Abstract
Natural ribozymes require metal ion cofactors that aid both in structural folding and in chemical catalysis. In contrast, many protein enzymes produce dramatic rate enhancements using only the chemical groups that are supplied by their constituent amino acids. This fact is widely viewed as the most important feature that makes protein a superior polymer for the construction of biological catalysts. Herein we report the in vitro selection of a catalytic DNA that uses histidine as an active component for an RNA cleavage reaction. An optimized deoxyribozyme from this selection requires l-histidine or a closely related analog to catalyze RNA phosphoester cleavage, producing a rate enhancement of ≈1-million-fold over the rate of substrate cleavage in the absence of enzyme. Kinetic analysis indicates that a DNA–histidine complex may perform a reaction that is analogous to the first step of the proposed catalytic mechanism of RNase A, in which the imidazole group of histidine serves as a general base catalyst. Similarly, ribozymes of the “RNA world” may have used amino acids and other small organic cofactors to expand their otherwise limited catalytic potential.
The “RNA world” theory for the origins of life holds that RNA was able to replicate in the absence of DNA and proteins (1). Some manifestations of this theory (1–3) include the view that, during the latter stages of the RNA world, ribozymes were not primitive and ineffectual but were directing a complex metabolic state replete with RNA and DNA synthesis machinery and a host of RNA enzymes necessary to catalyze supportive chemical reactions. Proponents of this view argue that a sophisticated metabolism mediated entirely by RNA would have been necessary for ribozymes to give rise to encoded protein synthesis—a precursor to the complex system of translation that is present in all extant organisms.
To achieve maximum catalytic rate enhancements, natural ribozymes require Mg2+ or other metal ion cofactors to induce structure formation or to participate directly in the catalytic process (4), perhaps defining a limitation to the catalytic potential for polynucleotide-based enzymes. When the limited catalytic repertoire of natural ribozymes is considered, the notion that complex networks of metabolic reactions could be guided entirely by RNA appears tenuous. Ancient ribozymes, however, may have created a larger catalytic repertoire by using various divalent metals (5) and by using nucleotide-like cofactors (6–8) to supplement the limited set of chemical groups that make up unmodified RNA. In recent years, a surprising number of artificial ribozymes have been created by using in vitro selection (9) that catalyze various chemical reactions including alkylation, acylation, phosphorylation, and ester and amide bond formation. In addition, the catalytic rates of ribozymes can be tightly regulated by using allosteric mechanisms (10), indicating that the potential for extensive and sophisticated catalysis by nucleic acids is large. Moreover, the full range of reactive groups found in protein enzymes becomes available to catalytic polynucleotides if amino acids could be used as cofactors. We set out to investigate whether nucleic acid enzymes could make use of amino acids or other organic compounds as cofactors to supplement their inherently meager chemical composition.
MATERIALS AND METHODS
In Vitro Selection and Reselection.
In vitro selection was carried out essentially as described (11–13). The initial DNA pool was prepared by PCR amplification of the template 5′-CTAATACGACTCACTATAGGAAGAGATGGCGACATCTC(N)40GTGAGGTTGGTGTGGTTG (50 pmol; N is any one of the four nucleotides occurring with an equal probability) in a 500-μl PCR mixture containing 400 pmol of primer B2 (5′-biotin-GAATTCTAATACGACTCACTATrA) and 400 pmol of primer 1 (5′-CAACCACACCAACCTCAC) with four thermocycles of 94°C (15 sec), 50°C (30 sec), and 72°C (30 sec). PCR mixture was prepared as described (13). Amplified DNA was precipitated with ethanol, resuspended in binding buffer (50 mM Hepes, pH 7.5 at 23°C/0.5 M NaCl/0.5 M KCl/0.5 mM EDTA), and the solution was passed through a streptavidin-derivatized affinity matrix to generate immobilized single-stranded DNA (12). The matrix displaying the pool DNA was repeatedly washed with binding buffer (1.5 ml over 30 min), and material was subsequently eluted over the course of 1 hr with three 20-μl aliquots of reaction buffer in which Hepes was replaced with 50 mM histidine (pH 7.5, 23°C). In rounds 8–11, reaction time was reduced to 25–15 min to favor those molecules that cleave more efficiently. Selected DNAs were precipitated with ethanol and amplified by PCR using primer 1 and primer 2 (5′-GAATTCTAATACGACTCACTATAGGAAGAGATGGCGAC), and the resulting PCR was reamplified as described above to reintroduce the biotin and embedded ribonucleotide moieties.
Reselection of the class II deoxyribozyme was initiated with a pool of 1013 DNAs, each carrying a 39-nucleotide core that had been mutagenized with a degeneracy of 0.21 per position. Similarly, HD2 reselections were conducted with an initial pool in which 26 nucleotides was mutagenized to a degeneracy of 0.33 per position. Individuals from the final selected pools were analyzed by cloning and sequencing as described (13). The DNA pools were prepared for this process by PCR amplification using primer 2 in place of primer B2. DNA populations and individual precursor DNAs were prepared for assays as described (12).
Deoxyribozyme Catalysis Assays.
All catalytic assays were conducted in the presence of 0.5 M NaCl, 0.5 M KCl, and 0.5 mM EDTA. Single-turnover assays contained a trace amount of (≈50 nM) substrate oligonucleotide and an excess (1–10 μM) of DNA catalyst as described for each assay. The cofactor used was l-histidine unless otherwise stated. Reactions were terminated by addition of an equal volume of 95% formamide containing 0.05% xylene cyanol and 0.05% bromophenol blue. Buffers containing both urea and EDTA were incapable of completely terminating deoxyribozyme activity.
Caged histidine experiments were conducted with intact dipeptides or with a concentration of hydrolyzed dipeptide products. Hydrolysis of dipeptides was achieved by incubating solutions containing 100 mM dipeptide and 6 M HCl in a sealed tube at 115°C for 23 hr. Samples were evaporated in vacuo and coevaporated with deionized water, and the resuspended samples were adjusted to neutral pH before use.
Kinetic Analyses.
All catalytic rate constants (kobs) reported were derived under self-cleavage or single-turnover conditions unless otherwise stated. Rate constants derived under self-cleavage or single-turnover conditions were established either by determining the initial velocity of the reaction (13) or by plotting the natural logarithm of the fraction of substrate remaining over time where the negative slope of the line obtained over several half-lives represents kobs. For these reactions, trace amounts of 5′-32P-labeled substrate were incubated with an excess of DNA enzyme at a concentration above the Kd for enzyme–substrate complex formation. The multiple turnover assay was conducted at 23°C with 1 μM HD2 and 12 μM substrate in 0.5 M NaCl/0.5 M KCl/50 mM Hepes, pH 7.5 at 23°C/5 mM histidine.
The rate constant for the uncatalyzed reaction was determined by incubating a trace amount of 5′-32P-labeled substrate under selection conditions in the absence of deoxyribozyme at 23°C for 20 days. Comparative analysis of RNA phosphoester cleavage indicates that the rate constant for uncatalyzed RNA cleavage in the presence of 50 mM histidine alone is ≈3.6 × 10−7 min−1. This rate is measurably faster than the rate of RNA phosphoester cleavage at 23°C in the absence of histidine (≈3.8 × 10−8 min−1). Rate constants reported for the uncatalyzed reactions are averaged from two replicate experiments.
RESULTS AND DISCUSSION
Isolation of a Histidine-Dependent Deoxyribozyme.
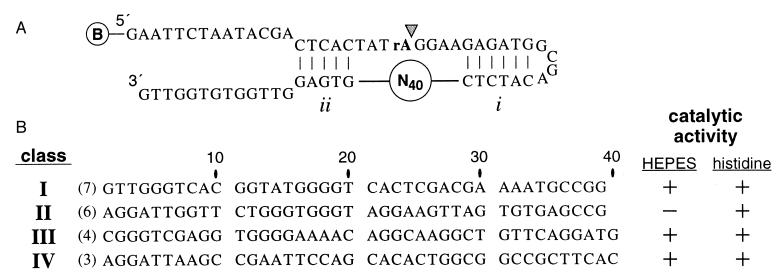
With in vitro selection (9) (Fig. 1A), we identified a class of deoxyribozymes (14) that catalyze the cleavage of an RNA phosphoester bond by using the amino acid histidine as a cofactor. The pool of DNA molecules used to initiate the selective-amplification process included a domain of 40 random-sequence nucleotides that is flanked on each side by regions of base complementation (Fig. 1A; pairing elements i and ii). The preengineered pairing elements were included in the design in an attempt to form a crude substrate binding site. To disfavor the isolation of metal-dependent deoxyribozymes from the random-sequence pool of DNAs, the divalent metal-chelating agent EDTA was included in a reaction mixture that was buffered with 50 mM l-histidine (pH 7.5). The DNA pool recovered after 11 rounds of selective amplification displayed RNA phosphoester-cleaving activity both under in vitro selection conditions and with a reaction buffer in which Hepes (50 mM, pH 7.5) was substituted for histidine. Individual molecules cloned from the DNA population at generation 11 were grouped into one of four sequence classes (Fig. 1B), and representative clones were tested for catalytic activity. Only class II DNAs demonstrate complete dependence on histidine, whereas the remaining classes appear to operate independently of any metal ion or small organic cofactor (unpublished observations). The latter examples are likely to be similar to other cofactor-independent deoxyribozymes reported recently (15, 16).
Figure 1.
In vitro selection of histidine-dependent deoxyribozymes. (A) A pool of 2 × 1013 biotin-modified DNAs was immobilized on a streptavidin-derivatized column matrix. Each DNA carries a single embedded RNA linkage (rA) and a 40-nucleotide random-sequence domain that is flanked by regions that are complementary to nucleotides that reside both 5′ and 3′ of the target phosphodiester (pairing elements i and ii). These preengineered substrate-binding interactions are expected to increase the probability of isolating active catalysts. DNAs that catalyze the cleavage of the RNA linkage upon incubation with a solution buffered with histidine were released from the matrix and amplified by PCR, and the amplification products were immobilized again to complete the selection cycle. Arrowhead identifies the expected site of cleavage. (B) The 40-nucleotide core of four classes of deoxyribozymes were identified by comparative sequence analysis. Variants within each group differed by no more than two mutations from the sequences shown. DNAs from classes I, III, and IV are active (+) with either Hepes or histidine buffers, whereas class II DNAs show no measurable activity (−) without histidine. Nucleotides are numbered to reflect their relationship to the original 40-nucleotide random-sequence domain.
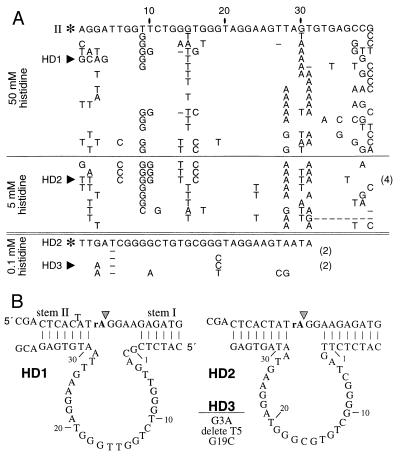
Although the original random-sequence domain included 40 nucleotides, all examples of class I and class II DNAs that were isolated carry only 39 nucleotides that correspond to this region. Presumably, these DNAs have undergone a single nucleotide deletion during the in vitro selection process. The rate of self-cleavage for the class II deoxyribozyme is ≈1000-fold slower (kobs = 1.5 × 10−3 min−1) than most natural self-cleaving ribozymes (17). Therefore, we subjected a mutagenized pool based on the original class II deoxyribozyme to additional rounds of in vitro selection to improve catalytic activity and to provide an artificial phylogeny of variant DNAs for comparative sequence analysis. A new DNA pool (sampling all possible variant DNAs with seven or fewer mutations relative to the class II sequence) was created such that the 39 nucleotides remaining from the original random sequence domain were mutagenized with a degeneracy of 0.21 (18). Parallel reselections were conducted by using reaction solutions buffered with either 50 mM histidine or with 5 mM histidine/50 mM Hepes. Individual DNAs isolated from the populations after five rounds of reselection (e.g., HD1 and HD2) are ≈100-fold more active than the original class II deoxyribozyme and show specific patterns of conserved sequence and mutation acquisition (Fig. 2A) that are characteristic of a relatively large and complex tertiary structure. Reselection with lower histidine concentrations using a mutagenized pool based on the sequence of HD2 gave rise to variant DNAs (e.g., HD3) in which most nucleotides in the putative catalytic domain are conserved (Fig. 2A).
Figure 2.
Sequences and secondary structures of variant deoxyribozymes. (A) Individual DNAs shown above and below the double line were isolated after reselection of mutagenized pools based on the sequence of the original class II deoxyribozyme (II) or the class II variant HD2, respectively. Depicted are the nucleotide sequences for the mutagenized core of the parent DNAs (asterisks) and the number (in parentheses) and type of nucleotide changes for each variant deoxyribozyme. HD1 and HD2 were recovered from a mutagenized pool based on II after five rounds of reselection with 50 or 5 mM histidine, respectively. HD3 was isolated from a mutagenized pool of truncated HD2 deoxyribozymes. (B) Each deoxyribozyme was reorganized to create a bimolecular complex, whereby separate substrate molecules are recognized by two regions of base complementation (stems I and II) with the enzyme domain. Nucleotides corresponding to the mutagenized core are numbered as in Fig. 1 and A.
We speculated that engineered pairing element i (Fig. 1A) was being used by class II deoxyribozymes. In contrast, we recognized that a conserved sequence domain near the 3′ end of the original random-sequence domain (Fig. 2A, nucleotides 32–36) was identical to pairing element ii, indicating that this second engineered pairing element possibly could be deleted. Based on these observations, the deoxyribozymes HD1, HD2, and HD3 were designed to operate with separate substrate and enzyme domains (Fig. 2B), where specificity for substrate is defined by Watson–Crick base complementation with the two pairing arms of the enzyme. In each case, the bimolecular complexes remain fully active and were used in this study to further characterize deoxyribozyme function.
Cofactor Requirements for Deoxyribozyme Function.
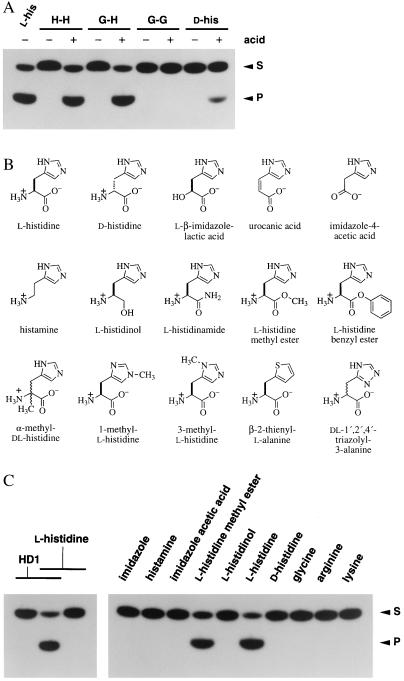
To establish definitively that histidine is required for catalysis, we exploited the inability of HD1 to accept dipeptides as cofactors. HD1 remains inactive in the presence of “caged” histidine that is delivered in the context of dipeptides but displays full activity when the constituent amino acids are liberated from each dipeptide by acid hydrolysis (Fig. 3A). HD1 is active only when incubated with acid-hydrolyzed dipeptides that contain at least one histidine residue. In addition, HD1 accepts l- but not d-histidine as a cofactor, indicating that the deoxyribozyme forms an amino acid binding site that is stereoselective. Interestingly, HD1 exhibits activity in the presence of acid-treated d-histidine, in accordance with the accelerated rate of interconversion between the two isomeric forms of the amino acids under acidic conditions (19).
Figure 3.
Cofactor recognition by a deoxyribozyme. (A) Catalytic activity of HD1 with l-histidine, d-histidine, and various dipeptides that did (+) or did not (−) receive pretreatment with hydrochloric acid. HD1 (10 μM) was incubated in the presence of trace amounts of 5′-32P-labeled substrate oligonucleotide (Fig. 2B) at 23°C for 2.5 hr in a mixture containing 50 mM l-histidine, d-histidine, or the dipeptides histidylhistidine (H-H), glycylhistidine (G-H), or glycylglycine (G-G) as indicated. Reaction products were analyzed by denaturing (8 M urea) PAGE and imaged by autoradiography. S and P identify substrate and product (5′ cleavage fragment) bands, respectively. (b) Chemical structures of l-histidine and the histidine analogs used to probe deoxyribozyme cofactor specificity. (C) Representative deoxyribozyme assays for HD1 catalytic activity with selected amino acids and histidine analogs. Reactions and analyses were conducted as described in A.
We examined a series of histidine analogs (Fig. 3B) to determine more carefully the chemical groups that are important for catalytic activity. HD1 discriminates against a variety of analogs, but shows full activity with the methyl ester of l-histidine (Fig. 3C). Neither 1-methyl- nor 3-methyl-l-histidine supports HD1 activity (data not shown), suggesting that an unaltered imidazole ring is important for deoxyribozyme function. As expected, HD2 has a similar pattern of cofactor discrimination (Table 1). These catalysts demonstrate stereospecific recognition of histidine and may maximize cofactor binding by using interactions with the carboxyl oxygens, the α-amino group, and the imidazole side chain. These characteristics also are consistent with the presence of a specific binding site for histidine. Although a number of analogs (e.g., dl-1′,2′,4′-triazolyl-3-alanine) fail to support deoxyribozyme activity, no compound was found to function as a competitive inhibitor, likely due to the inability of each analog to form a tight complex with the deoxyribozyme.
Table 1.
Relative kobs values for the bimolecular HD2 deoxyribozyme with 25 mM l-histidine or various analogs (kobs for l-histidine = 0.11 min−1)
| Cofactor | Fold discrimination |
|---|---|
| l-Histidine methyl ester | 1.1 |
| l-Histidine benzyl ester | 1.3 |
| α-Methyl-dl-histidine | 24 |
| l-Histidinamide | 41 |
| Glycylhistidine | 185 |
| l-Histidinol | 357 |
| 3-Methyl-l-histidine | 455 |
| d-Histidine | 714 |
| l-β-Imidazolelactic acid | 1,235 |
| 1-Methyl-l-histidine | 1,250 |
| Histamine | 5,263 |
| Imidazole | 6,667 |
| Urocanic acid | >10,000 |
| dl-1′,2′,4′-Triazolyl-3-alanine | >10,000 |
| β-2-Thienyl-l-alanine | >10,000 |
| Imidazole-4-acetic acid | >10,000 |
In general, deoxyribozyme activity diminishes in proportion to the level of disruption of the carboxyl and amino groups of l-histidine. Samples of 1- and 3-methyl-l-histidine may contain as much as 10 μM unmodified l-histidine as a contaminant. This concentration of preferred cofactor is more than sufficient to produce the level of HD2 activity seen with these analogs.
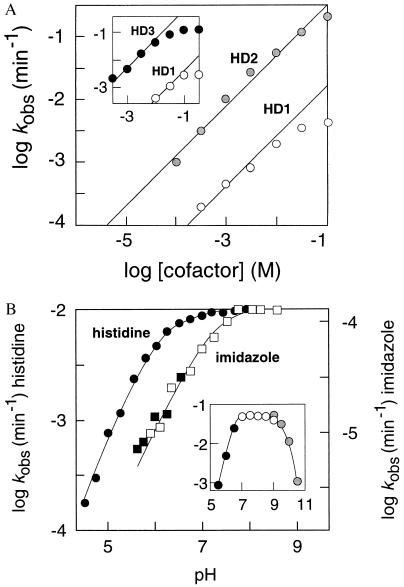
Although the cofactor binding site of HD1 can be saturated with histidine at high concentrations (apparent Kd ≈ 25 mM; max. kobs = 4.7 × 10−3 min−1 at pH 7.5), the putative binding site of HD2 is not saturated even at a cofactor concentration of 100 mM (Fig. 4A). Histidine becomes insoluble at higher concentrations due to its zwitterionic configuration, which prohibits examining catalytic activity in the presence of additional histidine. The zwitterion is disrupted with the methyl ester of histidine, thereby facilitating the testing of higher concentrations of cofactor. A plot of histidine methyl ester concentration versus HD1 and HD3 activity (Fig. 4A Inset) is also consistent with the presence of a saturable binding site with an apparent Kd of ≈25 mM. HD2 (kobs of 0.2 min−1, 100 mM histidine) demonstrates greater catalytic activity than HD1 despite the fact that the cofactor-binding site is not saturated. This indicates that although HD2 binds the cofactor with poorer affinity than does HD1, HD2 may position bound cofactor more favorably for the subsequent catalytic process. Reselection of a mutagenized DNA pool based on HD2 (Fig. 2A) yielded variant deoxyribozymes that display only a small improvement in catalytic activity despite selection using 100 μM cofactor. Therefore, HD2 and HD3 may represent near optimal versions of this class of deoxyribozymes.
Figure 4.
Involvement of histidine in deoxyribozyme function. (A) Concentration-dependent induction of deoxyribozyme function by histidine. HD1 and HD2 activities increase linearly with increasing concentrations of histidine. Unlike HD1, the concentration-dependent values for the rate constant of HD2 do not vary from a line depicting a slope of 1, indicating that the cofactor binding site has not been saturated with histidine. A similar plot for HD1 and HD3 (Inset) shows a plateau of kobs values at high concentrations of l-histidine methyl ester, indicating that the binding site approaches saturation at cofactor concentrations above 25 mM. (B) Dependence of deoxyribozyme function on pH. Data represented in the main plot were obtained with 1 mM histidine (circles) or 100 mM imidazole (squares) and data in the Inset were obtained with 5 mM histidine. Data depicted in B with solid, open, and shaded symbols were collected by using Mes-, Tris- (or imidazole-), and Caps-buffered solutions, respectively.
The rate constant for HD2-promoted catalysis is similar to that of natural self-cleaving ribozymes and under optimal conditions corresponds to a rate enhancement of ≈1 million-fold over the reaction rate in the absence of enzyme. HD2 exhibits multiple turnover kinetics, with an average of 7.5 reactions per enzyme per 2.5 hr (5 mM l-histidine). The activity of this deoxyribozyme is neither inhibited by the addition of EDTA nor enhanced by the addition of a number of divalent metal ions. Ni2+ completely inhibits deoxyribozyme function when added in equimolar amounts with histidine. The mechanism of this inhibition presumably involves the formation of Ni2+–histidine chelates at the expense of deoxyribozyme–amino acid complexes. HD2 requires K+ but not Na+ for catalytic function, indicating that the active DNA conformation may include a potassium-sensitive structure such as a G quartet (20).
Evidence for a Catalytic Role for the Histidine Cofactor.
To investigate the functional role that histidine might serve in the catalytic process, we examined the pH-dependent activity profile of the deoxyribozyme. The rate constant of HD2 is independent of pH between pH 7 and pH 9 (Fig. 4B). A significant loss of catalytic activity was expected due to DNA denaturation at pH values greater than 9 or less than 5 caused by deprotonation of thymidine and guanosine residues or protonation of cytidine and adenosine residues, respectively. However, most revealing is the response of HD2 to low pH conditions. The kobs values increase linearly with increasing pH between pH 4.5 and 5.5, giving a slope of 1. This result would be expected if the protonation state of a single functional group influences catalytic rate. A rate constant that is half the maximum is obtained at pH 6, suggesting a pKa of 6 for this putative catalytic group. This pKa value corresponds precisely with that for the imidazole group of histidine, indicating that if the chemical step of this enzymatic process is rate limiting, the deprotonated form of the amino acid side chain may serve as the catalytic moiety.
HD2 also displays activity with free imidazole as a cofactor, albeit with catalytic rates that are >6,000-fold slower than with histidine (Table 1). The corresponding pH profile of HD2 with imidazole is shifted toward the basic range by 1 pH unit compared with histidine (Fig. 4B). This shift in pKa of the putative catalytic group corresponds with the pKa of 7 for the free imidazole group. Although we cannot rule out a purely structural role for the amino acid, these data are consistent with a mechanism in which the imidazole group of histidine serves as a general base catalyst for the deprotonation of the 2′-hydroxyl group, thereby activating the 2′ oxygen for nucleophilic attack on the neighboring phosphorus atom. These pH-dependent activity profiles implicate the direct involvement of histidine in the chemical step of the reaction.
CONCLUSIONS
Histidine was chosen as a candidate cofactor because of the potential for the imidazole side chain to function in both general acid and general base catalysis near neutral pH. This property is inherent neither to the standard nucleotides nor to the remaining natural amino acids. As a consequence, histidine is one of the residues most frequently used to form the active sites of protein enzymes. For example, two histidines form a critical portion of the active site of bovine pancreatic ribonuclease A. Like this protein enzyme, the deoxyribozymes reported here cleave RNA phosphoesters via a cyclizing mechanism to produce free 5′ hydroxyl and 2′,3′-cyclic phosphate groups (data not shown). Although ribonuclease A has long served as a model for the study of enzyme action, the specific roles that each active-site residue plays in the catalytic process are still vigorously debated (21). The classical view holds that the histidine at position 12 acts as a general base for the deprotonation of the 2′ hydroxyl, whereas the histidine at position 119 acts as a general acid and protonates the 5′ oxyanion leaving group. Breslow (22) and others (23, 24) have proposed that the role for histidine 119 instead may be to protonate the phosphorane intermediate, thereby implicating general acid catalysis by the imidazole group as a preeminent step during the catalytic process. Our data indicate that the histidine cofactor for class II deoxyribozymes is not involved in a protonation step but is functioning exclusively as a general base catalyst.
Recently, several divalent metal-dependent deoxyribozymes that catalyze RNA phosphoester cleavage have been isolated by in vitro selection (11, 12, 15, 25). Like these metal-dependent deoxyribozymes, the optimized class II DNAs display rate constants that approach 1 min−1 despite the use of an amino acid cofactor rather than a metal ion. In addition, HD2 is capable of cleaving a substrate oligonucleotide that is made entirely of RNA, although with a rate that is ≈1,000-fold slower than with the substrate containing a single embedded ribonucleotide. The histidine-dependent deoxyribozyme might serve as a starting point for the further development of a DNA enzyme that more closely mimics the function of RNase A, including the simultaneous use of several mechanistic approaches to enhance the rate of RNA cleavage beyond that seen with existing ribozymes or deoxyribozymes. Specifically, a histidine-dependent deoxyribozyme might be made that matches the proficiency of a recently described Mg2+-dependent deoxyribozyme that also cleaves RNA (25).
In comparison to proteins, the more repetitive nature of the monomeric units that make up nucleic acids limits both the formation of fine structure in folded polynucleotides and the chemical reactivity of RNA and DNA. The observation that a nucleic acid enzyme can co-opt a favorite chemical unit of protein-based enzymes supports the notion that early ribozymes could have augmented their limited catalytic potential with organic as well as metal ion cofactors. Indeed, ribozymes may have been the first biocatalysts to use the chemical diversity of modern proteins using cofactors in the form of free amino acids, before giving up command over the larger share of biological catalysis. Perhaps it is the protein enzymes of contemporary biology that have borrowed some of their tools and tactics for catalytic function from the long-extinct ribozymes of the RNA world.
Acknowledgments
We thank Jeff Bada, Venkat Gopalan, and members of the Breaker laboratory for helpful discussions. This work was supported by a Young Investigator Award to R.R.B. from the Arnold and Mabel Beckman Foundation.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
A commentary on this article begins on page 5845.
References
- 1.Gilbert W. Nature (London) 1986;319:618. [Google Scholar]
- 2.Benner S A, Ellington A D, Tauer A. Proc Natl Acad Sci USA. 1989;86:7054–7058. doi: 10.1073/pnas.86.18.7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hirao I, Ellington A D. Curr Biol. 1995;5:1017–1022. doi: 10.1016/s0960-9822(95)00205-3. [DOI] [PubMed] [Google Scholar]
- 4.Pyle A M. Science. 1993;261:709–714. doi: 10.1126/science.7688142. [DOI] [PubMed] [Google Scholar]
- 5.Yarus M. FASEB J. 1993;7:31–39. doi: 10.1096/fasebj.7.1.8422972. [DOI] [PubMed] [Google Scholar]
- 6.White H B. J Mol Evol. 1976;7:101–104. doi: 10.1007/BF01732468. [DOI] [PubMed] [Google Scholar]
- 7.Connell G J, Christian E L. Origins of Life. 1993;23:291–297. doi: 10.1007/BF01582079. [DOI] [PubMed] [Google Scholar]
- 8.Breaker R R, Joyce G F. J Mol Evol. 1995;40:551–558. doi: 10.1007/BF00160500. [DOI] [PubMed] [Google Scholar]
- 9.Breaker R R. Chem Rev. 1997;97:371–390. doi: 10.1021/cr960008k. [DOI] [PubMed] [Google Scholar]
- 10.Tang J, Breaker R R. Chem Biol. 1997;4:453–459. doi: 10.1016/s1074-5521(97)90197-6. [DOI] [PubMed] [Google Scholar]
- 11.Breaker R R, Joyce G F. Chem Biol. 1994;1:223–229. doi: 10.1016/1074-5521(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 12.Breaker R R, Joyce G F. Chem Biol. 1995;2:655–660. doi: 10.1016/1074-5521(95)90028-4. [DOI] [PubMed] [Google Scholar]
- 13.Carmi N, Shultz L A, Breaker R R. Chem Biol. 1996;3:1039–1046. doi: 10.1016/s1074-5521(96)90170-2. [DOI] [PubMed] [Google Scholar]
- 14.Breaker R R. Nat Biotechnol. 1997;15:427–431. doi: 10.1038/nbt0597-427. [DOI] [PubMed] [Google Scholar]
- 15.Faulhammer D, Famulok M. J Mol Biol. 1997;269:188–202. doi: 10.1006/jmbi.1997.1036. [DOI] [PubMed] [Google Scholar]
- 16.Geyer C R, Sen D. Chem Biol. 1997;4:579–593. doi: 10.1016/s1074-5521(97)90244-1. [DOI] [PubMed] [Google Scholar]
- 17.Symons R H. Annu Rev Biochem. 1992;61:641–671. doi: 10.1146/annurev.bi.61.070192.003233. [DOI] [PubMed] [Google Scholar]
- 18.Breaker R R, Joyce G F. Trends Biotechnol. 1994;12:268–275. doi: 10.1016/0167-7799(94)90138-4. [DOI] [PubMed] [Google Scholar]
- 19.Engel M H, Hare P E. Year Book Carnegie Inst Washington. 1982;81:422–425. [Google Scholar]
- 20.Williamson J R. Annu Rev Biophys Biomol Struct. 1994;23:703–730. doi: 10.1146/annurev.bb.23.060194.003415. [DOI] [PubMed] [Google Scholar]
- 21.Perreault D M, Anslyn E V. Angew Chem Int Ed Engl. 1997;36:432–450. [Google Scholar]
- 22.Breslow R. Acc Chem Res. 1991;24:317–324. [Google Scholar]
- 23.Lim C, Tole P. J Am Chem Soc. 1992;114:7245–7252. [Google Scholar]
- 24.Wladkowski B D, Krauss M, Stevens W J. J Am Chem Soc. 1995;117:10537–10545. [Google Scholar]
- 25.Santoro S W, Joyce G F. Proc Natl Acad Sci USA. 1997;94:4262–4266. doi: 10.1073/pnas.94.9.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]