Abstract
Large numbers of genetic disorders are caused by nonsense mutations for which compound-induced readthrough of premature termination codons (PTCs) might be exploited as a potential treatment strategy. We have successfully developed a sensitive and quantitative high-throughput screening (HTS) assay, protein transcription/translation (PTT)–enzyme-linked immunosorbent assay (ELISA), for identifying novel PTC-readthrough compounds using ataxia-telangiectasia (A-T) as a genetic disease model. This HTS PTT-ELISA assay is based on a coupled PTT that uses plasmid templates containing prototypic A-T mutated (ATM) mutations for HTS. The assay is luciferase independent. We screened ∼34,000 compounds and identified 12 low-molecular-mass nonaminoglycosides with potential PTC-readthrough activity. From these, two leading compounds consistently induced functional ATM protein in ATM-deficient cells containing disease-causing nonsense mutations, as demonstrated by direct measurement of ATM protein, restored ATM kinase activity, and colony survival assays for cellular radiosensitivity. The two compounds also demonstrated readthrough activity in mdx mouse myotube cells carrying a nonsense mutation and induced significant amounts of dystrophin protein.
Translation termination is signaled by three stop codons: UAA, UAG, and UGA. This mechanism is highly conserved, although each stop codon has a different efficiency for terminating translation. UGA is considered to be a “leaky” stop codon with the highest intrinsic readthrough potential. UAA shows high fidelity and little intrinsic readthrough potential, whereas UAG has intermediate fidelity (Weiner and Weber, 1973; Lovett et al., 1991). Nonsense mutations create primary premature termination codons (PTCs) and result in either no formation of the target protein or truncated protein with impaired stability.
Certain compounds influence the fidelity of stop codon recognition and induce readthrough of primary PTCs, which allows translation of some full-length protein. In many cases, the readthrough-induced protein is functional, even when it contains a wrongly incorporated amino acid (Keeling and Bedwell, 2005; Zingman et al., 2007).
It is estimated that 30% of human disease-causing alleles are nonsense mutations (Mendell and Dietz, 2001). Other types of mutation, such as frameshift and splicing mutations, lead to secondary PTCs; however, these are not therapeutic targets for readthrough compounds (RTCs). Considering that >1,800 distinct genetic disorders are caused by nonsense mutations, the readthrough of primary PTCs has treatment potential for large numbers of patients.
To date, most reported PTC-RTCs that are active in mammalian cells have belonged to the aminoglycoside antibiotics class (Keeling and Bedwell, 2005; Zingman et al., 2007). Certain types of aminoglycosides can induce ribosomes to read through PTC mutations via insertion of a random amino acid by near-cognate transfer RNA. The therapeutic potential of aminoglycosides has been evaluated in the laboratory for different genetic models, such as cystic fibrosis (Howard et al., 1996; Bedwell et al., 1997; Du et al., 2002), muscular dystrophy (Howard et al., 2000; Wagner et al., 2001; Dunant et al., 2003; Loufrani et al., 2004), Hurler syndrome (Keeling et al., 2001), cystinosis (Helip-Wooley et al., 2002), spinal muscular atrophy (Sossi et al., 2001), ataxia-telangiectasia (A-T; Lai et al., 2004), and type 1 Usher syndrome (Rebibo-Sabbah et al., 2007). Clinical trials also indicate that aminoglycosides can induce some functional protein production; however, the therapeutic benefits remain uncertain (Wilschanski et al., 2000; Clancy et al., 2001; Wagner et al., 2001; Politano et al., 2003). Furthermore, the toxicity of most commercial aminoglycosides in mammals has greatly diminished their potential for successful readthrough therapy (Mingeot-Leclercq and Tulkens, 1999; Guan et al., 2000). Therefore, efforts are underway to develop better aminoglycoside derivatives with reduced toxicity and enhanced activity (Nudelman et al., 2006; Rebibo-Sabbah et al., 2007). Recently, PTC Therapeutics (South Plainfield, NJ) described a more efficient nonaminoglycoside RTC, PTC124, which was developed synthetically by screening >800,000 chemicals and analogues using a luciferase-based high-throughput screening (HTS) assay (Welch et al., 2007; Hirawat et al., 2007; M. Du et al., 2008). A phase-I clinical study in cystic fibrosis confirmed that PTC124 is generally well tolerated and appears to have more efficient readthrough activity than aminoglycosides (Hirawat et al., 2007). Moreover, PTC124 does not induce ribosomal readthrough of normal stop codons. A phase-II clinical trial is underway (Kerem et al., 2008). However, a recent study indicates that the initial discovery of PTC124 by HTS may have been biased by its direct effect on the FLuc (firefly luciferase) reporter used (Auld et al., 2009), indicating the importance of a luciferase-independent HTS assay for future drug screening.
In an effort to discover new RTCs, we developed a sensitive and quantitative luciferase-independent HTS assay, protein transcription/translation (PTT)–ELISA. The PTT-ELISA assay was validated for a fully automated 384-well robotic platform and used to screen ∼34,000 compounds. We focused followup efforts on 12 low-molecular-mass nonaminoglycoside compounds. From there, we identified two compounds that induced low levels of full-length functional A-T mutated (ATM) protein in A-T cells carrying ATM nonsense mutations, as demonstrated by direct measurement of ATM protein using ATM-ELISA, ATM-Ser1981 autophosphorylation, trans-phosphorylation of structural maintenance of chromosome (SMC) 1–Ser966, and colony survival assay (CSA). Both compounds also showed PTC-readthrough activity in mdx mouse myotube cells carrying a nonsense mutation and induced significant amounts of dystrophin protein.
Collectively, these studies provide the first robust luciferase-independent HTS assay for identifying RTCs and proof of principle for PTC readthrough by nonaminoglycoside compounds. They further establish that enhanced PTC readthrough can be considered as a therapeutic strategy for correcting nonsense mutations in many genetic diseases.
RESULTS
Development and validation of HTS assay PTT-ELISA
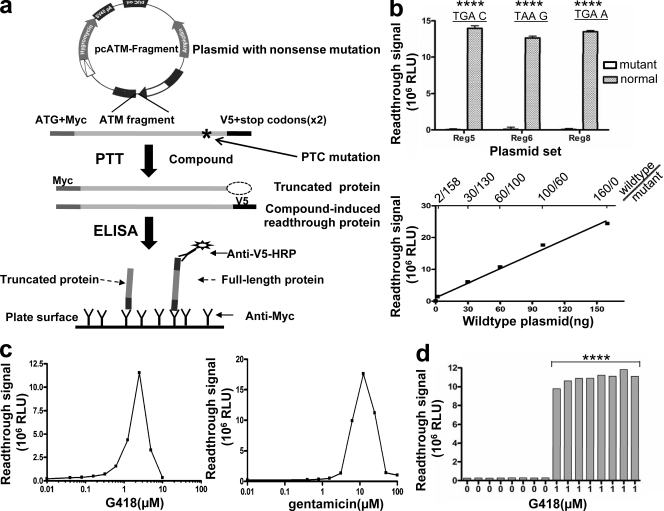
In vitro PTT was originally designed to detect truncating mutations (Roest et al., 1993; Telatar et al., 1996). ELISA was coupled into PTT to improve throughput for the detection of these mutations (Gite et al., 2003; L. Du et al., 2008). In a previous study, we used a PTT-PAGE gel approach to evaluate various aminoglycosides for readthrough activity of ATM nonsense mutations (Lai et al., 2004). However, the gel-based PTT assay was time consuming and involved the use of radioactive material; thus, it was difficult to automate for a high-throughput format. Herein, we have developed a plasmid-driven PTT-ELISA assay for screening large numbers of compounds for PTC readthrough activity. The assay uses plasmid templates containing prototypic ATM mutations, patterned after specific disease-causing ATM mutations. So as to work in a mammalian system, rabbit reticulocytes were chosen to drive the PTT reaction. Various fragments of mutated ATM alleles from cells of A-T patients were cloned into the plasmids and were N- and C-terminally tagged with the epitopes myc and V5, respectively. Anti-myc antibody was used to capture the translated protein onto an ELISA plate. If compounds induce PTC-readthrough in the assay, the plasmid-driven PTT results in a full-length ATM fragment including the V5 tag, which is detected with anti–V5–horseradish peroxidase (HRP) antibody (Fig. 1 a).
Figure 1.
HTS PTT-ELISA assay for RTCs screening. (a) Schematic of PTT-ELISA HTS. ATM regions containing a PTC mutation were cloned into plasmids and tagged with c-myc and V5 epitopes at each end. Compounds with PTC readthrough activity induce full-length protein, which can be identified by anti–V5-HRP. (b) Specificity and sensitivity of PTT-ELISA. All three mutant plasmids showed only background readings, whereas wild-type plasmids showed signals >200-fold over background (P < 0.0001), indicating that the assay specifically detects full-length proteins. The sample containing 2 ng of wild-type plasmid still gave signals that were twofold higher than those of mutant plasmid alone (P < 0.01), indicating that the sensitivity of the assay is ∼1% (2/158 ng). Error bars indicate the variation of duplicate samples. (c) Assay evaluation using aminoglycosides. Both G418 and gentamicin induced significant PTC readthrough over a large dose range. (d) Readings for a sample row from a 384-well plate. Samples containing G418 exhibited significantly different signal (Z′ factor > 0.6) over the no-drug wells (P < 0.0001). ****, P < 0.0001, as compared with control. Experiments were repeated three times for b and c and five times for d.
We constructed three mutant plasmids that contain prototypic PTC mutations in the ATM gene from three different A-T patients. The plasmid used for preliminary screening, plasmid-TAT51, contains ATM region 5 fragment (codons 1403–1886) and harbors a nonsense (PTC) mutation (c.5623C→T) that leads to a TGA C stop codon. The second mutant plasmid, plasmid-AT153LA, contains the same TGA stop codon but at a different position within the gene (c.8977C→T) in region 8 (codons 2550–3050) and also has a different +4 nt (TGA A). This was used to monitor the effect of surrounding sequences of PTCs on readthrough ability. A third mutant plasmid, plasmid-AT185LA, contains a different stop codon, TAA G, resulting from a nonsense mutation (c.3673C→T) in region 4 (codons 1041–1531). Plasmids containing the same fragments but without mutations were constructed by in vitro mutagenesis of patient-derived complementary DNA and were used as wild-type controls.
We first evaluated the specificity and sensitivity of PTT-ELISA to detect the in vitro–translated full-length protein fragment. All three mutant plasmids gave only background signal, whereas the comparable wild-type plasmids showed signals greater than ∼200-fold over background (Fig. 1 b, top), indicating that the assay specifically recognized full-length protein fragments. We next used TAT51 wild-type plasmid to evaluate the sensitivity of the assay. The TAT51 wild-type plasmid was serially diluted with TAT51 mutant plasmid and used to drive PTT reactions. The sample containing 1.2% of wild-type plasmid (2/158 ng) still gave signal that was twofold higher than that of mutant plasmid (Fig. 1 b, bottom), establishing the sensitivity of the assay at ∼1%. The sensitivity of an HTS assay is especially important for RTC screening because all RTCs to date have been only weak PTC readthrough inducers, and new classes of RTCs are expected only to be identified with a highly sensitive screening assay.
Next, we used two well known RTCs, G418 and gentamicin, to test the efficiency of the PTT-ELISA assay. Both compounds showed significant PTC readthrough activity with TAT51 mutant plasmid (TGA C) over a large dose range (40 nM–10 µM; Fig. 1 c). Moreover, G418 showed obvious toxicity for the PTT reaction at concentrations >2.5 µM, whereas gentamicin toxicity was not observed until 12.5 µM. These results are consistent with our previously published S35-PTT gel data (Lai et al., 2004) and demonstrate that the PTT-ELISA readily identifies compounds with readthrough activity.
We next validated the assay for use in a fully automated 384-well robotic platform, using plasmid-TAT51 (TGA C) as readthrough template and 1 µM G418 as the positive readthrough control. To further reduce the cost of the assay, we optimized a decrease in the PTT reaction volume from 25 to 5 µl. As shown in Fig. 1 d, the wells containing G418 exhibited significantly different signals (Z′ factor > 0.8) over the no-drug wells. The fully automated assay also showed consistent plate-to-plate accuracy and efficiency, indicating the suitability of the assay for HTS (unpublished data).
Screening of chemical libraries for RTCs
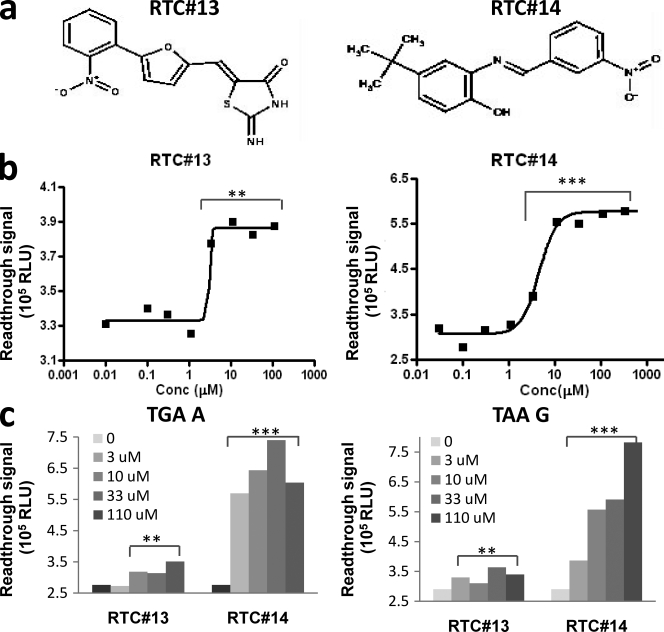
About 34,000 compounds were screened to discover novel RTCs. Plasmid-TAT51 (TGA C) was used for the initial screening. Each compound was screened at a final concentration of 10 µM in the assay mixture. For each screening plate, we included both positive (with 1 µM G418) and negative (with DMSO) samples as quality controls. Samples with signal >2-fold over the negative control were scored as potential “hits.” Our initial screens yielded 12 low-molecular-mass RTCs with appreciable readthrough activity. All 12 hits were then confirmed by manual PTT-ELISA at multiple concentrations (chemical names shown in Table S1). None of them were aminoglycosides and none of them had been previously reported. The chemical names and structures of the two leading compounds, RTC#13 and #14, are shown in Fig. 2 a; the names and structures for the remaining 10 RTCs are given in Fig. S1 and Table S1.
Figure 2.
Two leading RTCs were identified by HTS. (a) Molecular structures of RTC#13 and #14 (2-imino-5-{[5-(2-nitrophenyl)-2-furyl]methylene}-1,3-thiazolidin-4-one and 4-tert-butyl-2-[(3-nitrobenzylidene)amino]pheno). (b) TGA C readthrough activity by PTT-ELISA. (c) TGA A and TAA G readthrough activity. **, P < 0.01; ***, P < 0.001, as compared with untreated control. Experiments were repeated four times for b and three times for c.
The EC50 of five compounds (RTC#4, #11, #13, #14, and #16) were <10 µM (Fig. 2 b, RTC#13 and #14), implying therapeutic potential of those compounds. Notably, in these initial cell-free experiments, the maximum in vitro readthrough effect for the new compounds was not as favorable as that of G418 or gentamicin. For example, the maximum readthrough activity of RTC#13 and #14, detected by the cell-free PTT-ELISA assay, was ∼10% of the maximum activity of G418 and gentamicin in the same assay (Fig. 1 c vs. Fig. 2 b). This may be related to the solubility, permeability, or toxicity of the compounds. In contrast, both RTC#13 and #14 had an EC50 <10 µM, they were less toxic, and they did not show obvious inhibition of PTT at high concentrations (>50 µM), unlike both G418 and gentamicin (Fig. 1 c).
The readthrough efficiency of these compounds was also tested on two other stop codon contexts, TGA A and TAA G. For TGA A, all 12 compounds showed various extents of readthrough activity (unpublished data). For TAA G, six compounds (RTC#10, #11, #13, #14, #16, and #17) showed appreciable activity (unpublished data). The data for RTC#13 and #14 are shown in Fig. 2 c.
RTC-induced ATM protein and ATM-Ser1981 phosphorylation in A-T lymphoblastoid cell lines (LCLs)
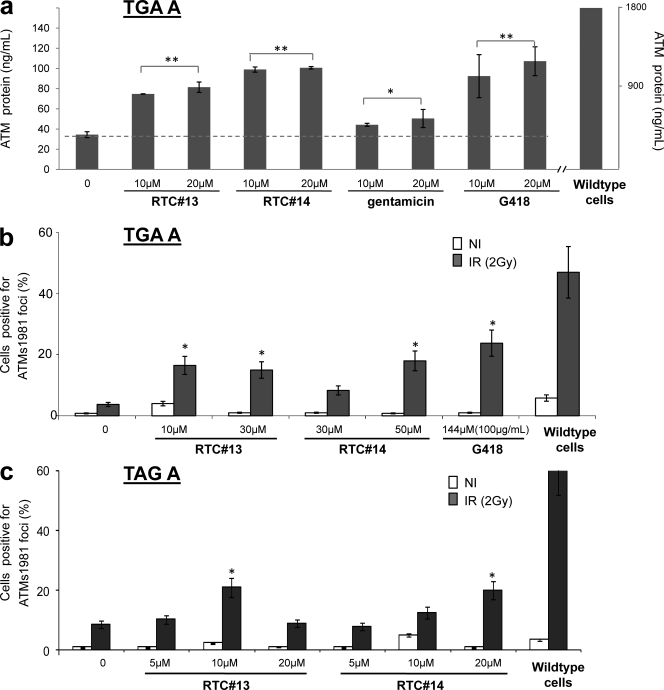
To test whether the newly identified compounds could induce ATM protein in A-T cells with PTC mutations, we exposed A-T LCLs to each compound for 4 d before harvesting the cells. Western blotting of nuclear lysates was generally of insufficient sensitivity for monitoring RTC-induced ATM protein (Lai et al., 2004); therefore, we used an ATM-ELISA method to measure intranuclear ATM protein in cells (Butch et al., 2004). Concentrations of individual RTCs used to treat AT153LA LCL with a homozygous TGA A mutation were based on their cytotoxicity profile (Fig. S2, RTC#13 and #14). Cells treated with the highest RTC concentrations still had >70% viability as compared with untreated cells. RTC#13 and #14 consistently induced low but detectable levels of ATM protein in A-T LCLs (Fig. 3 a). The restored ATM protein was ≤5% of the wild-type LCLs. Gentamicin and G418 also induced small amounts of ATM protein (Fig. 3 a). We were encouraged by these results because A-T patients with some residual ATM protein levels can have substantial ATM kinase activity, and this has been sometimes associated with a later onset and slower progression of symptoms (Gilad et al., 1998; Chun et al., 2003). Thus, we believe that even modest increases in ATM protein levels have therapeutic potential.
Figure 3.
RTCs induced intranuclear ATM protein and post-IR ATM-Ser1981 foci in A-T LCLs. LCLs were treated with compound for 4 d before harvesting. Induced ATM protein was assessed by ATM-ELISA and ATM-Ser1981 IRIF. For IRIF test, cells were irradiated with 2 Gy and IRIFs were scored after 30 min. All experiments were repeated three times. (a) Cells treated with various doses of compound RTC#13 and #14 showed a significantly increased ATM protein level, as compared with nontreated A-T samples (P < 0.05). The dashed line indicates the basal ATM protein level in untreated A-T cells. (b) RTC-induced ATM-Ser1981 IRIF in AT153LA (TGA A) cells. (c) RTC-induced ATM-Ser1981 IRIF in AT229LA (TAG A) cells. *, P < 0.05; **, P < 0.01, as compared with untreated sample. Error bars indicate the variation of two independent experiments.
To evaluate the function of RTC-induced ATM protein, we measured irradiation-induced foci (IRIF) formation of ATM-Ser1981 in the same AT153LA cells (TGA A). A-T cells do not form post–ionizing radiation (IR) ATM-Ser1981 foci because ATM protein is either absent or functionally impaired (usually the former). We found that RTC#13 and #14 induced considerable numbers of ATM-Ser1981 IRIFs in A-T cells with TGA A, whereas only a background level of IRIFs was observed in the untreated controls (Fig. 3 b). At same time, ∼51% of wild-type cells showed distinct post-IR foci. The maximum IRIF induction achieved in A-T cells by 10 µM RTC#13 was ∼40% of wild-type level. As a positive readthrough control, 144 µM of G418 (100 µg/ml) was used to treat cells. Our previous studies had shown that, at this concentration, G418 induced a maximum level of ATMs1981 IRIF (Lai et al., 2004). As anticipated, significant IRIFs were induced in G418-treated cells. The level in A-T cells was about half that in wild-type cells. The compounds were also tested in another A-T cell line, AT229LA, with homozygous TAG A mutations, and both RTC#13 and #14 induced significant numbers of ATM-Ser1981 IRIF, as compared with nontreated A-T cells (Fig. 3 c).
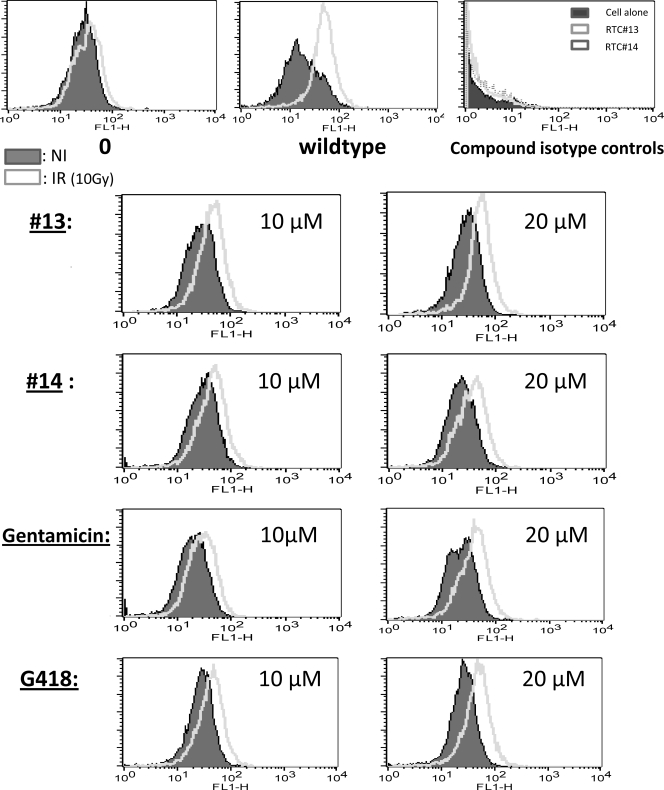
To further compare the readthrough activity of RTC#13 and #14 with gentamicin and G418, we used a flow cytometry (FC)–based ATM-Ser1981 phosphorylation assay to assess readthrough activity in AT229LA cells (TAG A; Fig. 4). Both RTC#13 and #14 induced increased ATM-Ser1981 autophosphorylation, as indicated by the right shift of fluorescence intensity (FI). This was consistent with previous ATM IRIF data (Fig. 3 c). Encouragingly, ATM-Ser1981 phosphorylation levels induced by RTC#13 and #14 were similar to those induced by the same concentrations of gentamicin and G418 in the same cell line (Fig. 4). Neither of the two compounds produces green autofluorescence (isotype controls; Fig. 4, top right histogram). Similar readthrough effects were also observed in AT153LA cells (TGA A) using cytometry-based ATM-Ser1981 phosphorylation assay (unpublished data).
Figure 4.
RTCs induced FC-ATM-Ser1981 phosphorylation in A-T LCLs. AT229LA cells (TAG A) were treated with RTC#13 and #14 for 4 d and analyzed for ATM-Ser1981 phosphorylation using FC. Gentamicin and G418 were used as positive readthrough controls. All compounds induced ATM phosphorylation in A-T cells, as indicated by a right FI shift. Neither compound produced autofluorescence, as shown in the top right histogram. Results were consistent in three independent experiments.
RTC-induced restoration of SMC1-Ser966 phosphorylation in A-T LCLs
As an alternative assay to assess ATM kinase function, we used a recently developed FC-based assay to measure the trans-phosphorylation of SMC1 in A-T LCLs (Fig. 5; Nahas et al., 2009). ATM phosphorylates SMC1 at Ser966 and Ser957 after IR-induced double-strand breaks (Yazdi et al., 2002; Kitagawa et al., 2004), and IR-induced SMC1 phosphorylation is deficient in A-T cells. In untreated AT153LA cells (TGA A), we observed no phosphorylation of SMC1 (Fig. 5 a, top, left). After treatment with either RTC#13 or #14, a right shift in FI was observed, indicating restored ATM kinase activity. Both G418 and gentamicin induced SMC1 phosphorylation in A-T cells at similar levels. We also tested the same compounds on TAG A mutation using AT229LA cells. They all induced detectable SMC1 phosphorylation (Fig. 5 b). RTC-induced correction of SMC1 phosphorylation was dose dependent in AT153LA cells (TGA A; Fig. S3).
Figure 5.
RTCs induced FC-SMC1 pSer966 phosphorylation in A-T LCLs. (a) RTC#13 and #14 restored SMC Ser966 phosphorylation in AT153LA cells (TGA A). (b) RTC#13 and #14 restored SMC1 Ser966 phosphorylation in AT229LA cells (TAG A). All experiments were repeated three times.
RTC-induced restoration of ATM kinase in A-T fibroblast cells
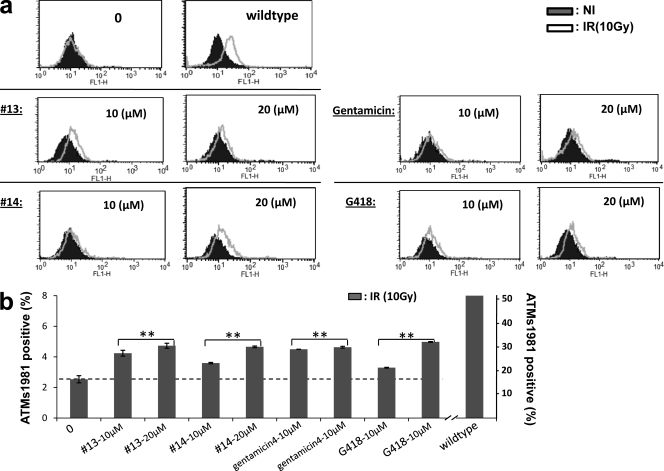
To determine whether the two lead RTCs were also active in other types of cells, we tested them against the A-T fibroblast cell line GM02052, which contains a homozygous TGA G mutation (c. 103C→T). RTC-treated cells showed slightly increased IR-induced SMC1-Ser966 (Fig. 6 a) and ATM-Ser1981 phosphorylation (Fig. 6 b), as compared with untreated cells. As positive readthrough controls, gentamicin and G418 showed readthrough activities similar to RTC#13 and #14 at the concentrations compared, indicating that both RTC#13 and #14 were also active on A-T fibroblasts.
Figure 6.
RTCs restored ATM kinase activity in A-T fibroblast cells. GM02052 cells with a homozygous c.103C→T mutation (TGA G) were treated with RTC#13 and #14 for 4 d, and ATM kinase activity was assessed using FC-based SMC1-Ser966 phosphorylation and ATM-Ser1981 phosphorylation. All experiments were repeated three times. (a) Histograms of FC-SMC1-Ser966. (b) Cell population positive for ATM-Ser1981 staining. The dashed line indicates the basal ATM s1981 phosphorylation in the nontreated A-T cells after radiation (10 Gy). **, P < 0.01, as compared with untreated sample. Error bars indicate the variation of two independent experiments.
RTC-induced cell survival correction in A-T LCLs
Because the radiosensitivity of A-T cells results from the ATM deficiency, we next investigated whether RTC#13 and #14 can abrogate A-T cell’s radiosensitivity of A-T cells. AT153LA cells (TGA A) were treated with compounds for 4 d and followed by CSA. One wild-type cell line and one different A-T cell line were used as assay quality controls. As expected, the wild-type and A-T controls showed 49 and 11% survival fraction, respectively. These were within the normal (>36%) and radiosensitive (<21%) range, respectively (Fig. 7, right two bars). Untreated AT153LA cells showed a 13.5% survival fraction, which is radiosensitive (Fig. 7, left bar). Encouragingly, we found that both RTC#13 and #14 (at 10 and 20 µM) abrogated AT153LA cell radiosensitivity, from radiosensitive (13.5%) to intermediate (21–36%) range. 100 µM G418 also abrogated radiosensitivity to intermediate range, whereas gentamicin did not show a significant effect in this assay at the tested concentrations (10 and 20 µM). The definition of CSA range used was previously established in our laboratory and is used for clinical A-T diagnosis (see Materials and methods).
Figure 7.
RTCs abrogated the radiosensitivity of A-T LCLs. AT153LA cells (TGA A) were treated with compounds and the CSA was measured. RTC#13, RTC#14, and G418 increased cell survival fractions to intermediate range. Gentamicin did not show an effect at tested concentrations. Results were consistent in three independent experiments.
RTCs induced mdx PTC readthrough in mouse myotube cells
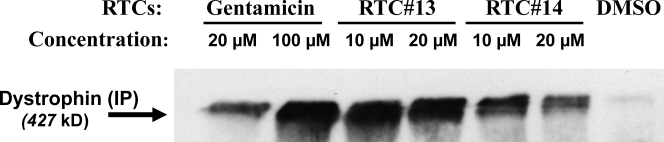
To investigate the ability of RTC#13 and #14 to readthrough a PTC mutation in genes other than ATM, we used mdx myotubes. The mdx mouse has been widely used as a model for Duchenne muscular dystrophy. It carries a C to T transition in exon 23 of the dystrophin gene that creates a premature stop codon (TAA), resulting in an absence of dystrophin protein (Sicinski, et al., 1989). Because gentamicin has been shown to induce readthrough of the mdx PTC mutation and to restore dystrophin expression in mdx mice both in vitro and in vivo (Barton-Davis, et al., 1999), we used it as a positive readthrough control in the experiments. Muscle progenitor cells isolated from mdx mice were induced to differentiate for 24 h before addition of gentamicin or compounds RTC#13 and #14. Myotube cultures were exposed to compounds for 72 h and then analyzed for dystrophin by Western blotting. Full-length dystrophin was clearly detected in cells treated with RTC#13 and #14 at concentrations of 10 and 20 µM but not in cultures treated with gentamicin at concentrations of 20 µM (unpublished data). We next used immunoprecipitation to increase the sensitivity of the assay (Fig. 8). Dystrophin expression was present in cells treated with gentamicin at concentrations of 20 and 100 µM. Both of our lead compounds showed readthrough activity, inducing significant amount of dystrophin protein. Results were consistent in multiple experiments performed.
Figure 8.
RTCs restored full-length dystrophin protein in mouse mdx myotubes (TAA). Cells were prepared and treated with RTCs for 3 d. Dystrophin proteins were detected by Western blot analysis using immunoprecipitation. Wild-type cells were used in the experiment to localize dystrophin protein. Gentamicin was used as a positive readthrough control. Both RTC#13 and #14 induced a significant amount of dystrophin protein. Results were consistent in three independent experiments.
DISCUSSION
We believe that HTS can identify new nonaminoglycoside RTCs with therapeutic potential. In this study, we used A-T as a disease model to identify new RTCs. A-T is a progressive autosomal recessive neurodegenerative disorder resulting from mutations in the ATM gene (Perlman et al., 2003; Chun and Gatti, 2004). ATM protein plays a very important role in cell cycle control, DNA damage repair, the oxidative stress response, and apoptosis (Shiloh, 2006). The A-T disorder provides an appropriate laboratory model for demonstrating novel principles of mutation-targeted therapy. In the ATM mutation spectrum, primary nonsense mutations account for ∼15% of the unique mutations detected in A-T patients (www.LOVD.nl/ATM). A well characterized spectrum of ATM mutations, supported by an extensive library of LCLs derived from patients with those mutations, allowed us to investigate the effect of nonaminoglycoside RTCs on various primary premature stop codons. We have previously used aminoglycosides and antisense oligonucleotides to correct nonsense and splicing ATM mutations, respectively, and to restore functional ATM protein in A-T LCLs (Lai et al., 2004; Du et al., 2007). These studies suggest that therapeutic benefits might be achieved if even modest increases in functional ATM protein levels can be induced. In these limited studies, we saw no significant effects on efficiency of readthrough from the fourth nucleotide of each tested stop codon.
Herein, we successfully developed a sensitive luciferase-independent HTS assay by coupling PTT and ELISA. PTT-ELISA shows high specificity for detecting readthrough products and, thus, minimizes false positives in the initial large-scale library screening. The assay is also very sensitive; the minimum detection threshold is ∼1%, which ensures its efficiency as a HTS assay. The efficiency of PTT-ELISA was further evaluated using two well known RTCs, G418 and gentamicin. The assay was able to detect their readthrough activity over a very large concentration range (G418, 40 nM–10 µM; gentamicin, 40 nM–100 µM). Furthermore, PTT-ELISA has been validated for a fully automated robotic platform, with consistent accuracy between plates. The 384-well format dramatically reduced the workload for screening thousands of compounds and also saved time, costs, and reagents. Moreover, this assay has the potential to be validated for a 1,536-well format. For these reasons, we believe that the PTT-ELISA HTS assay provides a powerful new tool for identifying new RTCs.
From a library of ∼34,000 compounds, we identified 12 low-molecular-mass compounds (between ∼300 and 450 daltons) with PTC readthrough activity. None of these new compounds were aminoglycosides. Several compounds showed EC50 values <10 µM, implying their potential for further development. To further assess these compounds in cell systems, we tested their readthrough activity in A-T LCLs with a variety of nonsense mutations. We used ATM-ELISA to directly detect ATM protein levels in treated cells. Subsequently, more sensitive cell-based assays, such as FC-based SMC1-Ser966, ATM-Ser1981 phosphorylation, and ATM-Ser1981-IRIF, were used to assess restored ATM kinase activity. Among the 12 compounds, RTC#13 and #14 showed PTC readthrough activity in A-T cells, both in LCLs and fibroblasts, as demonstrated by ATM-ELISA, ATM kinase activity (autophosphorylation of ATM and trans-phosphorylation of SMC1), and CSA. To determine whether the compounds can read through PTC mutations in other genes, we selected mouse mdx myotube cells and tested their ability to induce readthrough in a different species, a different cell type (nondividing muscular cells), and a different premature stop codon (TAA). In other studies, the TAA codon has proven the most difficult to read through (Kimura et al., 2005; Welch et al., 2007). Both RTC#13 and #14 induced PTC readthrough of the mouse mdx dystrophin gene.
Collectively, our data showed that both RTC#13 and #14 had comparable readthrough activity in cell-based assays, even though their activities in cell-free PTT-ELISA assay were much lower than gentamicin and G418. This may be related to differences in cell-based metabolism, drug solubility, and permeability of compounds.
To assess the impact of the compounds on readthrough of normal stop codons of other proteins, we performed two-dimensional gel electrophoresis. Neither compound significantly interfered with protein expression patterns (Fig. S4), implying that these compounds have potential for further development. However, we anticipate that significant therapeutic effects may not be discernable in A-T patients if the RTC-induced ATM level is <15% of normal, and this very likely also applies to other genetic disorders. Therefore, further structural modifications to improve pharmodynamics will be necessary. Structural optimization has improved the readthrough activity and lowered the toxicity of aminoglycosides (Nudelman et al., 2006; Rebibo-Sabbah et al., 2007). Therefore, it is expected to be effective for developing better nonaminoglycoside readthrough analogues as well.
The underlying mechanisms of PTC readthrough activity for these newly identified compounds remain unknown. It has been demonstrated that all of the known PTC-RTCs function by interfering with ribosomal translation. Certainly, aminoglycosides interact with the decoding center of the ribosomal 16S subunit and cause misincorporation of an amino acid at the PTC site which allows translation to continue (Keeling and Bedwell, 2005; Zingman et al., 2007). PTC124 is believed to act at a different location on the ribosome (Linde and Kerem, 2008). It will be interesting to learn whether RTC#13 and #14 interact with the ribosome.
It has been reported that nonsense-mediated messenger RNA (mRNA) decay (NMD) can significantly affect RTC-induced PTC-readthrough because mRNA transcripts carrying nonsense mutations are degraded by this pathway (Wilkinson and Shyu, 2002; Holbrook et al., 2004). Therefore, inhibition of NMD may stabilize mutant mRNA transcripts and increase RTC-induced readthrough output (Linde et al., 2007; Linde and Kerem, 2008). We have examined this in our laboratory using many different ATM nonsense mutations, and we have failed to detect a clear pattern of NMD or inhibition of NMD, suggesting that NMD in A-T cells may be a matter of degree. NMD efficiency may also vary between different mutations and different genes. The role of NMD in RTC-induced treatment remains to be clarified.
MATERIALS AND METHODS
Plasmids construction.
To construct plasmids containing stop codon mutations in their own context, we selected ATM nonsense mutations that resulted directly from disease-causing point mutations in A-T patients. The LCLs used in this study carried the following mutations: TAT51, homozygous 5623C→T (TGA C); AT185LA, homozygous 3673C→T (TAA G); and AT153LA, homozygous 8977C→T (TGA A). Reverse-transcription PCR was performed using custom designed primers which introduced N- and C-terminal epitopes (c-myc and V5, respectively) into the PCR products (Du et al., 2008). PCR products were then cloned into pcDNA5/FRT/TO TOPO plasmids according to the manufacturer’s protocols (Invitrogen). The PCR products for each mutant were mutagenized back to normal and used as paired normal control plasmids. DNA sequencing confirmed the PTT fragments in all constructs.
High-throughput PTT-ELISA.
Three chemical libraries, ChemBridge, Prestwick, and MicroSource, were used in our primary screening. The final concentration for each compound tested was 10 µM. Screening was performed on a fully integrated CORE System (Beckman Coulter). PTT was performed in a reaction volume of 5 µl. The TNT T7 PCR Quick Master Mix, containing 20 µM methionine, was aliquoted into a 384-well low-volume plate (MatriCal Bioscience). Compounds were added using a 384-well pin tool (V&P Scientific, Inc.). The PTT reaction was started by the addition of purified plasmid at 50 ng/well, which was incubated for 2 h at 30°C. PTT samples were subsequently stored at 4°C for ELISA analysis. For each plate, G418 at a final concentration of 1 µM served as positive control and reactions with 1% DMSO served as negative control. ELISA was performed in a 384-well MaxiSorp ELISA plate (Nunc). The plate was coated with 20 µl of 5-µg/ml mouse anti-Myc antibody (Invitrogen) overnight at 4°C, followed by washing with PBS and blocking with 50 µl PBSTM (PBS-containing 0.05% Tween-20 and 5% milk) for 30 min at 37°C. 20 µl H2O was added in PTT plate and 15 µl of reaction solution was transferred into ELISA plate, followed by overnight incubation at room temperature. After washing the plate, 20 µl of 1:500 mouse anti-V5 HRP (Invitrogen) was added and incubated for 2 h at 37°C. The plate was washed, incubated with 30 µl SuperSignal ELISA Pico working solution (Thermo Fisher Scientific), and measured on the Victor-3V using “top read” and an integration time of 0.5 s.
Immunoassay for measurement of intranuclear ATM protein.
Readthrough-induced full-length ATM protein in A-T cells was measured by ATM immunoassay (Butch et al., 2004). Cell nuclear extracts were prepared using NE-PER protocol (Thermo Fisher Scientific). Then ATM-ELISA was performed using 200-µg nuclear extracts. ATM concentrations of tested samples were calculated from the standard calibration curve using purified ATM protein (Chun et al., 2004).
Immunofluorescence of ATM-Ser1981 IRIF.
Immunostaining of nuclear foci of ATM-Ser1981was performed as previously reported (Du et al., 2007). In brief, after being treated with compound for 4 d, cells were irradiated with 2 Gy and incubated at 37°C for 30 min. The cells were dropped onto coverslips, fixed with 4% paraformaldehyde, and permeabilized. Coverslips were blocked for 1 h and incubated with mouse anti-ATM pSer1981 for 1 h (1:500; Rockland Immunochemicals, Inc.). After a second blocking, cells were stained with FITC-conjugated anti–mouse IgG (1:150; Jackson ImmunoResearch Laboratories) for 1 h and mounted onto slides.
FC analysis of ATM-Ser1981 and SMC1-Ser966 phosphorylation.
FC-SMC1 assay was performed as recently described (Nahas et al., 2009). FC-ATM-Ser1981 assay was based on Honda’s assay (Honda et al., 2009) with modifications. In brief, cells were resuspended in PBS and radiated for 10 Gy. After 1 h, the cells were fixed and permeabilized using the FIX & PERM cell permeabilization kit (Invitrogen). The cells were then incubated with 1 µl of mouse ATM-Ser1981 antibody (Cell Signaling Technology) for 2 h at room temperature. Cells were washed and resuspended in 100 µl PBS with Alexa Fluor 488 anti–mouse IgG (Invitrogen) for 45 min. Cells were next washed and resuspended in PBS with 0.2% paraformaldehyde and analyzed using a FACSCalibur (BD).
CSA.
CSA was performed as previously described (Sun et al., 2002). After 4 d of incubation with compounds, LCLs were plated, in duplicate, in 96-well plates at 100 and 200 cells per well. One plate was exposed to 1.0 Gy radiation, whereas the other was left unirradiated. The cells were incubated for 10–13 d and then stained with MTT. The presence of a colony of 32 cells was scored as a positive well, and survival fractions were calculated.
Mdx myotubes treatment and Western blot analysis of dystrophin.
Cells were derived from limb muscle of neonatal mdx and C57 mice, as previously described (Bertoni and Rando, 2002). For growth, cells were plated on dishes coated with 5 g/ml laminin (Invitrogen) and maintained in growth medium consisting of Ham’s F10 nutrient mixture (Mediatech, Inc.) supplemented with 20% fetal bovine serum, penicillin, and streptomycin. Cell differentiation was induced by maintaining the cells in low serum medium (differentiation medium) consisting of DME supplemented with 2% horse serum, penicillin, and streptomycin. Myoblasts were plated in wells of 6-well dishes and were allowed to differentiate for 24 h before adding the compounds. Media was replaced every 24 h with fresh differentiation media containing the compounds. Cells were lysed 96 h after induction of differentiation (72 h after addition of the compounds). Total protein was determined and dystrophin immunoblot analysis was performed as previously described (Sicinski et al., 1989). 250 µg of total protein from each sample was immunoprecipitated using a monoclonal antibody directed toward the rod domain (MANDYS -8; 1:40; Sigma-Aldrich) of the dystrophin protein and detected by Western blotting (Barton-Davis et al., 1999; Bertoni and Rando, 2002).
Statistical analysis.
Analysis of variance was performed for comparison of multiple means, and a two-sample Student’s t-test was used for comparison of two means. Statistical significance (P < 0.05) was assessed using Prism 4 (GraphPad Software, Inc.).
Online supplemental material.
Table S1 lists the chemical names of all 12 RTCs. Fig. S1 provides the chemical structures of 10 RTCs identified by HTS. Fig. S2 shows the cytotoxicity of RTC#13 and #14 in A-T LCLs. Fig. S3 shows the dose-dependent SMC1-Ser966 phosphorylation induced by RTC#13 in AT153LA cells. Fig. S4 provides the two-dimensional gel electrophoresis comparisons for before and after exposure to RTCs. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20081940/DC1.
Acknowledgments
We thank Dr. Joseph Loo for liquid chromatography–mass spectrometry confirmation of chemical identities.
This work was supported by National Institutes of Health grants 1R01NS052528 and 5U19AI067769, the A-T Medical Research Foundation, and the Muscular Dystrophy Association USE (C. Bertoni) . FC was performed in the University of California, Los Angeles (UCLA) Jonsson Comprehensive Cancer Center (JCCC) and Center for AIDS Research Flow Cytometry Core Facility, which is supported by National Institutes of Health awards CA-16042 and AI-28697, and by the JCCC, the UCLA AIDS Institute, and the UCLA David Geffen School of Medicine. Two-dimensional gel electrophoresis was performed in the UCLA Proteomics Core Facility.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- A-T
- ataxia-telangiectasia
- ATM
- A-T mutated
- CSA
- colony survival assay
- FC
- flow cytometry
- FI
- fluorescence intensity
- HRP
- horseradish peroxidase
- HTS
- high-throughput screening
- IR
- ionizing radiation
- IRIF
- irradiation-induced foci
- LCL
- lymphoblastoid cell line
- mRNA
- messenger RNA
- NMD
- nonsense-mediated mRNA decay
- PTC
- premature termination codon
- PTT
- protein transcription/translation
- RTC
- readthrough compound
- SMC
- structural maintenance of chromosome
References
- Auld D.S., Thorne N., Maguire W.F., Inglese J. 2009. Mechanism of PTC124 activity in cell-based luciferase assays of nonsense codon suppression.Proc. Natl. Acad. Sci. USA. 106:3585–3590 doi:10.1073/pnas.0813345106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton-Davis E.R., Cordier L., Shoturma D.I., Leland S.E., Sweeney H.L. 1999. Aminoglycoside antibiotics restore dystrophin function to skeletal muscles of mdx mice.J. Clin. Invest. 104:375–381 doi:10.1172/JCI7866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedwell D.M., Kaenjak A., Benos D.J., Bebok Z., Bubien J.K., Hong J., Tousson A., Clancy J.P., Sorscher E.J. 1997. Suppression of a CFTR premature stop mutation in a bronchial epithelial cell line.Nat. Med. 3:1280–1284 doi:10.1038/nm1197-1280 [DOI] [PubMed] [Google Scholar]
- Bertoni C., Rando T.A. 2002. Dystrophin gene repair in mdx muscle precursor cells in vitro and in vivo mediated by RNA-DNA chimeric oligonucleotides.Hum. Gene Ther. 13:707–718 doi:10.1089/104303402317322276 [DOI] [PubMed] [Google Scholar]
- Butch A.W., Chun H.H., Nahas S.A., Gatti R.A. 2004. Immunoassay to measure ataxia-telangiectasia mutated protein in cellular lysates.Clin. Chem. 50:2302–2308 doi:10.1373/clinchem.2004.039461 [DOI] [PubMed] [Google Scholar]
- Chun H.H., Gatti R.A. 2004. Ataxia-telangiectasia, an evolving phenotype.DNA Repair (Amst.). 3:1187–1196 doi:10.1016/j.dnarep.2004.04.010 [DOI] [PubMed] [Google Scholar]
- Chun H.H., Sun X., Nahas S.A., Teraoka S., Lai C.H., Concannon P., Gatti R.A. 2003. Improved diagnostic testing for ataxia-telangiectasia by immunoblotting of nuclear lysates for ATM protein expression.Mol. Genet. Metab. 80:437–443 doi:10.1016/j.ymgme.2003.09.008 [DOI] [PubMed] [Google Scholar]
- Chun H.H., Cary R.B., Lansigan F., Whitelegge J., Rawlings D.J., Gatti R.A. 2004. ATM protein purified from vaccinia virus expression system: DNA binding requirements for kinase activation.Biochem. Biophys. Res. Commun. 322:74–81 doi:10.1016/j.bbrc.2004.07.085 [DOI] [PubMed] [Google Scholar]
- Clancy J.P., Bebök Z., Ruiz F., King C., Jones J., Walker L., Greer H., Hong J., Wing L., Macaluso M., et al. 2001. Evidence that systemic gentamicin suppresses premature stop mutations in patients with cystic fibrosis.Am. J. Respir. Crit. Care Med. 163:1683–1692 [DOI] [PubMed] [Google Scholar]
- Du L., Pollard J.M., Gatti R.A. 2007. Correction of prototypic ATM splicing mutations and aberrant ATM function with antisense morpholino oligonucleotides.Proc. Natl. Acad. Sci. USA. 104:6007–6012 doi:10.1073/pnas.0608616104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Lai C.H., Concannon P., Gatti R.A. 2008. Rapid screen for truncating ATM mutations by PTT-ELISA.Mutat. Res. 640:139–144 [DOI] [PubMed] [Google Scholar]
- Du M., Jones J.R., Lanier J., Keeling K.M., Lindsey J.R., Tousson A., Bebök Z., Whitsett J.A., Dey C.R., Colledge W.H., et al. 2002. Aminoglycoside suppression of a premature stop mutation in a Cftr−/− mouse carrying a human CFTR-G542X transgene.J. Mol. Med. 80:595–604 doi:10.1007/s00109-002-0363-1 [DOI] [PubMed] [Google Scholar]
- Du M., Liu X., Welch E.M., Hirawat S., Peltz S.W., Bedwell D.M. 2008. PTC124 is an orally bioavailable compound that promotes suppression of the human CFTR-G542X nonsense allele in a CF mouse model.Proc. Natl. Acad. Sci. USA. 105:2064–2069 doi:10.1073/pnas.0711795105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunant P., Walter M.C., Karpati G., Lochmüller H. 2003. Gentamicin fails to increase dystrophin expression in dystrophin-deficient muscle.Muscle Nerve. 27:624–627 doi:10.1002/mus.10341 [DOI] [PubMed] [Google Scholar]
- Gilad S., Chessa L., Khosravi R., Russell P., Galanty Y., Piane M., Gatti R.A., Jorgensen T.J., Shiloh Y., Bar-Shira A. 1998. Genotype-phenotype relationships in ataxia-telangiectasia and variants.Am. J. Hum. Genet. 62:551–561 doi:10.1086/301755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gite S., Lim M., Carlson R., Olejnik J., Zehnbauer B., Rothschild K. 2003. A high-throughput nonisotopic protein truncation test.Nat. Biotechnol. 21:194–197 doi:10.1038/nbt779 [DOI] [PubMed] [Google Scholar]
- Guan M.X., Fischel-Ghodsian N., Attardi G. 2000. A biochemical basis for the inherited susceptibility to aminoglycoside ototoxicity.Hum. Mol. Genet. 9:1787–1793 doi:10.1093/hmg/9.12.1787 [DOI] [PubMed] [Google Scholar]
- Helip-Wooley A., Park M.A., Lemons R.M., Thoene J.G. 2002. Expression of CTNS alleles: subcellular localization and aminoglycoside correction in vitro.Mol. Genet. Metab. 75:128–133 doi:10.1006/mgme.2001.3272 [DOI] [PubMed] [Google Scholar]
- Hirawat S., Welch E.M., Elfring G.L., Northcutt V.J., Paushkin S., Hwang S., Leonard E.M., Almstead N.G., Ju W., Peltz S.W., Miller L.L. 2007. Safety, tolerability, and pharmacokinetics of PTC124, a nonaminoglycoside nonsense mutation suppressor, following single- and multiple-dose administration to healthy male and female adult volunteers.J. Clin. Pharmacol. 47:430–444 doi:10.1177/0091270006297140 [DOI] [PubMed] [Google Scholar]
- Holbrook J.A., Neu-Yilik G., Hentze M.W., Kulozik A.E. 2004. Nonsense-mediated decay approaches the clinic.Nat. Genet. 36:801–808 doi:10.1038/ng1403 [DOI] [PubMed] [Google Scholar]
- Honda M., Takagi M., Chessa L., Morio T., Mizuatni S. 2009. Rapid diagnosis of ataxia-telangiectasia by flow cytometric monitoring of DNA damage-dependent ATM phosphorylation.Leukemia. 23:409–414 doi:10.1038/leu.2008.195 [DOI] [PubMed] [Google Scholar]
- Howard M., Frizzell R.A., Bedwell D.M. 1996. Aminoglycoside antibiotics restore CFTR function by overcoming premature stop mutations.Nat. Med. 2:467–469 doi:10.1038/nm0496-467 [DOI] [PubMed] [Google Scholar]
- Howard M.T., Shirts B.H., Petros L.M., Flanigan K.M., Gesteland R.F., Atkins J.F. 2000. Sequence specificity of aminoglycoside-induced stop condon readthrough: potential implications for treatment of Duchenne muscular dystrophy.Ann. Neurol. 48:164–169 doi:10.1002/1531-8249(200008)48:2<164::AID-ANA5>3.0.CO;2-B [PubMed] [Google Scholar]
- Keeling K.M., Bedwell D.M. 2005. Pharmacological suppression of premature stop mutations that cause genetic diseases.Current Pharmacogenomics. 3:259–269 doi:10.2174/157016005774913149 [Google Scholar]
- Keeling K.M., Brooks D.A., Hopwood J.J., Li P., Thompson J.N., Bedwell D.M. 2001. Gentamicin-mediated suppression of Hurler syndrome stop mutations restores a low level of alpha-L-iduronidase activity and reduces lysosomal glycosaminoglycan accumulation.Hum. Mol. Genet. 10:291–299 doi:10.1093/hmg/10.3.291 [DOI] [PubMed] [Google Scholar]
- Kerem E., Hirawat S., Armoni S., Yaakov Y., Shoseyov D., Cohen M., Nissim-Rafinia M., Blau H., Rivlin J., Aviram M., et al. 2008. Effectiveness of PTC124 treatment of cystic fibrosis caused by nonsense mutations: a prospective phase II trial.Lancet. 372:719–727 doi:10.1016/S0140-6736(08)61168-X [DOI] [PubMed] [Google Scholar]
- Kimura S., Ito K., Miyagi T., Hiranuma T., Yoshioka K., Ozasa S., Matsukura M., Ikezawa M., Matsuo M., Takeshima Y., Miike T. 2005. A novel approach to identify Duchenne muscular dystrophy patients for aminoglycoside antibiotics therapy.Brain Dev. 27:400–405 doi:10.1016/j.braindev.2004.09.014 [DOI] [PubMed] [Google Scholar]
- Kitagawa R., Bakkenist C.J., McKinnon P.J., Kastan M.B. 2004. Phosphorylation of SMC1 is a critical downstream event in the ATM-NBS1-BRCA1 pathway.Genes Dev. 18:1423–1438 doi:10.1101/gad.1200304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C.H., Chun H.H., Nahas S.A., Mitui M., Gamo K.M., Du L., Gatti R.A. 2004. Correction of ATM gene function by aminoglycoside-induced read-through of premature termination codons.Proc. Natl. Acad. Sci. USA. 101:15676–15681 doi:10.1073/pnas.0405155101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linde L., Kerem B. 2008. Introducing sense into nonsense in treatments of human genetic diseases.Trends Genet. 24:552–563 doi:10.1016/j.tig.2008.08.010 [DOI] [PubMed] [Google Scholar]
- Linde L., Boelz S., Nissim-Rafinia M., Oren Y.S., Wilschanski M., Yaacov Y., Virgilis D., Neu-Yilik G., Kulozik A.E., Kerem E., Kerem B. 2007. Nonsense-mediated mRNA decay affects nonsense transcript levels and governs response of cystic fibrosis patients to gentamicin.J. Clin. Invest. 117:683–692 doi:10.1172/JCI28523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loufrani L., Dubroca C., You D., Li Z., Levy B., Paulin D., Henrion D. 2004. Absence of dystrophin in mice reduces NO-dependent vascular function and vascular density: total recovery after a treatment with the aminoglycoside gentamicin.Arterioscler. Thromb. Vasc. Biol. 24:671–676 doi:10.1161/01.ATV.0000118683.99628.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett P.S., Ambulos N.P., Jr., Mulbry W., Noguchi N., Rogers E.J. 1991. UGA can be decoded as tryptophan at low efficiency in Bacillus subtilis.J. Bacteriol. 173:1810–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell J.T., Dietz H.C. 2001. When the message goes awry: disease-producing mutations that influence mRNA content and performance.Cell. 107:411–414 doi:10.1016/S0092-8674(01)00583-9 [DOI] [PubMed] [Google Scholar]
- Mingeot-Leclercq M.P., Tulkens P.M. 1999. Aminoglycosides: nephrotoxicity.Antimicrob. Agents Chemother. 43:1003–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahas S.A., Butch A.W., Du L., Gatti R.A. 2009. Rapid flow cytometry-based structural maintenance of chromosomes 1 (SMC1) phosphorylation assay for identification of ataxia-telangiectasia homozygotes and heterozygotes.Clin. Chem. 55:463–472 doi:10.1373/clinchem.2008.107128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudelman I., Rebibo-Sabbah A., Shallom-Shezifi D., Hainrichson M., Stahl I., Ben-Yosef T., Baasov T. 2006. Redesign of aminoglycosides for treatment of human genetic diseases caused by premature stop mutations.Bioorg. Med. Chem. Lett. 16:6310–6315 doi:10.1016/j.bmcl.2006.09.013 [DOI] [PubMed] [Google Scholar]
- Perlman S., Becker-Catania S., Gatti R.A. 2003. Ataxia-telangiectasia: diagnosis and treatment.Semin. Pediatr. Neurol. 10:173–182 doi:10.1016/S1071-9091(03)00026-3 [DOI] [PubMed] [Google Scholar]
- Politano L., Nigro G., Nigro V., Piluso G., Papparella S., Paciello O., Comi L.I. 2003. Gentamicin administration in Duchenne patients with premature stop codon. Preliminary results.Acta Myol. 22:15–21 [PubMed] [Google Scholar]
- Rebibo-Sabbah A., Nudelman I., Ahmed Z.M., Baasov T., Ben-Yosef T. 2007. In vitro and ex vivo suppression by aminoglycosides of PCDH15 nonsense mutations underlying type 1 Usher syndrome.Hum. Genet. 122:373–381 doi:10.1007/s00439-007-0410-7 [DOI] [PubMed] [Google Scholar]
- Roest P.A., Roberts R.G., Sugino S., van Ommen G.J., den Dunnen J.T. 1993. Protein truncation test (PTT) for rapid detection of translation-terminating mutations.Hum. Mol. Genet. 2:1719–1721 doi:10.1093/hmg/2.10.1719 [DOI] [PubMed] [Google Scholar]
- Shiloh Y. 2006. The ATM-mediated DNA-damage response: taking shape.Trends Biochem. Sci. 31:402–410 doi:10.1016/j.tibs.2006.05.004 [DOI] [PubMed] [Google Scholar]
- Sicinski P., Geng Y., Ryder-Cook A.S., Barnard E.A., Darlison M.G., Barnard P.J. 1989. The molecular basis of muscular dystrophy in the mdx mouse: a point mutation.Science. 244:1578–1580 doi:10.1126/science.2662404 [DOI] [PubMed] [Google Scholar]
- Sossi V., Giuli A., Vitali T., Tiziano F., Mirabella M., Antonelli A., Neri G., Brahe C. 2001. Premature termination mutations in exon 3 of the SMN1 gene are associated with exon skipping and a relatively mild SMA phenotype.Eur. J. Hum. Genet. 9:113–120 doi:10.1038/sj.ejhg.5200599 [DOI] [PubMed] [Google Scholar]
- Sun X., Becker-Catania S.G., Chun H.H., Hwang M.J., Huo Y., Wang Z., Mitui M., Sanal O., Chessa L., Crandall B., Gatti R.A. 2002. Early diagnosis of ataxia-telangiectasia using radiosensitivity testing.J. Pediatr. 140:724–731 doi:10.1067/mpd.2002.123879 [DOI] [PubMed] [Google Scholar]
- Telatar M., Wang Z., Udar N., Liang T., Bernatowska-Matuszkiewicz E., Lavin M., Shiloh Y., Concannon P., Good R.A., Gatti R.A. 1996. Ataxia-telangiectasia: mutations in ATM cDNA detected by protein-truncation screening.Am. J. Hum. Genet. 59:40–44 [PMC free article] [PubMed] [Google Scholar]
- Wagner K.R., Hamed S., Hadley D.W., Gropman A.L., Burstein A.H., Escolar D.M., Hoffman E.P., Fischbeck K.H. 2001. Gentamicin treatment of Duchenne and Becker muscular dystrophy due to nonsense mutations.Ann. Neurol. 49:706–711 doi:10.1002/ana.1023 [PubMed] [Google Scholar]
- Weiner A.M., Weber K. 1973. A single UGA codon functions as a natural termination signal in the coliphage q beta coat protein cistron.J. Mol. Biol. 80:837–855 doi:10.1016/0022-2836(73)90213-1 [DOI] [PubMed] [Google Scholar]
- Welch E.M., Barton E.R., Zhuo J., Tomizawa Y., Friesen W.J., Trifillis P., Paushkin S., Patel M., Trotta C.R., Hwang S., et al. 2007. PTC124 targets genetic disorders caused by nonsense mutations.Nature. 447:87–91 doi:10.1038/nature05756 [DOI] [PubMed] [Google Scholar]
- Wilkinson M.F., Shyu A.B. 2002. RNA surveillance by nuclear scanning? Nat. Cell Biol. 4:E144–E147 doi:10.1038/ncb0602-e144 [DOI] [PubMed] [Google Scholar]
- Wilschanski M., Famini C., Blau H., Rivlin J., Augarten A., Avital A., Kerem B., Kerem E. 2000. A pilot study of the effect of gentamicin on nasal potential difference measurements in cystic fibrosis patients carrying stop mutations.Am. J. Respir. Crit. Care Med. 161:860–865 [DOI] [PubMed] [Google Scholar]
- Yazdi P.T., Wang Y., Zhao S., Patel N., Lee E.Y., Qin J. 2002. SMC1 is a downstream effector in the ATM/NBS1 branch of the human S-phase checkpoint.Genes Dev. 16:571–582 doi:10.1101/gad.970702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zingman L.V., Park S., Olson T.M., Alekseev A.E., Terzic A. 2007. Aminoglycoside-induced translational read-through in disease: overcoming nonsense mutations by pharmacogenetic therapy.Clin. Pharmacol. Ther. 81:99–103 doi:10.1038/sj.clpt.6100012 [DOI] [PubMed] [Google Scholar]