Abstract
Androgen receptor (AR) is phosphorylated at multiple sites in response to ligand binding, but the functional consequences and mechanisms regulating AR phosphorylation remain to be established. We observed initially that okadaic acid, an inhibitor of the major PPP family serine/threonine phosphatases PP2A and protein phosphatase 1 (PP1), had cell type-dependent effects on AR expression. More specific inhibitors of PP2A (fostriecin) and PP1 (tautomycin and siRNA against the PP1α catalytic subunit) demonstrated that PP1 and protein phosphatase 2A had opposite effects on AR protein and transcriptional activity. PP1 inhibition enhanced proteasome-mediated AR degradation, while PP1α overexpression increased AR expression and markedly enhanced AR transcriptional activity. Coprecipitation experiments demonstrated an AR-PP1 interaction, while immunofluorescence and nuclear-cytoplasmic fractionation showed androgen-stimulated nuclear translocation of both AR and PP1 in prostate cancer cells. Studies with phosphospecific AR antibodies showed that PP1 inhibition dramatically increased phosphorylation of Ser-650, a site in the AR hinge region shown to mediate nuclear export. Significantly, PP1 inhibition caused a marked decrease in nuclear localization of the wild-type AR, but did not alter total or nuclear levels of a S650A mutant AR. These findings reveal a critical role of PP1 in regulating AR protein stability and nuclear localization through dephosphorylation of Ser-650. Moreover, AR may function as a PP1 regulatory subunit and mediate PP1 recruitment to chromatin, where it can modulate transcription and splicing.
Androgen receptor (AR)3 plays a central role in prostate cancer (PCa) development and progression, with androgen deprivation therapy being the standard systemic treatment for PCa (1). Unliganded AR associates with an Hsp90 chaperone complex and is rapidly degraded. Ligand binding stabilizes AR, enhances nuclear entry, and allows AR to recruit coactivator proteins to androgen-regulated genes. AR is phosphorylated at Ser-94 and Ser-650 in the absence of androgen, and androgen treatment further stimulates AR phosphorylation, primarily at multiple serine-proline sites (2–5). Similar to other steroid receptors, AR transcriptional activity and sensitivity to low levels of androgen can be enhanced by multiple kinases or kinase signaling pathways, which may contribute to tumor progression subsequent to androgen deprivation therapy. However, the kinases mediating AR phosphorylation at specific sites, and the functional importance of AR phosphorylation at particular residues, remain to be clearly defined.
AR transcriptional activity may also be modulated directly or indirectly by serine/threonine phosphatases (6). A recent study demonstrated SV40 small T-antigen-dependent loading of protein phosphatase 2A (PP2A) onto AR, with subsequent AR dephosphorylation (7, 8). AR also interacts with small C-terminal domain phosphatase 2 (SCP2), which is recruited by AR to the androgen-regulated PSA promoter and negatively regulates AR transcriptional activity, possibly by dephosphorylation of RNA polymerase II (9). We report here that protein phosphatase 1 (PP1) increases AR protein stability and markedly enhances AR-mediated transcription. We show that AR binds the catalytic subunit of PP1, PP1α, and that androgens stimulate nuclear translocation of PP1α in conjunction with AR. Moreover, we determine that PP1 selectively dephosphorylates a specific site on the AR, Ser-650. Finally, consistent with a recent report that Ser-650 phosphorylation mediates AR nuclear export (10), we demonstrate that PP1 inhibition markedly decreases nuclear AR. These findings demonstrate that PP1 is a direct positive regulator of AR nuclear expression and transcriptional activity and identifies the AR-PP1 interaction as a potential therapeutic target for PCa drug development.
EXPERIMENTAL PROCEDURES
Reagents
Sources were as follows: steroids, MG115, MG132, CHX, and anti-FLAG M2 beads (Sigma); OA (Roche Applied Science); tautomycin, and fostriecin (Calbiochem); anti-AR(PG21), anti-pAR-S81, anti-PP1α, anti-PP2A, and microcystin-agarose (Upstate Biotechnology); anti-AR (N441) (Lab Vision); anti-PSA (Biodesign); anti-β-tubulin (Chemicon); normal mouse serum (NMS), normal rabbit serum (NRS), protein G, and NE-PER kit (Pierce); goat anti-rabbit Alexa 594 (Molecular Probes); serum (FBS and CDS) (Hyclone); PP1α plasmid (Origene); control and AR RNAi (Dharmacon). Two separate PP1α RNAi were from Dharmacon (SMARTpool, M-008927-00) and Santa Cruz (sc-36299).
Transient Transfection Reporter Assays and Real-time RT-PCR
AR expression and reporter plasmids, reporter assays, and real-time RT-PCR analyses have been described (12). Cells were grown in RPMI 1640 with 10% FBS (LNCaP and C4-2) or Dulbecco's modified Eagle's medium with 5% FBS. For androgen starvation, cells were grown in medium containing 5% charcoal dextran stripped FBS (CDS). For transfection, plasmid DNA or RNAi was transfected with Lipofectamine 2000. Empty pCDNA3.1 vector (Invitrogen) was used for equalization and a CMV-Renilla luciferase reporter (Promega) was an internal control. Luciferase was measured with a Dual-Luciferase assay kit (Promega). The ratio between firefly and Renilla luciferase is relative light unit (RLU), and the results are mean and standard deviation from triplicate samples.
DNA Mutagenesis and Generation of Stable Lines
The Flag-AR-S650A mutant was generated with the Site-directed Mutagenesis kit (Stratagene). LNCaP cell stable lines expressing Flag-AR or Flag-AR-S650A were established by selection of cells grown in RPMI 1640 with 10% FBS and 1 μg/ml puromycin.
Immunoblotting, Nuclear Fractionation, Coprecipitation, and Immunofluorescence
Total proteins were isolated with 2% SDS, quantified with BCA reagent (Pierce), and equal amounts were analyzed on prepoured gels (Invitrogen). Cytoplasmic-nuclear fractionation was with the NE-PER kit (Pierce). For coprecipitation, cells were harvested in Triton lysis buffer (0.5% Triton X-100, 20 mm Tris-HCl, 150 mm NaCl, 5 mm EDTA, and 2 mm dithiothreitol) with protease and phosphatase inhibitors. Supernatants were incubated with 40 μl of microcystin-agarose, anti-Flag M2, or anti-AR (N441) versus NMS cross-linked to protein G beads.
To assess AR phosphorylation, AR was first precipitated by anti-AR (N441), and equal amounts of total AR were immunoblotted with a panel of phosphospecific AR Abs (7, 10). For immunofluorescence, cells were fixed in 3.7% formaldehyde for 10 min, washed in PBS, and permeabilized in 0.1% Triton X-100 for 3 min. After washing in PBS, cells were blocked in 1% bovine serum albumin for 30 min and incubated in primary antibody (1:200) for 2 h. After washing in PBS, cells were incubated in goat anti-rabbit Alexa 594 (1:400) for 2 h, washed in PBS, and stained with 4′,6-diamidino-2-phenylindole (DAPI) (1:2000) for 5 min. After washing in PBS, cells were mounted on slides and observed with the ZEISS ApoTome-HAL100 Microscope.
RESULTS
Protein Phosphatase Inhibitor Okadaic Acid Has Cell Type-specific Effects on AR Protein Expression
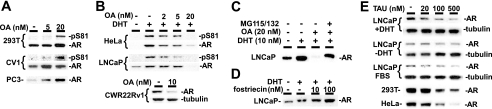
We initially examined protein levels of transfected AR in a series of cell lines after treatment with okadaic acid (OA), a nonspecific inhibitor of multiple phosphatases including PP1 and PP2A (11). OA increased AR protein levels in transfected 293T, CV1, and PC3 cells, which was accompanied by substantial increases in phosphorylation of Ser-81 (as detected by a pS81 antibody) (Fig. 1A). The Ser-81 site is phosphorylated in response to androgen stimulation by proline-directed serine/threonine kinases, including Cdk1, and is associated with AR stabilization and transcriptional activity (4, 12). These results were consistent with a previous report showing that PP2A can dephosphorylate several proline-directed serine sites in the AR N-terminal domain (including Ser-81), although PP2A interaction with AR in this previous study was dependent on loading by SV40 small T antigen (7, 8). However, OA had the opposite effect on AR protein expression and Ser-81 phosphorylation in AR-transfected HeLa cells, where the androgen (DHT)-stimulated increase in AR protein expression and Ser-81 phosphorylation were both markedly decreased by OA (Fig. 1B). Significantly, OA similarly caused a dose-dependent decrease in expression of the endogenous AR in the LNCaP and CWR22Rv1 PCa cell lines (Fig. 1B). This OA-induced decrease in endogenous AR protein was blocked by MG115/132, indicating that it was proteasome-mediated (Fig. 1C). The decreased mobility of the MG115/132 rescued AR is consistent with increased phosphorylation.
FIGURE 1.
Inhibition of PP1, but not PP2A, leads to reduction in AR protein expression. A, 293T, CV1, and PC3 cells were transfected with 100 ng of pCIneo-hAR plasmid and grown in CDS medium with 10 nm DHT and different doses of OA for 24 h, as indicated. In all experiments, cells were harvested in 2% SDS and equal amounts of total proteins were immunoblotted. B, HeLa cells were AR transfected as above, while endogenous AR was monitored in LNCaP and CWR22Rv1 cells. C and D, LNCaP cells in CDS medium were treated for 24 h with 10 nm DHT, 20 nm OA, proteasome inhibitors MG(MG115/132, 5 μm each), or fostriecin, as indicated. E, cells with endogenous (LNCaP) or transfected (293T and HeLa) AR were cultured in CDS medium ± 10 nm DHT (LNCaP) or in FBS medium (LNCaP, 293T, HeLa), with tautomycin (TAU) as indicated for 24 h.
PP1 Enhances AR Protein Expression-independent of Cell Type
The cell-specific effects of OA led us to examine more selective inhibitors of PP1 and PP2A, the major cellular phosphatases that are inhibited by OA. Fostriecin, a PP2A specific inhibitor, increased endogenous AR protein expression in LNCaP cells (Fig. 1D) and increased the expression of transfected AR in HeLa, COS1, and 293T cells (data not shown), indicating that PP2A negatively regulates AR expression in cells that do not express SV40 T antigen. In contrast to fostriecin, the PP1 inhibitor tautomycin gave a dose-dependent decrease in endogenous AR protein expression in LNCaP cells cultured in medium with steroid-depleted serum (charcoal dextran-stripped, CDS), with or without DHT, or in medium with nondepleted serum (FBS) (Fig. 1E). Tautomycin similarly decreased expression of transfected AR in 293T and HeLa cells (Fig. 1E).
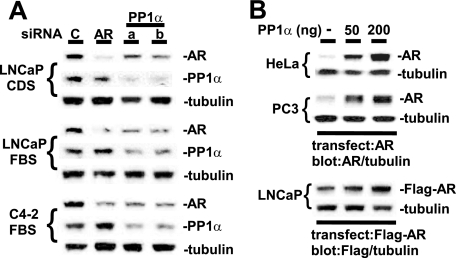
To confirm that the effect of tautomycin on endogenous AR in LNCaP cells was due to PP1 inhibition, we used siRNA to down-regulate endogenous PP1 activity. Mammalian cells have three homologous genes that encode PP1 catalytic subunits, PPP1CA, PPP1CB, and PPP1CC, which are the direct targets of tautomycin (13, 14). Affymetrix oligonucleotide microarray studies indicate that LNCaP cells express all three catalytic subunits, with expression of PPP1CA and PPP1CC being greater than PPP1CB (data not shown). By immunoblotting we could readily detect the protein product of PPP1CA, PP1α. Therefore, we used two different siRNA pools against PPP1CA to determine whether endogenous PP1α was enhancing AR expression in LNCaP cells. Both PPP1CA siRNA, but not a control siRNA or an AR siRNA, decreased the levels of endogenous PP1α in LNCaP cells and in the LNCaP-derived C4-2B cell line (Fig. 2A). Significantly, this PP1α down-regulation also caused a decrease in AR levels in the LNCaP cells cultured in the presence or absence of DHT and in the C4-2B cells.
FIGURE 2.
AR protein expression is increased by PP1. A, 10 nm of control nonspecific RNAi (lane C), AR RNAi (lane AR), or two separate PP1α RNAi (lanes a and b) were transfected into LNCaP or C4-2B cells for 3 days. B, HeLa and PC3 cells were transfected with 100 ng of pCIneo-hAR plasmid and LNCaP cells were transfected with 100 ng of Flag epitope-tagged AR (Flag-AR) plasmid, together with 0, 50, or 200 ng of PP1α vector. The medium was then changed to CDS medium with 10 nm DHT for 24 h.
As a further approach, we determined whether overexpression of PP1α would increase expression of AR protein. Indeed, co-transfection of PP1α markedly increased the expression of AR in both HeLa and PC3 cells, and these increases were observed in the absence and presence of DHT (Fig. 2B and data not shown). To study the effect of PP1 on AR expression in PCa cells expressing endogenous AR, LNCaP cells were cotransfected with PP1α and Flag epitope-tagged AR (Flag-AR) to distinguish exogenous AR from endogenous AR in nontransfected cells. As shown in Fig. 2B, PP1α also increased Flag-AR in LNCaP cells, although the effect was not as marked as in HeLa and PC3 cells (possibly reflecting a higher level of endogenous PP1 activity in LNCaP cells). Taken together, these results confirmed that PP1 was enhancing AR expression and indicated that PP1α was making a major contribution to this effect.
PP1 Inhibition Increases AR Protein Degradation via Proteasome-dependent Pathway
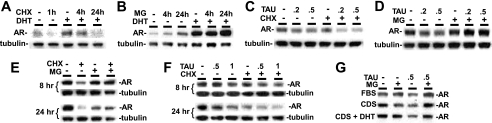
Previous studies have shown that the unliganded AR is rapidly degraded, while the liganded AR is more stable but still has a relatively short half-life (15). In agreement with these previous studies, cycloheximide treatment to block new protein synthesis in AR-transfected HeLa cells caused a substantial decline in AR protein levels over 4 h, with a more rapid decline in the absence of androgen (Fig. 3A). Proteasome inhibitors (MG115/132) increased transfected AR protein expression in the absence and presence of ligand (Fig. 3B), consistent with previous studies showing that AR is degraded through a proteasome-dependent pathway (16).
FIGURE 3.
PP1 inhibition promotes AR protein degradation via the proteasome pathway. A–D, HeLa cells were transfected with 100 ng of pCIneo-hAR plasmid and then placed into CDS medium with 10 nm DHT as indicated for 24 h. A, cycloheximide (CHX, 5 μg/ml) was added during the last 1 h, 4 h, or during entire 24 h. B, MG (MG115/132, 5 μm each) were added for last 4 h or entire 24 h. C, cells were treated with DHT and tautomycin (TAU) as indicated (0.2 or 0.5 μm) for 24 h, with cycloheximide added during last 4 h. D, cells were treated for 24 h with DHT, with compounds added during last 8 h. E and F, LNCaP cells were cultured in CDS for 2 days, followed by 10 nm DHT for 24 h. Cycloheximide, MG115/132 (5 μm each), and tautomycin (0.5 or 1.0 μm) were then added for the last 8 h or 24 h as indicated. G, LNCaP cells were grown in FBS, CDS, or CDS+10 nm DHT medium for 2 days, with MG115/132 (5 μm each) and tautomycin (0.5 μm) added during last 4 h.
To determine whether tautomycin reduces AR protein expression by increased degradation, we examined the effect of tautomycin after new protein synthesis was blocked with cycloheximide. As shown in Fig. 3C, the addition of tautomycin to cells treated with cycloheximide caused a further decrease in transfected AR protein levels, showing that PP1 inhibition was increasing AR protein degradation. To determine whether this degradation was through the proteasome, AR-transfected HeLa cells were subjected to tautomycin treatment in combination with MG115/132. As shown in Fig. 3D, the tautomycin-mediated decrease in AR expression could be blocked by MG115/132, indicating that tautomycin was increasing proteasome-dependent degradation of AR protein.
Similar experiments were carried out in LNCaP cells to examine the effects of PP1 inhibition on endogenous AR. Treatment of LNCaP cells with cycloheximide for 8 or 24 h caused a marked decrease in AR protein expression that was prevented by the addition of MG115/132, indicating that degradation of endogenous AR in LNCaP was through the proteasome (Fig. 3E). It should be noted that MG115/132 alone causes a decline in AR after 24 h, which reflects a decrease in AR message levels (data not shown). As observed above in HeLa cells, tautomycin for 8 h and 24 h caused a further decrease in AR levels in cycloheximide-treated LNCaP cells, showing that PP1 inhibition was increasing degradation of endogenous AR protein (Fig. 3F). This tautomycin-induced AR degradation was not observed in LNCaP cells treated with MG115/132, indicating that tautomycin was enhancing proteasome-dependent degradation of the endogenous AR (Fig. 3G). Similar results were obtained in LNCaP cells cultured in steroid hormone-depleted medium (charcoal-dextran-stripped FBS medium, CDS), CDS medium supplemented with DHT, or FBS medium (Fig. 3G).
PP1 Interacts with AR
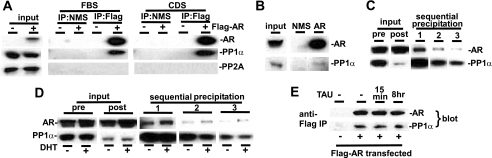
To assess a potential direct interaction between PP1 and AR, we carried out coprecipitation experiments in 293T cells cotransfected with Flag-AR and PP1α. Significantly, substantial levels of PP1α, but no detectable PP2A, were precipitated specifically by anti-Flag from the Flag-AR-transfected cells cultured in FBS or CDS medium (Fig. 4A). Importantly, PP1α could also be coimmunoprecipitated with endogenous AR from LNCaP cells (Fig. 4B). To further verify the interaction between endogenous PP1α and AR, we used microcystin, which binds tightly to PPP family phosphatases and has been widely used to precipitate PP1 and PP2A and associated proteins (17). For these experiments we first used microcystin-conjugated beads to efficiently precipitate PP1 from LNCaP cells grown in FBS medium. By immunoblotting, we readily detected AR as well as high levels of PP1α associated with the microcystin beads (Fig. 4C).
FIGURE 4.
AR binds to PP1α. A, 293T cells were transfected without or with 2 μg of Flag-AR vector and then cultured for 24 h in FBS or CDS medium. Lysates were precipitated with anti-Flag M2 beads or with normal mouse serum (NMS) beads as control. B, lysates from LNCaP cells in FBS medium were precipitated with anti-AR antibody (N441) or normal mouse serum (NMS), both cross-linked to protein G beads. C, lysates from LNCaP cells in FBS medium were precipitated sequentially three times with microcystin-agarose beads. Inputs are 2% of the lysate prior to the first precipitation (pre) and after the third precipitation (post). D, lysates from LNCaP cells in CDS medium, without or with 10 nm DHT for 24 h, were precipitated as in C. E, as in A, 293T cells in FBS medium were transfected with 1 μg of PP1CA, together with 2 μg of CMV-Flag vector or Flag-AR vector. Cells were then treated with 500 nm tautomycin for 15 min or 8 h as indicated. Lysates were precipitated with anti-Flag M2 beads.
To address whether AR was associating nonspecifically with the microcystin beads, we carried out serial precipitations from the lysates to deplete PP1 and determine whether this abrogated the AR association. As shown in Fig. 4C, three rounds of microcystin precipitation substantially depleted PP1α from the lysates, without markedly affecting AR levels (compare AR and PP1α inputs before the first immunoprecipitation, pre, and after the third precipitation, post). Importantly, AR precipitation by microcystin beads was markedly decreased in the PP1-depleted lysates, indicating that AR was not associating nonspecifically with microcystin beads. Similar results were obtained using lysates from LNCaP cells cultured in CDS medium, minus or plus DHT (Fig. 4D). The amount of PP1α-associated AR was increased in the presence of DHT, but total input AR levels were also enhanced by DHT, so the binding does not appear to be strongly ligand-dependent. Finally, the AR-PP1α interaction is not dependent on PP1 catalytic activity, as it was not altered by PP1 inhibition with tautomycin (Fig. 4E). Taken together, these findings support the conclusion that there is a direct or indirect (mediated by a PP1 regulatory subunit) interaction between AR and PP1α, and that this interaction does not require androgen.
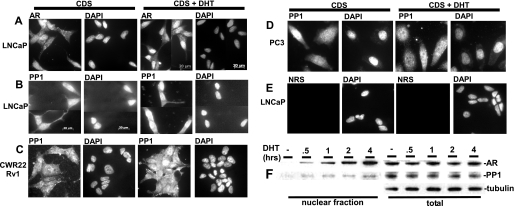
Androgens Stimulate PP1 Nuclear Translocation in AR-expressing Cells
The unliganded AR is distributed diffusely in the cell, and ligand binding stimulates rapid nuclear accumulation of AR in LNCaP cells (Fig. 5A). Therefore, to further assess whether there is a physiological AR-PP1 interaction, we next determined whether there was any detectable nuclear translocation of PP1α in response to androgen. Significantly, DHT induced the rapid nuclear accumulation of a fraction of endogenous PP1α in LNCaP and CWR22Rv1 PCa cells, but not in AR-negative PC3 cells (Fig. 5, B–D). Fractionation of nuclear proteins from DHT-treated LNCaP cells similarly showed a time-dependent nuclear entry of AR, accompanied by nuclear import of PP1α (Fig. 5F). These results provide further support for an AR-PP1 interaction and suggest that PP1α may associate with AR in the nucleus and participate in regulation of AR nuclear functions.
FIGURE 5.
Androgens stimulate PP1α nuclear translocation in AR-expressing cells. A–E, LNCaP, CWR22Rv1, and PC3 cells were grown on glass coverslips in CDS medium for 2 days, treated without or with 10 nm DHT for 15 min, and then fixed in formalin. Cells were then stained with anti-AR, anti-PP1, normal rabbit serum (NRS), or DAPI, and representative fields were photographed. F, LNCaP cells grown in CDS medium for 2 days were treated with 10 nm DHT for 30 min, 1, 2, and 4 h, and nuclear versus whole cell protein extracts were blotted.
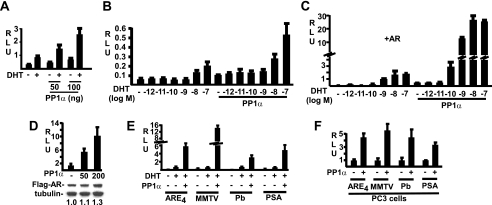
PP1 Enhances AR Transcriptional Activity
The data demonstrating an AR-PP1α interaction and coordinated nuclear translocation indicated that PP1 may participate in regulation of AR nuclear functions. To test this hypothesis, we first examined whether PP1α can modulate AR transcriptional activity. As shown in Fig. 6A, co-expression of PP1α enhanced the basal and DHT-stimulated transcriptional activity of the endogenous AR in LNCaP cells. To determine whether PP1 sensitizes AR to low levels of androgen, PP1α-transfected LNCaP cells were stimulated over a range of DHT concentrations. As shown in Fig. 6B, AR activity could be stimulated ∼4-fold by DHT in the absence of PP1α. This was enhanced ∼2.5-fold further when cells were cotransfected with PP1α, although there was a similar about 2-fold increase in basal activity in the absence of DHT. This stimulation did not appear to be a general effect on transcription, as there was no effect on a control CMV-regulated Renilla luciferase reporter gene (data not shown). Significantly, PP1α did not detectably lower the level of DHT required for stimulation (∼10 nm DHT).
FIGURE 6.
PP1 stimulates AR transcriptional activity. A, LNCaP cells were transfected with 2.5 ng of control Renilla reporter, 50 ng of ARE-luciferase reporter, and PP1α plasmids as indicated. The medium was changed to CDS without or with 10 nm DHT for 24 h, and relative light units (RLU, the ratio of firefly/Renilla luciferase) were assessed. B and C, LNCaP cells were transfected with Renilla control, ARE4-luciferase, 50 ng of PP1α plasmids, and cells in C were also cotransfected with 50 ng of pCIneo-hAR plasmid. The medium was changed to CDS plus different doses of DHT for 24 h. D, LNCaP cells were transfected with reporters and PP1α as in A, in addition to 100 ng of Flag-AR plasmid. Replicate wells were analyzed for luciferase activity or Flag-AR expression by anti-Flag immunoblotting. The band intensities for Flag antibody were normalized to β-tubulin, and the ratio in the absence of PP1 was set at 1. E and F, LNCaP (E) and PC3 (F) cells were transfected with AR (100 ng), PP1α (100 ng), Renilla luciferase control reporter, and the indicated AR-driven luciferase reporters (Pb: Probasin), and were then cultured for 24 h in CDS medium without or with 10 nm DHT as indicated. RLU is shown normalized to the non-PP1α samples.
The modest induction of AR activity by DHT in LNCaP cells transiently transfected with reporter genes appears to reflect limiting amounts of endogenous AR, and can be increased by cotransfection with AR. Therefore, we next examined LNCaP cells transfected with AR and an ARE4-luciferase reporter, minus or plus PP1α. Transfection of AR, minus PP1α, increased the DHT response to ∼13-fold induction at 10 nm DHT (Fig. 6C). Cotransfection with PP1α again modestly increased basal activity, but dramatically enhanced DHT-stimulated reporter gene activity by ∼14-fold over the level in the absence of PP1 (Fig. 6C). Because of this amplified activity a response could be detected clearly at 10-fold lower DHT (0.1 nm DHT), but half-maximal stimulation was observed at ∼1 nm DHT in the absence and presence of co-transfected PP1α.
One mechanism by which PP1 may be enhancing androgen-stimulated transcription is by increasing AR protein expression (although, as shown in Fig. 2B, PP1α transfection does not markedly increase total AR levels in LNCaP cells). Nonetheless, to address this mechanism, LNCaP cells were cotransfected with PP1α, ARE4-luciferase reporter gene, and Flag-AR. As shown in Fig. 6D, coexpression of PP1α and AR in LNCaP led to a marked increase in AR reporter gene activity. In contrast, in the same experiment there was a more modest increase in expression of the Flag-AR, suggesting that increased AR protein is not the sole basis for enhanced reporter gene expression. Finally, to study whether the effects of PP1 are promoter/enhancer-specific, LNCaP and PC3 cells (an AR-negative PCa cell line) in steroid-depleted medium were transiently transfected with AR and a panel of AR-regulated luciferase reporter genes, minus or plus PP1α, and DHT-stimulated activity was assessed. As shown in Fig. 6E (LNCaP) and 6F (PC3), co-transfection of PP1α strongly enhanced AR transactivation of all four androgen-regulated reporter genes. Taken together, the marked increases in DHT-stimulated AR reporter gene expression in AR and PP1α-cotransfected LNCaP cells, in conjunction with more modest increases in AR protein in these cells, indicate that PP1α can enhance AR transcriptional activity in addition to its effects on AR protein expression.
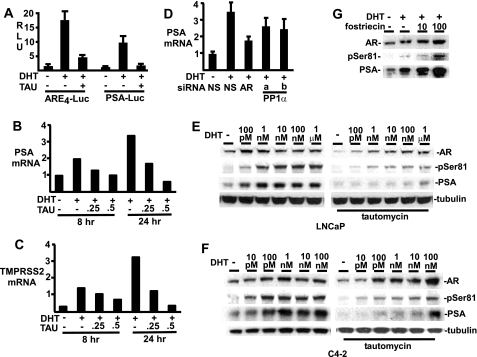
PP1 Inhibitor Tautomycin Impairs AR Transcriptional Activity
Consistent with the above results, transient transfection with AR-regulated reporter genes showed that the PP1 inhibitor tautomycin could markedly decrease DHT-stimulated transcriptional activity (Fig. 7A). Moreover, using quantitative real time RT-PCR, we found that tautomycin treatment of LNCaP cells could suppress DHT-stimulated expression of the endogenous androgen-regulated PSA and TMPRSS2 genes (Fig. 7, B and C). Consistent with these results, we also found that two different PP1α siRNA decreased DHT-stimulated PSA mRNA levels in LNCaP cells (Fig. 7D).
FIGURE 7.
PP1 inhibition reduces endogenous AR-mediated transcriptional activity. A, LNCaP cells were transfected with 2.5 ng of Renilla control, 50 ng of ARE4- or PSA-luciferase reporter plasmids. The medium was then changed to CDS without or with 10 nm DHT and 500 nm tautomycin as indicated for 24 h. B and C, LNCaP grown in CDS medium for 2 days were treated with DHT and tautomycin as indicated. Total RNA was isolated for real-time RT-PCR analysis of PSA (B) and TMPRSS2 (C) gene expression. Values shown are means of duplicate samples and are representative of two experiments. D, LNCaP were transfected with 20 nm of RNAi for nonspecific control (NS), AR, or PP1, incubated in CDS medium for 2 days, and then treated with 10 nm DHT for 24 h as indicated. Total RNA was isolated for real-time RT-PCR analysis of PSA gene expression. E and F, LNCaP (E) and C4-2 (F) cells were split in CDS medium for 2 days, and treated for 24 h with different doses of DHT, without or with 250 nm tautomycin. G, LNCaP cells split in CDS medium for 2 days were treated with 10 nm DHT 24 h, together with different doses (nm) of PP2A-specific inhibitor fostriecin.
To assess the effects of tautomycin on endogenous AR protein versus AR transcriptional activity, we examined a DHT dose response in LNCaP cells. In the absence of tautomycin, AR protein levels were increased, and PSA expression was strongly stimulated at 0.1–1 nm DHT (Fig. 7E). Tautomycin caused a decrease in total AR protein at lower DHT concentrations, but AR levels partially recovered at higher DHT concentrations. AR phosphorylation at Ser-81 also partially recovered at higher DHT concentrations, although the level of pS81 relative to total AR was still decreased at high DHT concentrations. In contrast, PSA levels were markedly decreased by tautomycin at low DHT concentrations, and did not recover at DHT concentrations up to 100 nm, despite the increases in total AR (Fig. 7E). Similar results were obtained in C4-2 cells, which have substantial basal AR activity in the absence of added DHT and respond to lower levels of DHT (Fig. 7F). Taken together, the PP1α transfection and tautomycin data demonstrate that endogenous PP1α can markedly enhance AR transcriptional activity, and that mechanisms in addition to AR protein stabilization may contribute to this effect.
In contrast to tautomycin, the PP2A inhibitor fostriecin increased AR protein and markedly increased both pS81 and PSA in LNCaP cells (Fig. 7G). This result confirms that PP2A negatively regulates AR in cells that do not express SV40 T antigen, and shows that protein phosphatases have distinct effects on AR expression and phosphorylation (Fig. 7G).
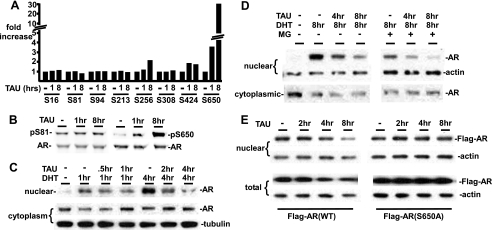
PP1 Inhibition Enhances AR Phosphorylation at Ser-650 and Decreases AR Nuclear Localization
The above results showed that PP1α could both stabilize AR and enhance it transcriptional activity, but it was not clear whether these were direct effects of PP1 on AR versus indirect effects of PP1 on other proteins that regulate AR stability and activity. Therefore, we next used a panel of phosphospecific antibodies directed at previously identified AR phosphorylation sites to determine whether AR phosphorylation at specific sites was increased by PP1 inhibition (7, 10). The AR was precipitated from tautomycin or control treated LNCaP cells, and equal amounts of total AR protein were then immunoblotted with phosphospecific AR antibodies. Tautomycin treatment resulted in a marked increase in the phosphorylation of Ser-650, and a modest increase in Ser-256 and Ser-424, with no clear change at Ser-16, Ser-81, Ser-94, Ser-213, or Ser-308 (Fig. 8, A and B and data not shown). This result indicates that PP1 may selectively dephosphorylate AR at Ser-650, although we cannot yet rule out dephosphorylation of other sites or the possibility that PP1 inhibition is strongly activating kinases that target Ser-650 (4, 10). Interestingly, a previous study of sites targeted by PP2A in the AR N terminus found that Ser-650 phosphorylation was not decreased by PP2A, consistent with Ser-650 being a site targeted specifically by PP1 (7).
FIGURE 8.
PP1 mediates AR dephosphorylation at Ser-650 and nuclear retention. A, LNCaP cells were split in FBS medium for 2 days, and 500 nm tautomycin was added for 1 or 8 h. AR was precipitated with anti-AR(N441), and equal amounts of total AR were immunoblotted with a panel of phospho-AR-specific antibodies and a total AR antibody. The band intensities for each phospho-antibody were normalized to the total AR, and the ratio in the absence of tautomycin was set at 1. Fold increase after 1 or 8 h of tautomycin were then determined, and the graph shows the average fold change from three independent experiments. B, representative blot showing results with pS81 and pS650 antibodies. C, LNCaP cells were grown in CDS medium for 2 days, and 10 nm DHT was added for 1 and 4 h. Tautomycin (500 nm) was added as indicated, and cells were harvested with the NE-PER kit for nuclear and cytoplasmic proteins. D, as in C, LNCaP cells grown in CDS medium were treated with 10 nm DHT and tautomycin, with and without proteasome inhibitor (MG115/132, 5 μm each) as indicated, and nuclear versus cytoplasmic AR were assessed. E, LNCaP stable lines expressing Flag-tagged AR wild-type or S650A mutant were grown in FBS medium and treated with tautomycin (500 nm) as indicated. Nuclear and total protein extracts were then immunoblotted with an anti-Flag Ab.
The functional significance of AR phosphorylation at most sites has not been established, but recent results indicate that AR nuclear export can be enhanced by Ser-650 phosphorylation and by OA treatment (10, 18). Therefore, we determined whether tautomycin treatment altered the nuclear localization of endogenous AR in LNCaP cells. As expected, DHT treatment of LNCaP cells in androgen-depleted medium stimulated a marked increase in nuclear AR and a corresponding decrease in cytoplasmic AR (Fig. 8C). The addition of tautomycin reduced this nuclear localization, with a marked decrease in the ratio of nuclear to cytoplasmic AR. Tautomycin also caused a loss in nuclear AR in cells that were co-treated with proteasome inhibitors (MG115 + MG132), consistent with a direct effect on AR cellular localization that is independent increased degradation (Fig. 8D). Taken together, these results indicate that PP1 inhibition is enhancing AR Ser-650 phosphorylation, nuclear export, and degradation.
Finally, to determine whether these effects of PP1 inhibition were dependent on increased Ser-650 phosphorylation, we examined an S650A mutant AR. For these experiments we generated LNCaP cells stably transfected with N-terminal Flag epitope-tagged wild-type or S650A AR. As observed with the endogenous AR, tautomycin treatment of cells expressing the Flag-tagged wild-type AR caused a marked loss of nuclear Flag-AR and a decrease in total Flag-AR expression (Fig. 8E). In contrast, tautomycin did not decrease levels of the Flag-AR(S650A) in the nucleus or decrease total cellular levels (Fig. 8E), indicating that these effects of PP1 inhibition are mediated by increased Ser-650 phosphorylation.
DISCUSSION
We initially observed cell type-specific effects on AR of a nonspecific phosphatase inhibitor, OA. Using a more specific PP1 inhibitor (tautomycin) and PP1 siRNA, we found that proteasome-mediated AR degradation was increased in response to PP1 suppression in all cell types examined. Conversely, overexpression of PP1α increased AR expression and transcriptional activity. In contrast, the PP2A specific inhibitor fostriecin enhanced AR protein expression and transactivation, supporting a physiological role for PP2A in negative AR regulation in cells that do not express SV40 small T-antigen (7). While an association between AR and PP2A could not be detected, presumably reflecting a weak and transient interaction, we readily detected an AR-PP1 interaction by coprecipitation that was independent of androgen. DHT treatment also stimulated the nuclear translocation of both endogenous AR and PP1α, supporting a physiological interaction and indicating that AR may function as a PP1 regulatory subunit for nuclear targeting of PP1. Significantly, PP1 inhibition caused a dramatic and specific increase in Ser-650 phosphorylation. Moreover, consistent with recent data showing that Ser-650 phosphorylation stimulates AR nuclear export (10), we found that PP1 inhibition markedly decreased nuclear AR. Finally, these effects of PP1 inhibition on AR levels and nuclear localization were abrogated by a S650A mutation, indicating that they are due to phosphorylation at this site. Taken together, these results demonstrate that PP1 interacts with AR and enhances its stability, that AR contributes to PP1 nuclear recruitment, and that PP1 enhances AR nuclear expression by selective dephosphorylation of Ser-650.
The increase in proteasome-mediated degradation of AR in response to PP1 inhibition by tautomycin may be secondary to increased nuclear export and subsequent exposure to ubiquitin ligases in the cytoplasm. However, Ser-650 is also imbedded in a PEST sequence (residues 638–658) that may directly target AR for degradation through proteasome- or caspase-3-dependent pathways (19–21). Significantly, one report indicates that Ser-650 phosphorylation enhances the interaction between AR and the E3 ubiquitin-ligase CHIP (22). While PP1 binding is not androgen-dependent, conformational changes in AR mediated by ligand binding or interactions with other proteins may nonetheless regulate PP1 accessibility to pS650, and thereby indirectly regulate AR localization or degradation (8).
In this study, coprecipitation experiments indicated that there was a direct interaction between AR and PP1α, although an indirect interaction mediated by a PP1 regulatory protein cannot yet be excluded. Binding of PP1 catalytic subunits to many regulatory subunits is mediated in part by a short motif, (R/K)VXF (13, 14, 23, 24). The AR has one site (KVFF) that fits this motif, which is located in the DNA binding domain and is part of the DNA recognition helix. Our preliminary studies indicate that this site is not required for AR-PP1 binding (data not shown). However, further studies are needed to determine whether AR is interacting directly with the PP1 catalytic subunit through this site or other motifs (25). Alternatively, it remains possible that binding is mediated through a PP1 regulatory protein.
In prostate cancer cells, overexpression of PP1 markedly stimulated AR-mediated transcription, while PP1 inhibition significantly impaired AR transcriptional activity. The magnitude of variations in total AR protein expression were less dramatic and unlikely to fully account for these effects on transcription. Increased nuclear AR clearly provides a further mechanism for the stimulation of AR transcriptional activity by PP1. Additionally, PP1 is known to interact with multiple proteins that regulate chromatin remodeling, transcription, and mRNA splicing (26–31). Therefore, in addition to mediating Ser-650 dephosphorylation and increasing nuclear AR, PP1 may be recruited to AR target genes and function as a coactivator of AR transcriptional activity. Moreover, nuclear recruitment of PP1 by AR would provide a mechanism by which AR could both enhance transcription and regulate splicing of specific genes. These hypotheses are currently under investigation.
Acknowledgments
We thank Dr. Daniel Gioeli (University of Virginia) for generously providing the pS650 AR antibody. We thank Tao Zhang and Dr. Xin Yuan for generating and providing the Flag-AR vector, Dr. Howard Shen for providing Probasin-Luc, and Robert Borgesi for technical assistance.
This work was supported by Department of Defense (DOD) Grant PC060807 and National Institutes of Health Grants R01CA111803 and P50 CA90381 (to S. P. B.). This work was also supported by a SPORE Career Development Award, DOD Postdoctoral Award PC040499, and NIH K99CA135592 (to S. C.), by the Hershey Family Prostate Cancer Research Fund, and by a Challenge Award from the Prostate Cancer Foundation.
- AR
- androgen receptor
- FBS
- fetal bovine serum
- PBS
- phosphate-buffered saline
- OA
- okadaic acid
- DAPI
- 4′,6-diamidino-2-phenylindole
- PP
- protein phosphatase
- PCa
- prostate cancer
- RLU
- relative light unit
- DHT
- dihydrotestosterone.
REFERENCES
- 1.Gelmann E. P. (2002) J. Clin. Oncol. 20, 3001–3015 [DOI] [PubMed] [Google Scholar]
- 2.Kuiper G. G., Brinkmann A. O. (1995) Biochemistry 34, 1851–1857 [DOI] [PubMed] [Google Scholar]
- 3.Zhou Z. X., Kemppainen J. A., Wilson E. M. (1995) Mol. Endocrinol. 9, 605–615 [DOI] [PubMed] [Google Scholar]
- 4.Gioeli D., Ficarro S. B., Kwiek J. J., Aaronson D., Hancock M., Catling A. D., White F. M., Christian R. E., Settlage R. E., Shabanowitz J., Hunt D. F., Weber M. J. (2002) J. Biol. Chem. 277, 29304–29314 [DOI] [PubMed] [Google Scholar]
- 5.Wong H. Y., Burghoorn J. A., Van Leeuwen M., De Ruiter P. E., Schippers E., Blok L. J., Li K. W., Dekker H. L., De Jong L., Trapman J., Grootegoed J. A., Brinkmann A. O. (2004) Biochem. J. 383, 267–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ikonen T., Palvimo J. J., Kallio P. J., Reinikainen P., Jänne O. A. (1994) Endocrinology 135, 1359–1366 [DOI] [PubMed] [Google Scholar]
- 7.Yang C. S., Vitto M. J., Busby S. A., Garcia B. A., Kesler C. T., Gioeli D., Shabanowitz J., Hunt D. F., Rundell K., Brautigan D. L., Paschal B. M. (2005) Mol. Cell. Biol. 25, 1298–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang C. S., Xin H. W., Kelley J. B., Spencer A., Brautigan D. L., Paschal B. M. (2007) Mol. Cell. Biol. 27, 3390–3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thompson J., Lepikhova T., Teixido-Travesa N., Whitehead M. A., Palvimo J. J., Jänne O. A. (2006) EMBO J. 25, 2757–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gioeli D., Black B. E., Gordon V., Spencer A., Kesler C. T., Eblen S. T., Paschal B. M., Weber M. J. (2006) Mol. Endocrinol. 20, 503–515 [DOI] [PubMed] [Google Scholar]
- 11.Dounay A. B., Forsyth C. J. (2002) Curr. Med. Chem. 9, 1939–1980 [DOI] [PubMed] [Google Scholar]
- 12.Chen S., Xu Y., Yuan X., Bubley G. J., Balk S. P. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 15969–15974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ceulemans H., Bollen M. (2004) Physiol. Rev. 84, 1–39 [DOI] [PubMed] [Google Scholar]
- 14.Cohen P. T. (2002) J. Cell Sci. 115, 241–256 [DOI] [PubMed] [Google Scholar]
- 15.Zhou Z. X., Lane M. V., Kemppainen J. A., French F. S., Wilson E. M. (1995) Mol. Endocrinol. 9, 208–218 [DOI] [PubMed] [Google Scholar]
- 16.Sheflin L., Keegan B., Zhang W., Spaulding S. W. (2000) Biochem. Biophys. Res. Commun. 276, 144–150 [DOI] [PubMed] [Google Scholar]
- 17.Moorhead G., MacKintosh R. W., Morrice N., Gallagher T., MacKintosh C. (1994) FEBS Lett. 356, 46–50 [DOI] [PubMed] [Google Scholar]
- 18.Shank L. C., Kelley J. B., Gioeli D., Yang C. S., Spencer A., Allison L. A., Paschal B. M. (2008) J. Biol. Chem. 283, 10568–10580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanner T., Claessens F., Haelens A. (2004) Ann. N.Y. Acad. Sci. 1030, 587–592 [DOI] [PubMed] [Google Scholar]
- 20.Lin H. K., Altuwaijri S., Lin W. J., Kan P. Y., Collins L. L., Chang C. (2002) J. Biol. Chem. 277, 36570–36576 [DOI] [PubMed] [Google Scholar]
- 21.Kang Z., Pirskanen A., Jänne O. A., Palvimo J. J. (2002) J. Biol. Chem. 277, 48366–48371 [DOI] [PubMed] [Google Scholar]
- 22.Rees I., Lee S., Kim H., Tsai F. T. (2006) Biochim. Biophys. Acta 1764, 1073–1079 [DOI] [PubMed] [Google Scholar]
- 23.Egloff M. P., Johnson D. F., Moorhead G., Cohen P. T., Cohen P., Barford D. (1997) EMBO J. 16, 1876–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wakula P., Beullens M., Ceulemans H., Stalmans W., Bollen M. (2003) J. Biol. Chem. 278, 18817–18823 [DOI] [PubMed] [Google Scholar]
- 25.Garcia A., Cayla X., Caudron B., Deveaud E., Roncal F., Rebollo A. (2004) C.R. Biol. 327, 93–97 [DOI] [PubMed] [Google Scholar]
- 26.Washington K., Ammosova T., Beullens M., Jerebtsova M., Kumar A., Bollen M., Nekhai S. (2002) J. Biol. Chem. 277, 40442–40448 [DOI] [PubMed] [Google Scholar]
- 27.Wu D. Y., Tkachuck D. C., Roberson R. S., Schubach W. H. (2002) J. Biol. Chem. 277, 27706–27715 [DOI] [PubMed] [Google Scholar]
- 28.Canettieri G., Morantte I., Guzmán E., Asahara H., Herzig S., Anderson S. D., Yates J. R., 3rd, Montminy M. (2003) Nat. Struct. Biol. 10, 175–181 [DOI] [PubMed] [Google Scholar]
- 29.Boudrez A., Beullens M., Groenen P., Van Eynde A., Vulsteke V., Jagiello I., Murray M., Krainer A. R., Stalmans W., Bollen M. (2000) J. Biol. Chem. 275, 25411–25417 [DOI] [PubMed] [Google Scholar]
- 30.Chen C. S., Weng S. C., Tseng P. H., Lin H. P., Chen C. S. (2005) J. Biol. Chem. 280, 38879–38887 [DOI] [PubMed] [Google Scholar]
- 31.Li C., Liang Y. Y., Feng X. H., Tsai S. Y., Tsai M. J., O'Malley B. W. (2008) Mol. Cell 31, 835–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deleted in proof