000. E2A and EBF act in synergy with the V(D)J recombinase to generate a diverse immunoglobulin repertoire in nonlymphoid cells.Mol. Cell. 5:343–353 doi:10.1016/S1097-2765(00)80429-3 [DOI] [PubMed] [Google Scholar]
- Rudin C.M., Storb U. 1992. Two conserved essential motifs of the murine immunoglobulin lambda enhancers bind B-cell-specific factors.Mol. Cell. Biol. 12:309–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schebesta M., Pfeffer P.L., Busslinger M. 2002. Control of pre-BCR signaling by Pax5-dependent activation of the BLNK gene.Immunity. 17:473–485 doi:10.1016/S1074-7613(02)00418-1 [DOI] [PubMed] [Google Scholar]
- Seet C.S., Brumbaugh R.L., Kee B.L. 2004. Early B cell factor promotes B lymphopoiesis with reduced interleukin 7 responsiveness in the absence of E2A.J. Exp. Med. 199:1689–1700 doi:10.1084/jem.20032202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semerad C.L., Mercer E.M., Inlay M.A., Weissman I.L., Murre C. 2009. E2A proteins maintain the hematopoietic stem cell pool and promote the maturation of myelolymphoid and myeloerythroid progenitors.Proc. Natl. Acad. Sci. USA. 106:1930–1935 doi:10.1073/pnas.0808866106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh H., Pongubala J.M., Medina K.L. 2007. Gene regulatory networks that orchestrate the development of B lymphocyte precursors.Adv. Exp. Med. Biol. 596:57–62 doi:10.1007/0-387-46530-8_5 [DOI] [PubMed] [Google Scholar]
- Sun X.H., Baltimore D. 1991. An inhibitory domain of E12 transcription factor prevents DNA binding in E12 homodimers but not in E12 heterodimers.Cell. 64:459–470 doi:10.1016/0092-8674(91)90653-G [DOI] [PubMed] [Google Scholar]
- Tiegs S.L., Russell D.M., Nemazee D. 1993. Receptor editing in self-reactive bone marrow B cells.J. Exp. Med. 177:1009–1020 doi:10.1084/jem.177.4.1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vela J.L., Aït-Azzouzene D., Duong B.H., Ota T., Nemazee D. 2008. Rearrangement of mouse immunoglobulin kappa deleting element recombining sequence promotes immune tolerance and lambda B cell production.Immunity. 28:161–170 doi:10.1016/j.immuni.2007.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Claus C.L., Vaccarelli G., Braunstein M., Schmitt T.M., Zúñiga-Pflücker J.C., Rothenberg E.V., Anderson M.K. 2006. The basic helix-loop-helix transcription factor HEBAlt is expressed in pro-T cells and enhances the generation of T cell precursors.J. Immunol. 177:109–119 [DOI] [PubMed] [Google Scholar]
- Xu C.R., Feeney A.J. 2009. The epigenetic profile of Ig genes is dynamically regulated during B cell differentiation and is modulated by pre-B cell receptor signaling.J. Immunol. 182:1362–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Kardava L., St Leger A., Martincic K., Varnum-Finney B., Bernstein I.D., Milcarek C., Borghesi L. 2008. E47 controls the developmental integrity and cell cycle quiescence of multipotential hematopoietic progenitors.J. Immunol. 181:5885–5894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y., Soriano P., Weintraub H. 1994. The helix-loop-helix gene E2A is required for B cell formation.Cell. 79:875–884 doi:10.1016/0092-8674(94)90076-0 [DOI] [PubMed] [Google Scholar]
- Zhuang Y., Jackson A., Pan L., Shen K., Dai M. 2004. Regulation of E2A gene expression in B-lymphocyte development.Mol. Immunol. 40:1165–1177 doi:10.1016/j.molimm.2003.11.031 [DOI] [PubMed] [Google Scholar]