Abstract
Copper plays an essential role in human physiology. It is required for respiration, radical defense, neuronal myelination, angiogenesis, and many other processes. Copper has distinct physicochemical properties that pose uncommon challenges for its transport across biological membranes. Only small amounts of copper are present in biological fluids, and essentially none of it exists in a free ion form. These properties and the low redox potential of copper dictate special structural and mechanistic features in copper transporters. This minireview discusses molecular mechanisms through which copper enters and exits human cells.
Dietary Acquisition and Excretion of Copper
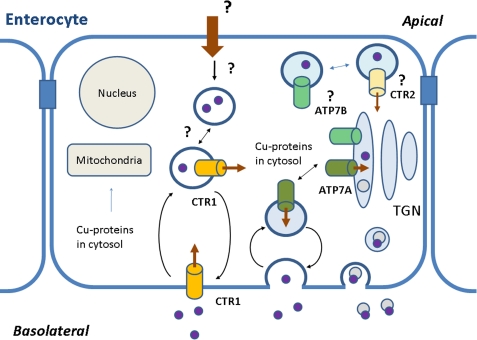
Dietary copper is acquired via the small intestine through a process that is not fully understood. Earlier studies showing the major involvement of the mammalian transporter CTR1 in cellular copper uptake (1) led to the assumption that in enterocytes CTR1 mediates the acquisition of dietary copper at the apical membrane. Recent studies questioned this assumption. Cell-surface labeling revealed that the majority (and perhaps all) of surface hCTR13 in Caco-2 cells, a model for enterocytes, is located at the basolateral border, i.e. the blood side, and thus cannot mediate apical copper entry (2). Basolateral CTR1 is also observed in renal cells and hepatocytes. Furthermore, in mice with an intestine-specific deletion of CTR1, enterocyte copper levels are higher (and not lower) than in normal mice, arguing against the direct role of CTR1 in apical uptake (3). It seems most likely that in enterocytes some other transporter plays a major role in copper uptake from the fluid that exits the stomach (Fig. 1). The existence of such pathways is supported by the observation that fibroblasts from CTR1 knock-out mice retain ∼25–30% of their copper uptake (4). Divalent metal transporter DMT1 was suggested as a candidate for copper uptake (5). Other systems not thought of as major copper transporters may accept copper as a surrogate substrate and contribute to the overall copper absorption. Endocytic mechanisms may also contribute to apical copper entry. It is not yet clear how the copper pool from apical dietary uptake in enterocytes interacts with the copper that enters from the blood (Fig. 1).
FIGURE 1.
Copper transport in enterocytes. Copper is shown to enter the enterocyte from the blood via hCTR1 and from the intestine through an unidentified mechanism. Copper then binds to copper chaperones, which deliver it to the target proteins, including ATP7A and ATP7B. CTR1 and CTR2 may also be involved in copper release from the intracellular vesicles. TGN, trans-Golgi network.
In intestinal cells and tissue, CTR1 was detected at the plasma membrane and in vesicles (2, 6). Thus, CTR1 in enterocytes may take up copper from the blood (as in other cell types) and also from the vesicles in which dietary copper has been sequestered (Fig. 1). This latter role is suggested by the finding that, although the intestinal deletion of CTR1 results in high copper levels in enterocytes, this copper is not available (3). It is tempting to speculate that CTR1 is uniquely positioned to make copper bioavailable either because it has a specific recognition mechanism and/or it is directly coupled to the copper chaperones that distribute copper for further utilization.
After copper crosses the membrane, it is delivered to acceptor proteins by cytosolic metallochaperones. The Cu-ATPase ATP7A accepts copper from Atox1 and facilitates copper export from enterocytes to blood (and hence to other tissues). Mutations of ATP7A (in Menkes disease patients) result in intestinal accumulation of copper, impaired efflux of copper from enterocytes, and pathologies due to diminished activity of copper-dependent enzymes. Recently, the second Cu-ATPase, ATP7B, was detected in mouse intestine and in Caco-2 cells (7). The role of ATP7B in the intestine is unclear (Fig. 1); however, based on the vesicular localization in high copper, ATP7B is thought to play a role in copper sequestration (7), similar to its proposed role in kidneys (8).
The major homeostatic control of copper takes place in the liver. Copper enters the liver from the blood via CTR1; the liver-specific inactivation of CTR1 disrupts copper uptake into the liver (9). Copper is then used in the liver for many purposes, one of which is to synthesize holo-ceruloplasmin, an abundant secreted copper ferroxidase. Excess copper is exported from hepatocytes by ATP7B. The ATPase-driven copper export is unlikely to be mediated directly across the plasma membrane; rather, Cu-ATPases sequester copper into vesicles that subsequently fuse with the plasma membrane, and copper is extruded.
Special Characteristics of Copper Transport
Essentially all intracellular copper is bound to cellular constituents. Hence, the definition of cellular copper concentration (rather than content) is imprecise. Because in the extracellular fluid all copper is also bound, the electrochemical potential and concentration gradients, essential in understanding the transport mechanisms of ions such as sodium and potassium, have been less useful in the characterization of copper transport. Furthermore, copper exists as Cu(I) and Cu(II), and many important copper-dependent biological processes employ the redox chemistry to cycle copper between these two states. This raises interesting questions related to the redox state in which copper is bound, transported, and released. In yeast, copper uptake via Ctr1p depends upon a Cu(II)/Fe(III) metalloreductase, Fre1, and Cu(I) is the presumed transported species. In agreement with this, in human cells, divalent metal ions do not inhibit hCTR1, whereas silver ions that are isoelectronic and of similar size to Cu(I) block the hCTR1-mediated copper uptake. Ascorbate converts Cu(II) to Cu(I) and stimulates copper uptake in some (but not all) cell lines. This could be due to different amounts of oxidoreductases such as the STEAP family of proteins (10) at the cell surface.
Cu(I) is also the form for intracellular distribution. Atox1 binds Cu(I) (11) and transfers Cu(I) to the MBSs in Cu-ATPases. In which form copper is released into the blood (from the intestine) or into the bile (from the liver) is not known. We also lack quantitative measurements of copper transport. The indications are that the transport rates are low (12, 13). It is assumed that when copper transport across membranes is assayed, it is the permeation step that is rate-limiting. This may not be the case. Protein-protein interactions that facilitate transfer to and from the transporter or the reduction of Cu(II) to Cu(I) (or oxidation upon release) may be slow steps in the pathway.
Structural Requirements for Copper Uptake
Channel-like Design of CTR1 Could Be the Key to Its Transport Mechanism
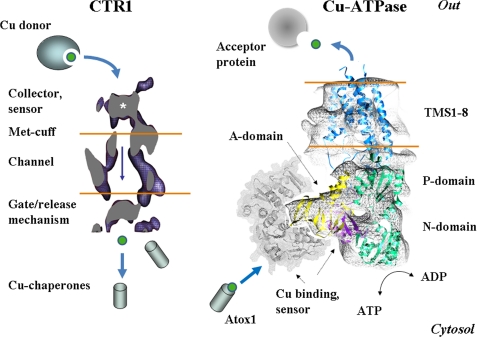
Recent studies yielded the first insights into the general organization and operating principles of copper transporters (Fig. 2). Despite their important role in cell physiology, the CTR1 orthologs vary greatly in primary sequence and length. Thus, the overall design of CTR proteins rather than sequence identity is the key to their transport mechanism. hCTR1 has only 190 amino acids and, like other CTR proteins, forms a stable trimer (1, 14, 15). The monomer has three TMSs with an extracellular N terminus and cytosolic C terminus (16). The N terminus of hCTR1 is N- and O-glycosylated (16, 17). The latter may serve to protect the protein against proteolysis because its absence leads to intracellular proteolytic removal of the N terminus (17). Whether the sugar chains influence the in vivo delivery of copper to hCTR1 or the trafficking of hCTR1 to its cellular destinations is not known.
FIGURE 2.
Structural model of hCTR1 from De Feo et al. (15) and the hypothetical structure of human Cu-ATPase based on the structure of archaeal CopA from Wu et al. (31) and modeling predictions for the N-terminal MBS1–4 (gray). The A-domain (yellow) and N- and P-domains (green) interact with the MBS in the N-terminal domain (purple and gray).
The three-dimensional structure of hCTR1 has recently been described (14, 15). It strongly supports the proposal (18) that the translocation pathway is formed at the center of the hCTR1 trimer. How copper is delivered from the serum to the pore remains unclear. Serum copper is largely associated with a high molecular weight fraction enriched in α2-macroglobulin, which may transfer copper directly to hCTR1 or may act as a source of exchangeable copper (19). The N terminus of CTR1, rich in His and Met residues, is likely to play a role in detecting copper carriers and in guiding copper toward the permeation pathway. The MxxMxM motif indeed binds copper with μm affinity (20). In vitro, a large portion of the N terminus can be deleted without significant loss of transport function (18). This suggests that the N terminus is important in vivo, where free copper is not available.
In addition to its possible role as a copper acceptor, the N terminus may act as a sensor. When fully loaded with copper, it may signal the need for endocytosis and removal of hCTR1 from the plasma membrane (21). From this perspective, it is interesting to compare the roles of the N termini in CTR1 and Cu-ATPases. Although very different structurally, both may perform an important role in receiving copper from specific donors and regulating transporter activity and intracellular localization (Ref. 21 and below).
Molecular Events at the Extracellular and Cytosolic Portions of CTR1 Are Coupled
Current data suggest that copper is guided to the permeation pathway through a narrow structure (Fig. 2) that includes highly conserved Met residues (Met150 and Met154 in hCTR1). Mutation of these Met residues abolishes copper transport in all examined CTR1 proteins. In addition to selecting and directing Cu(I) toward the pore, the conserved Met residues may couple the copper entry and exit from the pore. The binding of copper to hCTR1 at the extracellular surface leads to the conformational changes in the cytosolic loop (16). It seems that copper binding to conserved Met residues allows copper entry into the pathway that in turn facilitates opening of a “gate” at the exit of the pore (Fig. 2).
Mutagenesis data provide strong evidence that tight interaction with specific residues within the pathway is not required for copper transport (18). This explains poor conservation of intramembrane residues (with the notable exception of the GxxxG motif, which is required for oligomer stability) (22). The mechanism of copper release from CTR1 is a fascinating problem. Does copper diffuse out of the permeation pathway and bind to accessible copper-binding proteins based on their affinity, proximity, or abundance, or is the opening of the gate facilitated by intracellular copper acceptors such as the copper chaperones, making copper release a more targeted process? Whatever the answer, it is clear that as an ion transport mechanism, this process is quite unique.
CTR2
CTR2, a second mammalian copper transporter (23), is smaller than hCTR1 (143 amino acids), lacks most of the N terminus, and has limited sequence homology with hCTR1. In cells, CTR2 is detected in lysosomes and late endosomes (24); a fraction of overexpressed hCTR2 appears at the plasma membrane, where it can mediate copper uptake (25). By analogy with yeast Ctr2p, which has a role in mobilizing copper from intracellular stores (26), hCTR2 may be involved in retrieving copper from intracellular compartments (Fig. 1).
Structure and Mechanism of Human Cu-ATPases
Exporting copper from the cytosol poses challenges that are similar to those faced by the copper uptake machinery. Free cytosolic copper is negligible and specific copper capture prior to the transport step is necessary. The absence of free copper also implies that the metal status of donor and acceptor proteins (holo or apo) may have a profound effect on the kinetic properties of the copper pumps. Mutation in the acceptor (ceruloplasmin) affects the cellular location of the Cu-ATPase ATP7B (27); however, the effect on transport kinetics has not yet been explored. It is also important that the human Cu-ATPases translocate copper from the cytosol into the lumen of the trans-Golgi network or endocytic vesicles, where the low luminal pH may have a marked effect on copper release and ATPase turnover. This is in contrast to prokaryotic systems, where Cu-ATPases are located at the plasma membrane and extrude copper directly into the outside milieu.
Studies of ATP7A and ATP7B produced direct evidence of their function as P-type ATPases and yielded many details of their regulation (28). The analysis of bacterial homologs has further enriched our understanding of their enzymatic characteristics (29, 30). Recent electron microscopic reconstructions of Cu-ATPases yielded the overall shape of the molecule and the predicted orientation of functional domains (Fig. 2) (31). Human Cu-ATPases are 165–170-kDa proteins with eight TMSs and intracellular N and C termini. The Cu-ATPases couple the energy of ATP hydrolysis with the transfer of copper from the cytosol into the intracellular compartments. ATP hydrolysis is performed by the cytosolic nucleotide-binding (N-domain) and phosphorylation (P-domain) domains (Fig. 2), connected by a short linker. The hydrolysis is accompanied by a transient phosphorylation of the invariant Asp residue in the DKTG signature motif. The sequence of the N-domain and the nucleotide coordination environment in Cu-ATPases are unique compared with other P-type ATPases (32, 33), whereas the three-dimensional fold of the domain is common for all P-type pumps. These data suggest that the domain architecture (rather than the details of ligand binding) is central to the function of these transporters. That the P-type ATPases share general operating principles is apparent from the similarity in conformational transitions between archaeal CopA and mammalian Ca-ATPase (34).
At the same time, Cu-ATPases have distinct mechanistic properties. Some Cu-ATPases can form a phosphorylated intermediate in the absence of copper (29, 35). Human Cu-ATPases do not form measurable intermediate in the absence of copper; however, the well known chemotherapeutic drug cisplatin stimulates formation of phospho-intermediate (12, 35). The effect of clinically important drugs on Cu-ATPase function may have important practical implications.
Copper Delivery to Cu-ATPases
The transport cycle of human Cu-ATPases is initiated by the cytosolic copper chaperone Atox1, which interacts with the N-terminal domain of Cu-ATPases and transfers copper initially to one of the N-terminal MBSs (MBS2 in ATP7B) (36) and subsequently to other MBSs (37). Human Cu-ATPases have six MBSs in the N terminus; the first four sites are regulatory, whereas the two sites close to TMS1 are necessary for the transport function. All N-terminal MBSs are formed by the GMTCxxC sequences that provide a linear coordination environment for Cu(I) via conserved Cys residues. By analogy with archaeal CopA (38), the membrane portion of human Cu-ATPases may have two additional copper-binding sites.
Recent studies on archaeal CopA provided convincing evidence for the transfer of copper from the metallochaperone to the membrane sites in the absence of cytosolic MBSs (39). Unlike CopA, human Cu-ATPases require at least one N-terminal MBS for their activity. In addition, the inactivation of the N-terminal MBS2 disrupts copper transfer from Atox1 to the membrane sites (36), arguing in favor of a less direct, stepwise process of copper delivery to the membrane in the human system. Such a transfer process may include copper transfer to the N-terminal MBSs, which alters the interdomain interactions in the ATPase and increases the affinity or accessibility of the membrane sites for copper. Atox1 then transfers copper to the membrane sites either directly or, more likely, via the “intermediary” sites in proximity to the membrane.
Copper Release
Structurally diverse proteins receive copper in the secretory pathway from ATP7A or ATP7B, and tight interactions between the acceptor proteins and transporter are not necessary for copper release (40). Copper could be released from the ATPases as a free ion and then diffuse to the acceptor molecule. Such a mechanism would require the acceptor protein to have a higher affinity for copper than for Cu-ATPases. Because some of the well known acceptor proteins such as peptidyl α-monooxygenase have a relatively low affinity for copper, the binding of copper to intramembrane sites of the Cu-ATPase when exposed to the lumen must be weak. In CopA, the intramembrane copper-binding sites have a 3-fold coordination environment (38). One site appears to be formed by the CPC motif in TMS6 and a conserved Tyr in TMS7. A conserved Asn in TMS7 with Met and Ser in TMS8 may form the second site. The affinity of each site for Cu(I) was estimated as 1 fm, indicating that the trigonal coordination must be disrupted prior to copper exit from the ATPase. Because Asn, Ser, or Met alone is not a very good ligand for Cu(I), whereas Cys is, it is likely that the distortion of the unusual “second” site is a trigger for copper release.
Alternatively, transient interaction of the acceptor protein with the Cu-ATPase may facilitate copper transfer from the intramembrane sites of the transporter to the binding site in the acceptor. It is interesting that many Cu-ATPases, including ATP7A and ATP7B, have Met residues at the luminal rim of TMSs. This location suggests that methionines may “guide” the released copper from the transport sides toward the extracellular surface. The role of Met residues here is reminiscent of the predicted role of Met residues in hCTR1, except for the directionality of copper movement (Fig. 2).
Regulation of Copper Transport via Trafficking
Human copper transporters change their cellular location in response to various stimuli (41–43). The trafficking to/from specific compartments represents an important mechanism through which mammalian cells allocate copper and modulate intracellular copper levels. Trafficking in response to changing copper levels is the best characterized. In basal growth medium (containing 0.5–1.5 μm copper) or in cells treated with a copper chelator, Cu-ATPases are found in the trans-Golgi network. When copper increases, Cu-ATPases relocalize to endocytic vesicles in proximity to basolateral (ATP7A) or apical (ATP7B) membranes (43). Cu-ATPases sequester copper in the vesicles, which then fuse with the plasma membrane, and copper is released to the extracellular medium. Only a small fraction of the total Cu-ATPases is found at the plasma membrane (44), likely due to rapid endocytosis (45). Recent studies indicate that the trafficking of Cu-ATPase could also be coupled to hormone-mediated signaling, changes in Ca2+ fluxes, and kinase-mediated phosphorylation (46). These data provide the first mechanistic evidence of the tight link between copper homeostasis and such complex physiological responses as lactation (41), neuronal activity (47), hypoxia (48), and others.
hCTR1 has been detected at the plasma membrane and intracellular sites, and recent data suggest functional roles for CTR1 in both locations (see “Dietary Acquisition and Excretion of Copper”). Experiments in cell cultures showed that in elevated copper hCTR1 is internalized; however, results diverge on the extent of internalization and the fate of internalized CTR1. Some reports suggest that internalized CTR1 is completely degraded, and others suggest that internalization is a reversible process and that hCTR1 returns promptly to the membrane when copper is lowered. As with Cu-ATPases, recycling of hCTR1 may provide a mechanism for an acute regulatory response to changes in copper levels. However, many steps of this process remain poorly understood. Does endocytosis require saturation of the N-terminal copper-binding sites? (Does CTR1 sense high extracellular copper, or does the signal sensing high levels of copper come from inside the cell?) What is the mechanism for return of hCTR1 to the membrane? Is there a cell-specific storage pool? Do copper chaperones play a role in “reporting” the cellular copper status? These and many other questions remain to be addressed.
Concluding Remarks
Our knowledge of the mechanisms of human copper homeostasis is at best incomplete. We are close to obtaining important mechanistic information on the isolated transporters. However, an adequate understanding will come only from probing the physiological interactions among the several players at cellular and higher levels of integration.
Acknowledgments
We thank Drs. Stokes and Unger for providing recent data.
This work was supported, in whole or in part, by National Institutes of Health Grants P01 067166 (to J. H. K.) and DK071865 (to S. L.). This minireview will be reprinted in the 2009 Minireview Compendium, which will be available in January, 2010.
- hCTR1
- human CTR1
- MBS
- metal-binding site
- TMS
- transmembrane segment.
REFERENCES
- 1.Lee J., Peña M. M., Nose Y., Thiele D. J. (2002) J. Biol. Chem. 277, 4380–4387 [DOI] [PubMed] [Google Scholar]
- 2.Zimnicka A. M., Maryon E. B., Kaplan J. H. (2007) J. Biol. Chem. 282, 26471–26480 [DOI] [PubMed] [Google Scholar]
- 3.Nose Y., Kim B. E., Thiele D. J. (2006) Cell Metab. 4, 235–244 [DOI] [PubMed] [Google Scholar]
- 4.Lee J., Petris M. J., Thiele D. J. (2002) J. Biol. Chem. 277, 40253–40259 [DOI] [PubMed] [Google Scholar]
- 5.Arredondo M., Munoz P., Mura C. V., Nunez M. T. (2003) Am. J. Physiol. Cell Physiol. 284, C1525–C1515 [DOI] [PubMed] [Google Scholar]
- 6.Klomp A. E., Tops B. B., Van Denberg I. E., Berger R., Klomp L. W. (2002) Biochem. J. 364, 497–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weiss K. H., Wurz J., Gotthardt D., Merle U., Stremmel W., Füllekrug J. (2008) J. Anat. 213, 232–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barnes N., Bartee M. Y., Braiterman L., Gupta A., Ustiyan V., Zuzel V., Kaplan J. H., Hubbard A. L., Lutsenko S. (2009) Traffic 10, 767–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim H., Son H. Y., Bailey S. M., Lee J. (2009) Am. J. Physiol. Gastrointest. Liver Physiol. 296, G356–G364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohgami R. S., Campagna D. R., McDonald A., Fleming M. D. (2006) Blood 108, 1388–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ralle M., Lutsenko S., Blackburn N. J. (2003) J. Biol. Chem. 278, 23163–23170 [DOI] [PubMed] [Google Scholar]
- 12.Safaei R., Otani S., Larson B. J., Rasmussen M. L., Howell S. B. (2008) Mol. Pharmacol. 73, 461–468 [DOI] [PubMed] [Google Scholar]
- 13.Hung Y. H., Layton M. J., Voskoboinik I., Mercer J. F., Camakaris J. (2007) Biochem. J. 401, 569–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aller S. G., Unger V. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 3627–3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Feo C. J., Aller S. G., Siluvai G. S., Blackburn N. J., Unger V. M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 4237–4242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eisses J. F., Kaplan J. H. (2002) J. Biol. Chem. 277, 29162–29171 [DOI] [PubMed] [Google Scholar]
- 17.Maryon E. B., Molloy S. A., Kaplan J. H. (2007) J. Biol. Chem. 282, 20376–20387 [DOI] [PubMed] [Google Scholar]
- 18.Eisses J. F., Kaplan J. H. (2005) J. Biol. Chem. 280, 37159–37168 [DOI] [PubMed] [Google Scholar]
- 19.Moriya M., Ho Y. H., Grana A., Nguyen L., Alvarez A., Jamil R., Ackland M. L., Michalczyk A., Hamer P., Ramos D., Kim S., Mercer J. F., Linder M. C. (2008) Am. J. Physiol. Cell Physiol. 295, C708–C721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang J., Nadas I. A., Kim M. A., Franz K. J. (2005) Inorg. Chem. 44, 9787–9794 [DOI] [PubMed] [Google Scholar]
- 21.Guo Y., Smith K., Lee J., Thiele D. J., Petris M. J. (2004) J. Biol. Chem. 279, 17428–17433 [DOI] [PubMed] [Google Scholar]
- 22.Aller S. G., Eng E. T., De Feo C. J., Unger V. M. (2004) J. Biol. Chem. 279, 53435–53441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou B., Gitschier J. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 7481–7486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van den Berghe P. V., Folmer D. E., Malingré H. E., van Beurden E., Klomp A. E., van de Sluis B., Merkx M., Berger R., Klomp L. W. (2007) Biochem. J. 407, 49–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bertinato J., Swist E., Plouffe L. J., Brooks S. P., L'Abbé M. R. (2008) Biochem. J. 409, 731–740 [DOI] [PubMed] [Google Scholar]
- 26.Rees E. M., Lee J., Thiele D. J. (2004) J. Biol. Chem. 279, 54221–54229 [DOI] [PubMed] [Google Scholar]
- 27.di Patti M. C., Maio N., Rizzo G., De Francesco G., Persichini T., Colasanti M., Polticelli F., Musci G. (2009) J. Biol. Chem. 284, 4545–4554 [DOI] [PubMed] [Google Scholar]
- 28.Lutsenko S., Barnes N. L., Bartee M. Y., Dmitriev O. Y. (2007) Physiol. Rev. 87, 1011–1046 [DOI] [PubMed] [Google Scholar]
- 29.Hatori Y., Hirata A., Toyoshima C., Lewis D., Pilankatta R., Inesi G. (2008) J. Biol. Chem. 283, 22541–22549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mandal A. K., Cheung W. D., Argüello J. M. (2002) J. Biol. Chem. 277, 7201–7208 [DOI] [PubMed] [Google Scholar]
- 31.Wu C. C., Rice W. J., Stokes D. L. (2008) Structure 16, 976–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dmitriev O., Tsivkovskii R., Abildgaard F., Morgan C. T., Markley J. L., Lutsenko S. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 5302–5307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsuda T., Toyoshima C. (2009) EMBO J. 28, 1782–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hatori Y., Lewis D., Toyoshima C., Inesi G. (2009) Biochemistry 48, 4871–4880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leonhardt K., Gebhardt R., Mössner J., Lutsenko S., Huster D. (2009) J. Biol. Chem. 284, 7793–7802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walker J. M., Huster D., Ralle M., Morgan C. T., Blackburn N. J., Lutsenko S. (2004) J. Biol. Chem. 279, 15376–15384 [DOI] [PubMed] [Google Scholar]
- 37.Walker J. M., Tsivkovskii R., Lutsenko S. (2002) J. Biol. Chem. 277, 27953–27959 [DOI] [PubMed] [Google Scholar]
- 38.González-Guerrero M., Eren E., Rawat S., Stemmler T. L., Argüello J. M. (2008) J. Biol. Chem. 283, 29753–29759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.González-Guerrero M., Argüello J. M. (2008) Proc. Natl. Acad. Sci. U.S.A. 22, 105, 5992–5997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.El Meskini R., Culotta V. C., Mains R. E., Eipper B. A. (2003) J. Biol. Chem. 278, 12278–12284 [DOI] [PubMed] [Google Scholar]
- 41.Michalczyk A., Bastow E., Greenough M., Camakaris J., Freestone D., Taylor P., Linder M., Mercer J., Ackland M. L. (2008) J. Histochem. Cytochem. 56, 389–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hardman B., Michalczyk A., Greenough M., Camakaris J., Mercer J., Ackland L. (2007) Cell Physiol. Biochem. 20, 1073–1084 [DOI] [PubMed] [Google Scholar]
- 43.Mercer J. F., Barnes N., Stevenson J., Strausak D., Llanos R. M. (2003) Biometals 16, 175–184 [DOI] [PubMed] [Google Scholar]
- 44.Nyasae L., Bustos R., Braiterman L., Eipper B., Hubbard A. (2007) Am. J. Physiol. Gastrointest. Liver Physiol. 292, G1181–G1194 [DOI] [PubMed] [Google Scholar]
- 45.Pase L., Voskoboinik I., Greenough M., Camakaris J. (2004) Biochem. J. 378, 1031–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Veldhuis N. A., Gaeth A. P., Pearson R. B., Gabriel K., Camakaris J. (2009) Biometals 22, 177–190 [DOI] [PubMed] [Google Scholar]
- 47.Schlief M. L., Craig A. M., Gitlin J. D. (2005) J. Neurosci. 25, 239–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.White C., Kambe T., Fulcher Y. G., Sachdev S. W., Bush A. I., Fritsche K., Lee J., Quinn T. P., Petris M. J. (2009) J. Cell Sci. 122, 1315–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]