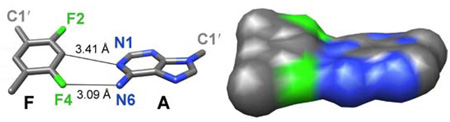
Abstract
Certain DNA polymerases were found to efficiently insert A opposite the hydrophobic T isostere 2,4-difluorotoluene (F) and vice versa, resulting in the widely held belief that some pols rely on shape rather than H-bonding for accurate replication. Using X-ray crystallography we have analyzed the geometry of F:A pairs in duplex DNA and observed a distance between fluorine and the exocyclic amino group of A that is consistent with a H-bond, thus challenging the assumption that the F analog is unable to engage in H-bonding as well as the steric hypothesis of DNA replication. Therefore, shape and H-bonding are inherently related and steric constraints at a pol active site, or conferred by stacking or the DNA backbone conformation may enable H-bonding by F.
The 2′-deoxyribo-2,4-difluorotoluene nucleoside analog (dF, Figure 1) was created as an isostere of 2′-deoxythymidine (dT) to investigate the role of Watson-Crick hydrogen bonds (W-C H-bonds) in DNA duplex stability and the fidelity of replication by DNA polymerases (pols).1 Despite strengthening stacking significantly relative to dT,2 incorporation of dF leads to a net destabilization of the duplex (ΔΔTm = −14°C and ΔΔG = −3.5 kcalmol−1 between the F:A-and T:A-containing DNAs).3 Molecular dynamics (MD) simulations indicated an increased local flexibility at sites of dF incorporation,4 although an initial NMR solution structure of a DNA duplex containing a single F:A pair provided support for similar shapes of the F:A and T:A pairs and limited conformational perturbations of the helical geometry.3
Figure 1.
Structures of dT, dF and dA and a hypothetical dF:dA pair. Putative H-bonds are dashed lines; arrows designate distances d1 [F4 (dF)…N6(dA)], d2 [C3(dF)…N1(dA)] and d3 [F2(dF)…C2(dA)].
DNA pol I Klenow fragment (Kf exo-) inserts dATP opposite template dF with surprisingly high efficiency (Vmax/Km reduced 40-fold) and fidelity compared with template dT.5 However, incorporation of dFTP opposite template dA by the same pol is inhibited more significantly relative to dTTP by comparison (>500-fold reduction in efficiency),6 likely due to some extent to different syn/anti and sugar conformational equilibria between dF and dT. Extensive studies of the kinetics of both replicative7 and lesion bypass (Y-class) DNA pols,8 involving among other hydrophobic analogs dF, have led to the steric hypothesis of DNA replication, i.e. that particularly the former class of pols appears to rely on shape rather than W-C H-bonding for accurate replication (reviewed in ref. 1c). However, by determining crystal structures of the Y-class DNA pol Dpo4 from S. solfataricus in complex with DNA duplexes containing dF in the template strand, we recently found that the shapes of F:A and F:G pairs at the pol active site differ significantly from those of the canonical T:A and wobble T:G pairs.9 The steric hypothesis of replication hinges on the assumption that dF lacks the ability to form H-bonds. This is indeed supported by semi-empirical calculations that indicated distances (d1 and d2, Figure 1) between F and A that were increased by between 0.5 and 0.7 Å relative to the T:A pair.10 However, to date no accurate experimental model of the F:A pair in a DNA duplex environment has been presented.
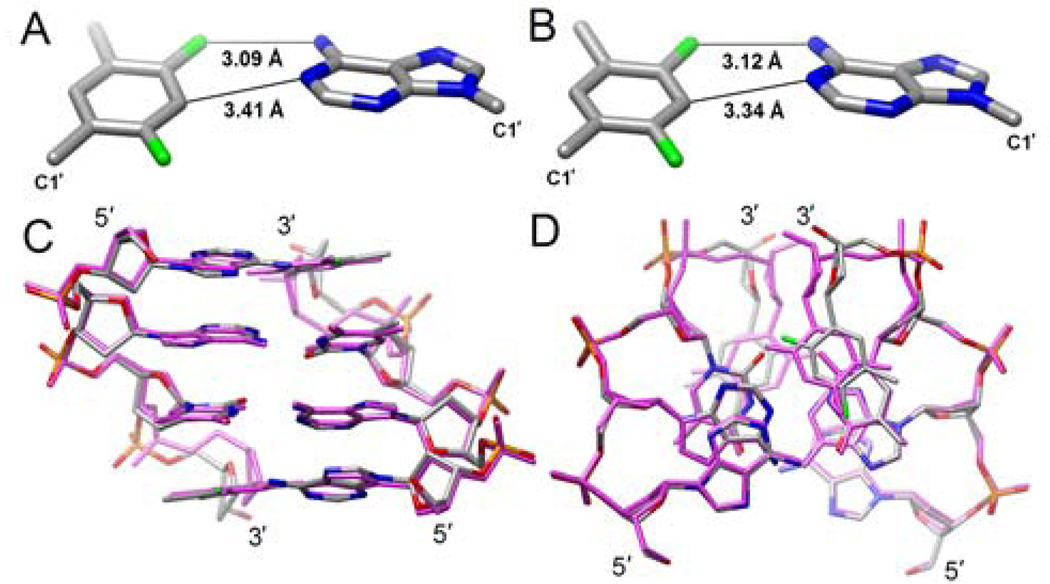
We have determined the crystal structure of a Dickerson-Drew Dodecamer (DDD) DNA duplex with a single F nucleotide [d(CGCGAATFCGCG)]2 bound to Bacillus halodurans ribonuclease H (BhRNase H) at 1.6 Å resolution. For experimental procedures, selected crystal and refinement parameters (Table S1) and the quality of the final electron density (Figure S1), please see the si file. Although crystals of the modified DDD alone could not be grown, protein-DNA contacts in the complex are limited to the CG portion and the conformation of the central tetramer including F:A pairs is unlikely to be distorted as a result of protein binding (Figure S2). Like the structure of the complex with the native DDD,11 the asymmetric unit of the complex with the F-modified DDD contains two independent 12mer strands (both duplexes are located on a dyad) and RNase H molecules. This allowed us to analyze the geometry of two independent F:A pairs and compare them with the geometries of the corresponding T:A pairs in the native complex as well as in crystal structures at high-resolution of the DDD alone (Table 1).12
Table 1.
Watson-Crick H-bond distances d1 and d2 and distance d3 (O2/F2…C2) for T:A and F:A pairs in crystal structures of B-form DNA.
| B-form DNA duplex |
Base pair |
d1 [Å] |
d2 [Å] |
d3 [Å] |
Resol. [Å] |
PDB code |
Ref. |
|---|---|---|---|---|---|---|---|
| Native DDDa | A5:T20 T8:A17 |
2.98 2.96 |
2.77 2.82 |
3.54 3.62 |
1.1 | 436D | 12a |
| Native DDDa | A5:T20 T8:A17 |
3.01 3.11 |
2.83 2.77 |
3.55 3.46 |
1.4 | 355D | 12b |
| Native DDD : BhRNase Hb |
F8:A17 F8:A17 |
2.95 2.99 |
2.81 2.76 |
3.57 3.47 |
1.8 | 3D0P | 11 |
| F8-DDD : BhRNase Hb |
F8:A17 F8:A17 |
3.09 3.12 |
3.41 3.34 |
4.40 4.34 |
1.6 | 3I8D | this work |
The duplex [d(CGCGAATTCGCG)]2 (nucleotides in one strand are numbered 1–12 and 13–24 in the other) lies in a general position, and the A5:T20 and T8:A17 base pairs (bold font) exhibit different geometries.
The first base pair is from duplex 1 and the second from duplex 2. Both duplexes sit on a dyad and F8:A17 and A5:F20 are symmetry-equivalent.
As expected, due to replacement of N3 in T by C3 in F (Figure 1), d2 is increased in the F:A relative to the T:A pair (by about 0.5 to 0.6 Å). However, at 3.34 Å as seen in the F:A pair of duplex 2, the separation of C3(F) and N1(A) is still below the sum of the van der Waals (vdW) radii (ca. 3.7 Å). Surprisingly, d1 is not significantly longer in F:A than in T:A (Figure 2AB, Table 1), and well below the distance consistent with a vdW contact of around 3.55 Å (assuming a radius of 1.35 Å for F). Even if we consider a vdW radius of F that equals that of H (1.2 Å),13 the observed separations between F4 and NH2 are still suggestive of an attractive interaction. At the other edge of the base pair, in the minor groove, d3 is increased by about 1 Å in F:A relative to T:A due to opening (Table 1).
Figure 2.
Geometries of the F:A pairs in (A) duplex 1 and (B) in duplex 2 with d1 and d2 indicated. Conformational consequences at the duplex level due to replacement of T with F. Superimposition of the central tetramer duplexes (AATF):(AATF) and (AATT):(AATT) from the crystal structures of the modified and native DDD in complex with BhRNase H, resp., viewed (C) along the dyad and into the major groove, and (D) rotated around the horizontal by 90° and viewed along the helical axis. Duplex DNA atoms are colored gray (C; pink, native DDD), red (O), blue (N), orange (P) and green (F).
Comparison between the conformations of the native and F-modified duplexes in the structures of the complexes reveals only deviations near the sites of modifications (Figure 2CD). As a result of the longer d2 distance in F:A relative to T:A, the former base pair is stretched. This pushes the backbones outwards, and goes along with changes in the 10 to 20° range in backbone torsion angles ε and α of F and the preceding T. In addition to stretching, F:A pairs exhibit increased stagger (a shift of F and A relative to one other along the helical axis) and the aforementioned opening. However, propeller twisting that is quite pronounced in T:A pairs does not appear to be increased in F:A. Similarly, local helical twist and rise are virtually unaffected by the replacement of T with F (Figure 2).
In crystal structures of B-form DNA, the H-bond distance between O4 of thymine and N6 of adenine (d1) is typically longer than that between the N3(T) and N1(A) atoms12 (d2, 3.04 ± 0.17 Å vs. 2.83 ± 0.13 Å, respectively,14 Table 1) and computational simulations paint a similar picture.14 This means that the loss of the (T)N3-H…N1(A) H-bond in the F:A pair results in a considerable loss of stability relative to T:A, even if we attribute a minor stabilizing effect to the C3-H…N1 contact in F:A. On the other hand, there is a surprisingly small difference between the lengths of d1 in the T:A and F:A pairs and it is reasonable to postulate formation of a H-bond between F4 and N6 based on our structural data. Although we don’t observe the positions of hydrogen atoms at 1.6 Å resolution, the distance between the calculated position of the N6(A) hydrogen atom and F4 amounts to ca. 2.1 Å and is thus comparable to the shortest distances found between fluorine and N-H donors in the crystal structures of small molecules.15 The pairing mode seen here between dF and dA in B-DNA is much tighter than that between rF and rA previously analyzed in the crystal structure of an RNA duplex,16 but comparable to the pairing of rF and rG that was consistent with H-bond formation between F2(rF) and N1(rG) (min. dist. = 3.03 Å).17
Our structural data at high resolution contradict the earlier assumption that the pairing of F and A does not involve H-bonding (i.e. refs. 1,10). But the structural data accumulated to date also indicate a considerable plasticity of the F:A pair, with different geometries observed in DNA here, RNA,16,17 and at the post-replicative site of a Y-class DNA pol.9 As far as the steric hypothesis and the reliance on shape rather than H-bonding by certain DNA pols for accurate replication are concerned, a more complicated picture is emerging. Shape and H-bonding cannot be separated readily and steric constraints such as backbone geometry, stacking or enzyme active sites should be considered enablers of H-bonding, in line with earlier theoretical work by Guerra and Bickelhaupt.18 The finding here that dF engages in a H-bond to dA also raises the possibility that some DNA pols may probe the minor groove of dF:dATP or dA:dFTP pairs at the active site with H-bonds.
Supplementary Material
Acknowlegdment
This work was supported by NIH grant R01 GM55237. Vanderbilt University is a member institution of LS-CAT at the Advanced Photon Source (Argonne, IL). Use of the APS was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under Contract No. W-31-109-Eng-38.
Footnotes
Supporting Information Available: Experimental procedures and Figure S1 and S2 and Table S1. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.(a) Schweitzer BA, Kool ET. J. Org. Chem. 1994;59:7238–7242. doi: 10.1021/jo00103a013. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Kool ET. Annu. Rev. Biochem. 2002;71:191–219. doi: 10.1146/annurev.biochem.71.110601.135453. [DOI] [PubMed] [Google Scholar]; (c) Kool ET, Sintim HO. Chem. Comm. 2006:3665–3675. doi: 10.1039/b605414e. [DOI] [PubMed] [Google Scholar]
- 2.Guckian KM, Schweitzer BA, Ren RX, Sheils C, Paris PL, Tahmassebi DC, Kool ET. J. Am. Chem. Soc. 1996;118:8182–8183. doi: 10.1021/ja961733f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guckian KM, Krugh TR, Kool ET. Nat. Struct. Biol. 1998;5:954–959. doi: 10.1038/2930. [DOI] [PubMed] [Google Scholar]
- 4.Cubero E, Laughton CA, Luque FJ, Orozco M. J. Am. Chem. Soc. 2000;122:6891–6899. [Google Scholar]
- 5.Moran S, Ren RX-F, Rumney S, IV, Kool ET. J. Am. Chem. Soc. 1997;119:2056–2057. doi: 10.1021/ja963718g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.(a) Moran S, Ren RX, Kool ET. Proc. Natl. Acad. Sci. USA. 1997;94:10506–10511. doi: 10.1073/pnas.94.20.10506. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Morales JC, Kool ET. Nat. Struct. Biol. 1998;5:950–954. doi: 10.1038/2925. [DOI] [PubMed] [Google Scholar]
- 7.(a) Kim TW, Delaney JC, Essigmann JM, Kool ET. Proc. Natl. Acad. Sci. USA. 2005;102:15803–15808. doi: 10.1073/pnas.0505113102. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Potapova O, Chan C, DeLucia AM, Helquist SA, Kool ET, Grindley NDF, Joyce CM. Biochem. 2006;45:890–898. doi: 10.1021/bi051792i. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Kim TW, Brieba LG, Ellenberger T, Kool ET. J. Biol. Chem. 2006;281:2289–2295. doi: 10.1074/jbc.M510744200. [DOI] [PubMed] [Google Scholar]
- 8.(a) Washington MT, Helquist SA, Kool ET, Prakash L, Prakash S. Mol. Cell. Biol. 2003;23:5107–5112. doi: 10.1128/MCB.23.14.5107-5112.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Wolfle WT, Washington MT, Kool ET, Spratt TE, Helquist SA, Prakash L, Prakash S. Mol. Cell. Biol. 2005;25:7137–7143. doi: 10.1128/MCB.25.16.7137-7143.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Mizukami S, Kim TW, Helquist SA, Kool ET. Biochem. 2006;45:2772–2778. doi: 10.1021/bi051961z. [DOI] [PubMed] [Google Scholar]
- 9.Irimia A, Eoff RL, Pallan PS, Guengerich FP, Egli M. J. Biol. Chem. 2007;282:36421–36433. doi: 10.1074/jbc.M707267200. [DOI] [PubMed] [Google Scholar]
- 10.(a) Santhosh C, Mishra PC. Int. J. Quantum Chem. 1998;68:351–355. [Google Scholar]; (b) Wang X, Houk KN. Chem. Comm. 1998:2631–2632. [Google Scholar]
- 11.Pallan PS, Egli M. Cell Cycle. 2008;7:2562–2569. doi: 10.4161/cc.7.16.6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.(a) Tereshko V, Minasov G, Egli M. J. Am. Chem. Soc. 1999;121:470–471. [Google Scholar]; (b) Shui X, McFail-Isom L, Hu GG, Williams LD. Biochem. 1998;37:8341–8355. doi: 10.1021/bi973073c. [DOI] [PubMed] [Google Scholar]
- 13.Dunitz JD, Schweizer WB. Chem. Europ. J. 2006;12:6804–6815. doi: 10.1002/chem.200600092. [DOI] [PubMed] [Google Scholar]
- 14.Srinivasan AR, Sauers RR, Fenley MO, Boschitsch AH, Matsumoto A, Colasanti AV, Olson WK. Biophys. Rev. 2009;1:13–20. doi: 10.1007/s12551-008-0003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.(a) Howard JAK, Hoy VJ, O’Hagan D, Smith GT. Tetrahedron. 1996;52:12613–12622. [Google Scholar]; (b) Dunitz JD. ChemBioChem. 2004;5:614–621. doi: 10.1002/cbic.200300801. [DOI] [PubMed] [Google Scholar]
- 16.Xia J, Noronha A, Li F, Rajeev KG, Akinc A, Braich R, Egli M, Manoharan M. Am. Chem. Soc. Chem. Biol. 2006;1:176–183. doi: 10.1021/cb600063p. [DOI] [PubMed] [Google Scholar]
- 17.Li F, Pallan PS, Maier MA, Rajeev KG, Mathieu SL, Kreutz C, Fan Y, Sanghvi J, Micura R, Rozners E, Manoharan M, Egli M. Nucleic Acids Res. 2007;35:6424–6438. doi: 10.1093/nar/gkm664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guerra CF, Bickelhaupt FM. Angew. Chem. Int. Ed. 2002;41:2092–2095. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.