Abstract
Functional absence of fragile X mental retardation protein (FMRP) causes the fragile X syndrome, a hereditary form of mental retardation characterized by a change in dendritic spine morphology. The RNA-binding protein FMRP has been implicated in regulating postsynaptic protein synthesis. Here we have analyzed whether the abundance of scaffold proteins and neurotransmitter receptor subunits in postsynaptic densities (PSDs) is altered in the neocortex and hippocampus of FMRP-deficient mice. Whereas the levels of several PSD components are unchanged, concentrations of Shank1 and SAPAP scaffold proteins and various glutamate receptor subunits are altered in both adult and juvenile knock-out mice. With the exception of slightly increased hippocampal SAPAP2 mRNA levels in adult animals, altered postsynaptic protein concentrations do not correlate with similar changes in total and synaptic levels of corresponding mRNAs. Thus, loss of FMRP in neurons appears to mainly affect the translation and not the abundance of particular brain transcripts. Semi-quantitative analysis of RNA levels in FMRP immunoprecipitates showed that in the mouse brain mRNAs encoding PSD components, such as Shank1, SAPAP1–3, PSD-95, and the glutamate receptor subunits NR1 and NR2B, are associated with FMRP. Luciferase reporter assays performed in primary cortical neurons from knock-out and wild-type mice indicate that FMRP silences translation of Shank1 mRNAs via their 3′-untranslated region. Activation of metabotropic glutamate receptors relieves translational suppression. As Shank1 controls dendritic spine morphology, our data suggest that dysregulation of Shank1 synthesis may significantly contribute to the abnormal spine development and function observed in brains of fragile X syndrome patients.
In humans, the functional loss of the fragile X mental retardation protein (FMRP)2 causes the fragile X syndrome (FXS), a severe form of inherited mental retardation (1–4). In the brain of both humans and mice, FMRP deficiency results in a significant change in both dendritic spine morphology and synaptic function (5–9). FMRP is an RNA-binding protein that is thought to act primarily as a repressor of mRNA translation. Among other subcellular regions in neurons, FMRP appears to exercise this control function at postsynaptic sites. It has been hypothesized that in dendrites FMRP locally controls the synthesis of proteins, such as components of the postsynaptic density (PSD), which regulate both dendritic spine morphology and synaptic function (2, 9, 10). The PSD is a complex protein network lying underneath the postsynaptic membrane of excitatory synapses (11–13). It serves to cluster glutamate receptors and cell adhesion molecules, recruit signaling proteins, and anchor these components to the microfilament-based cytoskeleton in dendritic spines. To combine these functions, the central layers of the PSD consist of several scaffold proteins, such as members of the PSD-95, SAPAP/GKAP, and Shank/ProSAP families. Because of their capacity to directly interact with many different PSD components and to regulate the size and shape of dendritic spines, Shanks in particular are assumed to represent master scaffold proteins of the PSD (11). Activity-dependent changes in the PSD composition are thought to represent a molecular basis for most principal brain functions, including learning and memory. Several of these long term synaptic changes and learning paradigms critically depend on dendritic protein synthesis (14–17). Interestingly, mRNAs encoding some of the central components of the PSD, such as Shank1–3, SAPAP3, PSD-95, and α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid-type glutamate receptor subunits (GluR), are present in dendrites (18–23).
As FMRP has been implicated in the local regulation of mRNA translation at synapses, one crucial question is as follows: which postsynaptic proteins are affected by the loss of FMRP in a quantitative manner and may thus contribute to abnormal dendritic spine morphology and impaired synaptic plasticity? To specifically address this question, we took advantage of the possibility to isolate PSDs. In PSD fractions prepared from two major brain areas of wild-type and FMRP-deficient mice, we compared the levels of major scaffold proteins and glutamate receptor subunits. Thereby, we identified a select group of postsynaptic proteins, including the central scaffold protein Shank1, that are enriched in PSDs of FMRP-deficient mice. Functional data further suggest that FMRP represses translation of Shank1 transcripts in neurons via an interaction with its 3′-untranslated region (3′UTR). This translation block is abolished upon the activation of metabotropic glutamate receptors (mGluR). Thus, a deregulated postsynaptic synthesis of Shank1, a master scaffold protein of the PSD, may significantly contribute to the aberrant dendritic spine morphology caused by the absence of FMRP.
EXPERIMENTAL PROCEDURES
Animals, Cell Culture, Transfection, and Luciferase Assays
Fmr1−/− mice (B6.129P2-Fmr1tmlCgr/J strain, The Jackson Laboratory, Bar Harbor, ME) were maintained on a C57BL/6J background. Tissue was dissected from different brain regions of adult male Fmr1−/− and congenic C57BL/6J wild-type mice raised in the animal facility of the University Medical Center Hamburg-Eppendorf. Cortical neurons were prepared from mouse embryos (embryonic day 18) and grown in Neurobasal medium (Invitrogen). For luciferase assays with unstimulated neurons, cells were transfected 7 days after plating and harvested 1 day after transfection (24). Transfection efficiencies were in the range of 3–5%. Neurons transfected 14 days after plating were treated 1 day later with 0.1 mm (S)-3,5-dihydroxyphenylglycine (DHPG; Sigma) for 10 min at 37 °C and harvested after stimulation. The dual luciferase reporter assay system (Promega GmbH, Mannheim, Germany) was used according to the manufacturer's recommendations with cell extracts prepared 24 h after transfection.
Biochemical Cell Fractionation, Antibodies, and Western Blot Analysis
From mouse cortices and hippocampi, cytosolic, synaptosome, and PSD fractions were prepared as essentially described (25), snap-frozen in liquid nitrogen, and stored at −80 °C. Glutathione S-transferase fusion proteins containing amino acid residues 722–776, 636–699, and 694–747 of rat SAPAP1 (GenBankTM accession number NP_075235), SAPAP2 (P97837), and SAPAP3 (AAS90634) were used to raise and affinity-purify custom rabbit polyclonal antisera 5280, 5281, and 5269 (Pineda, Berlin, Germany), respectively. Antibodies directed against PSD-95 (26), SAP97 (27), SAP102 (28), Chapsyn-110/PSD-93 (29), Shank1 (30), Shank3 (obtained from Ref. 31), IRSp53 (32), and poly(A)-binding protein (33) have been previously described. For commercially available antibodies, antibody sources, and dilutions used for Western blotting see Table 1. Western blots were performed as described (34). Immune complexes were visualized with the Lumi-Light Western blotting substrate (Roche Diagnostics) on x-ray films (Eastman Kodak Co.). After scanning of the films, the intensity of individual signals was determined by densitometric measurement using ImageJ software.
TABLE 1.
Antibodies, antibody sources, and dilutions used for Western blotting
| Antibody | Antibody source | Dilution for Western blotting |
|---|---|---|
| Anti-FMRP, mouse monoclonal, clone 1C3-1a | Euromedex, Mundolsheim, France | As recommended by supplier |
| Anti-FMRP, rabbit polyclonal, H-120 | Santa Cruz Biotechnology, Heidelberg, Germany | |
| Anti-β-tubulin III, rabbit polyclonal | ||
| Anti-RGS4, rabbit polyclonal | Abcam, Cambridge, UK | |
| Anti-NR1, mouse monoclonal, mAb363 | CHEMICON Europe, Hofheim, Germany | |
| Anti-NR2A, rabbit polyclonal, AB1555P | ||
| Anti-NR2B, rabbit polyclonal, AB1557P | ||
| Anti-GluR1, rabbit polyclonal, AB1504 | ||
| Anti-GluR2/3, rabbit polyclonal, AB1506 | ||
| Anti-GluR4, rabbit polyclonal | Dianova, Hamburg, Germany | |
| Anti-SAPAP1, affinity-purified rabbit polyclonal 5280 | Stefan Kindler | 1:33 |
| Anti-SAPAP2, affinity-purified rabbit polyclonal 5281 | 1:83 | |
| Anti-SAPAP3, affinity-purified rabbit polyclonal 5269 | 1:1000 | |
| Anti-SAP97, rabbit polyclonal | See Valtschanoff et al. (27) | 1:250 |
| Anti-SAP102, rabbit polyclonal | See Müller et al. (28) | 1.500 |
| Anti-SAP90/PSD-95, rabbit polyclonal | See Kistner et al. (26) | 1:1000 |
| Anti-Chapsyn-110, rabbit polyclonal | See Kim et al. (29) | 1:1000 |
| Anti-Shank1, rabbit polyclonal | See Zitzer et al. (30) | 1:2000 |
| Anti-Shank3, rabbit polyclonal | See Redecker et al. (31) | 1:3000 |
| Anti-IRSp53, rabbit polyclonal | See Soltau et al. (32) | 1:2000 |
| Anti-PABP, rabbit polyclonal | See Brendel et al. (33) | 1:2000 |
Statistical Data Analysis
Data generated in entirely independent experiments were analyzed with a two-sided paired Student's t test (level of significance α = 0.05). Results obtained from clustered experiments, such as those using identical PSD or RNA preparations, were analyzed with a linear mixed models application that considers the clustered structure of the experiment (SPSS 15.0.1, SPSS Inc., Chicago, 2006; level of significance α = 0.05).
RNA Preparation, Northern Blotting, Immunoprecipitation, and Real Time Reverse Transcription-PCR
Total RNA from mouse brain cytosolic and synaptosome fractions was isolated using TRIzol reagent (Invitrogen), and Northern blots were generated using the formaldehyde method (35). cDNA fragments containing nucleotides 259–408 and 23–1128 of the mouse PSD-95 (GenBankTM accession number NM_001109752) and human β-actin cDNA (DQ890960.2), respectively, were labeled with 32P-containing nucleotides using the Rediprime II labeling system (GE Healthcare) and used to probe blotted RNAs. Labeled bands were visualized using a BAS-1800II PhosphorImager (Fujifilm, Düsseldorf, Germany) and x-ray films (Kodak). Immunoprecipitations from brains of adult Fmr1−/− and wild-type mice with antibodies against FMRP and PABP as well as irrelevant IgG, extraction of RNA from immunoprecipitates, and real time RT-PCR were performed as described (36). Reverse transcription (50 °C for 50 min) was directly followed by gene-specific amplification (initial strand separation at 94 °C for 15 min; 40 cycles of each at 94 °C for 15 s, 58 °C for 30 s, and 72 °C for 30 s). Transcript levels of hypoxanthine-guanine phosphoribosyltransferase were used for normalization as described (37). Levels of PSD-95, SAP97, SAPAP1, SAPAP2, SAPAP3, Shank1, NR1, and GluR1 mRNAs were quantified using gene-specific primers as follows: PSD-95-fw (5′-GGCACCGACTACCCCACAG-3′) and PSD-95-rev (5′-AACACCATTGACCGACAGGA-3′); SAP97-fw (5′-AGTGACGAAGTCGGAGTGATT-3′) and SAP97-rev (5′-GTCAGGGATCTCCCCTTTATCT-3′); SAPAP1-fw (5′-CGTAAGTGAAGTCTCCATCAAC-3′) and SAPAP1-rev (5′-CTCGCTCACCTGACTTATGG-3′); SAPAP2-fw (5′-GACAGACAGACTGCGGATCG-3′) and SAPAP2-rev (5′-GCAGACATCCTTGGGCTTTC-3′); SAPAP3-fw (5′-ACTATTTGCAGGTGCCGCAAG-3′) and SAPAP3-rev (5′-GGGCTACCATCTGAGTCTCC-3′); Shank1-fw (5′-AGCCTGCAGCAGTGCCCAGCA-3′) and Shank1-rev (5′-ATGCGAGGCCGCCAGGCCCA-3′); NR1-fw (5′-AGCCAGGTCTACGCTATCC-3′) and NR1-rev (5′-TAGGGTGGTACGGTGCGAAG-3′); NR2B-fw (5′-AGAGGTGGTTGACTTCTCTGTGCC-3′) and NR2B-rev (5′-TGAAGTATTCAAAGACAAAGACAGC-3′); and GluR1-fw (5′-ACCACTACATCCTCGCCAAC-3′) and GluR1-rev (5′-TCACTTGTCCTCCACTGCTG-3′), respectively. The concentration of IRSp53 mRNA was quantified using the Mm_Baiap2_1_SG QuantiTect primer assay (QT01061431, Qiagen, Hilden, Germany). Data analysis was performed using REST-2005 software (38).
Eukaryotic Expression Vectors
pFiRe-basic, a derivative of pBicFire (39), contains two recombinant genes, both of which are controlled by independent cytomegalovirus immediate-early promoters and encode Photinus (PhoLuc) and Renilla luciferase (RenLuc), respectively. To create pFiRe-Shank1, the cDNA sequence in the PhoLuc gene of pFiRe-basic that encodes a short synthetic 3′UTR was exchanged for a cDNA region corresponding to nucleotides 6593–7180 of the rat Shank1 3′UTR (GenBankTM accession number NM_031751).
RESULTS
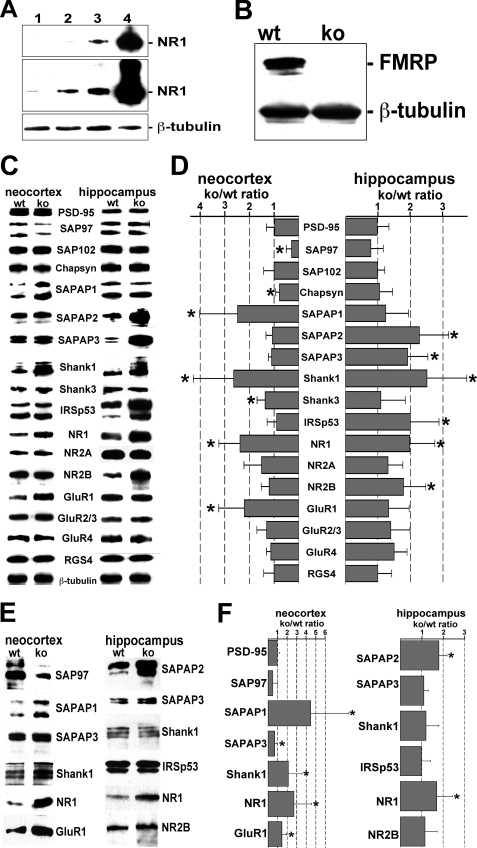
To identify changes in the molecular framework of the PSD occurring in Fmr1−/− (knock-out) mice, we compared the levels of different scaffold proteins in PSD fractions prepared from neocortices and hippocampi of knock-out and wild-type animals. Successful enrichment of PSDs was documented by a strong increase in the levels of the N-methyl-d-aspartate-type glutamate receptor subunit NR1, a known PSD component, in successive steps of the purification protocol as compared with β-tubulin (a known contaminant of PSD fractions; Fig. 1A). Also, FMRP is clearly detectable in brain homogenates of wild-type but not FMRP knock-out animals (Fig. 1B). For the comparative analysis of PSD fractions, equal protein amounts were analyzed by Western blotting with specific antibodies (Fig. 1, C and E). Relative levels of individual proteins in the PSD of FMRP-deficient versus wild-type mice were determined by densitometric measurement of the respective immunochemical signal normalized to the β-tubulin signal intensity (Fig. 1, D and F). In neocortical and hippocampal PSD fractions obtained from 2-month-old animals, levels of PSD-95 and SAP102 were similar in FMRP-deficient and wild-type mice. The concentration of Shank1, however, was strongly increased in both the neocortex and hippocampus of FMRP-deficient as compared with wild-type animals. In contrast, SAPAP1 and Shank3 levels were only elevated in neocortical PSD preparations, whereas SAPAP2, SAPAP3, and IRSp53 concentrations were selectively increased in hippocampal PSD fractions prepared from knock-out animals. Finally, levels of SAP97 and Chapsyn were significantly decreased in neocortical but not hippocampal PSD preparations obtained from FMRP-deficient mice. In PSD fractions prepared from juvenile mouse brains (2 weeks after birth), some of these changes were already apparent, such as increases in neocortical levels of SAPAP1 and Shank1, an elevated concentration of SAPAP2 in the hippocampus, and a decrease in neocortical SAP97 levels. However, increases in hippocampal SAPAP3, Shank1, and IRSp53 concentrations were only observed in 2-month-old animals.
FIGURE 1.
In the mouse neocortex and hippocampus, FMRP loss leads to altered levels of select scaffold proteins and receptor subunits in the PSD. A, proteins (15 μg per lane) from soluble protein (lane 1), crude membrane P2 (lane 2), synaptosomal (lane 3), and PSD fractions (lane 4) of adult wild-type mice were analyzed via Western blotting with antibodies directed against NR1 and β-tubulin. Levels of the PSD marker protein NR1 increase in consecutive fractions obtained during PSD purification. The upper panel represents a shorter exposure of the blot shown in the middle panel. B, Western blot analysis of proteins from the soluble brain protein fraction (15 μg per lane) of 2-month-old wild-type (wt) and Fmr1−/− (ko) mice. Although β-tubulin is detected in both protein extracts, FMRP is only present in lysates obtained from wild-type animals. C, in Western blots performed with PSD fractions from the neocortex and hippocampus of 2-month-old wild-type and Fmr1−/− mice (7 μg per lane), different proteins (as indicated) were detected with specific antibodies. D, bar graph indicating the knock-out to wild-type ratio (ko/wt ratio) in the levels of individual proteins detected in Western blot experiments as shown in C. For each PSD component with an altered level in knock-out as compared with wild-type mice, the final value is based on at least three different PSD preparations and three or more independent Western blot experiments per preparation. For proteins with unchanged levels, at least two independently prepared PSD fractions were investigated and each by two or more immunoblotting experiments. Protein levels were determined via densitometric evaluation of the immunochemical signal and normalized to the signal intensity obtained for β-tubulin. A bar value of 1 indicates that the normalized levels of the respective protein in PSD preparations from knock-out and wild-type animals are identical. Simple horizontal lines specify standard deviations. Altered knock-out to wild-type ratios as observed for SAP97 (neocortex, p = 0.001, linear mixed models application), Chapsyn-110 (neocortex, p = 0.031), SAPAP1 (neocortex, p = 0.001), SAPAP2 (hippocampus, p < 0.001), SAPAP3 (hippocampus, p < 0.001), Shank1 (neocortex, p < 0.001; hippocampus, p < 0.001), Shank3 (neocortex, p < 0.04), IRSp53 (hippocampus, p = 0.005), NR1 (neocortex, p = 0.013; hippocampus, p < 0.001), NR2B (hippocampus, p = 0.016), and GluR1 (neocortex, p = 0.001) are statistically significant (marked by asterisk). E, Western blots were performed with PSD fractions from the neocortex and hippocampus of 2-week-old wild-type (wt) and Fmr1−/− (ko) mice (7 μg per lane) and antibodies directed against different postsynaptic components (as indicated). F, bar graph indicating the knock-out to wild-type ratios of the postsynaptic protein levels determined in Western blot experiments as shown in E. Altered knock-out to wild-type ratios as observed for SAPAP1 (neocortex, p = 0.01, linear mixed models application), SAPAP2 (hippocampus, p = 0.028), SAPAP3 (neocortex, p = 0.03), Shank1 (neocortex, p = 0.012), NR1 (neocortex, p < 0.001; hippocampus, p = 0.008), and GluR1 (neocortex, p = 0.024) are statistically significant (marked by asterisk). For further details see text.
One crucial function of scaffold proteins in the PSD is to anchor neurotransmitter receptors in the postsynaptic membrane. Thus, we also analyzed whether the observed changes in scaffold protein levels affect the abundance of glutamate receptors in the PSD (Fig. 1, C and D). As compared with wild-type mice, glutamate receptor subunits NR2A, GluR2/3, and GluR4 were slightly but not significantly enriched in PSD fractions of FMRP-deficient mice. In contrast, the N-methyl-d-aspartate receptor subunit NR1 was increased in both the neocortex and hippocampus of knock-out animals, whereas elevated levels of NR2B and GluR1 were only observed in the hippocampus and neocortex, respectively. Increases in NR1 and GluR1 levels were already apparent in juvenile FMRP-deficient mice, whereas postsynaptic NR2B concentrations appear to rise thereafter. Taken together, the loss of FMRP results in an increased abundance of the scaffold proteins SAPAP1, SAPAP2, SAPAP3, Shank1, and IRSp53 as well as the glutamate receptor subunits NR1, NR2B, and GluR1 at synapses. Although most changes become manifest until the end of the 2nd week after birth, others occur rather late during postnatal development. Moreover, the absence of FMRP affects some PSD components differentially in neocortical and hippocampal neurons, respectively.
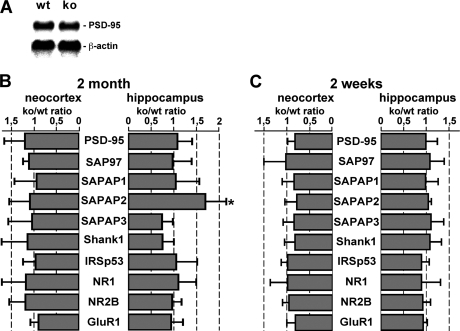
Next we wanted to determine whether altered levels of individual proteins at synapses of FMRP knock-out mice correlate with changes in the concentration of their corresponding mRNAs. Thus, we first sought to establish a control mRNA whose levels are identical in the brains of FMRP-deficient and wild-type mice. A Northern blot analysis carried out with total RNA extracted from the neocortex showed that PSD-95 mRNA levels are identical in the cortex of FMRP-deficient and wild-type mice (Fig. 2A). This result was confirmed by real time RT-PCR analysis with template RNA extracted from both the neocortex and hippocampus of knock-out and wild-type animals (Fig. 2B) and is consistent with stable postsynaptic PSD-95 levels in both brain regions as described above (Fig. 1, C–F). Real time RT-PCR data further showed that the total levels of SAP97, SAPAP1, SAPAP3, Shank1, IRSp53, NR1, NR2B, and GluR1 mRNAs in both neocortex and hippocampus of 2-week- and 2-month-old animals are essentially identical in FMRP-deficient and wild-type mice (Fig. 2, B and C). Only SAPAP2 transcript levels in the adult hippocampus were significantly increased in knock-out animals (1.69-fold; p = 0.001, mixed models). Adult neocortical and juvenile hippocampal and neocortical SAPAP2 mRNA concentrations were unchanged in FMRP-deficient mice as compared with wild-type mice. Thus, although an increase in hippocampal SAPAP2 transcript levels may contribute to a similar rise in SAPAP2 concentrations in Fmr1−/− mice as compared with wild-type mice, the changes in the levels of other postsynaptic proteins in knock-out animals described herein do not appear to result from an altered abundance of the corresponding mRNAs in the absence of FMRP.
FIGURE 2.
Total levels of most mRNAs encoding postsynaptic scaffold proteins and glutamate receptor subunits are unchanged in Fmr1−/− mice as compared with wild-type animals. A, Northern blots were performed with total RNA (40 μg per lane) prepared from the neocortex of 2-month-old wild-type (wt) and knock-out (ko) animals and radioactively labeled cDNA probes against PSD-95 and β-actin mRNAs. B and C, bar graphs summarizing real time RT-PCR data obtained with 2-month-old (B) and 2-week-old mice (C). Individual bars indicate the knock-out to wild-type (ko/wt) ratio in the levels of PSD-95, SAP97, SAPAP1, SAPAP2, SAPAP3, Shank1, IRSp53, NR1, NR2B, and GluR1 mRNAs detected in the neocortex and hippocampus, respectively. For all transcripts the final value is based on at least two different experiments using two separate tissue preparations. Values close to 1 indicate that the levels of both mRNAs are similar in wild-type and knock-out animals. Only hippocampal SAPAP2 mRNA levels in 2-month-old mice are significantly increased (1.69-fold ± 0.46; p = 0.001, linear mixed models application; marked by asterisk). Simple horizontal lines represent S.D. For more details see text.
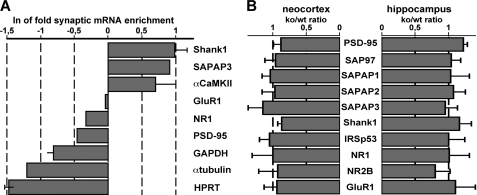
To assess whether local mRNA concentrations at synapses may be affected by the loss of FMRP, we used total RNA extracted from both neocortical and hippocampal synaptosome fractions as a template for real time RT-PCR. The observation that well established dendritic transcripts, such as Shank1, SAPAP3, and αCaMKII mRNAs (18, 20, 40–42), are strongly enriched in synaptosome fractions as compared with total brain lysates, and known somatic transcripts, including glyceraldehyde-3-phosphate dehydrogenase, α-tubulin, and hypoxanthine-guanine phosphoribosyltransferase mRNAs (24, 41, 43), are highly enriched in total brain homogenates in comparison with synaptosome fractions (Fig. 3A) shows that this method represents a valid approach to determine synaptic mRNA levels. Real time RT-PCR analysis further revealed that synaptic levels of PSD-95, SAP97, SAPAP1, SAPAP2, SAPAP3, Shank1, IRSp53, NR1, NR2B, and GluR1 mRNAs in both the neocortex and the hippocampus are essentially identical in FMRP-deficient and wild-type mice (Fig. 3B). Taken together, these data suggest that the loss of FMRP does not primarily alter the total and synaptic abundance of mRNAs encoding postsynaptic scaffold proteins and glutamate receptor subunits, whereas it selectively affects the synaptic levels of particular PSD components in cortical and hippocampal neurons.
FIGURE 3.
Synaptosomal levels of mRNAs encoding postsynaptic scaffold proteins and glutamate receptor subunits are identical in Fmr1−/− and wild-type mice. A, real time RT-PCR analysis was used to determine mRNA concentrations in total brain lysates and synaptosome fractions prepared from 2-month-old mice. The bar graph depicts the natural logarithms of the enrichment coefficients determined for synaptic compared with total mRNA levels. Known dendritic transcripts encoding Shank1, SAPAP3, and αCaMKII are strongly enriched in synaptosome fractions. B, bar graph summarizing real time RT-PCR data obtained with RNA extracted from synaptosome fractions of 2-month-old mice. Individual bars indicate the knock-out to wild-type (ko/wt) ratio in the levels of PSD-95, SAP97, SAPAP1, SAPAP2, SAPAP3, Shank1, IRSp53, NR1, NR2B, and GluR1 mRNAs detected in the neocortex and hippocampus, respectively. For all transcripts, the final value is based on at least two different experiments using two separate tissue preparations. Values close to 1 indicate that the levels of mRNAs are similar in both wild-type and knock-out animals. Simple horizontal lines represent standard deviations. For more details see text.
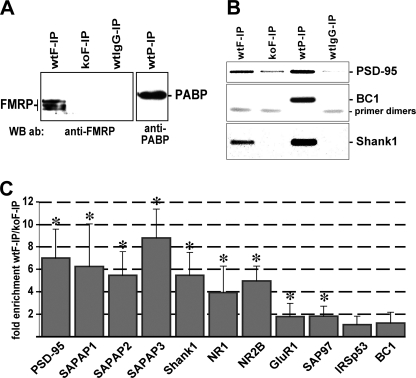
Over the recent years, consensus has been developed that FMRP most likely acts as a regulator of translation (1, 3, 5–7). Therefore, we hypothesized that altered postsynaptic levels of scaffold proteins and glutamate receptors described herein result from a translational dysregulation of the respective mRNAs in the absence of FMRP. To evaluate this hypothesis, we first performed immunoprecipitation assays with whole mouse brain homogenates to identify FMRP-associated mRNAs. Western blot analysis confirmed that immunoprecipitates obtained from brain homogenates of wild-type mice using antibodies directed against FMRP (wtF-IP) and the poly(A)-binding protein (PABP, wtP-IP) indeed contain the respective antigens (Fig. 4A). FMRP was not present in anti-FMRP precipitates from knock-out brains (koF-IP) and material precipitated from wild-type homogenates utilizing irrelevant IgGs (wtIgG-IP). From all four immunoprecipitates, total RNA was extracted and analyzed by RT-PCR using primer pairs specific for PSD-95, BC1, and Shank1 RNAs. RT-PCR products were separated and visualized in an agarose gel (Fig. 4B). All three RNAs were present in the wtP-IP that served as a positive control as PABP is known to interact with all polyadenylated mRNAs and BC1 RNA (36, 44–46). In contrast, the wtF-IP contained only Shank1 and PSD-95 transcripts but not the BC1 RNA. Although this observation is consistent with previous reports showing an association of FMRP with PSD-95 transcripts (21, 22, 36) but not BC1 RNA (36), it also shows that Shank1 mRNA is an in vivo FMRP target. Moreover, in the wtIgG-IP none of the three RNAs was detectable, thus excluding the possibility of unspecific RNA precipitation. Similarly, in the koF-IP negative control, both Shank1 transcripts and BC1 RNA were absent, and the PSD-95 mRNA-specific amplification resulted in only a very faint product. To further assess a putative association of other RNAs with FMRP, we performed real time RT-PCR analysis with template RNA extracted from wtF-IP and koF-IP material. Similar to the negative control BC1 RNA, IRSp53 transcripts were not enriched in wtF-IP as compared with koF-IP (Fig. 4C). In contrast, PSD-95, SAPAP1, SAPAP2, SAPAP3, Shank1, NR1, and NR2B mRNA levels were about 4–9-fold higher in wtF-IP in comparison with koF-IP. These data suggest that in the mouse brain these transcripts represent in vivo FMRP targets whose translation is dysregulated in the absence of the RNA-binding protein. SAP97 and GluR1 mRNA concentrations were only moderately (less than 2-fold) yet significantly increased in wtF-IP, and thus these transcripts do not appear to represent major FMRP-associated mRNAs.
FIGURE 4.
mRNAs encoding PSD-95, SAPAP1, SAPAP2, SAPAP3, Shank1, NR1, and NR2B associate with FMRP in vivo. A, in a Western blot (WB) experiment, immunoprecipitates from wild-type (wt) and knock-out (ko) lysates obtained with antibodies (ab) directed against FMRP (F-IP) and PABP (P-IP) as well as irrelevant antibodies (IgG-IP) were probed for FMRP and PABP, respectively. B, from the input material and precipitates described in A, total RNA was extracted and used as template for RT-PCR experiments performed with primers specific for Shank1, PSD-95, and BC1 RNA, respectively. PCR products were separated and visualized in an agarose gel. C, real time RT-PCR analysis was used to determine relative mRNA concentrations in immunoprecipitates obtained with anti-FMRP antibodies. The bar graph depicts the enrichment coefficients determined for wtF-IP as compared with koF-IP mRNA levels. Simple vertical lines represent standard deviations. (PSD-95, p < 0.001; SAPAP1, p < 0.001; SAPAP2, p < 0.001; SAPAP3, p < 0.001; Shank1, p < 0.001; NR1, p = 0.002; NR2B, p = 0.003; GluR1, p = 0.021; SAP97, p < 0.001; IRSp53, p = 0.38; BC1, p = 0.93).
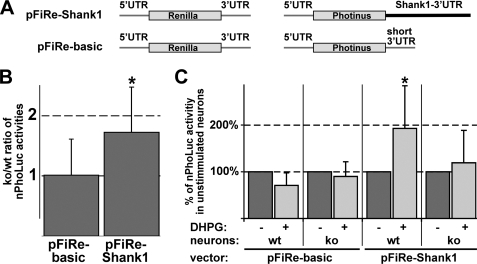
The hypothesis that FMRP regulates the translation of Shank1 mRNAs was further evaluated by performing luciferase assays in primary cortical neurons derived from Fmr1−/− and wild-type mice. For this purpose we constructed two eukaryotic expression vectors each containing two separate genes encoding either Photinus (PhoLuc) or Renilla luciferase (RenLuc) (Fig. 5A). RenLuc-encoding genes are identical in both vectors, and the respective enzyme activity was used to normalize for differences in transfection rates. In contrast, PhoLuc mRNAs synthesized from both plasmids differ in their 3′UTR sequence. While pFiRe-basic derived transcripts contain a short recombinant 3′UTR, pFiRe-Shank1-encoded mRNAs instead possess the first 588 nucleotides of the Shank1–3′UTR, including a cis-acting dendritic targeting element (18). Eukaryotic expression vectors were introduced into 7-day-old differentiated neurons. One day after transfection, the primary neurons were harvested, and the activity of both luciferases was determined in cell homogenates. The ratio of normalized PhoLuc (nPhoLuc) activities observed in knock-out and wild-type neurons was 1.01 ± 0.6 and 1.72 ± 0.75 (Fig. 5B; p = 0.0004; two-sided paired t test; n = 25 and 29, respectively) for pFiRe-basic and pFiRe-Shank1-transfected cells, respectively, suggesting that FMRP represses Shank1 mRNA translation via an interaction with the 3′UTR. This interpretation provides a molecular explanation for the increased Shank1 levels in PSD fractions obtained from FMRP-deficient mice.
FIGURE 5.
FMRP mediates mGluR-regulated Shank1 mRNA translation via an interaction with the 3′UTR. Primary cortical neurons derived from wild-type (wt) and knock-out (ko) mice were transfected with the indicated eukaryotic expression vectors. Cell lysates were assayed for Photinus and Renilla luciferase activities. A, two eukaryotic expression vectors, pFiRe-Shank1 and pFiRe-basic, each contain two separate genes encoding either Photinus (PhoLuc) or Renilla luciferase (RenLuc). RenLuc mRNAs encoded by both vectors are identical. In contrast, PhoLuc transcripts synthesized from both plasmids differ in their 3′UTR sequence. While pFiRe-basic derived mRNAs possess a short recombinant 3′UTR, pFiRe-Shank1-encoded transcripts instead contain the first 588 nucleotides of the Shank1–3′UTR. B, bar graph shows the knock-out to wild-type ratios (ko/wt) of normalized PhoLuc activities (nPhoLuc) determined in unstimulated 8-day-old neurons. C, bar graph depicts the percentile nPhoLuc activity in transfected 15-day-old cells before and after DHPG-mediated mGluR activation (dark and light gray bars, respectively). For each experimental condition, the respective normalized PhoLuc activity measured without DHPG treatment is arbitrarily set to 100%. Simple vertical lines represent standard deviations. Statistically significant alterations are marked by an asterisk.
It has been implicated that FMRP-mediated translation control is regulated by metabotropic glutamate receptors (mGluR) (8–10, 47–50). Thus, 2-week-old primary neurons were transfected with either pFiRe-basic or pFiRe-Shank1, incubated with the mGluR agonist DHPG for 10 min, and harvested. Whereas DHPG treatment led to a strongly increased nPhoLuc activity in pFiRe-Shank1-transfected wild-type neurons (Fig. 5C; 192 ± 91%; p = 0.0078; two-sided paired t test; n = 13), it did not alter the nPhoLuc activity in corresponding knock-out cells (119 ± 69%; p = 0.603; n = 10). Moreover, the wild-type-specific increase in enzyme activity upon mGluR stimulation was not observed in pFiRe-basic transfected neurons (71 ± 27%; p = 0.084; n = 9). Taken together, these data suggest that FMRP interacts with the 3′UTR of Shank1 mRNAs to repress translation, a block that is abolished after mGluR activation.
DISCUSSION
In humans and mice, loss of FMRP causes changes in dendritic spine morphology and synaptic function (1, 2, 51, 52). It has been hypothesized that in dendrites FMRP locally controls the synthesis of proteins, such as components of the PSD, which regulate both cellular events (1–9). A crucial question is as follows: which postsynaptic proteins are affected by the loss of FMRP in a quantitative manner? Here we specifically focused our analysis on the postsynaptic pool of proteins by investigating PSD preparations derived from the mouse brain. PSD fractions were selectively prepared from the neocortex and the hippocampus, respectively, brain regions in which the loss of FMRP causes abnormal dendritic spine development (51, 53–55) and impaired synaptic plasticity (8–10, 50, 56–59).
Alterations in the composition of synaptic glutamate receptors have been suggested as a principal cellular mechanism for cognitive impairment (60, 61). Accordingly, we show here that in FMRP-deficient mice the postsynaptic levels of the glutamate receptor subunits NR1 (neocortex and hippocampus), NR2B (hippocampus), and GluR1 (neocortex) are increased. Our data further indicate that NR1, NR2B, and GluR1 mRNAs are in vivo mRNA targets of FMRP. However, as the loss of FMRP affects neither the total nor the synaptic concentration of these transcripts, FMRP may rather regulate their translation.
In contrast to the glutamate receptor subunits, the postsynaptic levels of SAP97 were found to be strongly decreased in the neocortex of Fmr1−/− mice as compared with wild-type animals. Yet both cytoplasmic and synaptic levels of the respective mRNAs are unchanged in the absence of FMRP, suggesting that FMRP enhances the translation of SAP97 transcripts. Consistently, it was recently shown that FMRP can also act as a translational activator (62, 63). In both the neocortex and the hippocampus, postsynaptic levels of two scaffold proteins related to SAP97, namely PSD-95 and SAP102, were found to be identical in wild-type and FMRP-deficient mice. Despite the fact that PSD-95- and SAP102-deficient mice exhibit impaired learning (64, 65) and functional loss of SAP102 results in mental retardation in humans (66), our data suggest that both proteins do not significantly contribute to the learning disabilities of FXS patients. Interestingly, we and others have identified PSD-95 mRNA as an in vivo FMRP target (21, 22, 36, 67). Zalfa et al. (22) recently reported that through this interaction FMRP selectively stabilizes the PSD-95 mRNA in the hippocampus but not the neocortex, thus causing reduced hippocampal levels of PSD-95 mRNA and protein in FMRP-deficient mice. However, here we did not observe any quantitative changes in both cytoplasmic and synaptic PSD-95 mRNA concentrations and postsynaptic protein levels in the absence of FMRP, neither in the neocortex nor the hippocampus. In accordance with this observation, Muddashetty et al. (21) described identical dendritic and synaptic PSD-95 mRNA levels in primary neurons from Fmr1−/− and wild-type mice. Thus, although several reports, including this study, strongly support an in vivo interaction between FMRP and the PSD-95 mRNA (21, 22, 36, 67), the cellular consequences of this association remain ambiguous.
Increased postsynaptic SAPAP2 levels in the hippocampus of adult FMRP-deficient mice appear to at least in part be due to a slight but significant increase in the concentration of the respective mRNA, indicating that FMRP may enhance SAPAP2 mRNA turnover. Furthermore, despite unchanged cytoplasmic and synaptic levels of the corresponding transcripts, SAPAP1 and SAPAP3 are more abundant at neocortical and hippocampal synapses of adult Fmr1−/− mice, respectively. Consistently, an elevated SAPAP3 concentration was observed in hippocampal lysates of FMRP-deficient mice (68). Here we also show that mRNAs encoding SAPAP1–3 are in vivo brain targets of FMRP. Noteworthy, another study ranks SAPAP3 and SAPAP4 transcripts among the 11 most prominent mouse brain mRNAs associated with FMRP (69). Also, very recently FMRP was shown to regulate activity-dependent transport of SAPAP4 mRNAs into dendrites (70). In conjunction with our observations, these data suggest that FMRP regulates the translation and/or dendritic trafficking of all four SAPAP mRNAs. The recent finding that SAPAP3-deficient mice exhibit a phenotypic behavior reminiscent of obsessive-compulsive disorder in humans (71) underscores the particular significance of these scaffold proteins for the correct functionality of the synapse and further supports the hypothesis that deregulated levels of SAPAPs contribute to FXS-associated synaptic dysfunctions. An increased postsynaptic abundance of IRSp53 in FMRP-deficient brains may play a similar role in FXS pathogenesis as both IRSp53 and SAPAPs are able to connect PSD-95 to Shank scaffold proteins (32). In accordance with this hypothesis, IRSp53-deficient mice exhibit impaired learning (72, 73).
We further identified Shank1 mRNAs as additional in vivo targets of FMRP. Again, neither the cytoplasmic nor the synaptic Shank1 transcript levels are altered in the absence of FMRP in both the neocortex and the hippocampus. These results are consistent with the finding that Shank1 mRNA levels are identical in primary hippocampal neurons prepared from Fmr1−/− and wild-type mice (22). FMRP therefore does not appear to regulate the stability of Shank1 transcripts. The prevalent view in FXS is that excess protein synthesis at synapses causes synaptic dysfunction and cognitive impairment (2, 8, 9). This hypothesis is consistent with our observation that the postsynaptic levels of proteins investigated herein, if altered, are primarily increased. Moreover, we show that via an interaction with the proximal 588 nucleotides of the Shank1 3′UTR FMRP diminishes mRNA translation in primary neurons at basal state. Consistently, we observed increased Shank1 levels at both cortical and hippocampal synapses of FMRP-deficient mice. In addition to excess translation of FMRP target mRNAs at basal state, it is generally assumed that FMRP deficiency impairs the activity-dependent regulation of synaptic protein synthesis (2, 8, 9). Accordingly, mGluR stimulation enhanced the translation of reporter mRNAs containing the proximal part of the Shank1 3′UTR in wild-type but not in FMRP-deficient primary neurons. Taken together, our data suggest that although FMRP does not regulate the abundance of Shank1 transcripts in neurons, its deficiency results in both an excess basal and a loss of mGluR-induced Shank1 mRNA translation. Altered translational regulation of Shank1 transcripts in particular may contribute to the FXS pathology, as Shank1 strongly affects spine morphology (74). In fact, when overexpressed, Shank1 can induce aberrant spine formation along otherwise spineless dendrites (75). In FXS patients, elevated PSD levels of Shank1 may thus stabilize spines, which are normally lost during maturation and pruning of synapses. The view that Shank1 levels need to be maintained within a very narrow range to ensure physiological synapse function is supported by the observation that loss of only one copy of the SHANK3 gene in humans causes mental retardation as observed in conjunction with the 22q13 deletion syndrome (76, 77). Shank proteins regulate the size and shape of dendritic spines because of their capacity to directly interact with many different PSD components and are assumed to represent master scaffold proteins of the PSD (11). Thus, we hypothesize that the observed increase of Shank1 levels at synapses resulting from the cellular loss of FMRP strongly interferes with the mental capacities of FXS patients.
Acknowledgments
We thank Claudia Schob and Evita Mohr (University Medical Center Hamburg-Eppendorf) for generously providing pFiRe-basic and PABP antibodies, respectively, and Susanne Sehner (University Medical Center Hamburg-Eppendorf) for providing assistance with the statistical data analysis.
This work was supported by the Deutsche Forschungsgemeinschaft Grants Ki488/2-6 (to S. K.) and KR 1321/4-1 (to H.-J. K. and S. K.) and Thyssen-Stiftung Az. 10.05.2.185 (to D. R. and S. K.). This article is based in part on a doctoral study by J. S. at the Faculty of Biology, University of Hamburg.
- FMRP
- fragile X mental retardation protein
- CMV
- cytomegalovirus
- DHPG
- (S)-3,5-dihydroxyphenylglycine
- FXS
- fragile X syndrome
- GluR
- α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid-type glutamate receptor subunit
- mGluR
- metabotropic glutamate receptors
- NR
- N-methyl-d-aspartate type glutamate receptor subunit
- PABP
- poly(A)-binding protein
- PhoLuc
- Photinus luciferase
- RGS4
- regulator of G-protein signaling
- PSD
- postsynaptic density
- RenLuc
- Renilla luciferase
- RT-PCR
- reverse transcription-initiated PCR
- UTR
- untranslated region
- fw
- forward
- rev
- reverse
- IP
- immunoprecipitate
- wtF-IP
- wild-type mice using antibodies directed against FMRP
- koF-IP
- FMRP not present in anti-FMRP precipitates from knock-out brains
- wtIgG-IP
- wild-type homogenates utilizing irrelevant IgG.
REFERENCES
- 1.Bardoni B., Davidovic L., Bensaid M., Khandjian E. W. (2006) Expert Rev. Mol. Med. 8, 1–16 [DOI] [PubMed] [Google Scholar]
- 2.Bassell G. J., Warren S. T. (2008) Neuron 60, 201–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Darnell J. C., Mostovetsky O., Darnell R. B. (2005) Genes Brain Behav. 4, 341–349 [DOI] [PubMed] [Google Scholar]
- 4.Oostra B. A., Willemsen R. (2009) Biochim. Biophys. Acta 1790, 467–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bagni C., Greenough W. T. (2005) Nat. Rev. Neurosci. 6, 376–387 [DOI] [PubMed] [Google Scholar]
- 6.Garber K. B., Visootsak J., Warren S. T. (2008) Eur. J. Hum. Genet. 16, 666–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grossman A. W., Aldridge G. M., Weiler I. J., Greenough W. T. (2006) J. Neurosci. 26, 7151–7155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bear M. F., Dölen G., Osterweil E., Nagarajan N. (2008) Neuropsychopharmacology 33, 84–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pfeiffer B. E., Huber K. M. (2009) Neuroscientist 10.1177/1073858409333075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bassell G. J., Gross C. (2008) Nat. Med. 14, 249–250 [DOI] [PubMed] [Google Scholar]
- 11.Gundelfinger E. D., Boeckers T. M., Baron M. K., Bowie J. U. (2006) Trends Biochem. Sci. 31, 366–373 [DOI] [PubMed] [Google Scholar]
- 12.Okabe S. (2007) Mol. Cell. Neurosci. 34, 503–518 [DOI] [PubMed] [Google Scholar]
- 13.Sheng M., Hoogenraad C. C. (2007) Annu. Rev. Biochem. 76, 823–847 [DOI] [PubMed] [Google Scholar]
- 14.Sutton M. A., Schuman E. M. (2006) Cell 127, 49–58 [DOI] [PubMed] [Google Scholar]
- 15.Kindler S., Wang H., Richter D., Tiedge H. (2005) Annu. Rev. Cell Dev. Biol. 21, 223–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang Y. S., Richter J. D. (2007) Methods Enzymol. 431, 143–162 [DOI] [PubMed] [Google Scholar]
- 17.Bramham C. R., Wells D. G. (2007) Nat. Rev. Neurosci. 8, 776–789 [DOI] [PubMed] [Google Scholar]
- 18.Böckers T. M., Segger-Junius M., Iglauer P., Bockmann J., Gundelfinger E. D., Kreutz M. R., Richter D., Kindler S., Kreienkamp H. J. (2004) Mol. Cell. Neurosci. 26, 182–190 [DOI] [PubMed] [Google Scholar]
- 19.Ju W., Morishita W., Tsui J., Gaietta G., Deerinck T. J., Adams S. R., Garner C. C., Tsien R. Y., Ellisman M. H., Malenka R. C. (2004) Nat. Neurosci. 7, 244–253 [DOI] [PubMed] [Google Scholar]
- 20.Kindler S., Rehbein M., Classen B., Richter D., Böckers T. M. (2004) Brain Res. Mol. Brain Res. 126, 14–21 [DOI] [PubMed] [Google Scholar]
- 21.Muddashetty R. S., Keliæ S., Gross C., Xu M., Bassell G. J. (2007) J. Neurosci. 27, 5338–5348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zalfa F., Eleuteri B., Dickson K. S., Mercaldo V., De Rubeis S., di Penta A., Tabolacci E., Chiurazzi P., Neri G., Grant S. G., Bagni C. (2007) Nat. Neurosci. 10, 578–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Falley K., Schütt J., Iglauer P., Menke K., Maas C., Kneussel M., Kindler S., Wouters F. S., Richter D., Kreienkamp H. J. (2009) Traffic 10, 844–857 [DOI] [PubMed] [Google Scholar]
- 24.Blichenberg A., Schwanke B., Rehbein M., Garner C. C., Richter D., Kindler S. (1999) J. Neurosci. 19, 8818–8829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carlin R. K., Grab D. J., Cohen R. S., Siekevitz P. (1980) J. Cell Biol. 86, 831–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kistner U., Wenzel B. M., Veh R. W., Cases-Langhoff C., Garner A. M., Appeltauer U., Voss B., Gundelfinger E. D., Garner C. C. (1993) J. Biol. Chem. 268, 4580–4583 [PubMed] [Google Scholar]
- 27.Valtschanoff J. G., Burette A., Davare M. A., Leonard A. S., Hell J. W., Weinberg R. J. (2000) Eur. J. Neurosci. 12, 3605–3614 [DOI] [PubMed] [Google Scholar]
- 28.Müller B. M., Kistner U., Kindler S., Chung W. J., Kuhlendahl S., Fenster S. D., Lau L. F., Veh R. W., Huganir R. L., Gundelfinger E. D., Garner C. C. (1996) Neuron 17, 255–265 [DOI] [PubMed] [Google Scholar]
- 29.Kim E., Cho K. O., Rothschild A., Sheng M. (1996) Neuron 17, 103–113 [DOI] [PubMed] [Google Scholar]
- 30.Zitzer H., Hönck H. H., Bächner D., Richter D., Kreienkamp H. J. (1999) J. Biol. Chem. 274, 32997–33001 [DOI] [PubMed] [Google Scholar]
- 31.Redecker P., Bockmann J., Böckers T. M. (2006) Histochem. Cell Biol. 126, 679–685 [DOI] [PubMed] [Google Scholar]
- 32.Soltau M., Berhörster K., Kindler S., Buck F., Richter D., Kreienkamp H. J. (2004) J. Neurochem. 90, 659–665 [DOI] [PubMed] [Google Scholar]
- 33.Brendel C., Rehbein M., Kreienkamp H. J., Buck F., Richter D., Kindler S. (2004) Biochem. J. 384, 239–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Monshausen M., Rehbein M., Richter D., Kindler S. (2002) J. Neurochem. 81, 557–564 [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 36.Iacoangeli A., Rozhdestvensky T. S., Dolzhanskaya N., Tournier B., Schütt J., Brosius J., Denman R. B., Khandjian E. W., Kindler S., Tiedge H. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 734–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kurth I., Thompson D. A., Rüther K., Feathers K. L., Chrispell J. D., Schroth J., McHenry C. L., Schweizer M., Skosyrski S., Gal A., Hübner C. A. (2007) Mol. Cell. Biol. 27, 1370–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pfaffl M. W., Horgan G. W., Dempfle L. (2002) Nucleic Acids Res. 30, e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pedersen S. K., Christiansen J., Hansen T. O., Larsen M. R., Nielsen F. C. (2002) Biochem. J. 363, 37–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blichenberg A., Rehbein M., Müller R., Garner C. C., Richter D., Kindler S. (2001) Eur. J. Neurosci. 13, 1881–1888 [DOI] [PubMed] [Google Scholar]
- 41.Burgin K. E., Waxham M. N., Rickling S., Westgate S. A., Mobley W. C., Kelly P. T. (1990) J. Neurosci. 10, 1788–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Welch J. M., Wang D., Feng G. (2004) J. Comp. Neurol. 472, 24–39 [DOI] [PubMed] [Google Scholar]
- 43.Garner C. C., Tucker R. P., Matus A. (1988) Nature 336, 674–677 [DOI] [PubMed] [Google Scholar]
- 44.Wang H., Iacoangeli A., Popp S., Muslimov I. A., Imataka H., Sonenberg N., Lomakin I. B., Tiedge H. (2002) J. Neurosci. 22, 10232–10241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.West N., Roy-Engel A. M., Imataka H., Sonenberg N., Deininger P. (2002) J. Mol. Biol. 321, 423–432 [DOI] [PubMed] [Google Scholar]
- 46.Muddashetty R., Khanam T., Kondrashov A., Bundman M., Iacoangeli A., Kremerskothen J., Duning K., Barnekow A., Hüttenhofer A., Tiedge H., Brosius J. (2002) J. Mol. Biol. 321, 433–445 [DOI] [PubMed] [Google Scholar]
- 47.Weiler I. J., Irwin S. A., Klintsova A. Y., Spencer C. M., Brazelton A. D., Miyashiro K., Comery T. A., Patel B., Eberwine J., Greenough W. T. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 5395–5400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Waung M. W., Huber K. M. (2009) Curr. Opin. Neurobiol. 10.1016/j.conb.2009.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Narayanan U., Nalavadi V., Nakamoto M., Pallas D. C., Ceman S., Bassell G. J., Warren S. T. (2007) J. Neurosci. 27, 14349–14357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dölen G., Osterweil E., Rao B. S., Smith G. B., Auerbach B. D., Chattarji S., Bear M. F. (2007) Neuron 56, 955–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grossman A. W., Elisseou N. M., McKinney B. C., Greenough W. T. (2006) Brain Res. 1084, 158–164 [DOI] [PubMed] [Google Scholar]
- 52.Jin P., Warren S. T. (2003) Trends Biochem. Sci. 28, 152–158 [DOI] [PubMed] [Google Scholar]
- 53.Irwin S. A., Patel B., Idupulapati M., Harris J. B., Crisostomo R. A., Larsen B. P., Kooy F., Willems P. J., Cras P., Kozlowski P. B., Swain R. A., Weiler I. J., Greenough W. T. (2001) Am. J. Med. Genet. 98, 161–167 [DOI] [PubMed] [Google Scholar]
- 54.Irwin S. A., Idupulapati M., Gilbert M. E., Harris J. B., Chakravarti A. B., Rogers E. J., Crisostomo R. A., Larsen B. P., Mehta A., Alcantara C. J., Patel B., Swain R. A., Weiler I. J., Oostra B. A., Greenough W. T. (2002) Am. J. Med. Genet. 111, 140–146 [DOI] [PubMed] [Google Scholar]
- 55.Nimchinsky E. A., Oberlander A. M., Svoboda K. (2001) J. Neurosci. 21, 5139–5146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lauterborn J. C., Rex C. S., Kramár E., Chen L. Y., Pandyarajan V., Lynch G., Gall C. M. (2007) J. Neurosci. 27, 10685–10694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park S., Park J. M., Kim S., Kim J. A., Shepherd J. D., Smith-Hicks C. L., Chowdhury S., Kaufmann W., Kuhl D., Ryazanov A. G., Huganir R. L., Linden D. J., Worley P. F. (2008) Neuron 59, 70–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilson B. M., Cox C. L. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 2454–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nosyreva E. D., Huber K. M. (2006) J. Neurophysiol. 95, 3291–3295 [DOI] [PubMed] [Google Scholar]
- 60.Groc L., Bard L., Choquet D. (2009) Neuroscience 158, 4–18 [DOI] [PubMed] [Google Scholar]
- 61.Lau C. G., Zukin R. S. (2007) Nat. Rev. Neurosci. 8, 413–426 [DOI] [PubMed] [Google Scholar]
- 62.Fähling M., Mrowka R., Steege A., Kirschner K. M., Benko E., Förstera B., Persson P. B., Thiele B. J., Meier J. C., Scholz H. (2009) J. Biol. Chem. 284, 4255–4266 [DOI] [PubMed] [Google Scholar]
- 63.Bechara E. G., Didiot M. C., Melko M., Davidovic L., Bensaid M., Martin P., Castets M., Pognonec P., Khandjian E. W., Moine H., Bardoni B. (2009) PLoS Biol. 7, e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cuthbert P. C., Stanford L. E., Coba M. P., Ainge J. A., Fink A. E., Opazo P., Delgado J. Y., Komiyama N. H., O'Dell T. J., Grant S. G. (2007) J. Neurosci. 27, 2673–2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Migaud M., Charlesworth P., Dempster M., Webster L. C., Watabe A. M., Makhinson M., He Y., Ramsay M. F., Morris R. G., Morrison J. H., O'Dell T. J., Grant S. G. (1998) Nature 396, 433–439 [DOI] [PubMed] [Google Scholar]
- 66.Tarpey P., Parnau J., Blow M., Woffendin H., Bignell G., Cox C., Cox J., Davies H., Edkins S., Holden S., Korny A., Mallya U., Moon J., O'Meara S., Parker A., Stephens P., Stevens C., Teague J., Donnelly A., Mangelsdorf M., Mulley J., Partington M., Turner G., Stevenson R., Schwartz C., Young I., Easton D., Bobrow M., Futreal P. A., Stratton M. R., Gecz J., Wooster R., Raymond F. L. (2004) Am. J. Hum. Genet. 75, 318–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Todd P. K., Mack K. J., Malter J. S. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 14374–14378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Narayanan U., Nalavadi V., Nakamoto M., Thomas G., Ceman S., Bassell G. J., Warren S. T. (2008) J. Biol. Chem. 283, 18478–18482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brown V., Jin P., Ceman S., Darnell J. C., O'Donnell W. T., Tenenbaum S. A., Jin X., Feng Y., Wilkinson K. D., Keene J. D., Darnell R. B., Warren S. T. (2001) Cell 107, 477–487 [DOI] [PubMed] [Google Scholar]
- 70.Dictenberg J. B., Swanger S. A., Antar L. N., Singer R. H., Bassell G. J. (2008) Dev. Cell 14, 926–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Welch J. M., Lu J., Rodriguiz R. M., Trotta N. C., Peca J., Ding J. D., Feliciano C., Chen M., Adams J. P., Luo J., Dudek S. M., Weinberg R. J., Calakos N., Wetsel W. C., Feng G. (2007) Nature 448, 894–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim M. H., Choi J., Yang J., Chung W., Kim J. H., Paik S. K., Kim K., Han S., Won H., Bae Y. S., Cho S. H., Seo J., Bae Y. C., Choi S. Y., Kim E. (2009) J. Neurosci. 29, 1586–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sawallisch C., Berhörster K., Disanza A., Mantoani S., Kintscher M., Stoenica L., Dityatev A., Sieber S., Kindler S., Morellini F., Schweizer M., Boeckers T. M., Korte M., Scita G., Kreienkamp H. J. (2009) J. Biol. Chem. 284, 9225–9236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sala C., Piëch V., Wilson N. R., Passafaro M., Liu G., Sheng M. (2001) Neuron 31, 115–130 [DOI] [PubMed] [Google Scholar]
- 75.Roussignol G., Ango F., Romorini S., Tu J. C., Sala C., Worley P. F., Bockaert J., Fagni L. (2005) J. Neurosci. 25, 3560–3570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wilson H. L., Wong A. C., Shaw S. R., Tse W. Y., Stapleton G. A., Phelan M. C., Hu S., Marshall J., McDermid H. E. (2003) J. Med. Genet. 40, 575–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bonaglia M. C., Giorda R., Borgatti R., Felisari G., Gagliardi C., Selicorni A., Zuffardi O. (2001) Am. J. Hum. Genet. 69, 261–268 [DOI] [PMC free article] [PubMed] [Google Scholar]