Abstract
Abnormal expression of CEACAM6 is observed at the apical surface of the ileal epithelium in Crohn's disease (CD) patients, and CD ileal lesions are colonized by pathogenic adherent-invasive Escherichia coli (AIEC). We investigated the ability of AIEC reference strain LF82 to colonize the intestinal mucosa and to induce inflammation in CEABAC10 transgenic mice expressing human CEACAMs. AIEC LF82 virulent bacteria, but not nonpathogenic E. coli K-12, were able to persist in the gut of CEABAC10 transgenic mice and to induce severe colitis with reduced survival rate, marked weight loss, increased rectal bleeding, presence of erosive lesions, mucosal inflammation, and increased proinflammatory cytokine expression. The colitis depended on type 1 pili expression by AIEC bacteria and on intestinal CEACAM expression because no sign of colitis was observed in transgenic mice infected with type 1 pili–negative LF82-ΔfimH isogenic mutant or in wild-type mice infected with AIEC LF82 bacteria. These findings strongly support the hypothesis that in CD patients having an abnormal intestinal expression of CEACAM6, AIEC bacteria via type 1 pili expression can colonize the intestinal mucosa and induce gut inflammation. Thus, targeting AIEC adhesion to gut mucosa represents a new strategy for clinicians to prevent and/or to treat ileal CD.
Inflammatory bowel disease (IBD) mainly consists of two disorders, ulcerative colitis and Crohn's disease (CD), with a combined prevalence of ∼150–200 cases per 100,000 in Western countries (Shanahan, 2002; Loftus, 2004). The abnormal inflammatory response observed in IBD requires interplay between host genetic factors and the intestinal microbiota (Podolsky, 2002; for review see Strober et al., 2007). The role of the microbiota in IBD development is highlighted with the following observations: in CD patients, postsurgical exposure of the terminal ileum to luminal contents is associated with increased inflammation, and diversion of the fecal stream is associated with improvement (Rutgeerts et al., 1991); some IBD patients improve upon antibiotic treatment (Sartor, 2004; Sands, 2007); the severity of colitis in multiple animal models is decreased by the administration of antibiotics; and no sign of colitis is observed when those animals are in germ-free conditions (for review see Sartor, 2008).
Two broad hypotheses have arisen regarding the role of the intestinal microbiota in the pathogenesis of IBD. Several lines of evidence support the notion that IBD results from an excessive immune response to gut commensal organisms (for review see Strober et al., 2007). However, the disease could result from a problem in the composition of the microflora leading to generalized or localized dysbiosis. Thus, a breakdown in the balance between putative species of “protective” versus “harmful” intestinal bacteria has been reported and may promote inflammation. A low proportion of Faecalibacterium prausnitzii on resected ileal Crohn mucosa is associated with endoscopic recurrence at 6 mo, and this bacteria has antiinflammatory properties (Tamboli et al., 2004; Sokol et al., 2008). In addition, host-mediated inflammation in response to a pathogen infection can disrupt the intestinal microbiota and shift the balance between the protective microbiota and the pathogen in favor of the pathogen, as seen with Citrobacter rodentium infection promoting the overgrowth of Enterobacteriaceae (Lupp et al., 2007) and with Salmonella Typhimurium infection (Stecher et al., 2007). In patients with CD and ulcerative colitis, high concentrations of bacteria forming a biofilm on the surface of the gut mucosa (Swidsinski et al., 2002) and increased numbers of mucosa-associated Escherichia coli are observed (Darfeuille-Michaud et al., 1998; Martin et al., 2004; Conte et al., 2006; Baumgart et al., 2007; Kotlowski et al., 2007; Sasaki et al., 2007). This overgrowth of E. coli can result from host-mediated inflammation or abnormal expression of molecules acting as receptors for bacterial adhesion. In CD patients with ileal involvement of the disease, we recently reported an abnormal ileal expression of carcinoembryonic antigen-related cell adhesion molecules (CEACAMs) 5 and 6 (Barnich et al., 2007) and showed that only CEACAM6 acts as a receptor for pathogenic E. coli. These bacteria, called adherent-invasive E. coli (AIEC), colonize the ileal mucosa of CD patients (Darfeuille-Michaud et al., 2004). They are able to adhere to and invade intestinal epithelial cells and to survive and highly replicate within macrophages, leading to the secretion of high amounts of TNF (Boudeau et al., 1999; Glasser et al., 2001). Interestingly, in vitro studies demonstrated that CEACAM6 expression is increased in cultured intestinal epithelial cells not only after IFN-γ or TNF stimulation (Fahlgren et al., 2003) but also after infection with AIEC bacteria, indicating that AIEC could promote their own colonization in CD patients (Barnich et al., 2007). In the present paper, using transgenic CEABAC10 mice harboring a bacterial artificial chromosome that contains part of the human CEA family gene cluster, including complete human CEACAM3, CEACAM5 (CEA), CEACAM6, and CEACAM7 genes (Chan and Stanners, 2004), we investigated whether LF82 bacteria isolated from a CD patient can colonize the intestinal mucosa as a result of CEACAM expression and whether AIEC LF82 colonization could lead to the development of gut inflammation.
RESULTS
Intestinal expression of human CEACAMs allowed AIEC LF82 gastrointestinal (GI) colonization and persistence
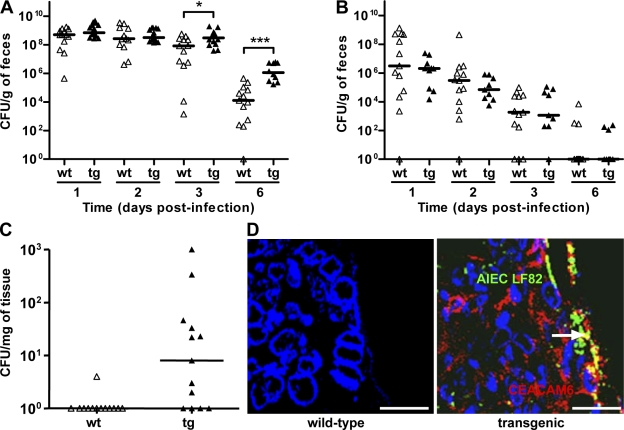
WT FVB/N and CEABAC10 mice were orally challenged with 109 bacteria, and the level of bacteria in the stools was followed to evaluate bacterial colonization rate. The analysis of the presence of AIEC bacteria in the stools revealed that there were no differences in bacterial counts between WT mice and transgenic mice until day 2 after infection. At day 3, significantly (P = 0.013) more AIEC LF82 bacteria were found in stools of mice expressing human CEACAMs (3.0 × 108 CFU/g of feces) compared with WT mice (8.5 × 107 CFU/g of feces; Fig. 1 A). At day 6, the differences between WT and transgenic mice were even higher and statistically more significant (P = 0.002) because the median number of AIEC LF82 bacteria found in stools of transgenic mice was 1.1 × 106 CFU/g of feces compared with 1.3 × 104 CFU/g of feces for WT mice (Fig. 1 A). It is of note that at day 6 after infection, 10/10 of the CEABAC10 mice still presented more than 105 CFU of AIEC LF82 per gram of feces compared with only 3/13 for WT mice. When experiments were performed with the nonpathogenic E. coli K-12, no differences in the numbers of bacteria in the stools were encountered between WT and transgenic mice (Fig. 1 B). Bacteria counts rapidly decreased in the feces of WT or transgenic mice because only 3.1 × 106 CFU/g of feces and 2.1 × 106 CFU/g of feces, respectively, were found at day 1 after infection. At day 3 after infection, the number of E. coli K-12 bacteria present in feces of infected mice was <105 CFU/g of feces.
Figure 1.
AIEC LF82 bacterial colonization and persistence in mouse intestine according to CEACAM expression. (A and B) Quantification of AIEC LF82 (A) or E. coli K-12 MG1655 (B) in the feces of WT (wt, white triangles) or CEABAC10 (tg, black triangles) mice receiving 0.25% DSS in drinking water after oral infection with 109 bacteria at day 0. (C) Quantification of colonic mucosal-associated AIEC LF82 bacteria at day 7 after oral infection. The quantification for each mouse (symbols) and the median (bars) is expressed as CFU/g of feces or CFU/mg of organ tissue. (D) Confocal microscopy analysis of colonic section at day 7 after infection from WT or transgenic mice infected with AIEC LF82. CEACAM6 expression was detected using anti-CEACAM6 monoclonal antibody and a Cy3-conjugated anti–mouse IgG. AIEC LF82 bacteria were detected using anti-O83 rabbit antibody and a FITC-conjugated anti–rabbit IgG. Arrow shows colocalization (yellow staining) between CEACAM6 and bacteria. WT FVB/N mice: E. coli K-12 MG1655 infected (n = 13), AIEC LF82 infected (n = 9); CEABAC10 transgenic FVB/N mice: E. coli K-12-MG1655 (n = 13), AIEC LF82-infected (n = 15). Results shown here are representative of three separate colonization experiments. *, P < 0.05; ***, P < 0.001. Bars, 50 µm.
To confirm AIEC LF82 colonization, we determined the number of colonic mucosa-associated bacteria to investigate whether the presence of AIEC LF82 bacteria in the feces correlated with mucosal colonization of the GI tract. At day 7 after infection, AIEC bacteria were not detected in 12 out of 13 colonic samples of WT mice (Fig. 1 C). In contrast, we found that a majority of transgenic mice (8/13) harbored colon-associated AIEC LF82 bacteria, and confocal microscopy examination of colonic sections stained for CEACAM6 showed that AIEC bacteria colocalized with CEACAM6 expression (Fig. 1 D).
AIEC LF82 GI colonization induced severe colitis in mice expressing human CEACAMs
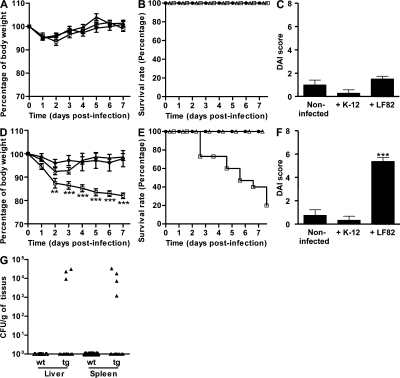
To analyze the consequences of AIEC LF82 colonization, we recorded the body weight loss, survival rate, and disease activity index (DAI) score of WT mice and CEABAC10 mice orally challenged with 109 bacteria. None of the WT mice challenged with AIEC LF82 or E. coli K-12 bacteria developed bloody diarrhea or appeared ill. There was a similar evolution in body weight in both infected and noninfected mice and no mortalities in either group (Fig. 2, A and B). None of the E. coli K-12– and LF82-infected WT mice presented clinical symptoms of colitis, and no significant difference in DAI scores was observed between noninfected, E. coli K-12–infected, and LF82-infected WT mice (Fig. 2 C). In contrast, clinical symptoms of colitis were observed in CEABAC10 mice orally challenged with AIEC LF82. At day 2, a significant (P < 0.01) decrease in body weight of mice infected with AIEC LF82 (87.4 ± 2.0%) was observed compared with noninfected transgenic mice (96.0 ± 1.6%) or mice infected with E. coli K-12 (92.5 ± 1.5%; Fig. 2 D), a difference which persisted until the end of the experiment. The severity of the disease induced by AIEC infection in transgenic mice was confirmed by a high mortality rate. Infection with AIEC LF82 bacteria induced a strongly reduced survival rate from day 2 (Fig. 2 E) and, at day 7 after infection, only 20% of mice survived, whereas no mortality was observed in noninfected transgenic mice or in those infected with E. coli K-12 bacteria. Increased clinical symptoms of colitis were observed in transgenic mice orally challenged with AIEC LF82 compared with E. coli K-12 bacteria. The DAI score of transgenic mice infected with AIEC LF82 bacteria was significantly higher at day 6 (5.4 ± 0.3) compared with noninfected mice (0.8 ± 0.5; P < 0.001) or those infected with E. coli K-12 (0.3 ± 0.3; P < 0.001; Fig. 2 F). AIEC colonization in WT mice over a 14-d period did not decrease body weight or result in mortality. Similar results were observed in CEABAC10 mice administered with K-12 bacteria (unpublished data). In addition, dissemination of AIEC LF82 bacteria was observed in the liver and the spleen of three and four transgenic mice, respectively (Fig. 2 G), but in no infected WT mice.
Figure 2.
AIEC LF82 bacteria induced clinical symptoms of colitis in CEABAC10 transgenic mice. (A–F) AIEC LF82 infections were performed in WT (A–C) and in CEABAC10 (D–F) mice. Shown is evolution of body weight (A and D), survival rate (B and E), and DAI score ascertained at day 7 after infection (C and F) of mice orally challenged at day 0 with CMC alone (black circles), with CMC containing 109 E. coli K-12 MG1655 bacteria (white triangles), or with CMC containing 109 AIEC LF82 bacteria (white squares). (G) Quantification of AIEC LF82 at day 7 after infection in the liver and spleen. WT FVB/N mice: CMC alone (n = 11), E. coli K-12 MG1655 infected (n = 7), AIEC LF82 infected (n = 14); CEABAC10 transgenic FVB/N mice: CMC alone (n = 9), E. coli K-12 MG1655 infected (n = 11), AIEC LF82 infected (n = 15). Shown here are representative body weight evolution, survival rate, and DAI of three separate experiments. Horizontal bars represent medians. Error bars represent SEM. **, P < 0.01; ***, P < 0.001 (compared with noninfected mice).
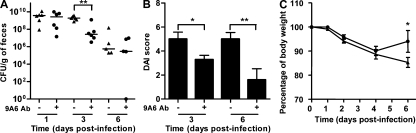
To confirm the involvement of human CEACAM6 in AIEC LF82 colonization and induction of inflammation in transgenic mice, AIEC-infected transgenic mice received intraperitoneal injections of anti-CEACAM6 monoclonal antibody 9A6 1 d before infection and at days 0, 1, and 3 after infection. The resulting blockade of CEACAM6 receptor significantly decreased AIEC LF82 colonization until the mice received anti-CEACAM6 monoclonal antibodies (Fig. 3 A). Significantly lower DAI scores and significant gains in body weight were also observed at day 6 after infection (Fig. 3, B and C). Overall, these results therefore indicate that among the various human CEACAMs expressed by transgenic mice, CEACAM6 is required to allow CD-associated AIEC bacteria to colonize the intestine and to induce clinical symptoms of colitis.
Figure 3.
Intraperitoneal administration of anti-CEACAM6 monoclonal antibody 24 h before infection and at days 0, 1, and 3 after infection prevents colonization and clinical symptoms of colitis in AIEC LF82-infected CEABAC10 transgenic mice. (A–C) Evolution of the number of AIEC LF82 in the feces (A), of DAI score ascertained at days 3 and 6 after infection (B), and of body weight (C) of transgenic mice orally challenged at day 0 with 109 AIEC LF82 bacteria. Horizontal bars represent medians. CEABAC10 transgenic FVB/N mice: AIEC LF82 infected (n = 5; black triangles); AIEC LF82 infected after intraperitoneal injection of anti-CEACAM6 antibody (n = 5; black circles). Two experiments were performed independently. Shown here are colonization, DAI, and body weight evolution of one representative experiment. Error bars represent SEM. *, P < 0.05; **, P < 0.01.
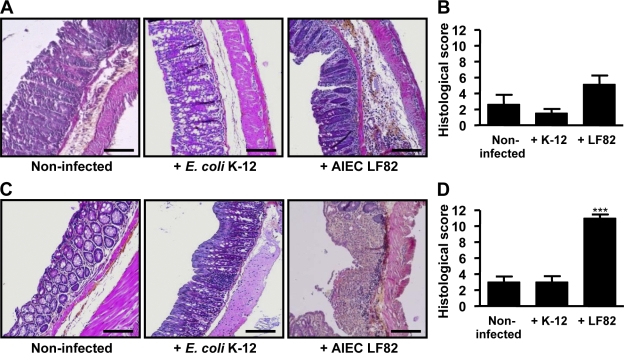
Histological analyses of colonic tissues were performed at day 7 after infection to evaluate the degree of inflammation. In WT mice infected with AIEC LF82 bacteria, a slight polynuclear infiltration was observed but colonic mucosa was normal, with the crypts being straight, well defined, and sitting on the muscularis mucosa (Fig. 4 A). In addition, the histological score was not significantly different between noninfected, E. coli K-12–infected, and AIEC LF82-infected mice (Fig. 4 B). In contrast, histological colonic sections of AIEC LF82-infected transgenic mice showed hemorrhagic walls with multiple ulcerations, mucosal edema, neutrophil infiltrations with transmural involvement, and presence of large erosion areas (Fig. 4 C). Also, the histological score was greatly increased in transgenic mice infected with LF82 (11.0 ± 0.4) compared with noninfected mice (3.0 ± 0.7) or mice infected with E. coli K-12 (3.0 ± 0.8), and the differences were highly statistically significant (P < 0.001; Fig. 4 D). Epithelial damage occurred in all areas of the colonic mucosa and the infiltrated immune cells were mostly polynuclear cells. In addition, some cryptic abscesses were observed in transgenic mice infected with AIEC LF82 but not in those infected with E. coli MG1655. These findings demonstrate that mice expressing human CEACAMs are susceptible to AIEC LF82 oral infection and develop severe clinical symptoms of colitis.
Figure 4.
Intestinal colonization by AIEC LF82 bacteria causes severe histopathological damage in colonic mucosa. (A–D) Hematoxylin/eosin/safran staining of colonic tissue sections obtained at day 7 after infection from WT mice (A) or CEABAC10 transgenic mice (C), noninfected or infected with E. coli K-12 MG1655 or with AIEC LF82 bacteria (magnification, 100×). Histopathological scoring for several parameters of colonic inflammation was performed for WT mice (B) or CEABAC10 transgenic mice (D), noninfected or infected with 109 E. coli K-12 MG1655 or with AIEC LF82 bacteria. WT FVB/N mice: CMC alone (n = 11), E. coli K-12 MG1655 infected (n = 12), AIEC LF82 infected (n = 14); CEABAC10 transgenic FVB/N mice: CMC alone (n = 9), E. coli K-12 MG1655 infected (n = 11), AIEC LF82 infected (n = 15). Shown here are representative histological scores of two separate experiments. Error bars represent SEM. ***, P < 0.001 compared with noninfected mice. Bars, 100 µm.
AIEC LF82 colonization and induced colitis is dependent on type 1 pili expression
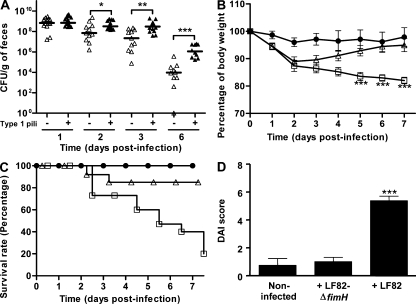
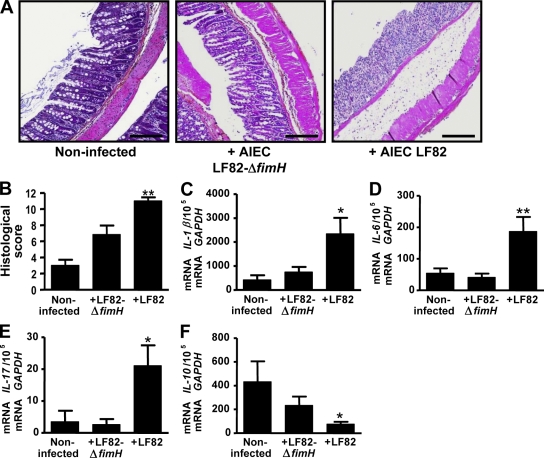
To investigate the role of type 1 pili in AIEC bacterial colonization, CEABAC10 transgenic mice were challenged with AIEC LF82-ΔfimH isogenic mutant, which did not produce type 1 pili but was still able to synthesize functional flagella (unpublished data). Analysis of the presence of AIEC LF82 bacteria or the LF82-ΔfimH mutant in the stools revealed that at day 2 after infection there was a statistically significant 4.5-fold decrease (P = 0.033) in LF82-ΔfimH mutant (7 × 107 CFU/g of feces) compared with WT AIEC LF82 bacteria (3.2 × 108 CFU/g of feces; Fig. 5 A). A 13.6-fold decrease in the LF82-ΔfimH mutant in the stools (2.2 × 107 CFU/g of feces) compared with WT AIEC LF82 bacteria (3.0 × 108 CFU/g of feces) was observed at day 3 after infection (P = 0.001). At day 6 after infection, a 2-log difference was observed between WT LF82 bacteria and type 1 pili–negative LF82-ΔfimH mutant. In agreement with these results, transgenic mice receiving the LF82-ΔfimH mutant, unlike those infected with WT AIEC LF82 bacteria, had a body weight similar to that observed for noninfected mice from day 5 after infection (Fig. 5 B). In addition, an 85% survival rate was observed for AIEC LF82-ΔfimH mutant infected mice, compared with only 20% for AIEC LF82 infected mice at day 7 after infection (Fig. 5 C). Also, mice infected with the AIEC LF82-ΔfimH isogenic mutant did not present any significant difference in the DAI score compared with noninfected mice (Fig. 5 D). Colonic examination revealed that histological damages observed in AIEC LF82-infected transgenic mice were no longer observed in mice infected with the LF82-ΔfimH mutant (Fig. 6 A). Indeed, these mice presented normal colonic histological structures with little infiltration of inflammatory cells; the colonic histological score was significantly (P = 0.003) decreased for mice infected with the LF82-ΔfimH isogenic mutant (6.8 ± 1.1) compared with mice infected with AIEC LF82 bacteria (11.0 ± 0.4; Fig. 6 B). Finally, significantly increased levels of IL-1β, IL-6, and IL-17 mRNAs (5.5-fold, 3.4-fold, and 6.1-fold, respectively) and significantly decreased levels of IL-10 mRNAs (5.6-fold) were observed in colonic specimens of CEABAC10 mice infected with AIEC LF82 bacteria compared with those of noninfected mice (Fig. 6, C–F). In contrast, such variations in cytokine levels were not observed after infection with the LF82-ΔfimH isogenic mutant. Altogether, these results reinforce the role of type 1 pili in AIEC LF82 colonization and reveal that only fully virulent AIEC LF82 bacteria can induce colitis in CEABAC10 mice by increasing the expression of proinflammatory cytokines.
Figure 5.
AIEC LF82 colonization via type 1 pili expression induce clinical symptoms of colitis in CEABAC10 transgenic mice. (A) Quantification of AIEC LF82 bacteria (black triangles) or type 1 pili–negative AIEC LF82-ΔfimH mutant (white triangles) in the feces of CEABAC10 mice after oral infection with 109 bacteria at day 0. Each symbol represents one mouse and the horizontal bars represent the medians. (B and C) Evolution of body weight (B) and survival rate (C) of CEABAC10 transgenic mice orally challenged at day 0 with CMC alone (black circles), CMC containing 109 AIEC LF82 bacteria (white squares), or CMC containing 109 AIEC LF82-ΔfimH bacteria (white triangles). (D) DAI score ascertained for CEABAC10 transgenic mice that were noninfected, infected with AIEC LF82-ΔfimH bacteria, or infected with AIEC LF82 bacteria at day 7 after infection. CEABAC10 transgenic FVB/N mice: CMC alone (n = 9), AIEC LF82-ΔfimH infected (n = 14), AIEC LF82 infected (n = 15). Shown here are representative body weight evolution, survival rate, and DAI of three separate experiments. Error bars represent SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (compared with mice infected with AIEC LF82-ΔfimH).
Figure 6.
Type 1 pili–dependent AIEC LF82 colonization is required to induce severe histopathological damage and inflammation of colonic mucosa. (A) Hematoxylin/eosin/safran staining of colonic tissue sections obtained at day 7 after infection from CEABAC10 transgenic mice noninfected, infected with AIEC LF82-ΔfimH isogenic mutant, or infected with AIEC LF82 bacteria (magnification, 100×). (B) Histopathological scoring for several parameters of colonic inflammation. (C–F) Total RNAs from mouse colon were isolated and IL-1β (C), IL-6 (D), IL-17 (E), and IL-10 (F) mRNA levels were measured by RT-PCR. CEABAC10 transgenic FVB/N mice: CMC alone (n = 9), AIEC LF82-ΔfimH infected (n = 14), AIEC LF82 infected (n = 15). Shown here are representative histological scores of two separate experiments. Error bars represent SEM. *, P < 0.05; **, P < 0.01 (compared with mice infected with AIEC LF82-ΔfimH). Bars, 100 µm.
DISCUSSION
Two broad hypotheses have emerged regarding the pathogenesis of IBD. One speculates that primary dysregulation of the mucosal immune system leads to excessive immunological responses to normal microflora and the other suggests that changes in the composition of gut microflora elicit pathological responses from the normal mucosal immune system (for review see Strober et al., 2007). In the ileal involvement of CD, changes in mucosa-associated microbiota have been observed, in particular a decrease in F. prausnitzii, bacteria with antiinflammatory properties (Sokol et al., 2008), and an increase in pathogenic E. coli (Neut et al., 2002; Swidsinski et al., 2002; Darfeuille-Michaud et al., 2004; Martin et al., 2004; Mylonaki et al., 2005; Conte et al., 2006; Kotlowski et al., 2007; Sasaki et al., 2007; Eaves-Pyles et al., 2008). The latter are able to adhere to and invade intestinal epithelial cells and to replicate within macrophages, eliciting a strong proinflammatory response through TNF secretion (Darfeuille-Michaud et al., 1998; Boudeau et al., 1999; Glasser et al., 2001). A possible explanation for the increased number of AIEC bacteria associated with ileal mucosa is the increased expression of CEACAM6 on the brush border of ileal enterocytes in CD patients because our previous in vitro studies suggested that CEACAM6 can act as a receptor for AIEC binding to the intestinal mucosa (Barnich et al., 2007). In this paper, we analyzed through in vivo experiments whether CEACAM expression can lead to abnormal colonization by AIEC bacteria and to the development of gut inflammation.
To mimic abnormal expression of CEACAM6 observed in CD patients, we used the transgenic CEABAC10 mouse model expressing human CEACAMs (Chan and Stanners, 2004). Oral challenge of these mice with the AIEC reference strain LF82 induced body weight loss, diarrhea, rectal bleeding, and a greatly reduced survival rate of only 20% at day 7 after infection. Infected transgenic CEABAC10 mice, but not WT mice, presented severe gut inflammation with increased proinflammatory cytokines IL-1β, IL-6, and IL-17 mRNA levels, decreased antiinflammatory cytokine IL-10 mRNA levels, and histopathological damage of gut mucosa. As the lesions were restricted to the colonic mucosa, we therefore further analyzed the discrepancy in the site of lesions between mice and CD patients because AIEC bacteria are associated with ileitis (Darfeuille-Michaud et al., 2004). Western blot analysis of CEACAMs expression in CEABAC10 transgenic mice indicated that CEACAM5 was highly expressed in both small intestine and colon, but that expression of CEACAM6 was restricted to the colonic mucosa (unpublished data). This confirmed the previously reported lack of ceacam6 mRNA in the ileal mucosa of CEABAC10 transgenic mice (Chan and Stanners, 2004). Thus, in this mouse model challenged with AIEC bacteria, the absence of ileal inflammation can be explained by the lack of ileal CEACAM6 expression. These results can be correlated with our previous study showing that ileal enterocytes from CD patients overexpressed both CEACAM5 and CEACAM6 and that the AIEC adhesion to the brush border of enterocytes was inhibited by CEACAM6 antibodies but not by CEACAM5 antibodies. This could be a result of the fact that mannose-rich sugars are more prevalent on CEACAM6 and that the adhesion of AIEC bacteria is mannose dependent (Barnich et al., 2007). Among the human CEACAM molecules expressed in the GI tract of CEABAC10 mice or CD patients, we show here that in transgenic mice expressing human CEACAMs, CEACAM6 plays a key role. Indeed, intraperitoneal administration of anti-CEACAM6 monoclonal antibodies, by blocking the CEACAM6 receptor, resulted in significant decreases in AIEC LF82 colonization and DAI scores.
As a general rule, colonization of the gut mucosa by bacteria can elicit gut inflammation, but this is not observed for Campylobacter jejuni, which failed to induce an inflammatory response (Rinella et al., 2006; Mansfield et al., 2007). In the present study, we observed for AIEC bacteria a strict correlation between colonization and development of inflammation. Indeed, in WT mice orally challenged with AIEC LF82, we observed neither colonization nor any sign of gut inflammation. In contrast, when CEABAC10 mice were challenged with AIEC bacteria, both bacterial colonization of the colonic mucosa and severe inflammation were observed. However, transgenic mice given nonpathogenic E. coli orally had no E. coli colonization or gut inflammation. A similar absence of colonization and inflammation was also observed when the nonpiliated mutant of AIEC LF82, which is unable to bind CEACAM6, was orally administered to transgenic mice. Overall, these results indicate that for AIEC involvement in CD the abnormal expression of CEACAM6 plays a crucial role by allowing the bacteria to colonize the gut mucosa and, subsequently, to trigger inflammation. It has been recently reported that the intrusion of enteropathogens into the gut ecosystem can result in the triggering of the host's inflammatory response, which disrupts colonization resistance, as reported for C. rodentium and Salmonella enterica serovar Typhimurium (Lupp et al., 2007; Stecher et al., 2007). Inflamed gut might offer altered conditions, such as a change in the adhesion sites, that can be exploited by the pathogen but not by the commensals. In the AIEC model, it would seem that it is not only inflammation that promotes intestinal colonization because we previously observed, using the DSS-injured colon model in WT BALB/c/J mice, that infection with AIEC LF82 bacteria significantly worsened colitis but this inflammation did not allow the AIEC bacteria to colonize the gut (Carvalho et al., 2008).
The inflammation induced by AIEC infection in CEABAC10 transgenic mice was not observed when mice were infected with the AIEC LF82 type 1 pili–negative mutant, indicating that the adhesion of AIEC bacteria to the gut mucosa via type 1 pili is essential for the development of inflammation. This result is of great interest because FimH could serve as a potent stimulator of the innate immune response via interaction with TLR4 independently of LPS (Mossman et al., 2008). Also, previous molecular dissections of virulence factor expression in the AIEC LF82 strain suggested that LF82 bacteria are highly flagellated and, therefore, highly piliated under GI tract conditions as a result of a coregulation between expression of flagella and type 1 pili (Barnich et al., 2003; Claret et al., 2007; Rolhion et al., 2007). The results reported in the present study showing that gut inflammation is linked to type 1 pili expression by pathogenic E. coli correlates with what was observed with uropathogenic E. coli and urinary tract infection (Cegelski et al., 2008).
We know from various models that the binding of adhesins expressed on the bacterial cell surface to defined glycosylated receptors on the host tissue surface is considered to be an initial and critical step in pathogenesis. As AIEC bacteria have the ability to colonize the intestinal mucosa, this opens a new avenue for therapy such as blocking the interaction between type 1 pili and CEACAMs. AIEC adhesion involving type 1 pili is mediated by recognition of mannose residues by the FimH adhesion site located at the tip of the pilus (Ofek et al., 2000). Recent crystal structure determination of FimH complexed with oligomannose-3 showed the feasibility of using natural and engineered mannose antagonists to block bacterial invasion (Wellens et al., 2008). Several strategies can be used to block AIEC adhesion and therefore colonization, such as use of probiotics and/or vaccination. Results of probiotic trials in CD are mixed (Sartor, 2004). E. coli Nissle 1917 was more effective than a placebo in preventing relapse of CD after induction of remission by standard medical therapy (Malchow, 1997), but no benefit of Lactobacillus GG administered for 1 yr could be demonstrated in preventing postoperative relapse of symptoms or endoscopic lesions in the neoterminal ileum (Prantera et al., 2002). However, the findings of the present study suggest that probiotics, such as yeasts or cell wall mannoprotein from yeasts, which are very rich in free mannose residues, would be a promising strategy to inhibit AIEC colonization in patients expressing CEACAM6 on the ileal mucosa and harboring AIEC bacteria and, therefore, to prevent recurrent intestinal inflammation and possibly to treat active CD. In addition, E. coli Nissle 1917, which has a strong and significant inhibitory effect on adherent-invasive E. coli adhesion and invasion in co-infection and preinfection experiments, could thus be effective for preventive or curative probiotic therapy in patients with CD (Boudeau et al., 2003). Adhesin-based vaccines may also be effective in the prevention of CD, as previously reported for the prevention of recurrent and acute infections of the urogenital mucosa (Langermann et al., 1997). It was reported for uropathogenic E. coli that antibodies specifically blocking the binding of FimH to its natural receptor prevent bacterial colonization and the subsequent inflammation of the urinary tract (Thankavel et al., 1997; Langermann et al., 2000). A vaccine containing a recombinant truncated form of FimH adhesin strongly reduced the in vivo colonization of the bladder by uropathogenic E. coli in a mouse cystitis model (Poggio et al., 2006), suggesting that a similar approach could be used to block gut AIEC colonization in CD patients expressing CEACAM6. Finally, another strategy could be to interrupt pilus assembly and thereby block pilus-mediated adhesion using pilicides, which are pilus inhibitors that target chaperone function by inhibiting pilus biogenesis (Svensson et al., 2001; Larsson et al., 2005). In conclusion, the results generated by the in vivo studies reported in this paper could lead to the development of specific therapies to treat patients with ileal involvement of CD by targeting AIEC bacterial adhesion to the gut mucosa.
MATERIALS AND METHODS
Bacterial strains and culture.
Ampicillin-erythromycin–resistant E. coli strain LF82, isolated from a chronic ileal lesion of a CD patient, was used as the AIEC reference strain (Darfeuille-Michaud et al., 1998). A nonpiliated isogenic mutant AIEC LF82 deleted for the fimH gene and the E. coli K-12 MG1655 rifampicin-resistant strain (laboratory stock) were also used in this study. Overnight bacterial cultures in Luria-Bertani (LB) broth without shaking at 37°C were harvested by centrifugation at 4,000 g for 15 min. The bacterial pellet was resuspended in carboxymethyl cellulose (CMC) at 0.5% (wt/vol) in distilled water.
Mice.
All mice were housed in specific pathogen-free conditions in the animal care facility at the University of Auvergne (Clermont-Ferrand, France). FVB/N WT mice were purchased from Charles River Laboratories and CEABAC10 transgenic mice (heterozygote; Chan and Stanners, 2004) were maintained in our animal facilities. WT and transgenic CEABAC10 mice were coupled to obtain 50% WT mice and 50% CEABAC10 mice. Mice from the same generation were used for experimentation. Animal protocols were approved by the Committee for Research and Ethical Issues of the International Association for the Study of Pain.
Infection of mice.
12-wk-old FVB/N WT or CEABAC10 transgenic male mice (body weight, ∼26–28 g) were pretreated by oral administration of the broad-spectrum antibiotic streptomycin (20 mg intragastric per mouse) to disrupt normal resident bacterial flora in the intestinal tract (Wadolkowski et al., 1988; Stecher et al., 2007) and were orally challenged with 109 bacteria 24 h later. Because we observed a very wide distribution range of the numbers of bacteria in the feces, as shown in Fig. S1 A, we adapted the protocol. Animals received the very low dose of 0.25% (wt/vol) of dextran sulfate sodium (DSS; molecular mass = 36,000-50,000 daltons; MP Biomedicals) in drinking water starting 3 d before infection to increase the accessibility of bacteria to the surface of the epithelial layer. The administration of 0.25% DSS did not affect the body weight of transgenic mice and did not induce clinical symptoms of colitis as assessed by the DAI score (Fig. S1, B and C). When mice attained 80% of their initial weight or 7 d after oral bacterial infection, they were anesthetized with isoflurane and then euthanized by cervical dislocation. Colon specimens were collected to quantify mucosal-associated bacteria, to analyze histological damages, and to perform quantitative RT-PCR. Spleen and liver were collected to quantify translocated bacteria. To perform anti-CEACAM6 antibody treatment, 500 µg of monoclonal anti-CEACAM6 9A6 (GENOVAC) were injected intraperitoneally in 200 µl of PBS 24 h before infection, and at days 0, 1, and 3 after infection.
Colonization evaluation.
1, 2, 3, and 6 d after bacterial infection, fresh fecal pellets (100–200 mg) were collected from individual mice and resuspended in PBS. After serial dilution, bacteria were enumerated by plating on LB agar medium containing 50 mg/µl ampicillin and 20 mg/µl erythromycin to isolate AIEC LF82 bacteria and isogenic mutants, 300 mg/µl or containing rifampicin to isolate K-12 MG1655 bacteria, and incubated at 37°C overnight. Colonization was confirmed by numbering AIEC-associated bacteria to the colonic mucosa at day 7 after infection. A 2-cm segment of colon, beginning at 0.5 cm from the cecal junction, was removed and opened longitudinally. The segment was placed in sterile PBS solution. The sample was homogenized and the resulting suspension was plated in duplicate onto LB agar plates containing appropriated antibiotics and incubated overnight at 37°C.
Clinical assessment of colitis and histological evaluation of colonic damage.
Colonic damage was ascertained by DAI as defined in Table I. Rectal bleeding was assessed by Hemoccult II test (SKD SARL). The scores range from 0 (healthy) to 12 (greatest activity of colitis). After mouse sacrifice, the entire colon was excised and rolls of the proximal colon were fixed in buffered 4% formalin, paraffin embedded, cut into 3-µm slices, and stained with hematoxylin/eosin/safranin. The histological severity of colitis was graded in a blinded fashion by a GI pathologist. The tissue samples were assessed for the extent and depth of inflammation and the extent of crypt damage as presented in Table II. The histology score corresponds to the sum of each items.
Table I.
DAI assessment
| Symptom/score | Characteristics |
| Body weight loss | |
| 0 | No loss |
| 1 | 1–5% loss of body weight |
| 2 | 5–10% loss of body weight |
| 3 | 10–20% loss of body weight |
| 4 | >20% loss of body weight |
| Stool consistency | |
| 0 | Normal feces |
| 1 | Loose stool |
| 2 | Watery diarrhea |
| 3 | Slimy diarrhea, little blood |
| 4 | Severe watery diarrhea with blood |
| Blood in stool | |
| 0 | No blood |
| 2 | Presence of blood assessed by Hemoccult II test |
| 4 | Visible bleeding |
Table II.
Histological grading of intestinal inflammation
| Symptom | Characteristics |
| Infiltration of inflammatory cells | |
| 0 | Rare inflammatory cells in the lamina propria |
| 1 | Increased numbers of inflammatory cells, including neutrophils in the lamina propria |
| 2 | Confluence of inflammatory cells extending into the submucosa |
| 3 | Transmural extension of the inflammatory cell infiltrate |
| Infiltration of lamina propria by mononuclear cells | |
| 0 | Normal |
| 1 | Moderate |
| 2 | Great |
| Infiltration of lamina propria by polynuclear cells | |
| 0 | Normal |
| 1 | Moderate |
| 2 | Great |
| Infiltration of epithelium by polynuclear cells | |
| 0 | No infiltration |
| 1 | Surface |
| 2 | Inside the crypt |
| 3 | Cryptic abscess |
| Severity of epithelial damage | |
| 0 | Absence of mucosal damage |
| 1 | Lymphoepithelial lesions |
| 2 | Mucosal erosion/ulceration |
| 3 | Extensive mucosal damage and extension through deeper structures of the bowel wall |
| Surface of epithelial damage | |
| 0 | Normal |
| 1 | Focal |
| 2 | Wide |
Confocal microscopy.
Colonic sections taken at day 7 after infection of WT or transgenic mice infected with AIEC LF82 were stained using mouse anti–human CEACAM6 monoclonal antibody clone 9A6 (GENOVAC) and Cy3-conjugated anti–mouse IgG (Vector Laboratories) as secondary antibody. AIEC LF82 bacteria were detected with rabbit anti-O83 antibody and FITC-conjugated anti–rabbit IgG (Vector Laboratories). Tissues were observed with a confocal microscope (LSM 510 Meta; Carl Zeiss, Inc.).
mRNA quantification.
Total RNA was isolated from colonic tissues using TRIzol (Invitrogen) according to the manufacturer's instructions. After treatment at 37°C for 30 min with 20–50 U RNase-free DNase I (Roche), complementary DNA were obtained using a Reverse transcription (Fermentas) and were quantified in realplex2 (Eppendorf) using SYBR green Taq ReadyMix (Sigma-Aldrich) with specific mouse oligonucleotides. The sense and antisense oligonucleotides used were, respectively, the following: GAPDH, 5′-ATGGCCTTCCGTGTTCCTAC-3′ and 5′-CAGATGCCTGCTTCACCAC-3′; IL-1β, 5′-ATGGCAACTGTTCCTGAACTCAACT-3′ and 5′-CAGGACAGGTATAGATTCTTTCCTTT-3′; IL-6, 5′-CTAGGTTTGCCGAGTAGATCT-3′ and 5′-CACAAAGCCAGAGTCCTTCAGAGA-3′; IL-17, 5′-ACCTCACACGAGGCACAAGTG-3′ and 5′-CTTCATTGCGGTGGAGAGTCC-3′; and IL-10, 5′-CCCTTTGCTATGGTGTCCTT-3′ and 5′-TGGTTTCTCTTCCCAAGACC-3′. Each sample was run in duplicate. All results were normalized to the unaffected housekeeping GAPDH gene.
Statistical analysis.
Statistical analysis was performed using a two-tailed Fisher's exact test. A p-value ≤ 0.05 was considered statistically significant. Data are expressed as the mean ± SEM. ANOVA was used for intergroup comparison.
Online supplemental material.
Fig. S1 presents data of AIEC LF82 bacterial colonization, evolution of body weight, and DAI according to CEACAM expression in mice receiving or not 0.25% of DSS in drinking water. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20090741/DC1.
Acknowledgments
We thank Dr Abdelkrim Alloui for animal care (Animal facilities, Clermont-Ferrand, France). The authors are grateful to Pierre Sauvanet and Frédéric Faure (JE2526, Clermont-Ferrand, France) and Monique Etienne (Institut National de la Santé et de la Recherche Médicale U766, Clermont-Ferrand, France) for technical assistance.
This study was supported by the Ministère de la Recherche et de la Technologie (JE2526), Institut National de la Recherche Agronomique (Unité Sous Contrat 2018), and by grants from the Association F. Aupetit, Institut de Recherche des Maladies de l'Appareil Digestif (Laboratoire Astra France), and European Commission through FP7 IBDase project.
The authors have no conflicting financials interests.
Footnotes
Abbreviations used:
- AIEC
- adherent-invasive E. coli
- CD
- Crohn' disease
- CEACAM
- carcinoembryonic antigen-related cell adhesion molecule
- CMC
- carboxymethyl cellulose
- DAI
- disease activity index
- GI
- gastrointestinal
- IBD
- inflammatory bowel disease
References
- Barnich N., Boudeau J., Claret L., Darfeuille-Michaud A. 2003. Regulatory and functional co-operation of flagella and type 1 pili in adhesive and invasive abilities of AIEC strain LF82 isolated from a patient with Crohn's disease.Mol. Microbiol. 48:781–794 doi:10.1046/j.1365-2958.2003.03468.x [DOI] [PubMed] [Google Scholar]
- Barnich N., Carvalho F.A., Glasser A.L., Darcha C., Jantscheff P., Allez M., Peeters H., Bommelaer G., Desreumaux P., Colombel J.F., Darfeuille-Michaud A. 2007. CEACAM6 acts as a receptor for adherent-invasive E. coli, supporting ileal mucosa colonization in Crohn disease.J. Clin. Invest. 117:1566–1574 doi:10.1172/JCI30504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgart M., Dogan B., Rishniw M., Weitzman G., Bosworth B., Yantiss R., Orsi R.H., Wiedmann M., McDonough P., Kim S.G., et al. 2007. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn's disease involving the ileum.ISME J. 1:403–418 doi:10.1038/ismej.2007.52 [DOI] [PubMed] [Google Scholar]
- Boudeau J., Glasser A.L., Masseret E., Joly B., Darfeuille-Michaud A. 1999. Invasive ability of an Escherichia coli strain isolated from the ileal mucosa of a patient with Crohn's disease.Infect. Immun. 67:4499–4509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudeau J., Glasser A.L., Julien S., Colombel J.F., Darfeuille-Michaud A. 2003. Inhibitory effect of probiotic Escherichia coli strain Nissle 1917 on adhesion to and invasion of intestinal epithelial cells by adherent-invasive E. coli strains isolated from patients with Crohn's disease.Aliment. Pharmacol. Ther. 18:45–56 doi:10.1046/j.1365-2036.2003.01638.x [DOI] [PubMed] [Google Scholar]
- Carvalho F.A., Barnich N., Sauvanet P., Darcha C., Gelot A., Darfeuille-Michaud A. 2008. Crohn's disease-associated Escherichia coli LF82 aggravates colitis in injured mouse colon via signaling by flagellin.Inflamm. Bowel Dis. 14:1051–1060 doi:10.1002/ibd.20423 [DOI] [PubMed] [Google Scholar]
- Cegelski L., Marshall G.R., Eldridge G.R., Hultgren S.J. 2008. The biology and future prospects of antivirulence therapies.Nat. Rev. Microbiol. 6:17–27 doi:10.1038/nrmicro1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C.H., Stanners C.P. 2004. Novel mouse model for carcinoembryonic antigen-based therapy.Mol. Ther. 9:775–785 doi:10.1016/j.ymthe.2004.03.009 [DOI] [PubMed] [Google Scholar]
- Claret L., Miquel S., Vieille N., Ryjenkov D.A., Gomelsky M., Darfeuille-Michaud A. 2007. The flagellar sigma factor FliA regulates adhesion and invasion of Crohn disease-associated Escherichia coli via a cyclic dimeric GMP-dependent pathway.J. Biol. Chem. 282:33275–33283 doi:10.1074/jbc.M702800200 [DOI] [PubMed] [Google Scholar]
- Conte M.P., Schippa S., Zamboni I., Penta M., Chiarini F., Seganti L., Osborn J., Falconieri P., Borrelli O., Cucchiara S. 2006. Gut-associated bacterial microbiota in paediatric patients with inflammatory bowel disease.Gut. 55:1760–1767 doi:10.1136/gut.2005.078824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darfeuille-Michaud A., Neut C., Barnich N., Lederman E., Di Martino P., Desreumaux P., Gambiez L., Joly B., Cortot A., Colombel J.F. 1998. Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn's disease.Gastroenterology. 115:1405–1413 doi:10.1016/S0016-5085(98)70019-8 [DOI] [PubMed] [Google Scholar]
- Darfeuille-Michaud A., Boudeau J., Bulois P., Neut C., Glasser A.L., Barnich N., Bringer M.A., Swidsinski A., Beaugerie L., Colombel J.F. 2004. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn's disease.Gastroenterology. 127:412–421 doi:10.1053/j.gastro.2004.04.061 [DOI] [PubMed] [Google Scholar]
- Eaves-Pyles T., Allen C.A., Taormina J., Swidsinski A., Tutt C.B., Jezek G.E., Islas-Islas M., Torres A.G. 2008. Escherichia coli isolated from a Crohn's disease patient adheres, invades, and induces inflammatory responses in polarized intestinal epithelial cells.Int. J. Med. Microbiol. 298:397–409 doi:10.1016/j.ijmm.2007.05.011 [DOI] [PubMed] [Google Scholar]
- Fahlgren A., Baranov V., Frängsmyr L., Zoubir F., Hammarström M.L., Hammarström S. 2003. Interferon-gamma tempers the expression of carcinoembryonic antigen family molecules in human colon cells: a possible role in innate mucosal defence.Scand. J. Immunol. 58:628–641 doi:10.1111/j.1365-3083.2003.01342.x [DOI] [PubMed] [Google Scholar]
- Glasser A.L., Boudeau J., Barnich N., Perruchot M.H., Colombel J.F., Darfeuille-Michaud A. 2001. Adherent invasive Escherichia coli strains from patients with Crohn's disease survive and replicate within macrophages without inducing host cell death.Infect. Immun. 69:5529–5537 doi:10.1128/IAI.69.9.5529-5537.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotlowski R., Bernstein C.N., Sepehri S., Krause D.O. 2007. High prevalence of Escherichia coli belonging to the B2+D phylogenetic group in inflammatory bowel disease.Gut. 56:669–675 doi:10.1136/gut.2006.099796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langermann S., Palaszynski S., Barnhart M., Auguste G., Pinkner J.S., Burlein J., Barren P., Koenig S., Leath S., Jones C.H., Hultgren S.J. 1997. Prevention of mucosal Escherichia coli infection by FimH-adhesin-based systemic vaccination.Science. 276:607–611 doi:10.1126/science.276.5312.607 [DOI] [PubMed] [Google Scholar]
- Langermann S., Möllby R., Burlein J.E., Palaszynski S.R., Auguste C.G., DeFusco A., Strouse R., Schenerman M.A., Hultgren S.J., Pinkner J.S., et al. 2000. Vaccination with FimH adhesin protects cynomolgus monkeys from colonization and infection by uropathogenic Escherichia coli.J. Infect. Dis. 181:774–778 doi:10.1086/315258 [DOI] [PubMed] [Google Scholar]
- Larsson A., Johansson S.M., Pinkner J.S., Hultgren S.J., Almqvist F., Kihlberg J., Linusson A. 2005. Multivariate design, synthesis, and biological evaluation of peptide inhibitors of FimC/FimH protein-protein interactions in uropathogenic Escherichia coli.J. Med. Chem. 48:935–945 doi:10.1021/jm040818l [DOI] [PubMed] [Google Scholar]
- Loftus E.V., Jr. 2004. Clinical epidemiology of inflammatory bowel disease: Incidence, prevalence, and environmental influences.Gastroenterology. 126:1504–1517 doi:10.1053/j.gastro.2004.01.063 [DOI] [PubMed] [Google Scholar]
- Lupp C., Robertson M.L., Wickham M.E., Sekirov I., Champion O.L., Gaynor E.C., Finlay B.B. 2007. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae.Cell Host Microbe. 2:204 doi:10.1016/j.chom.2007.08.002 [DOI] [PubMed] [Google Scholar]
- Malchow H.A. 1997. Crohn's disease and Escherichia coli. A new approach in therapy to maintain remission of colonic Crohn's disease? J. Clin. Gastroenterol. 25:653–658 doi:10.1097/00004836-199712000-00021 [DOI] [PubMed] [Google Scholar]
- Mansfield L.S., Bell J.A., Wilson D.L., Murphy A.J., Elsheikha H.M., Rathinam V.A., Fierro B.R., Linz J.E., Young V.B. 2007. C57BL/6 and congenic interleukin-10-deficient mice can serve as models of Campylobacter jejuni colonization and enteritis.Infect. Immun. 75:1099–1115 doi:10.1128/IAI.00833-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin H.M., Campbell B.J., Hart C.A., Mpofu C., Nayar M., Singh R., Englyst H., Williams H.F., Rhodes J.M. 2004. Enhanced Escherichia coli adherence and invasion in Crohn's disease and colon cancer.Gastroenterology. 127:80–93 doi:10.1053/j.gastro.2004.03.054 [DOI] [PubMed] [Google Scholar]
- Mossman K.L., Mian M.F., Lauzon N.M., Gyles C.L., Lichty B., Mackenzie R., Gill N., Ashkar A.A. 2008. Cutting edge: FimH adhesin of type 1 fimbriae is a novel TLR4 ligand.J. Immunol. 181:6702–6706 [DOI] [PubMed] [Google Scholar]
- Mylonaki M., Rayment N.B., Rampton D.S., Hudspith B.N., Brostoff J. 2005. Molecular characterization of rectal mucosa-associated bacterial flora in inflammatory bowel disease.Inflamm. Bowel Dis. 11:481–487 doi:10.1097/01.MIB.0000159663.62651.4f [DOI] [PubMed] [Google Scholar]
- Neut C., Bulois P., Desreumaux P., Membré J.M., Lederman E., Gambiez L., Cortot A., Quandalle P., van Kruiningen H., Colombel J.F. 2002. Changes in the bacterial flora of the neoterminal ileum after ileocolonic resection for Crohn's disease.Am. J. Gastroenterol. 97:939–946 doi:10.1111/j.1572-0241.2002.05613.x [DOI] [PubMed] [Google Scholar]
- Ofek I., Hasty D.L., Abraham S.N., Sharon N. 2000. Role of bacterial lectins in urinary tract infections. Molecular mechanisms for diversification of bacterial surface lectins.Adv. Exp. Med. Biol. 485:183–192 doi:10.1007/0-306-46840-9_25 [DOI] [PubMed] [Google Scholar]
- Podolsky D.K. 2002. Inflammatory bowel disease.N. Engl. J. Med. 347:417–429 doi:10.1056/NEJMra020831 [DOI] [PubMed] [Google Scholar]
- Poggio T.V., La Torre J.L., Scodeller E.A. 2006. Intranasal immunization with a recombinant truncated FimH adhesin adjuvanted with CpG oligodeoxynucleotides protects mice against uropathogenic Escherichia coli challenge.Can. J. Microbiol. 52:1093–1102 doi:10.1139/W06-065 [DOI] [PubMed] [Google Scholar]
- Prantera C., Scribano M.L., Falasco G., Andreoli A., Luzi C. 2002. Ineffectiveness of probiotics in preventing recurrence after curative resection for Crohn's disease: a randomised controlled trial with Lactobacillus GG.Gut. 51:405–409 doi:10.1136/gut.51.3.405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinella E.S., Eversley C.D., Carroll I.M., Andrus J.M., Threadgill D.W., Threadgill D.S. 2006. Human epithelial-specific response to pathogenic Campylobacter jejuni.FEMS Microbiol. Lett. 262:236–243 [DOI] [PubMed] [Google Scholar]
- Rolhion N., Carvalho F.A., Darfeuille-Michaud A. 2007. OmpC and the sigma(E) regulatory pathway are involved in adhesion and invasion of the Crohn's disease-associated Escherichia coli strain LF82.Mol. Microbiol. 63:1684–1700 doi:10.1111/j.1365-2958.2007.05638.x [DOI] [PubMed] [Google Scholar]
- Rutgeerts P., Goboes K., Peeters M., Hiele M., Penninckx F., Aerts R., Kerremans R., Vantrappen G. 1991. Effect of faecal stream diversion on recurrence of Crohn's disease in the neoterminal ileum.Lancet. 338:771–774 doi:10.1016/0140-6736(91)90663-A [DOI] [PubMed] [Google Scholar]
- Sands B.E. 2007. Inflammatory bowel disease: past, present, and future.J. Gastroenterol. 42:16–25 doi:10.1007/s00535-006-1995-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartor R.B. 2004. Therapeutic manipulation of the enteric microflora in inflammatory bowel diseases: antibiotics, probiotics, and prebiotics.Gastroenterology. 126:1620–1633 doi:10.1053/j.gastro.2004.03.024 [DOI] [PubMed] [Google Scholar]
- Sartor R.B. 2008. Microbial influences in inflammatory bowel diseases.Gastroenterology. 134:577–594 doi:10.1053/j.gastro.2007.11.059 [DOI] [PubMed] [Google Scholar]
- Sasaki M., Sitaraman S.V., Babbin B.A., Gerner-Smidt P., Ribot E.M., Garrett N., Alpern J.A., Akyildiz A., Theiss A.L., Nusrat A., Klapproth J.M. 2007. Invasive Escherichia coli are a feature of Crohn's disease.Lab. Invest. 87:1042–1054 doi:10.1038/labinvest.3700661 [DOI] [PubMed] [Google Scholar]
- Shanahan F. 2002. Crohn's disease.Lancet. 359:62–69 doi:10.1016/S0140-6736(02)07284-7 [DOI] [PubMed] [Google Scholar]
- Sokol H., Pigneur B., Watterlot L., Lakhdari O., Bermúdez-Humarán L.G., Gratadoux J.J., Blugeon S., Bridonneau C., Furet J.P., Corthier G., et al. 2008. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients.Proc. Natl. Acad. Sci. USA. 105:16731–16736 doi:10.1073/pnas.0804812105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher B., Robbiani R., Walker A.W., Westendorf A.M., Barthel M., Kremer M., Chaffron S., Macpherson A.J., Buer J., Parkhill J., et al. 2007. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota.PLoS Biol. 5:2177–2189 doi:10.1371/journal.pbio.0050244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strober W., Fuss I., Mannon P. 2007. The fundamental basis of inflammatory bowel disease.J. Clin. Invest. 117:514–521 doi:10.1172/JCI30587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson A., Larsson A., Emtenäs H., Hedenström M., Fex T., Hultgren S.J., Pinkner J.S., Almqvist F., Kihlberg J. 2001. Design and evaluation of pilicides: potential novel antibacterial agents directed against uropathogenic Escherichia coli.ChemBioChem. 2:915–918 doi:10.1002/1439-7633(20011203)2:12<915::AID-CBIC915>3.0.CO;2-M [DOI] [PubMed] [Google Scholar]
- Swidsinski A., Ladhoff A., Pernthaler A., Swidsinski S., Loening-Baucke V., Ortner M., Weber J., Hoffmann U., Schreiber S., Dietel M., Lochs H. 2002. Mucosal flora in inflammatory bowel disease.Gastroenterology. 122:44–54 doi:10.1053/gast.2002.30294 [DOI] [PubMed] [Google Scholar]
- Tamboli C.P., Neut C., Desreumaux P., Colombel J.F. 2004. Dysbiosis in inflammatory bowel disease.Gut. 53:1–4 doi:10.1136/gut.53.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thankavel K., Madison B., Ikeda T., Malaviya R., Shah A.H., Arumugam P.M., Abraham S.N. 1997. Localization of a domain in the FimH adhesin of Escherichia coli type 1 fimbriae capable of receptor recognition and use of a domain-specific antibody to confer protection against experimental urinary tract infection.J. Clin. Invest. 100:1123–1136 doi:10.1172/JCI119623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadolkowski E.A., Laux D.C., Cohen P.S. 1988. Colonization of the streptomycin-treated mouse large intestine by a human fecal Escherichia coli strain: role of growth in mucus.Infect. Immun. 56:1030–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellens A., Garofalo C., Nguyen H., Van Gerven N., Slättegård R., Hernalsteens J.P., Wyns L., Oscarson S., De Greve H., Hultgren S., Bouckaert J. 2008. Intervening with urinary tract infections using anti-adhesives based on the crystal structure of the FimH-oligomannose-3 complex.PLoS One. 3:e2040 doi:10.1371/journal.pone.0002040 [DOI] [PMC free article] [PubMed] [Google Scholar]