Abstract
The GM2 activator protein (GM2AP) is an 18 kDa non-enzymatic accessory protein involved in the degradation of neuronal gangliosides. Genetic mutations of GM2AP can disrupt ganglioside catabolism and lead to deadly lysosomal storage disorders. Crystallography of wild-type GM2AP reveals 4 disulfide bonds and multiple conformations of a flexible loop region that is thought to be involved in lipid binding. To extend the crystallography results, a cysteine construct (L126C) was expressed and modified with 4-maleimide TEMPO for electron paramagnetic resonance (EPR) studies. However, because a ninth cysteine has been added by site-directed mutagenesis and the protein was expressed in E. coli in the form of inclusion bodies, the protein could misfold during expression. To verify correct protein folding and labeling, a sequential multiple-protease digestion, nano-LC electrospray ionization 14.5 T Fourier transform ion cyclotron resonance mass spectrometry assay was developed. High-magnetic field and robust automatic gain control results in sub-ppm mass accuracy for location of the spin-labeled cysteine and verification of proper connectivity of the four disulfide bonds. The sequential multiple protease digestion strategy and ultra-high mass accuracy provided by FT-ICR MS allow for rapid and unequivocal assignment of relevant peptides and provide a simple pipeline for analyzing other GM2AP constructs.
INTRODUCTION
The GM2 Activator Protein (GM2AP) is an 18 kDa, non-enzymatic accessory protein involved in ganglioside catabolism. A specific function of GM2AP involves binding and extracting GM2 from intralysosomal vesicles that positions the terminal GalNAc head group of GM2 for cleavage by β-hexosaminidase A (Hex A) to produce GM3.1, 2 Genetic mutations to either GM2AP or Hex A prevent the degradation of GM2, thereby causing lysosomal storage disease, such as AB variant gangliosidosis and Tay Sachs disease.3–5 GM2AP X-ray crystallographic structures reveal 4 disulfide bonds and a β-cup topology that forms a hydrophobic pocket for lipid binding.6 Various X-ray structures with different lipid substrates reveal multiple conformations of a mobile loop region that is observed in either a "closed" or "open" conformation. These "open" and "closed" conformations suggest a functional conformation change that may occur upon lipid ligand or vesicle binding.6
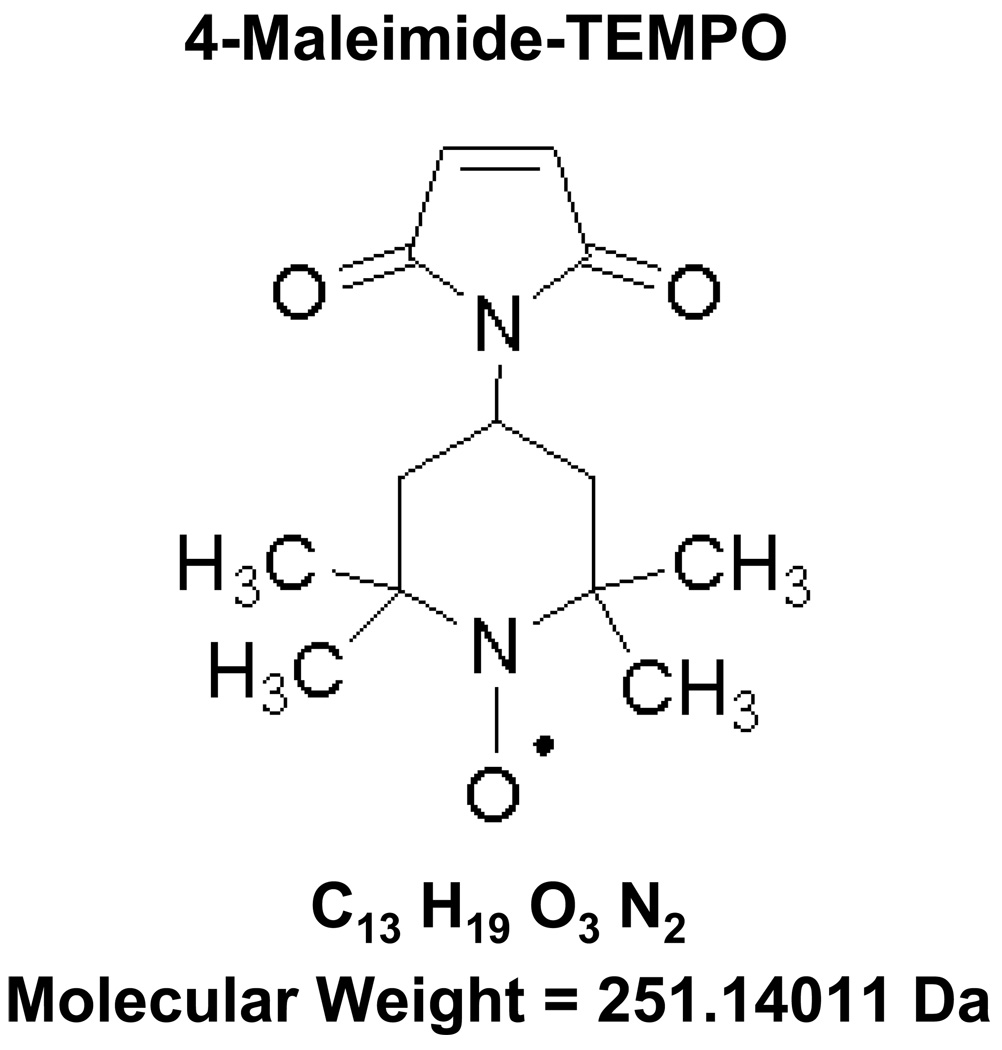
To further probe the "binding" conformational changes of the loop region, site-directed spin labeling electron paramagnetic resonance (EPR) spectroscopy7–10 was applied to GM2AP.11 In proteins, site-directed spin labeling typically proceeds by generating site-specific cysteine amino acid substitutions for modification with paramagnetic nitroxide probes, e.g., 4-maleimide-TEMPO (Figure 1). Reduced cysteine residues act as nucleophiles capabable of reacting with maleimide or thiosulfonate groups to form thioethers or disulfide bonds, respectively. Cysteine residues involved in disulfide bonds do not readily react with those functional groups at pH 6.8–7.4.12 To ensure that purified protein contained the proper connectivity of the 8 native cysteine residues and that only the introduced reporter cysteine (L126C) was labeled, a mass spectrometry based assay was developed.
Figure 1.
Structure, molecular weight, and elemental composition for 4-maleimide-TEMPO.
Mass spectrometry enables direct identification of post-translational modifications, including disulfide bonds.13–20 Because of the analytical challenge associated with verifying multiple disulfide bonds, a multi-tier assay was developed. First, the expressed protein was sequentially proteolytically digested with the enzymes, trypsin, endoproteinase GluC, and endoproteinase AspN. After each sequential digestion, the samples were analyzed by high-field Fourier transform ion cyclotron resonance (FT-ICR) MS.21 Although sequential proteolysis produces more complex mixtures for analysis, the resulting MS/MS spectra from less complex disulfide-bound peptide fragments result in simple verification of disulfide connectivity. High resolution mass measurement with our modified 14.5 T LTQ FT-ICR MS provides high mass accuracy and allows for quick peptide and fragment ion assignments with rms mass errors of less than 0.5 ppm.21 Here, we present a systematic approach to verify proper folding and labeling of a recombinantly over-expressed L126C GM2AP construct by combination of standard proteomic techniques and multiple scan functions with high resolution MS and/or low/high resolution MS/MS with our modified 14.5 T LTQ FT-ICR MS. The first scan function included the use of the typical top-5 data dependent (high resolution MS; low resolution MS/MS) to identify all proteolytic fragments, whereas the identified fragments were subsequently analyzed by directed high resolution MS/MS for verification of disulfide connectivity. Furthermore, electron capture dissociation (ECD) with our custom-built 9.4 T FT-ICR provided the last piece of information for verification of disulfide binding. The result is a high-confidence MS assay that will be easy to apply to other spin-labeled proteins containing multiple disulfide linkages.
EXPERIMENTAL PROCEDURES
Materials
All solvents, methanol and water, were purchased from J.T. Baker (Philipsburg, NJ) at HPLC grade. Trypsin, endoproteinase GluC, endoproteinase Asp N, formic acid, and ammonium bicarbonate were purchased from Sigma-Aldrich (St. Louis, MO).
Protein Expression and Purification of GM2AP Mutants
Wild-type and L126C GM2AP constructs were purified as previously described.22 The substituted cysteine residue was modified with 4-maleimide-TEMPO (4MT) overnight in the dark (4 °C) in 25 mM Tris, 2.5% glycerol, and 0.05% Tween 20 buffer (pH 7.0). The spin-labeled L126C-4MT GM2AP construct was then dialyzed against 25 mM Tris, 2.5% glycerol, and 0.05% Tween 20 buffer (pH 8.0) and concentrated to 200 µL. Concentrated spin-labeled protein was loaded onto a size-exclusion S-200 Sephacryl column (Amhersham Biosciences, Piscataway, NJ) and eluted with 25 mM ammonium bicarbonate (pH 8.0) to remove the salts and detergents.
Sequential Digestion with Trypsin, GluC, and AspN
The solvent-exchanged sample was diluted to 50 pmol/µL (i.e., 50 µM), with a final volume of 160 µL. To that solution, 100 ng of trypsin (final substrate:trypsin ratio ~1000:1) was added and incubated overnight at 36 °C. Next, a 100 µL aliquot of the trypsin digest was incubated overnight with 200 ng of endoproteinase GluC (substrate:GluC, 400:1) at 36 °C. Finally, 50 µL of the trypsin/Glu C digested solution was incubated with 100 ng of endoproteinase AspN (substrate:AspN, 400:1). Each sample was diluted to 1 pmol/µL prior to LC MS analysis.
Direct Infusion and Reversed-Phase Capillary Liquid Chromatography
Direct infusion of the intact proteins was performed with an Advion BioSystems Nanomate (Ithaca, NY).23 The intact proteins were diluted to 1–5 pmol/µL in H2O/methanol/formic acid, 43/50/2 (v/v/v). Digests were separated by reversed-phase nano-liquid chromatography (10 µL sample) with a C18 capillary column (New Objective, Woburn, MA] 5 cm× 75 µm, 15 µm i.d spray tip). An Eksigent NanoLC (Dublin, CA) was used to deliver a 35 minute gradient (5 to 95% B) at 400 nL/min with solution A as 0.5% formic acid (v/v) in 5% aqueous methanol and solution B as 0.5% formic acid (v/v) in 95% aqueous methanol.
Mass Spectrometry
Mass spectrometry was performed with either a modified hybrid linear quadrupole ion trap FT-ICR mass spectrometer (LTQ-FT, Thermo Fisher Corp., Bremen, Germany) equipped with an actively shielded 14.5 T superconducting magnet (Magnex, Oxford, U.K.),21, 24, 25 or a custom-built 9.4 T FT-ICR MS.26–28 The protease-treated samples were screened by on-line LC/MS with the 14.5 T LTQ FT-ICR instrument operated in top-5 data-dependent mode (high resolution FT-ICR, low resolution LTQ CID MS/MS. For each precursor ion measurement, one million charges were accumulated in the LTQ prior to transfer (~1 ms transfer period) through three octopole ions guides (2.2 MHz, 250 Vp-p) to a capacitively coupled29 open cylindrical ICR cell for analysis. The robust automatic gain control24 for transferring the same number of ions for each FT-ICR MS scan yielded less than 0.500 ppm rms mass error with external calibration. Each of the five most abundant ions were collisionally dissociated in the LTQ for low resolution MS/MS (3 microscans, 10,000 target ions, 2.0 Da isolation width, 35 % relative collision energy, 0.250 activation q, 30 ms activation period, and dynamic exclusion list size of 60 for 1 minute). Data was collected with Xcalibur 2.0 software (Thermo Fisher).
Following the identification of disulfide-containing peptides from the first pass low resolution LTQ MS/MS analysis (as described in Data analysis and Informatics section), the samples were re-analyzed by nano-LC, selected-ion high resolution MS/MS to confirm charge state and fragment ion assignments. The second experiment is needed for verification of disulfide connectivity. The scan type for the high resolution MS/MS was set at 7 to 10 scans per segment. The first scan was the same as the precursor ion scan found in the top-5 data-dependent experiment. The next 2 to 9 scans were set to select relevant ions (disulfide connected peptide fragments) for high resolution MS/MS. The 2 or 3 most abundant charge states (for relevant disulfide connected proteolytic fragment targets) were selected for high resolution MS/MS analysis because different charge states are known to fragment differently with CID.30–32 A selected-ion high resolution MS/MS scan consisted of a target ion accumulation target of 50,000 ions with a 5 Da mass selection window, CID fragmentation in the LTQ (35.0 scaled collision energy, 0.250 activation q, and 30 ms activation period), and transfer to the cell for measurement. Low resolution MS/MS spectra exhibiting ambiguous fragment assignments were easily assigned by high resolution MS/MS based on charge state validation and mass errors less than 0.500 ppm. High resolution MS/MS data were collected with both Xcalibur 2.0 and MIDAS software.
Electron capture dissociation (ECD) was performed with a custom-built 9.4 T FT-ICR and is known to specifically break disulfide bonds more readily than the polypeptide backbone.33–35 Thus, disulfide bonds not verified by high resolution CID MS/MS were analyzed by ECD FT-ICR MS. Ions were selected with a quadrupole mass filter and accumulated in a second octopole for 1 second.36 The ions were then transferred through multiple ion guides and trapped in an open-ended cylindrical Penning trap by gated trapping for ECD analysis.37 Accurate mass and time-tag38 knowledge of disulfide peptides from the top-5 data-dependent analysis of the different digest facilitated the use of "peak parking" with the Eksigent nano-LC. Namely, at the elution time for the peptide of interest, the flow rate was reduced from 400 nL/min to 50 nL/min so that 25 ECD spectra could be signal-averaged. Data acquisition was performed by use of a Predator data station and data analysis by MIDAS 3.4 software.39
Data Analysis and Informatics
Peaks from the Xcaliber files were extracted with a customized peak picking algorithm, thresholded at 10% of the maximum peak magnitude for low resolution MS/MS and 1% for high resolution MS. The resulting files were searched with MASCOT (Matrix Science, Cambridge, UK) against a custom-created database containing the wild-type and L126C mutant sequences. To aid in identification of the 4MT labeled cysteine, the mass of the 4-MT label (252.14739) was added to the MASCOT modification database. Results from MASCOT were visualized and validated with Scaffold (Proteomics Software, Portland, OR) software. After identification of the 4MT labeled peptide, the remaining disulfide peptides from each digest were identified by comparing high-resolution MS precursor ion measurements to values calculated from the elemental composition of the assigned peptides. High-resolution MS/MS and ECD fragment ion assignments were performed by hand.
RESULTS AND DISCUSSION
Intact Protein Analysis
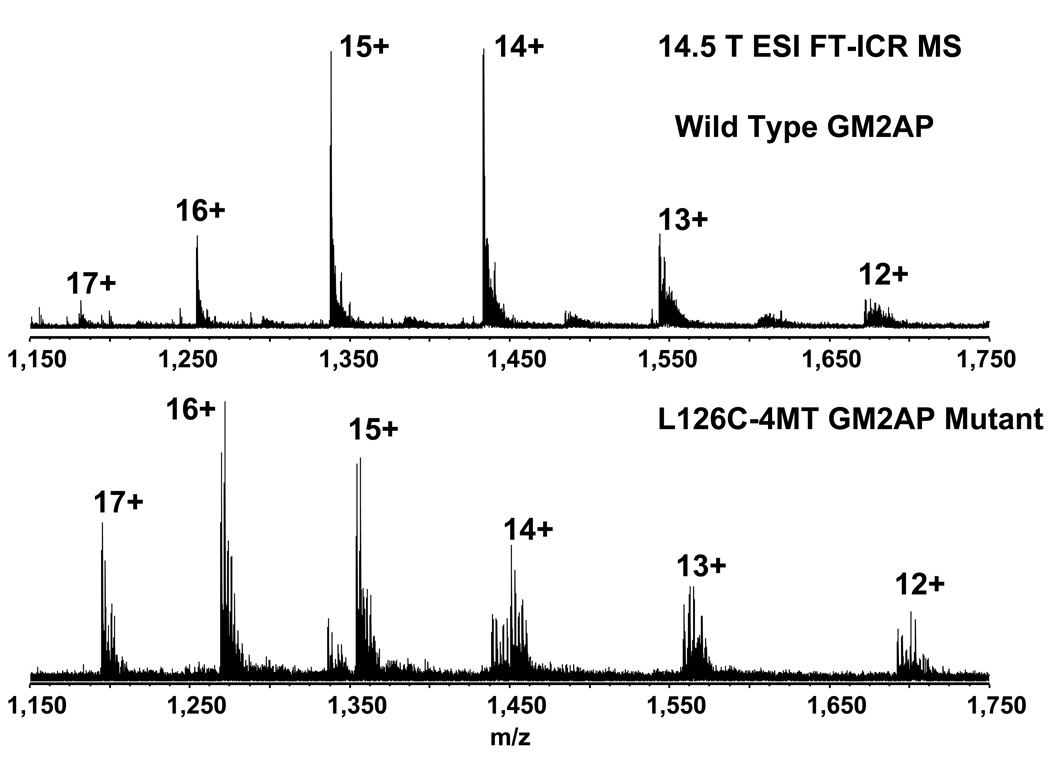
FT-ICR MS can resolve protein isotopic distributions40 and thus determine the charge state unambiguously based on the m/z separation between isotopic variants differing in elemental composition by 12Cn vs. 13C1 12Cn-1.41 Thus, before sequential proteolysis and LC MS analysis, the intact wild-type and L126C-4MT GM2AP were analyzed by direct infusion ESI FT-ICR MS (Figure 2). A 240 Da mass shift was observed between the two constructs, corresponding to the addition of one 4MT label to the substituted reporter cysteine (L126C). The spectra also show several salt and oxidation adducts, both common for intact protein analysis.
Figure 2.
Broadband 14.5 T electrospray positive-ion FT-ICR mass spectra of intact wild-type and L126C-4MT mutant GM2AP proteins. The L126C mutant was labeled with 4-maleimide-TEMPO. The shift in the m/z for the observed charge states indicates labeling of the protein.
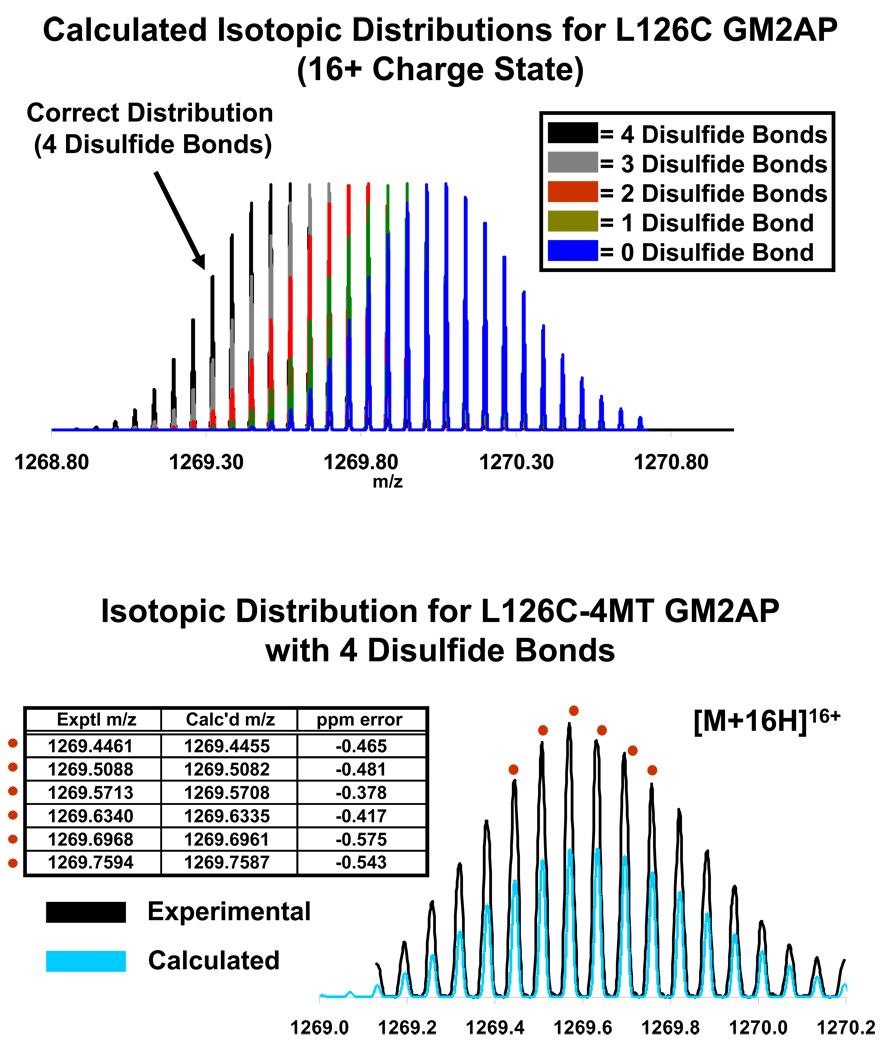
A second criterion for identification of the correct construct is presented in Figure 3, top. If there are less than four disulfide bonds, the 16+ charge state isotopic distribution, calculated from the molecular formula for the L126C-4MT GM2AP construct, will decrease by 0.12598 = (2)(1.007825)/16) for each addition of 2 hydrogens. Thus, a positive match between the calculated and experimental isotopic distributions precedes downstream analysis. For this case, Figure 2, bottom illustrates excellent agreement between the measured and experimental isotopic distributions for the 16+ charge state isotopic distribution of the L126C-4MT GM2AP construct. Also, the salt adducts and oxidized species match the correct isotopic distribution (data not shown). Thus, mass analysis of intact L126C-4MT GM2AP unambiguously confirms the presence of four disulfide bonds and a single 4MT modification, but not disulfide bonding arrangement.
Figure 3.
Top: Calculated isotopic distributions corresponding to 0, 1, 2, 3, and 4 disulfide bonds for L126C-4MT GM2AP [M+16H]16+. The calculated m/z distribution decreases by (2)(1.007825)/16 = 0.1260 for each additional disulfide linkage. Bottom: Calculated and experimental isotopic distributions for L126C-4MT GM2AP [M+16H]16+. The close fit of the calculated to the experimental distribution confirms that the protein has the correct amino acid sequence, with four disulfide bonds, and a single 4MT label on a cysteine residue. The five highest-magnitude in the isotopic distribution were chosen for comparison to calculated masses (red dots).
Tryptic Digest Peptide Analysis
Following tryptic digestion, the LC high-resolution precursor MS and low-resolution MS/MS data were searched with MASCOT against the known sequence of L126C GM2AP, allowing for the presence of 4MT. Low resolution MS/MS was chosen at first because of the slower duty cycle associated with high resolution MS/MS. Figure 4 (top) presents the sequence of L126C GM2AP, expected disulfide connectivity, location of the reporter cysteine, and sites of proteolytic cleavage for different proteases. MASCOT/Scaffold analysis identifies the tryptic digest fragments shown in blue in Figure 4, bottom), but cannot identify disulfide-containing peptides (red in Figure 4, bottom). MASCOT is typically used to analyze proteomic data sets in which the protein sample from cell lysate (including proteins containing disulfide bonds) has been reduced with dithiothreitol and alkylated with iodoacetamide. Thus, the peptides that contain cysteine will have reacted with iodoacetimide and can be identified with MASCOT. This experiment was designed to retain the disulfide connectivity. Our current version of MASCOT cannot identify peptides that are linked by disulfide bonds.
Figure 4.
Top: L126C GM2AP amino acid sequence with location of reported disulfide bonds and predicted cleavages for trypsin (blue: K,R), Glu C (dark green: E), and Asp N (orange: D). The location of the L126C amino acid substitution is shown in bright green. The HIS10-tag (light gray) is engineered to include a Factor Xa cleavage site. Bottom: Sequence coverage for the L126C-4MT GM2AP construct after trypsin digestion and nano-LC MS analysis. The blue sequence was identified by MASCOT; the red sequence was identified by hand, and corresponds to the peptides still containing disulfide bonds.
However, based on the known primary sequence, accurate mass measurement of the tryptic digest peptide precursor ions, and sequence tags from subsequent MS/MS, manual analysis revealed all of peptides containing one or more disulfide linkages, thus providing 100% sequence coverage. In addition, from Figure 4 (top), the connectivity for tryptic peptide Cys-1 (linking C8 to C152), containing a single disulfide bond (see Figure 5) could be uniquely determined (see below). Thus, from the predicted sequence and the results of the tryptic digest, the expected tryptic disulfide peptide sequences and masses could be generated. Because 100% sequence coverage identification was provided by trypsin digestion, other non-disulfide bound peptide fragments were not reported for the Trypsin/Glu C or Trypsin/Glu C/Asp N digestions. The need to verify the disulfide bound peptide fragments generated the need for the multiple digestion platform. Furthermore, high mass accuracy reduces the number of possible matches for a measured peptide to the possibilities found in a database (or protein) of known sequences. For example, if the tryptic peptide IESVLSSSGK, mass 1005.5342 Da, is searched against the possible peptide fragments from the sequence of GM2AP at 1, 20, and 2000 ppm mass error, the numbers of matched peptide sequences are 1, 4, and 7, respectively. Thus, high mass accuracy yields higher confidence in assignments by reducing the number of possible peptide matches.
Figure 5.
Scheme presenting the theoretical disulfide bound peptides after sequential proteolytic digestion. Various m/z values were calculated for charge states to aid during analysis and the selected ion monitoring MS/MS experiment. Other combinations were also calculated (not shown) but not identified.
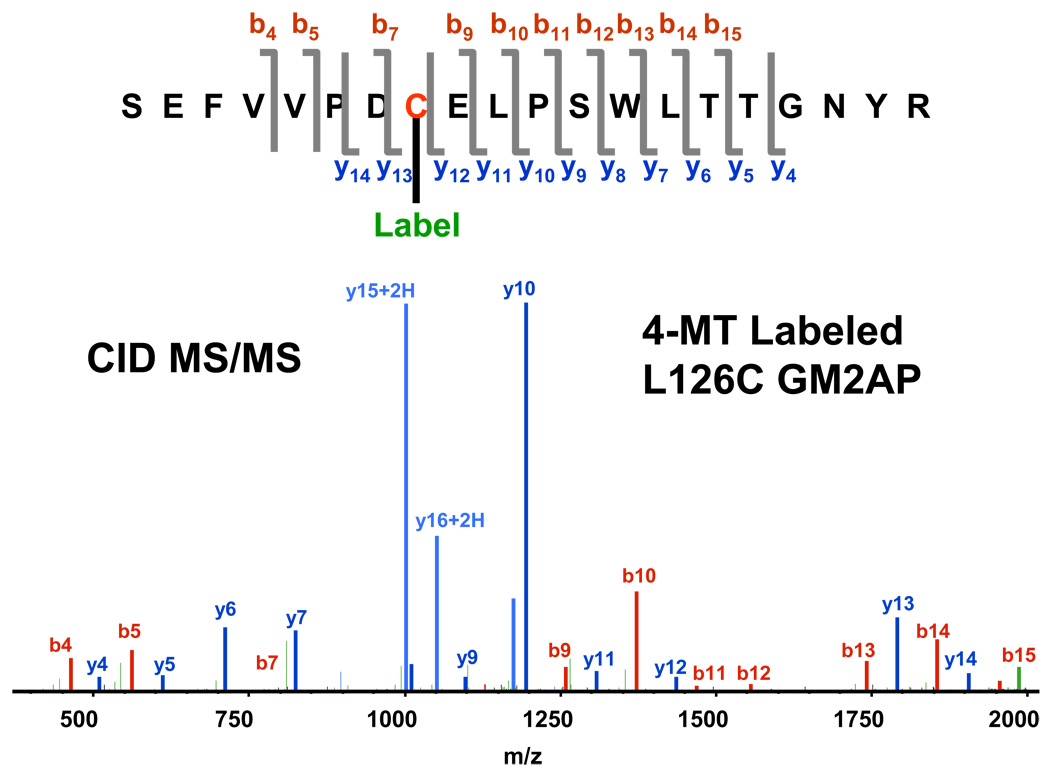
Location of Spin-Labeled Cysteine
Figure 6 shows the observed b- and y-ion series for the tryptic peptide containing the 4MT reporter cysteine. In contrast to other labile post-translational modifications,42–44 the present data illustrate that the 4MT thioester bond does not break during CID fragmentation, because the b9–15 and y13–14 ion series retain the mass associated with 4MT addition. Furthermore, if a population of the expressed protein were not properly folded, other non-reporter cysteine residues would be modified by the 4MT label. This is not the case because other 4MT modified non-reporter cysteine were not observed. Figure 6 thus establishes the sequence location of the 4MT spin-labeled cysteine.
Figure 6.
Identified fragments for low-resolution CID MS/MS of tryptic digested 4MT L126C reporter cysteine. The same 4MT labeled reporter cysteine was also observed from the other digests (data not shown).
Verification of Disulfide Bond Connectivity from Sequential Trypsin, Glu-C, and Asp-N Peptide Analysis
From the tryptic peptide assignments and the predicted subsequent Glu-C and AspN proteolytic cleavage sites, the remaining disulfide-connected peptides were generated and denoted as Cys-2a, Cys-2b, and Cys-2c (see Figure 5). Charge state m/z values are also calculated for nano-LC selected-ion high resolution MS/MS.
Figure 7 presents the fragment ions from the disulfide-containing peptides identified by high mass accuracy MS and CID MS/MS. From the trypsin digest, Cys-1 (Figure 7, top; Table 1) resulted in b- and y- ions that retained the mass increase associated with the second, shorter LGCIK sequence. High resolution MS/MS provided unambiguous assignment of the fragment ions for Cys-1; i.e., at low resolution/mass accuracy in the LTQ (close to unit resolution) many of the fragments had assignment ambiguities (data not shown). From the trypsin/Glu C digestion, Cys-2a was verified (Figure 7, bottom; Table 2) with a tripeptide sequence tag.45 The cysteine precursor ion mass measured to within 0.340 ppm mass error (Table 3) includes the loss of 2 hydrogens for the disulfide bond formed between C68 and C75.
Figure 7.
Disulfide bonds that were verified through CID MS/MS analysis. Top: High-resolution MS/MS of Cys-1. Several large peptide fragment masses verify the peptide sequence. Bottom: High-resolution MS/MS of Cys-2a (mass scale-expanded segment shown as an inset). The calculated mass for the intact peptide matches the experimental mass corresponding to one disulfide bond (Table 1). A sequence tag of three amino acids verifies the identity of the disulfide-containing peptide. As expected, the cyclic disulfide bond peptide does not yield fragments.
Table 1.
Peak list and mass errors from high-resolution CID MS/MS of Cys-1.
| Ion | Charge | Exptl m/z | Exptl Mass (Da) | Calc'd Mass (Da) | Error (ppm) |
|---|---|---|---|---|---|
| b8 | 1 | 978.3773 | 977.3700 | 977.3696 | 0.466 |
| b11 | 2 | 920.8761 | 1839.7377 | 1839.7383 | −0.348 |
| b11-H2O | 2 | 911.8707 | 1821.7269 | 1821.7277 | −0.478 |
| b12 | 2 | 985.3975 | 1968.7805 | 1968.7809 | −0.223 |
| b14 | 2 | 1077.9551 | 2153.8957 | 2153.8973 | −0.761 |
| b15 | 2 | 1135.4694 | 2268.9243 | 2268.9242 | 0.026 |
| b15 | 3 | 757.3150 | 2268.9232 | 2268.9242 | −0.447 |
| b15-H2O | 3 | 751.3115 | 2250.9127 | 2250.9136 | −0.420 |
| y5 | 1 | 555.3612 | 554.3539 | 554.3535 | 0.821 |
| y9 | 2 | 492.7772 | 983.5399 | 983.5395 | 0.386 |
| y9-H2O | 2 | 483.7719 | 965.5293 | 965.5295 | −0.212 |
| y12 | 2 | 923.9610 | 1845.9075 | 1845.9081 | −0.347 |
| y12 | 3 | 616.3098 | 1845.9076 | 1845.9081 | −0.279 |
| y13 | 2 | 981.4744 | 1960.9343 | 1960.9351 | −0.428 |
| y14 | 2 | 1074.5148 | 2147.0151 | 2147.0144 | 0.307 |
| y15 | 2 | 1118.0305 | 2234.0465 | 2234.0464 | 0.027 |
| y17 | 2 | 1235.0806 | 2468.1467 | 2468.1469 | −0.097 |
| y18 | 3 | 852.7333 | 2555.1781 | 2555.1789 | −0.319 |
| y19 | 3 | 896.4133 | 2686.2181 | 2686.2194 | −0.490 |
Table 2.
Peak list and mass errors from high-resolution CID MS/MS of Cys-2a.
| Ion | Charge | Exptl m/z | Exptl Mass (Da) | Calc'd Mass (Da) | Error (ppm) |
|---|---|---|---|---|---|
| M-2H2O | 2 | 705.7891 | 1409.5637 | 1409.5631 | 0.390 |
| b10 | 1 | 1051.4226 | 1050.4153 | 1050.4150 | 0.271 |
| b10-H2O | 1 | 1033.4120 | 1032.4047 | 1032.4044 | 0.286 |
| b11 | 1 | 1152.4700 | 1151.4627 | 1151.4627 | 0.004 |
| b11-H2O | 1 | 1134.4594 | 1133.4521 | 1133.4521 | 0.013 |
| b12 | 1 | 1299.5388 | 1298.5316 | 1298.5311 | 0.358 |
| b12-H2O | 1 | 1281.5282 | 1280.5209 | 1280.5205 | 0.308 |
Table 3.
Peak list and mass error for relevant peptides. The * indicates the spin-labeled L126C mutant peptide. Figure 4 lists the sequences for the different cysteine-containing peptides.
| Exptl m/z | Exptl Mass (Da) | Calc'd Mass (Da) | Error (ppm) | Peptide |
|---|---|---|---|---|
| 568.2611 | 2269.0151 | 2269.0148 | 0.138 | GHHHHHHHHHHSSGHIEGR |
| 670.3882 | 669.3810 | 669.3810 | 0.021 | DPAVIR |
| 1601.8937 | 3201.7728 | 3201.7747 | −0.596 | SLTLEPDPIVVPGNVTLSVVGSTSVPLSSPLK |
| 408.2472 | 814.4799 | 814.4800 | −0.110 | VDLVLEK |
| 458.2685 | 914.5224 | 914.5225 | −0.084 | EVAGLWIK |
| 447.7318 | 893.4490 | 893.4494 | −0.456 | EGTYSLPK |
| 855.7492 | 2564.2256 | 2564.2257 | −0.014 | SEFVVPDc*ELPSWLTTGNYR |
| 503.7744 | 1005.5342 | 1005.5342 | 0.015 | IESVLSSSGK |
| 706.8267 | 2823.2777 | 2823.2777 | 0.000 | Cys-1 |
| 845.8801 | 5069.2368 | 5069.2374 | −0.120 | Cys-2 |
| 723.7993 | 2562.2109 | 2562.2100 | 0.340 | Cys-2a |
| 911.4231 | 3641.6632 | 3641.6636 | −0.126 | Cys-2b |
| 869.3866 | 2605.1378 | 2605.1378 | −0.002 | Cys-2c |
As for Cys-2 and Cys-2b, high resolution CID MS/MS provided a sequence tag for identification, but did not reveal the disulfide bond connectivity. Cys-2 and Cys-2b are essentially "cyclic" peptides, and are thus expected to fragment poorly by low-energy CID.13, 14 Therefore, those peptides were fragmented by ECD. Both peptides resulted in only a charge-reduced species, not fragmentation (data not shown). The trypsin/Glu C sample was therefore digested with Asp N to release Cys-2c (Figure 5). Once again, high resolution MS/MS sequencing could not verify the final two disulfide bonds; however, peptide identity was confirmed. Figure 8 presents the spectrum after "peak parking" nano-LC ECD FT-ICR MS. Once peak parking is initiated, i.e., flow rate slowed from 200 nL/min to 50 nL/min, the separation will be less than optimal for the remainder of the analysis. However, the ECD experiment was designed for analysis of only Cys-2c, thus the loss in separation efficiency for the remainder of the gradient was irrelevant. ECD is known to specifically fragment disulfide bonds.33–35 In this case, ECD specifically broke the C94 – C105 disulfide bond to generate Frag2 (Figure 8) and EPC94PEPLR. ECD analysis thus provided the final piece of evidence to verify the last two disulfide bonds.
Figure 8.
"Peak-parked nano-LC" high-resolution ECD MS/MS (25 scans, 3 min). ECD typically breaks disulfide bonds prior to amino acid backbone fragmentation. Two fragments corresponding to the specific fragmentation of the C94-C105 disulfide bond were observed. The table lists the mass accuracy for ECD analysis.
Finally, Table 3 summarizes the tryptic peptides identified by MASCOT, along with the manually identified disulfide bonds. The data acquired from all three sequential digests by accurate mass measurement MS and MS/MS sequence tags resulted in quick identification of all four disulfide peptides as well as the sequence location of the 4MT-Cys.
CONCLUSIONS
A multi-tiered assay successfully validates protein folding for GM2AP mutants by use of conventional proteomic techniques and sequential digestion with a series of different proteases. Although sequential proteolysis digestion increased the compositional complexity of the sample (i.e., number of peptide fragments), the length of individual disulfide-containing peptides was reduced, thereby simplifying interpretation. The modified 14.5 T LTQ-FT ICR MS yielded mass errors of less than 1 ppm with automatic gain control (AGC). For external calibration coefficients to be applied on an LC time scale, the same number of ions must be transferred to the cell for each measurement.
High resolution MS combined with low-resolution MS/MS provides a quick screen for identification of the spin-labeled mutant peptide, protein sequence coverage, and disulfide bound peptide fragments. High resolution CID MS/MS allowed for validation of expected disulfide links. In cases for which high resolution CID MS/MS did not establish disulfide connectivity, ECD with the 9.4 T FT-ICR MS provided the necessary evidence. Finally, the present techniques can be easily applied to other GM2AP constructs produced for EPR studies, and more generally to other proteins with natural or artificial post-translational modifications and multiple disulfide linkages.
ACKNOWLEDGMENT
This work was supported by NIH (GM-78359), NIH (R01GM077232), NSF Division of Materials Research through DMR-0654118, and the State of Florida
Footnotes
Abbreviations: FT-ICR, Fourier transform ion cyclotron resonance; MS, mass spectrometry; LC, liquid chromatograph; ESI, electrospray ionization; LTQ, linear quadrupole ion trap; GM2AP, GM2 activator protein; CID, collision-induced dissociation; ECD, electron capture dissociation
REFERENCES
- 1.Meier EM, Schwarzmann G, Furst W, Sandhoff K. J. Biol. Chem. 1991;266:1879–1887. [PubMed] [Google Scholar]
- 2.Mchaourab HS, Lietzow MA, Hideg H, Hubbell WL. Biochemistry. 1996;35:7692–7704. doi: 10.1021/bi960482k. [DOI] [PubMed] [Google Scholar]
- 3.Bertoni C, Li YT, Li SC. J. Biol. Chem. 1999;274:28612–28618. doi: 10.1074/jbc.274.40.28612. [DOI] [PubMed] [Google Scholar]
- 4.Kolter T, Sandhoff K. J. Inherited Metabolic Disease. 1998;21:548–563. doi: 10.1023/a:1005419122018. [DOI] [PubMed] [Google Scholar]
- 5.Huang JQ, Trasler JM, Igdoura S, Michaud J, Hanal N, Gravel RA. Human Molecular Genetics. 1997;6:1879–1885. doi: 10.1093/hmg/6.11.1879. [DOI] [PubMed] [Google Scholar]
- 6.Wright CS, Li SC, Rastinejad F. J. Mol. Biol. 2000;304:411–422. doi: 10.1006/jmbi.2000.4225. [DOI] [PubMed] [Google Scholar]
- 7.Columbus L, Hubbell WL. Biochemistry. 2004;43:7273–7287. doi: 10.1021/bi0497906. [DOI] [PubMed] [Google Scholar]
- 8.Bordignon E, Klare JP, Doebber M, Wegener AA, Martell S, Engelhard M, Steinhoff HJ. J. Biol. Chem. 2005;280:38767–38775. doi: 10.1074/jbc.M509391200. [DOI] [PubMed] [Google Scholar]
- 9.Hubbell W, Cafiso D, Altenbach C. Nature Struct. Mol. Biol. 2000;7:735–739. doi: 10.1038/78956. [DOI] [PubMed] [Google Scholar]
- 10.Fanucci GE, Cafiso DS. Curr. Opin. Struct. Biol. 2006;16:644–653. doi: 10.1016/j.sbi.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 11.Mathias JD, Ran Y, Carter JD, Fanucci GE. Biophysical Journal. 2009;77 doi: 10.1016/j.bpj.2009.05.058. x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lundblad RL. Chemical Reagents for Protein Modification. 3 ed. CRC Press; 2004. [Google Scholar]
- 13.Bean MF, Carr SA. Anal. Biochem. 1992;201:216–226. doi: 10.1016/0003-2697(92)90331-z. [DOI] [PubMed] [Google Scholar]
- 14.Badock V, Raida M, Adermann K, Forssmann WG, Schrader M. Rapid Communications in Mass spectrometry. 1998;12:1952–1956. doi: 10.1002/(SICI)1097-0231(19981215)12:23<1952::AID-RCM420>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 15.Gorman JJ, Wallis TP, Pitt JJ. Mass Spectrom. Rev. 2002;21:183–216. doi: 10.1002/mas.10025. [DOI] [PubMed] [Google Scholar]
- 16.Xu H, Zhang L, Freitas MA. J. Proteome Res. 2008;138–144:1. doi: 10.1021/pr070363z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yen TY, Joshi RK, Yan H, Seto NOL, Palcic MM, Macher BA. J. Mass Spectrom. 2000;35:990–1002. doi: 10.1002/1096-9888(200008)35:8<990::AID-JMS27>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 18.Yen TY, Yan H, Macher BA. J. Mass Spectrom. 2002;31:15–30. doi: 10.1002/jms.241. [DOI] [PubMed] [Google Scholar]
- 19.Nair SS, Nilsson CL, Emmett MR, Schaub TM, Gowd KH, Thakur SS, Krishnan KS, Balaram P, Marshall AG. Anal. Chem. 2006;78:8082–8088. doi: 10.1021/ac0607764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Popolo L, Ragni E, Carotti C, Palomares O, Aardema R, Back JW, Dekker HL, de Koning LJ, de Jong L, de Koster CG. J. Biol. Chem. 2008;283:18553–18565. doi: 10.1074/jbc.M801562200. [DOI] [PubMed] [Google Scholar]
- 21.Schaub TM, Hendrickson CL, Horning S, Quinn JP, Senko MW, Marshall AG. Anal. Chem. 2008;80:3985–3990. doi: 10.1021/ac800386h. [DOI] [PubMed] [Google Scholar]
- 22.Ran Y, Fanucci GE. Anal. Biochem. 2008;382:132–134. doi: 10.1016/j.ab.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang S, Van Pelt CK, Henion JD. Electrophoresis. 2003;24:3620–3632. doi: 10.1002/elps.200305585. [DOI] [PubMed] [Google Scholar]
- 24.Schwartz JC, Senko MW, Syka JEP. J. Am. Soc. Mass Spectrom. 2002;13:659–669. doi: 10.1016/S1044-0305(02)00384-7. [DOI] [PubMed] [Google Scholar]
- 25.Syka JEP, Marto JA, Bai DL, S H, Senko MW, Schwartz JC, Ueberheide B, Garcia B, Busby S, Muratore T, Shabanowitz J, Hunt DF. J. Proteome Red. 2004;3:621–626. doi: 10.1021/pr0499794. [DOI] [PubMed] [Google Scholar]
- 26.Senko MW, Hendrickson CL, Pasa-Tolic L, Marto JA, White FM, Guan S, Marshall AG. Rapid Commun. Mass Spectrom. 1996;10:1824–1828. doi: 10.1002/(SICI)1097-0231(199611)10:14<1824::AID-RCM695>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 27.Håkansson K, Chalmers MJ, Quinn JP, McFarland MA, Hendrickson CL, Marshall AG. Anal. Chem. 2003;13:3256–3262. doi: 10.1021/ac030015q. [DOI] [PubMed] [Google Scholar]
- 28.Marshall AG, Guan S. Rapid Commun. Mass Spectrom. 1996;10:1829–1832. doi: 10.1002/(SICI)1097-0231(199611)10:14<1829::AID-RCM697>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 29.Beu SC, Laude DA. Anal. Chem. 1992;64:177–180. [Google Scholar]
- 30.Tang XJ, Thibault P, Boyd RK. Anal. Chem. 1993;65:2824–2834. doi: 10.1021/ac00068a020. [DOI] [PubMed] [Google Scholar]
- 31.Griffiths WJ, Jonsson AP. Proteomics. 2001;8:934–945. doi: 10.1002/1615-9861(200108)1:8<934::AID-PROT934>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 32.Tsapraili G, Nair H, Somogyi A, Wysocki VH, Zhong W, Futrell JH, Summerfield SG, Gaskell SJ. J. Am. Chem. Soc. 1999;121:5142–5154. [Google Scholar]
- 33.Zubarev RA, Krunger NA, Fridriksson EK, Lewis M, AH DM, Carpenter BK. Journal of the American Chemical Society. 1999;121:2857–2862. [Google Scholar]
- 34.Zubarev RA. Current Opinion in Biotechnol. 2004;15:12–16. doi: 10.1016/j.copbio.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 35.Uggerud E. Int. J. Mass Spectrom. 2004;234:45–50. [Google Scholar]
- 36.Senko MW, Hendrickson CL, Emmett MR, Shi SD, Marshall AG. J. Am. Soc. Mass Spectrom. 1997;8:970–976. [Google Scholar]
- 37.Tsybin Yl, Hendrickson CL, Beu SC, Marshall AG. Int. J. Mass Spectrom. 2006:144–149. [Google Scholar]
- 38.Pasa-Tolic L, Masselon CD, Barry RC, Shen Y, Smith RD. BioTechniques. 2004;37:621–636. doi: 10.2144/04374RV01. [DOI] [PubMed] [Google Scholar]
- 39.Blakney GT, Robinson DE, Ly NV, Kelleher NL, Hendrickson CL, Marshall AG. 53rd Amer. Soc. Mass Spectrom. Ann. Conf. on Mass Spectrom. & Allied Topics; June 4–9, 2005; San Antonio, TX. [Google Scholar]
- 40.Hendrickson CL, Emmett MR. Ann. Rev. Phys. Chem. 1990;50:517–513. doi: 10.1146/annurev.physchem.50.1.517. [DOI] [PubMed] [Google Scholar]
- 41.Senko MW, Beu SC, McLafferty FW. J. Am. Soc. Mass Spectrom. 1995;6:52–56. doi: 10.1016/1044-0305(94)00091-D. [DOI] [PubMed] [Google Scholar]
- 42.Cooper HJ, Håkansson K, Marshall AG. Mass Spectrom. Rev. 2005;24:201–222. doi: 10.1002/mas.20014. [DOI] [PubMed] [Google Scholar]
- 43.Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villén J, Li J, Cohn MA, Cantley LC, Gygi SP. Proc. Natl. Acad. Sci., U.S.A. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mørtz E, O'Connor PB, Roepstorff P, Kelleher NL, Wood TD, McLafferty FW, Mann M. Proc. Natl. Acad. Sci. U.S.A. 1996;93:8264–8267. doi: 10.1073/pnas.93.16.8264. [DOI] [PMC free article] [PubMed] [Google Scholar]